Abstract
Juvenile hormone (JH) is a key regulator of insect development and reproduction. In adult mosquitoes, it is essential for maturation of the ovary and normal male reproductive behavior, but how JH distribution and activity is regulated after secretion is unclear. Here, we report a new type of specific JH-binding protein, given the name mosquito juvenile hormone-binding protein (mJHBP), which circulates in the hemolymph of pupal and adult Aedes aegypti males and females. mJHBP is a member of the odorant-binding protein (OBP) family, and orthologs are present in the genomes of Aedes, Culex, and Anopheles mosquito species. Using isothermal titration calorimetry, we show that mJHBP specifically binds JH II and JH III but not eicosanoids or JH derivatives. mJHBP was crystallized in the presence of JH III and found to have a double OBP domain structure reminiscent of salivary “long” D7 proteins of mosquitoes. We observed that a single JH III molecule is contained in the N-terminal domain binding pocket that is closed in an apparent conformational change by a C-terminal domain-derived α-helix. The electron density for the ligand indicated a high occupancy of the natural 10R enantiomer of JH III. Of note, mJHBP is structurally unrelated to hemolymph JHBP from lepidopteran insects. A low level of expression of mJHBP in Ae. aegypti larvae suggests that it is primarily active during the adult stage where it could potentially influence the effects of JH on egg development, mating behavior, feeding, or other processes.
Keywords: development, insect, ligand-binding protein, reproduction, X-ray crystallography
Introduction
The involvement of juvenile hormone (JH)2 in metamorphosis, reproductive development, and aspects of mating behavior in both male and female insects is well established (1). A JH receptor protein, methoprene-tolerant (Met), was first identified in Drosophila melanogaster and has since been shown to be conserved in insects and other arthropods and to initiate signaling of JH-dependent pathways (2–4). The movement of JH within the insect is not completely understood, but in the insect order Lepidoptera (moths), hemolymph juvenile hormone-binding proteins (hJHBPs) are thought to distribute the hormone to target tissues, protect it from degradation, and regulate its bound concentration (5, 6). The lepidopteran hJHBPs belong to the invertebrate Takeout protein family and the TULIP superfamily of lipid-binding proteins (7). In the larvae of Manduca sexta, the hJHBP concentration is far higher than that of the hormone and is well above the dissociation constant for the complex, indicating that circulating JH is nearly all bound (8). The structure of hJHBP from Bombyx mori in complex with JH has been determined to contain a single binding site for the hormone (9). Structural comparison of ligand-bound and ligand-free forms of hJHBP has shown that JH binding induces a conformational change, resulting in burial of the hormone in the interior of the transport protein (9, 10). It has been suggested that contact of hJHBP with target tissues reverses this process, resulting in release of the ligand into cell membranes or cytoplasm. Outside of the Lepidoptera, direct evidence for circulating hJHBPs is scant. The presence of members of the Takeout protein family in other orders of insects has led to suggestions that hJHBPs of this type are broadly distributed and that the lepidopteran model of JH transport is widespread (11). However, Takeout proteins have relatively low sequence homology with known hJHBPs and have not been shown to bind JH, so the state of circulating JH in non-lepidopeteran insects has remained unclear.
The D7 proteins are insect odorant-binding protein (OBP) family members found in the saliva of female mosquitoes and other blood-feeding species in the dipteran suborder Nematocera (12–14). Long D7 proteins recognize hydrophobic ligands by means of two OBP domains that bind different types of small molecules. The N-terminal domain of the long D7s binds vertebrate eicosanoid mediators in Aedes and Anopheles mosquitoes (12, 14). In Aedes sp. the C-terminal domain binds biogenic amines including serotonin and histamine, whereas the Anopheles C-terminal domain does not appear to have a small-molecule binding site (12, 14). Binding of these ligands prevents the rapid inflammatory reactions in host skin that interfere with the feeding process, which is essential for the transmission of malaria parasites and pathogenic viruses to human hosts.
We have screened OBP sequences in the available mosquito genomic databases, including those of important disease vectors, in search of genes related to the salivary D7 proteins that may be expressed outside of the salivary gland. We hypothesized that proteins of this type would bind hydrophobic small molecules acting in essential physiological processes. In this study we identify a D7-like protein in the hemolymph of Ae. aegypti as a ligand-specific JH-binding protein. The X-ray crystal structure of the JH III–protein complex reveals a single binding site and suggests that the protein undergoes a conformational change upon hormone loading that stabilizes the ligand in the binding pocket. This protein, given the name mJHBP, is a novel form of circulating JHBP that is structurally unrelated to the lepidopteran hJHBP. Its restriction to the adult and pupal stages of mosquitoes suggests that it also plays a different physiological role than the lepidopteran binding protein.
Results
Characterization of AAEL008620
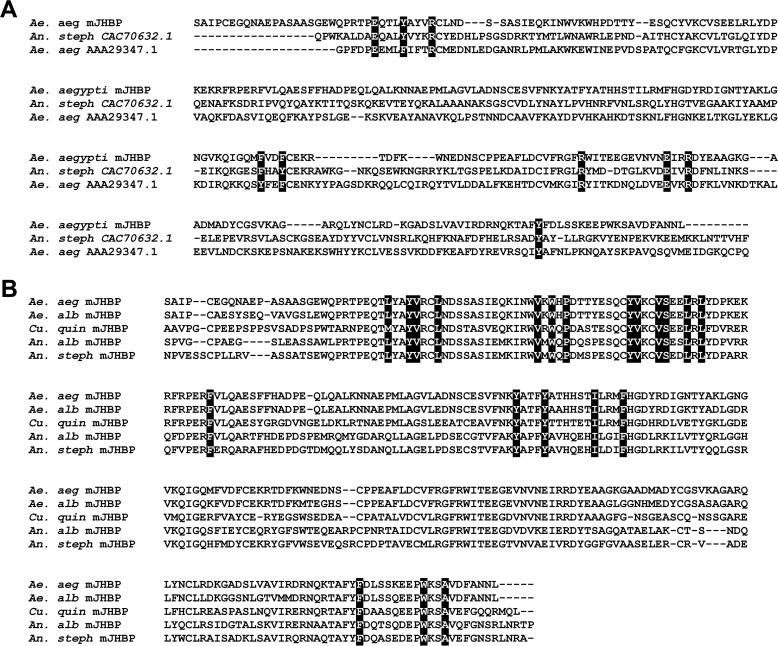
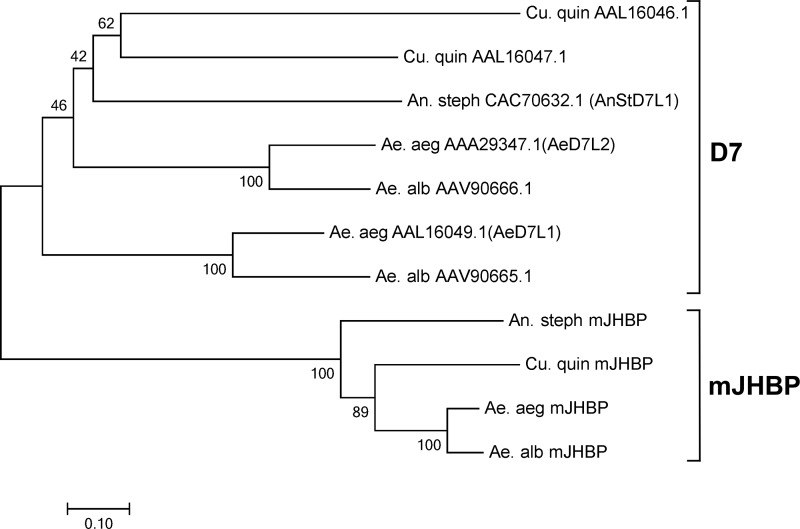
A transcript annotated as AAEL008620, containing a secretion signal sequence (as indicated using the program Signal-P; Ref. 15) and showing pronounced similarity to the salivary long D7 protein group, was identified in expressed sequence tag and genomic databases from Ae. aegypti but was present only at low frequency in the published salivary gland transcriptome (16). The D7s belong to the insect odorant-binding protein family and are secreted into the female saliva where they bind small-molecule mediators of vertebrate host hemostasis and inflammation during blood feeding. A broad search of sequence data detected apparent orthologs of AAEL008620 in the genomes of all Anopheles sp. (Fig. 1, supplemental Fig. S1). The available genomic data for Culex quinquefaciatus did not contain an orthologous sequence to AAEL008620, but partial sequences were found in a database of expressed sequence tags. The complete cDNA for the Culex ortholog of AAEL008620 was obtained using reverse-transcription PCR and 5′-RACE procedures with mRNA prepared from whole C. quinquefaciatus adults (Fig. 1B). The amino acid sequences of AAEL008620 and their orthologs show considerably more similarity to one another than to the salivary D7 sequences in the three genera, including those within the same species (Fig. 1, A and B). For example, AAEL008620 and SRS008484, their orthologs from Anopheles stephensi, show 58% amino acid identity in a pairwise alignment, whereas AAEL008620 and the Ae. aegypti salivary D7 AeD7L1 are only 24% identical. This group of D7-like proteins forms a distinct clade that is present in anopheline and culicine mosquito species (Fig. 2). Extensive database searches revealed no potential orthologs of AAEL008620 in the higher flies, including D. melanogaster or in any other order of insects.
Figure 1.
Sequence comparisons of Ae. aegypti mJHBP (AAEL008620) with salivary D7 proteins and orthologs from other mosquito species. A, amino acid sequence alignment, constructed with MUSCLE (37), of mJHBP with salivary D7 proteins from An. stephensi (An. steph) and Ae. aegypti (Ae. aeg). Highlighted residues correspond to structurally conserved positions in the interface between the N-terminal and C-terminal domains. B, amino acid sequence alignment of mJHBP with orthologs from Ae. albopictus (Ae. alb), C. quinquefaciatus (Cu. quin), An. albimanus (An. alb), and An. stephensi (An. steph). Highlighted residues indicate conserved binding pocket sites that contact the bound ligand in the Ae. aegypti mJHBP–JH III complex.
Figure 2.
Neighbor joining phylogenetic tree, constructed with MEGA (38), showing the relationship of mosquito salivary D7 proteins to members of the mJHBP (AAEL008620) clade. Salivary D7 and mJHBP clades are indicated. Abbreviations are as in Fig. 1. Values indicate percentage bootstrap support at each node.
The salivary long D7 proteins contain two odorant-binding protein domains, with the N-terminal domain functioning as a scavenger of vertebrate eicosanoid mediators including cysteinyl leukotrienes and thromboxane A2 (TXA2) (12, 14). The C-terminal domain of the culicine salivary D7s binds biogenic amines including serotonin and norepinephrine, whereas those of Anopheles sp. do not have a well-defined binding pocket. Clear evidence of a two-domain structure, including the conservation of amino acid residues present in the D7 domain interface, is seen in the AAEL008620 sequence (Fig. 1A). The region of AAEL008620 corresponding to the N-terminal domain binding pocket of the salivary D7s contains many conserved residues that contact the ligand in the D7 crystal structures, indicating that the protein has a pocket of similar shape and hydrophobicity to these forms and may bind a similar fatty acid-like molecule. The degree of amino acid identity with salivary D7s is lower in the C-terminal domain of AAEL008620 than in the N-terminal domain, and it is not completely clear from the sequences alone that that they are structurally similar in this portion of the protein.
Tissue distribution and expression of AAEL008620
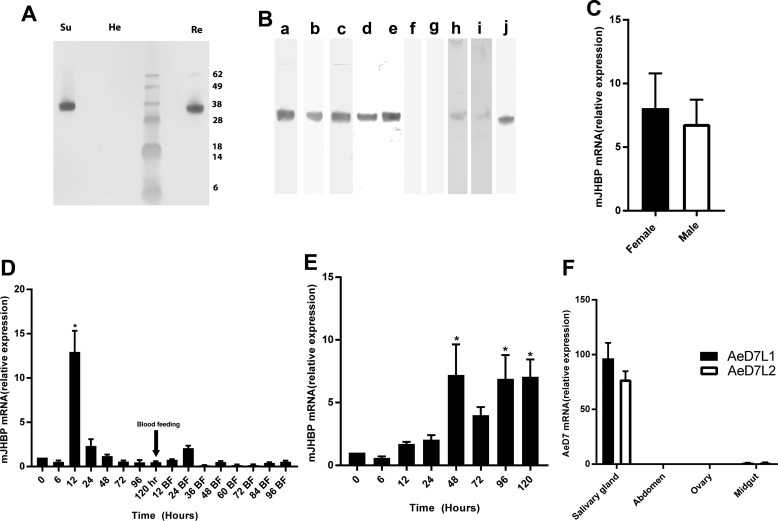
Recombinant Ae. aegypti AAEL008620 was produced in E. coli and purified to homogeneity. Antisera raised against the recombinant protein recognized both the recombinant protein itself and a protein of nearly identical size in whole-body homogenates of female adult mosquitoes when examined by immunoblotting. Hemolymph obtained by perfusion from male and female non-blood-fed mosquitoes also showed a strong signal in Western blots, indicating that AAEL008620 encodes a circulating hemolymph protein that is present in the adults of both sexes (Fig. 3, A and B). Fractionation of crude hemolymph by centrifugation at 10,000 × g followed by Western blotting of the supernatant and pellet fractions showed the protein to be present only in the cell-free fraction (Fig. 3A). Hemolymph samples obtained from pupae 1 and 3 days after pupation also showed the presence of AAEL008620, but hemolymph samples from third and fourth instar larvae showed no trace of the protein (Fig. 3B). Because D7s are secreted into the lumen of the adult female salivary gland, we also examined salivary gland extracts and detected small amounts of the AAEL008620 protein in 10 gland-pair equivalents, but the lack of detection in previous proteomic studies suggests that it is not a major component of the saliva (Fig. 3B) (17). Additionally, AAEL008620 has been previously detected by mass spectrometry in extracts of the male accessory gland of Ae. aegypti (18). We detected minute amounts of the protein by immunoblotting extracts equivalent to 30 pairs of male accessory gland dissected from unmated 4-day-old adults (Fig. 3B).
Figure 3.
Expression of mJHBP in Ae. aegypti. A, detection of mJHBP in cell-free hemolymph and hemocytes of female Ae. aegypti adults by Western blotting. Cell free hemolymph (Su), pelleted hemocytes (He), and recombinant mJHBP (Re) were analyzed on the same gel. Molecular mass standards are shown, and values for these are shown at the right edge of the panel. B. localization and stage-specific expression of mJHBP in Ae. aegypti as measured by Western blotting shown in blot strips prepared from different gels. Lane a, sugar fed female adult hemolymph (4 insect equivalents (ie); lane b, blood-fed female adult hemolymph (4 ie); lane c, male adult hemolymph (4 ie); lane d, pupal hemolymph, 1 day post molting (5 ie); lane e, pupal hemolymph, 3 days post molting (5 ie); lane f, larval hemolymph 3rd instar (5 ie); lane g, larval hemolymph 4th instar (5 ie); lane h, salivary gland homogenate (10 ie); lane i, male accessory gland homogenate (30 ie); j, recombinant mJHBP. C, quantitative PCR of mJHBP expression in male and female (3-days post eclosion) Ae. aegypti abdominal carcasses. The data are presented as enhancement of expression relative to samples of mRNA extracted from whole bodies (mean ± S.E.). D, time course of mJHBP expression in adult females as measured by quantitative PCR. mRNA samples were extracted from female abdominal carcasses and analyzed as in panel B. Data are displayed relative to a value of 1 at eclosion (time = 0). Females were fed on chickens at 120 h post eclosion. The bar marked * indicates statistical significance (p = 0.0001, one-way analysis of variance). E, time course of mJHBP expression in adult males measured as in panel C. (*, p ≤ 0.01). F, comparison of mJHBP and salivary D7 expression in different tissues by quantitative PCR. mRNA from dissected salivary glands, abdominal carcass, ovaries, and midgut was amplified using primers specific for two salivary D7 (AeD7L1 and AeD7L2) sequences and for mJHBP. The data are presented as salivary D7 expression relative to mJHBP for each tissue.
The AAEL008620 transcript was detected in mRNA preparations from both male and female adults (Fig. 3C). Preparations from adult abdomens after removal of digestive and reproductive organs (carcass) showed higher levels of transcript relative to control sequences than samples from whole bodies, suggesting that the fat body may be a major source of the protein (Fig. 3C). The time course of expression in female abdominal carcasses revealed a significant peak of transcription at ∼12 h post-eclosion, corresponding to a reported peak of JH III synthesis (Fig. 3D) (19). Transcript levels declined after this and remained at a similar level after blood feeding (Fig. 3D). In the male adult, transcript levels increased significantly 48 h after eclosion (Fig. 3E). Expression of AAEL008620 was also detected in the salivary gland, but the transcript levels were 70–100-fold lower than those for the salivary D7 proteins AeD7L1 and AeD7L2 (Fig. 3F).
Ligand binding by the AAEL008620 gene product
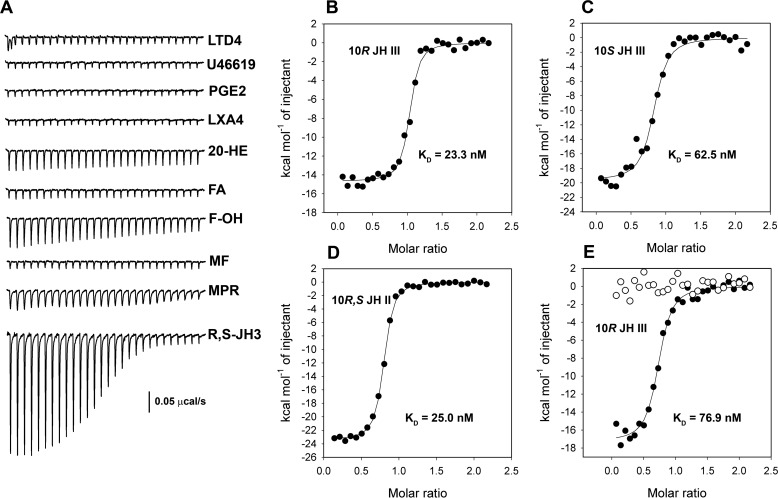
To identify binding partners for AAEL008620 we used the recombinant protein to screen potential ligands by isothermal titration calorimetry (ITC). We first tested several eicosanoid compounds to determine if the ligand selectivity profile resembled the salivary D7s. Neither leukotriene D4 (LTD4) nor the TXA2 analog U46619, two ligands known to bind with the N-terminal domains of D7 proteins from Aedes and Anopheles species (12, 14), showed any detectable binding (Fig. 4A). This indicates that the endogenously functioning protein does not recognize the vertebrate effectors bound by salivary D7s. Prostaglandins and lipoxins are eicosanoids known to be present in insect hemolymph where they serve as mediators of essential processes including antimicrobial responses (20). Recombinant AAEL008620 did not show detectable binding with either prostaglandin E2 (PGE2) or lipoxin A4 (LXA4), a known regulator of mosquito hemocyte function (Fig. 4A) (21).
Figure 4.
Ligand binding of mJHBP (AAEL008620) as measured by ITC. For all experiments, the mJHBP concentration in the cell was 5 μm, and the ligand concentration in the syringe was 50 μm. A, raw injection profiles for ligand screening. U46619, thromboxane A2 analog U46619; 20-HE, 20-hydroxyecdysone; FA, farnesoic acid; F-OH, 1-farnesol; MF, methyl farnesoate; MPR, methoprene; R,S-JH3, racemic JH III. B, injection enthalpies (filled circles) for binding of 10R-JH III with mJHBP fit to a single site-binding model (solid line). Thermodynamic parameters: ΔH = −14.7 ± 0.2 kcal mol−1, K = 4.3 ± 0.9 × 107 m−1, TΔS = −4.1 kcal mol−1, n = 1.0. C, injection enthalpies for binding of 10S-JH III with mJHBP fit to a single site-binding model. Thermodynamic parameters: ΔH = −19.7 ± 0.5 kcal mol−1, K = 1.6 ± 0.4 × 107 m−1, TΔS = −9.7 kcal mol−1, n = 0.8. D, injection enthalpies for binding of JH II with mJHBP fit to a single site-binding model. Thermodynamic parameters: ΔH = −23.4 ± 0.1 kcal mol−1; K = 4.0 ± 0.3 × 107 m−1, TΔS = −12.9 kcal mol−1, n = 0.8. E, injection enthalpies for binding of 10R-JH III with the An. stephensi mJHBP ortholog (filled circles) fit to a single site-binding model. Thermodynamic parameters: ΔH = −17.3 ± 0.3 kcal mol−1, K = 1.3 ± 0.2 × 107 m−1, TΔS = −7.4 kcal mol−1, n = 0.7. Also shown are injection enthalpies for binding of 10R-JH III with AnStD7L1, a salivary D7 protein from An. stephensi (open circles).
Because circulating JH-binding proteins have not been identified outside of the order Lepidoptera, we tested racemic JH III as a ligand and found that it bound tightly to the AAEL008620 gene product (Fig. 4A). The binding reaction was enthalpy-driven (ΔH = −20.0 ± 0.3 kcal/mol) with an equilibrium constant (K) of 9.11 × 106 m−1, corresponding to a dissociation constant (Kd) of 109 nm. Binding was found to be highly specific for the JH epoxy methyl ester, as methyl farnesoate showed no detectible interaction with the protein (Fig. 4A). Binding of farnesoic acid was also not detectible, whereas 1-farnesol and methoprene interacted only weakly with the protein (Fig. 4A). The insect-molting hormone, 20-hydroxyecdysone, which plays an important role in development of the vitellogenic ovary, displayed little or no detectable affinity for AAEL008620 (Fig. 4A).
Racemic JH III was chromatographically separated into its component optical isomers, and binding with each was measured. AAEL008620 bound the naturally occurring 10R enantiomer with approximately 3 times higher affinity as measured by ITC, indicating that the protein differentially recognizes the natural enantiomer of the hormone (Fig. 4, B and C). In Lepidoptera, the predominant JH forms are JH I, having ethyl substituents at C-7 and C-11, and JH II, having a methyl substituent at C-7 and an ethyl group at C-11. The AAEL008620 gene product was also found to bind racemic JH II with high affinity (Fig. 4D), indicating that there is sufficient space in the binding pocket for an additional methyl group. We conclude that AAEL008620 is a specific JH-binding protein that is circulating in the hemolymph of pupal and adult male and female mosquitoes. Because of this we have given this protein the name mJHBP, indicating that it is a mosquito juvenile hormone-binding protein functionally analogous to, but completely unrelated in structure to the previously described hJHBP of Lepidoptera.
The structure of mJHBP
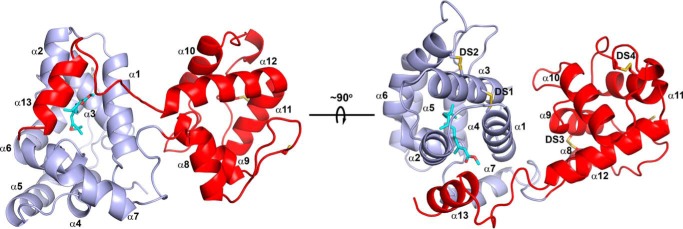
The crystal structure of mJHBP containing bound JH III was determined by molecular replacement using an unpublished polyalanine-substituted model of the N-terminal domain of a salivary long D7 from the sand fly Phlebotomus duboscqi to perform the search (Table 1). The structure of the protein is generally similar to that of the salivary long D7 proteins in that it contains two odorant-binding protein domains connected by a linker sequence and has a well-ordered domain interface that is structurally homologous with those of the salivary long D7 proteins from Ae. aegypti and An. stephensi (Fig. 5) (12, 14). The N-terminal domain is a bundle of seven α-helices and contains most of the residues included in the JH III-binding site. This domain is similar in its overall structure to the N-terminal domain of AeD7L1 and AnStD7L1, the structurally characterized salivary D7 proteins from Ae. aegypti and An. stephensi, with superpositions showing root mean square deviations of 1.88 Å (106 Cα atoms) and 1.52 Å (109 Cα atoms), respectively (12, 14). Two disulfide bonds are present in the N-terminal domain of mJHBP with Cys-36 of helix α1 being linked with Cys-67 of α3 and Cys-63 of α3 being linked with Cys-122 of α6. The linker peptide between the two domains extends from Gly-159 to Met-168 (Fig. 5).
Table 1.
Data collection and refinement statistics for the mJHBP–JH III complex
| Crystal | JH III complex |
|---|---|
| Resolution (Å) | 55.4-1.84 |
| Beamline | 22-ID |
| Wavelength (Å) | 0.979010 |
| Completeness (total/high resolution shell) | 98.5/99.8 |
| Average redundancy (total/high resolution shell) | 3.5/3.5 |
| Rmerge (total/high resolution shell, %) | 10.2/52.5 |
| CC1/2 (total/high resolution shell) | 0.99/0.80 |
| I/sigI (total/high resolution shell) | 10.8/2.7 |
| Observed reflections | 332,506 |
| Unique reflections | 78,904 |
| Space group | C2 |
| Unit cell dimensions (Å) | |
| a | 120.1 |
| b | 69.1 |
| c | 121.9 |
| β (°) | 112.8 |
| Refinement | |
| Total non-H protein atoms | 6532 |
| Total non-H solvent atoms | 672 |
| Total non-H ligand atoms | 57 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.828 |
| Wilson B factor (Å2) | 18.7 |
| Mean B factors (Å2) | |
| Protein | 25.4 |
| Solvent | 33.6 |
| Ligand | 18.2 |
| Molprobity analysis | |
| Ramachandran plot (allowed/total, %) | 97.7/100 |
| Clashscore | 0.94 |
| Rotamer outliers (%) | 0.58 |
| Coordinate error ML (Å, Phenix) | 0.18 |
| Rcryst/Rfree | 0.16/0.20 |
Figure 5.
Ribbon diagrams of the mJHBP structure. The N-terminal domain is colored light blue, and the C-terminal domain is colored red, and the right and left images are related by a ∼90° rotation around the horizontal axis shown. The helical elements making up the structure are designated α1-α13. The disulfide bonds are shown in stick representation with sulfur colored yellow and are designated DS1 to DS4 in the right-hand figure. The JH III ligand is also shown as a stick representation with carbon atoms colored cyan and oxygen red.
The C-terminal domain of mJHBP contains six helical segments with two disulfide bonds linking Cys-173 with Cys-195 of helix α9 and Cys-232 with Cys-245 of helix α11 (Fig. 5). The domain interface is stabilized by several interdomain interactions including salt bridges linking Glu-27 with Arg-201, Arg-73 with Glu-206, and Arg-35 with Glu-213 as well as a stacking interaction between Tyr-31 and Arg-201 supplemental Fig. S2). Although the sequence similarity of this domain with the salivary D7 proteins is lower than the N-terminal domain, the C-terminal domain contains a very similar arrangement of helical elements to AnStD7L1, with the exception of α13, which extends over the surface of the N-terminal domain in mJHBP and is discussed below in relation to JH III binding (Fig. 5). Notably, 14 residues at the N terminus of mJHBP are not visible in the structure and are presumably disordered. The signal peptide cleavage site was predicted using Signal-P and was not observed directly, but analogous predicted sites were found at approximately the same positions in putative mJHBP orthologs from various mosquito species. The slightly larger size of hemolymph mJHBP relative to recombinant could be due to glycosylation. An N-glycosylation site (NetNGlyc) is predicted at the solvent-exposed side chain of Asn-38 in helix α1 (22).
The JH III-binding pocket
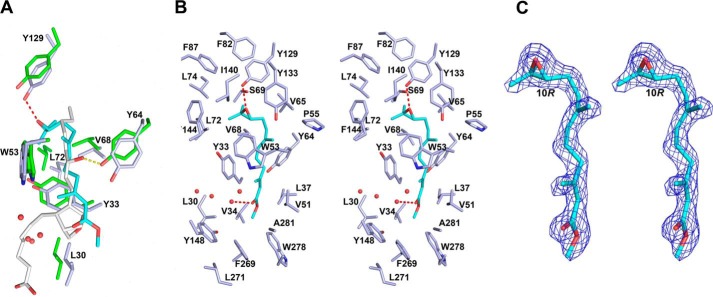
A single well-ordered molecule of JH III (refined occupancies 0.91–0.96) is present in the binding pocket of the N-terminal domain of mJHBP (Figs. 5, 6, and 7 and Table 1). Three molecules of the complex are present in the asymmetric unit of the crystal, and the ligand conformation is essentially identical in all three. The epoxy end of JH III is located deep in the core of the domain, whereas the methyl ester group of the hormone is oriented toward its surface (Figs. 5 and 7). The epoxy group forms a hydrogen bond with the phenolic hydroxyl of Tyr-129, and the remainder of the isoprenoid chain is surrounded by hydrophobic side chains including those of Phe-144, Tyr-64, Trp-53, Val-65, Val-68, Leu-72, Leu-74, Val-51, and Tyr-33 (Fig. 6, A and B). A hydrogen bond between an ordered water molecule and the carbonyl oxygen of the methyl ester moiety further stabilizes the ligand (Fig. 6, A and B). Although the protein was crystallized in the presence of a large excess of racemic JH III, the electron density for the ligand clearly indicates that the 10R enantiomer predominates in the binding pocket, reflecting the selectivity of mJHBP for the natural 10R form of JH III. (Fig. 6C).
Figure 6.
Ligand binding interactions in mJHBP and salivary D7. A, stick diagrams of conserved residues in the binding pockets of mJHBP (carbon atoms colored light blue) and An. stephensi salivary D7 protein AnStD7L1 (carbon atoms colored green). The JH III ligand in mJHBP is colored cyan with oxygen in red, and the U46619 ligand in AnStD7L1 is colored light gray, with oxygen colored red. The hydrogen bond between Tyr-129 of mJHBP and the JH III epoxide group is shown as a red dashed line, and the hydrogen bond between the equivalent of Tyr 64 in AnStD7L1 and the ω-6 hydroxyl is shown as a yellow dashed line. Numbering of the residues Is for mJHBP. B, stereoview of mJHBP binding pocket with JH III (cyan) bound. Hydrogen bonds are shown as red dashed lines, and ordered water molecules in the binding pocket are shown as red spheres. C, stereoview of simulated annealing omit Fo − Fc electron density (contoured at 3 σ) with the final JH III ligand model inserted. The map was calculated with all JH III ligand coordinates omitted prior to simulated annealing refinement in Phenix.refine (35). The C-10 position having an R configuration is labeled.
Figure 7.
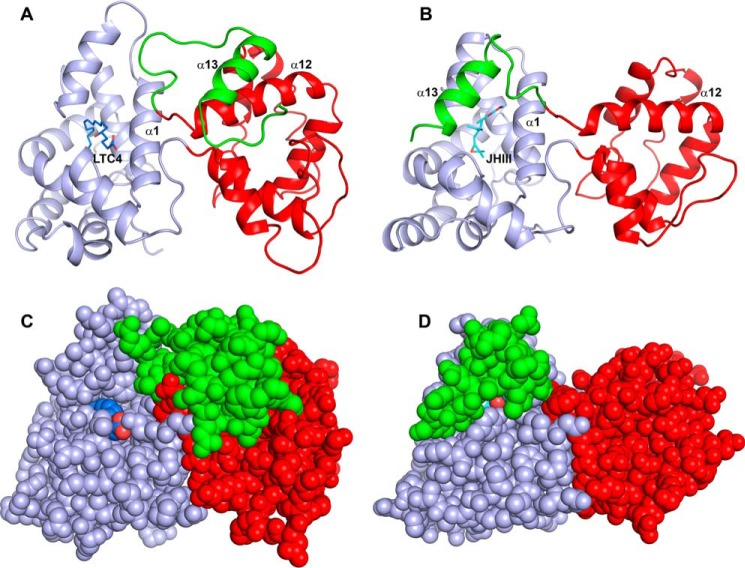
Structural comparison of a salivary D7 protein (AnStD7L1) and mJHBP. A, ribbon diagram of AnStD7L1 with the N-terminal domain colored light blue, the C-terminal domain colored red, and the extreme C terminus including helix α13 colored green. The leukotriene C4 ligand (LTC4) is shown in stick representation with carbon colored blue and oxygen red. B, mJHBP oriented and colored as in panel A with the JH III ligand shown in stick representation with carbon colored cyan and oxygen red. In the case of mJHBP, the C-terminal portion including helix α13 is positioned to cover the entry to the binding pocket. C, space filling representation of panel A to show the open entry to the N-terminal domain binding pocket that accommodates the 20-carbon fatty acid of leukotriene C4. D, the space filling model of mJHBP, oriented as in panel B, shows nearly complete burial of the bound ligand and occlusion of the entry channel.
No clear ligand entry or exit path is present in the mJHBP model, and the ligand is buried in its interior. The C terminus of the protein, including the coil region between α12 and α13 as well as helix α13, passes over the methyl ester end of the ligand and interacts with the surface of the N-terminal domain (Figs. 5 and 7B). Helix α13 is packed in an approximately parallel orientation against α2, and the facing surfaces of these structures are in van der Waals contact. One interdomain hydrogen bond, between the carbonyl oxygen of Ala-285 near the C terminus and the amide nitrogen of Tyr-148, is present. In the C-terminal region the side chains of Phe-269, Trp-278, and Ala-281 surround the methyl ester end of the JH III ligand. This closely packed arrangement of hydrophobic side chains explains the exclusion of JH III derivatives having hydrophilic groups in this part of the molecule as well as methoprene with its bulkier isopropyl ester moiety.
More than 20 amino acid residues were found to contact the ligand in the binding pocket of mJHBP (Fig. 6B). Examination of an amino acid sequence alignment of mJHBP with its apparent orthologs from Aedes albopictus and C. quinquefaciatus, and 20 species from the genus Anopheles show that the structure of the pocket is highly conserved (Fig. 1B, supplemental Fig. S1). Although the overall amino acid identity of the sequences between genera is ∼50%, nearly complete conservation of binding pocket residues is seen, including those contained in the C-terminal domain, strongly suggesting that JH III binding by mJHBP is functionally conserved in anopheline and culicine mosquitoes (Fig. 1B). To test this hypothesis we produced a recombinant version of the mJHBP ortholog from An. stephensi (ASTE000616) by the methods used for the Ae. aegypti protein and evaluated the binding of JH III by ITC. This protein bound 10R-JH III similarly to the Ae. aegypti mJHBP (Fig. 4E, Kd = 76 nm). In contrast, no detectable binding of JH III was seen with recombinant AnStD7L1, a salivary D7 protein from An. stephensi that is known to bind cysteinyl leukotrienes and TXA2 analogs in its N-terminal domain binding pocket (Fig. 4E).
Similarities of mJHBP and long D7 proteins
Comparison of the crystal structure of the salivary long D7 protein AnStD7L1 from An. stephensi (12) with hormone-bound mJHBP suggests that a conformational change may be important in the ligand binding/release mechanism (Fig. 6). In the salivary D7 protein, helix α13 is turned away from the N-terminal domain and interacts with the surface of the C-terminal domain (Fig. 7, A and C). This provides an entry path and binding surface for the longer eicosanoid fatty acid ligands LTC4 and the TXA2 analog U46619 that remains open (Fig. 7, A and C) (12). The D7 residue corresponding to Ile-165 in mJHBP is a lysine that forms a salt bridge with the carboxyl group of the fatty acid (Fig. 1A). In mJHBP, the area occupied by the carboxyl end of the fatty acid ligand is filled by the parts of the C-terminal domain including α13, which interacts with JH III, nearly occluding the apparent path of ligand entry seen in the salivary D7 proteins (Fig. 7, B and D). It appears likely that the C-terminal end of mJHBP forms its interaction with the N-terminal domain during ligand binding. This suggests a binding model in which the initial ligand entry occurs through the open channel seen in the N-terminal domain of salivary D7 proteins followed by ordering of the C-terminal region of the protein into a cap structure that covers the entry site in a conformational change that is perhaps driven by interaction with the ligand. Crystals of the ligand-free protein were not obtained after extensive screening, suggesting that there are significant structural differences between the ligand-bound and ligand-free forms.
In the channel of the N-terminal domain that accommodates the bulk of the JH III ligand, Leu-30, Val-48, Tyr-33, Trp-53, Tyr-64, and Tyr-129 are conserved between mJHBP and the salivary D7s (Fig. 6A) (12, 14). As discussed above, the phenolic hydroxyl group of the conserved residue Tyr-129 forms a hydrogen bond with the epoxide oxygen of JH III, whereas the rest of these residues surround the hydrocarbon portion of the molecule. The long eicosanoid chain of the D7 ligands extends further along the surface of the protein in the groove formed by helices α1 and α7 (Figs. 6A and 7B). This region is partially blocked by the C-terminal cap structure of mJHBP, but a cavity containing as many as four ordered water molecules is present in the corresponding space of mJHBP, suggesting that few amino acid changes would be necessary to form a functional eicosanoid binding pocket in an mJHBP ortholog.
Discussion
mJHBP belongs to the insect OBP family and is related to the mosquito salivary D7 proteins, which recognize vertebrate eicosanoid effectors of inflammation released during blood feeding (12–14). It does not detectably bind these vertebrate ligands but is specific for JH. Among compounds related to JH, only methyl ester epoxides bound tightly to mJHBP. Methyl farnesoate, differing from JH only in the absence of the epoxy group, showed little or no binding, nor did farnesoic acid, 1-farnesol, or the insecticidal JH analog methoprene. This spectrum of binding selectivity indicates a highly specific binding pocket that is more exclusive than that of the bHLH-PAS family of JH receptors (23). Moreover, it follows the same highly specific pattern of binding observed in lepidopteran hemolymph JHBPs (hJHBPs; Ref. 5).
The location of mJHBP in the hemolymph rather than the salivary gland is consistent with a JH binding or transport function for the protein. Unlike the salivary long D7 proteins, it is present in both male and female mosquitoes, suggesting that it acts in processes other than blood feeding and egg development. The dissociation constant (KD) of the mJHBP–JHIII complex (Fig. 4) is similar in magnitude to the circulating JH concentration in adult mosquitoes as would be expected for a transport protein whose function depends on binding and release of its ligand. Two recent studies have shown that JH levels in adult Ae. aegypti are in the range of 8–30 pg/individual female. If this was all contained in the hemolymph, the circulating JH III concentration range would be 85–322 nm, based on a reported hemolymph volume of ∼350 nl/individual in An. stephensi (24).
The structure of the mJHBP–JH III complex reveals a binding pocket that is exquisitely tailored in size, shape, and hydrophobicity to accommodate JH III. The ligand is enveloped on all sides by amino acid residues at a site homologous to the binding pocket for eicosanoids in salivary long D7 proteins. The C-terminal helical segment (α13, Fig. 5) lies along the surface of the N-terminal domain and closes the entry to the binding pocket. Protein side chains are packed around both the epoxy and ester ends of the linear JH III molecule, giving an arrangement that would exclude the binding of longer-chain lipid molecules in a similar mode. Notably, the crystallized complex contains predominantly 10R-JH III, whereas in solution the 10S enantiomer is also bound, albeit at lower affinity. This suggests that the 10R complex crystallizes preferentially under these conditions and that complexes of the two enantiomers may differ conformationally. Because only the R enantiomer is found in vivo we conclude that the observed structure represents the natural form of the complex.
Although mainly associated with olfactory or gustatory sensillae, insect OBPs have been found in salivary glands, hemolymph, eggshells, and male accessory glands of various insect species (25, 26). Sensory OBP ligands include sex pheromones as well volatile and non-volatile host compounds detected by olfactory and gustatory receptors. Before this study, no endogenous ligand for a non-sensory OBP had been described. Recently, the expression of an apparent single-domain OBP, OBP6, has been shown to be induced in the midgut of the tsetse fly, Glossina morsitans, by the presence of an endosymbiont and has been implicated in the regulation of immune responses (27). This result shows that essential physiological processes can be mediated by putatively non-sensory OBPs and that endogenous small molecule ligands may be involved in these activities.
The ancient origin and fundamental physiological importance of JH led us to expect a degree of taxonomic conservation of mJHBP at the family level or higher. We found that orthologs of mJHBP are present in the genomes of all available members of the Culicidae (mosquito family) but not outside of this group. Moreover, amino acid residues having contact with the JH ligand in the mJHBP structure show nearly complete conservation, indicating that all orthologs are likely to bind JH. It seems clear that an mJHBP was present in the ancestor of the culicine and anopheline mosquitoes, which diverged during the Jurassic period 150 million years ago (28, 29). The high degree of structural similarity between mJHBP and salivary D7s suggests that the latter may have evolved from an endogenous lipid transporting protein, perhaps a JH-binding protein. A relatively small number of structural changes, including movement of α13 to the position seen in AnStD7L1, could change the ligand selectivity of the protein to accommodate longer-chain vertebrate eicosanoids.
Here we have identified a novel, taxonomically conserved hemolymph-binding protein for JH III that is not related to any previously identified hormone-binding protein and whose existence was not predictable using only bioinformatic approaches. Our binding and structural analyses show stoichiometric, enantioselective, high-affinity incorporation of a single molecule of JH into a binding site that is partially formed through an apparent conformational change. These features are strongly suggestive of a true JH-binding function for this protein in vivo. The physiological activity of mJHBP has not yet been established, and it may act to regulate JH concentration in the hemolymph as is thought for the lepidopteran hJHBPs. However, it is restricted to the pupal and adult stages of Ae. aegypti and apparently does not play a role in the developmental activities of JH in the larval stage. Regulators of mosquito physiology are obvious targets in the development of control agents against these vectors of parasitic and viral diseases that threaten a large fraction of the human population. With this in mind, we are continuing to study the in vivo mechanism of mJHBP action and its role in JH function.
Experimental procedures
Materials
LTD4, U46619, PGE2, lipoxin A4, methoprene, and 20-hydroxyecdysone were obtained from Cayman chemical. Racemic JH III and 1-farnesol (2E,6E) were obtained from Sigma. Methyl farnesoate (2E,6E) and farnesoic acid (2E,6E) were obtained from Echelon Biosciences. Racemic JH II was obtained from Scitech. The enantiomers of JH III (10R and 10S) were purified as described by Cusson et al. (30).
Expression and purification of recombinant mJHBP
The recombinant mJHBP proteins were produced and purified as described previously with modifications (12). Codon-optimized synthetic Ae. aegypti and An. stephensi mJHBP (ASTE000616) cDNAs were cloned separately into pET17b vector and moved into Escherichia coli BL21(DE3) pLys S. The resulting strains were grown at 37 °C in LB broth and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Inclusion bodies were harvested, washed, and solubilized as described previously (12). The protein was refolded by adding the guanidine-solubilized protein solution dropwise into 4 liters of folding buffer (20 mm Tris-HCl, pH 8.0, 300 mm arginine, 2 mm cystamine HCl) and incubating with stirring at 4 °C overnight. The recombinant proteins were purified by gel filtration chromatography on Sephacryl S-200 and Superdex 75 (GE Healthcare), ion exchange chromatography on Q-Sepharose (GE Healthcare), and hydrophobic interaction chromatography on phenyl-Sepharose (GE Healthcare). Polyclonal antiserum was prepared by Spring Valley Laboratories (Sykesville, MD) after injection of a rabbit with refolded recombinant protein in Freund's complete adjuvant.
Hemolymph collection, tissue dissection, and immunoblotting
Hemolymph was obtained from adult Ae. aegypti (Liverpool strain) males and females by perfusion with phosphate-buffered saline (PBS). To fractionate crude hemolymph into cellular and cell-free fractions, perfusion was performed using an isotonic buffer composed of 41 mm sodium citrate, pH 4.5, 186 mm NaCl, 1.7 mm EDTA (31) Drawn capillary tubes were filled with buffer and used to pierce the abdomen. The buffer was then injected through the capillary using a syringe, and the hemolymph-buffer mixture was collected from an incision at the anterior end of the mosquito (21). Hemolymph was also collected from third and fourth instar larvae as well as pupae 1 and 3 days after the larval-pupal molt. In these cases an incision was made at the posterior end of each individual, and 5–10 individuals were placed in a Microcon centrifugal concentration device (100-kDa cutoff). The devices were centrifuged at 12,000–14,000 × g for 5 min, and hemolymph was collected as the filtrate. Salivary glands and male accessory glands were dissected into cold PBS and frozen at −80 °C until use. Before analysis the tissue samples were homogenized by sonication and centrifuged at 13,000 rpm. Samples were separated by SDS-PAGE and blotted to nitrocellulose. After incubation with anti-mJHBP IgG, blots were incubated with goat anti-rabbit alkaline phosphatase conjugate IgG, and bands were visualized using Western Blue substrate reagent (Promega).
Quantitative PCR
Ae. aegypti adult abdominal and tissue RNA were extracted using the TRIzol reagent (Life Technologies). Approximately 1 μg of total RNA was used to synthesize cDNA by using the QuantiTect reverse transcriptase kit (Qiagen). mJHBP-specific primers AE2 F and AE2 R were designed to amplify a 119-base pair amplicon for qPCR (supplemental Table 1). AeD7L1 and AeD7L2, Ae. aegypti salivary D7 proteins, gene-specific primers were designed to amplify 111 bp and 116 bp, respectively (supplemental Table 1). qPCR mixtures containing SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), 300 nm primers, and 100 ng of synthesized cDNA were analyzed for mJHBP RNA abundance with the CFX 96 Real-Time thermal cycler system (Bio-Rad) and normalized against the Ae. aegypti 60S ribosomal protein transcript as the reference gene (supplemental Table 1). A qPCR mixture containing non-template control was also tested in all experiments as a negative control. Six biological replicates were used, and each experiment was repeated twice. Processed data were inspected manually, and the -fold change values were calculated using the 2−ΔΔCt method (16). One-way analysis of variance and multiple comparison tests were used to determine statistical significance.
Cloning of the Culex quinquefasciatus mJHBP cDNA
A DNA sequence coding for a C. quinquefasciatus mJHBP ortholog was not detected in database searches of the C. quinquefasciatus genome (GCA_000209185.1), but a 5′-truncated apparent ortholog of Ae. aegypti mJHBP could be assembled from two publicly available EST/cDNAs (FF178189.1 and FF158205.1) accessed using Vectorbase. A complete cDNA encoding this mJHBP was obtained from C. quinquefasciatus total RNA, prepared from adult abdomens using the 5′ RACE method with the SMARTer RACE 5′/3′ kit (Clontech Laboratories) and the primers described in supplemental Table 1.
Isothermal titration calorimetry
ITC experiments were performed with a Microcal VPITC instrument at 30 °C. The proteins and ligands were dissolved in 20 mm Tris-HCl pH 7.5 at concentrations of 5 μm and 50 μm, respectively. Ligands were dissolved using sonication after evaporation of carrier solvent under a stream of nitrogen. The ligands were added as 10-μl injections to the protein samples contained in the calorimeter cell. Calculated injection enthalpies were fit to a single-site binding model in the Microcal data evaluation software.
Crystallization of mJHBP
Ae. aegypti mJHBP was crystallized using the hanging drop-vapor diffusion method from 30% w/v polyethylene glycol 6000 in 0.1 m MES pH 6.0 with 3.5% w/v sorbitol or d-(+)-glucose (Hampton Research) added to the drops. The protein sample contained a 10-fold molar excess of racemic JH III (Sigma), and the best crystals were obtained with 1 μl of protein and 1 μl of precipitant solution. The crystals were flash-cooled for data collection in the precipitant solution containing 10% glycerol.
Diffraction data collection and structure solution
Diffraction data were collected at beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, and images were processed using XDS (32). mJHBP crystallized in the space group C2 with three monomers contained in the asymmetric unit. The structure of JH-bound mJHBP was determined by molecular replacement using Phaser (Table 1) (33). An N-terminal domain polyalanine-substituted structure of an unpublished salivary D7 protein structure from the sand fly P. duboscqi (24.8% identity with mJHBP N-terminal domain) was used as the search model. After numerous cycles of manual rebuilding using Coot (34) and refinement of the N-terminal domain model using Phenix (35), some features of the C-terminal domain became visible in the electron density map. The structure could then be completed with additional cycles of manual rebuilding and refinement. Occupancy refinement of the JH III ligand in Phenix.refine gave values of 0.91–0.96 for the three copies. The quality of the structure was evaluated using Molprobity (36).
Data availability
Coordinates and structure factors for mJHBP were submitted to the PDB with the accession code 5V13.
Author contributions
I. H. K. performed the experiments and analyzed the data. V P. and W. J. performed the experiments. W. G. G. provided the reagents and performed the experiments. J. M. C. R. analyzed the data. J. F. A. conceived the study, performed the experiments, analyzed the data, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Andre Laughinghouse and Kevin Lee for mosquito rearing and the staff of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory for assistance with X-ray data collection.
This work was supported by the intramural research program of the NIAID, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 5V13) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- JH
- juvenile hormone
- Met
- methoprene-tolerant
- hJHBP
- hemolymph juvenile hormone-binding protein
- mJHBP
- mosquito juvenile hormone-binding protein
- OBP
- odorant-binding protein
- RACE
- rapid amplification of cDNA ends
- TXA2
- thromboxane A2
- ITC
- isothermal titration calorimetry
- LTD4
- leukotriene D4
- LXA4
- lipoxin A4
- PGE2
- prostaglandin E2
- qPCR
- quantitative PCR.
References
- 1. Riddiford L. M. (2008) Juvenile hormone action: a 2007 perspective. J. Insect. Physiol. 54, 895–901 [DOI] [PubMed] [Google Scholar]
- 2. Ashok M., Turner C., and Wilson T. G. (1998) Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 95, 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jindra M., Palli S. R., and Riddiford L. M. (2013) The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 [DOI] [PubMed] [Google Scholar]
- 4. Wilson T. G., and Fabian J. (1986) A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201 [DOI] [PubMed] [Google Scholar]
- 5. Goodman W., Schooley D. A., and Gilbert L. I. (1978) Specificity of the juvenile hormone binding protein: the geometrical isomers of juvenile hormone I. Proc. Natl. Acad. Sci. U.S.A. 75, 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Touhara K., Lerro K. A., Bonning B. C., Hammock B. D., and Prestwich G. D. (1993) Ligand binding by a recombinant insect juvenile hormone binding protein. Biochemistry 32, 2068–2075 [DOI] [PubMed] [Google Scholar]
- 7. Alva V., and Lupas A. N. (2016) The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim. Biophys. Acta 1861, 913–923 [DOI] [PubMed] [Google Scholar]
- 8. Goodman W., and Gilbert L. I. (1978) The hemolymph titer of juvenile hormone binding protein and binding sites during the fourth larval instar of Manduca sexta. Gen. Comp. Endocrinol. 35, 27–34 [DOI] [PubMed] [Google Scholar]
- 9. Suzuki R., Fujimoto Z., Shiotsuki T., Tsuchiya W., Momma M., Tase A., Miyazawa M., and Yamazaki T. (2011) Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori. Sci. Rep. 1, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolodziejczyk R., Bujacz G., Jakób M., Ozyhar A., Jaskolski M., and Kochman M. (2008) Insect juvenile hormone binding protein shows ancestral fold present in human lipid-binding proteins. J. Mol. Biol. 377, 870–881 [DOI] [PubMed] [Google Scholar]
- 11. Wei D., Li H. M., Tian C. B., Smagghe G., Jia F. X., Jiang H. B., Dou W., and Wang J. J. (2015) Proteome analysis of male accessory gland secretions in oriental fruit flies reveals juvenile hormone-binding protein, suggesting impact on female reproduction. Sci. Rep. 5, 16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alvarenga P. H., Francischetti I. M., Calvo E., Sá-Nunes A., Ribeiro J. M., and Andersen J. F. (2010) The function and three-dimensional structure of a thromboxane A2/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite. PLos Biol. 8, e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calvo E., Mans B. J., Andersen J. F., and Ribeiro J. M. (2006) Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 281, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 14. Calvo E., Mans B. J., Ribeiro J. M., and Andersen J. F. (2009) Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 3728–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 16. Ribeiro J. M., Martin-Martin I., Arcà B., and Calvo E. (2016) a deep insight into the sialome of male and female Aedes aegypti mosquitoes. PLoS ONE 11, e0151400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ribeiro J. M., Arcà B., Lombardo F., Calvo E., Phan V. M., Chandra P. K., and Wikel S. K. (2007) An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sirot L. K., Poulson R. L., McKenna M. C., Girnary H., Wolfner M. F., and Harrington L. C. (2008) Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect. Biochem. Mol. Biol. 38, 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hernández-Martínez S., Rivera-Perez C., Nouzova M., and Noriega F. G. (2015) Coordinated changes in JH biosynthesis and JH hemolymph titers in Aedes aegypti mosquitoes. J. Insect. Physiol. 72, 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanley D., and Kim Y. (2011) Prostaglandins and their receptors in insect biology. Front Endocrinol. (Lausanne) 2, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramirez J. L., de Almeida Oliveira G., Calvo E., Dalli J., Colas R. A., Serhan C. N., Ribeiro J. M., and Barillas-Mury C. (2015) A mosquito lipoxin/lipocalin complex mediates innate immune priming in Anopheles gambiae. Nat. Commun. 6, 7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta R., and Brunak S. (2002) Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 310–322 [PubMed] [Google Scholar]
- 23. Charles J. P., Iwema T., Epa V. C., Takaki K., Rynes J., and Jindra M. (2011) Ligand-binding properties of a juvenile hormone receptor, methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mack S. R., Foley D. A., and Vanderberg J. P. (1979) Hemolymph volume of noninfected and Plasmodium berghei-infected Anopheles stephensi. J. Invertebr. Pathol. 34, 105–109 [DOI] [PubMed] [Google Scholar]
- 25. Amenya D. A., Chou W., Li J., Yan G., Gershon P. D., James A. A., and Marinotti O. (2010) Proteomics reveals novel components of the Anopheles gambiae eggshell. J. Insect. Physiol. 56, 1414–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dottorini T., Nicolaides L., Ranson H., Rogers D. W., Crisanti A., and Catteruccia F. (2007) A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc. Natl. Acad. Sci. U.S.A. 104, 16215–16220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benoit J. B., Vigneron A., Broderick N. A., Wu Y., Sun J. S., Carlson J. R., Aksoy S., and Weiss B. L. (2017) Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. Elife 6, e19535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calvo E., Pham V. M., Marinotti O., Andersen J. F., and Ribeiro J. M. (2009) The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno M., Marinotti O., Krzywinski J., Tadei W. P., James A. A., Achee N. L., and Conn J. E. (2010) Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar. J. 9, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cusson M., Miller D., and Goodman W. G. (1997) Characterization of antibody 444 using chromatographically purified enantiomers of juvenile hormones I, II, and III: implications for radioimmunoassays. Anal. Biochem. 249, 83–87 [DOI] [PubMed] [Google Scholar]
- 31. Rodrigues J., Brayner F. A., Alves L. C., Dixit R., and Barillas-Mury C. (2010) Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for mJHBP were submitted to the PDB with the accession code 5V13.