Abstract
The long noncoding RNA highly up-regulated in liver cancer (HULC) is aberrantly elevated in hepatocellular carcinoma (HCC), and this up-regulation is crucial for HCC pathogenesis. However, the underlying mechanism in HULC up-regulation is poorly understood. We hypothesized that HULC might modulate the oncogene high mobility group A2 (HMGA2) to promote hepatocarcinogenesis. Quantitative real-time PCR analysis showed that the expression levels of HULC were positively correlated with those of HMGA2 in clinical HCC tissues. Interestingly, we also observed that HULC could up-regulate HMGA2 in HCC cells. Mechanistically, we found that the microRNA-186 inhibited HMGA2 expression by targeting the 3′-untranslated region (3′-UTR) of HMGA2 mRNA. Strikingly, HULC acted as a competing noncoding RNA to sequester miR-186 and thereby relieved miR-186–mediated HMGA2 repression. Functionally, HMGA2 knockdown decreased the HULC-enhanced growth of HCC cells both in vitro and in vivo. We conclude that the long noncoding RNA HULC increases HMGA2 expression by sequestering miR-186 post-transcriptionally and thereby promotes liver cancer growth, providing new insights into the mechanism by which HULC enhances hepatocarcinogenesis.
Keywords: cell proliferation; hepatocellular carcinoma; long noncoding RNA (long ncRNA, lncRNA); microRNA (miRNA); oncogene; up-regulated in liver cancer (HULC); high mobility group A2 (HMGA2); microRNA-186
Introduction
Growing evidence demonstrates that many regulatory noncoding RNAs (ncRNAs)3 play crucial roles in multiple biological cellular processes (1, 2). The dysregulation of ncRNAs contributes to the development of numerous human diseases (3, 4). At present, ncRNAs are operationally divided into two groups according to their sizes: small ncRNAs (<200 nt) and long ncRNAs (lncRNAs) (>200 nt). It has been reported that many identified lncRNAs can act as decoys, scaffold, guides, or microRNA sponges to affect tumorigenesis (5). Highly up-regulated in liver cancer (HULC) was identified as the specifically overexpressed long noncoding RNA in hepatocellular carcinoma (HCC) (6). Previously, our group reported that HULC could be up-regulated by hepatitis B virus X protein through transcription factor cAMP-response element-binding protein (7). HULC interacted with the 5′-UTR of CLOCK mRNA via complementary base pairing, resulting in the elevation of CLOCK expression (8). In addition, HULC could reduce the translational repression of PRKACB by sequestering miR-372 and up-regulated the expression of E2F1 by sequestering miR-107 (9, 10). Functionally, the elevated HULC increased abnormal lipid metabolism and promoted angiogenesis, proliferation, migration, and invasion of HCC (11–13). However, the underlying mechanism by which HULC promotes the development of HCC is not well documented.
MicroRNAs (miRNAs) are a class of small ncRNAs that contain ∼19–25 nucleotides. MiRNAs act as post-transcriptional regulators of gene expression through directly binding to the 3′-UTR of the target mRNAs (14). It has been reported that the overexpression of miR-186 can lead to a reduction of Twist family basic helix-loop-helix transcription factor 1 (Twist1) expression, resulting in the elevation of cell apoptosis and the arrest of cell cycle in ovarian cancer (15). Extensive evidence suggests that miR-186 is a novel tumor suppressor gene (16). In addition, miR-186 targets Yes-associated protein 1 (YAP1) to suppress the Hippo signaling pathway and tumorigenesis of HCC (17). However, the roles of miR-186 in HULC-mediated HCC remain unclear.
High-mobility group A protein 2 (HMGA2), as an architectural transcription factor, plays a crucial role in the development of numerous cancers, such as colorectal cancer, lung cancer, and gastric cancer (18–20). HMGA2 can influence the structure of chromatin by binding to AT-rich DNA sequences, leading to transcriptional regulation of genes by enhancing or suppressing transcription factors (21). In terms of functionality, it affects cell cycle, proliferation, epithelial-to-mesenchymal transition, and invasiveness of cancer cells (22, 23). More importantly, the expression of HMGA2 is correlated with metastatic potential (24, 25). HMGA2 expression is primarily increased in different cancers, and it can be regulated by different mechanisms. Up-regulation of HMGA2 often results from genetic alterations, such as gene amplification and translocation, which lengthen the promoter region or shorten the 3′-UTR of HMGA2 (26). It has been reported that HMGA2 is inhibited by let-7 in breast cancer and miR-107 in HCC (22, 27). It also can be increased by the Wnt/β-catenin pathway (28). However, whether HULC is involved in the elevation of HMGA2 in HCC is poorly understood.
In the present study, we are interested in whether lncRNA HULC enhances the development of HCC through HMGA2. Interestingly, our data showed that HULC increases HMGA2 expression by sequestering miR-186 post-transcriptionally and thereby promotes liver cancer growth. Our finding provides new insights into the mechanism of hepatocarcinogenesis mediated by lncRNA HULC.
Results
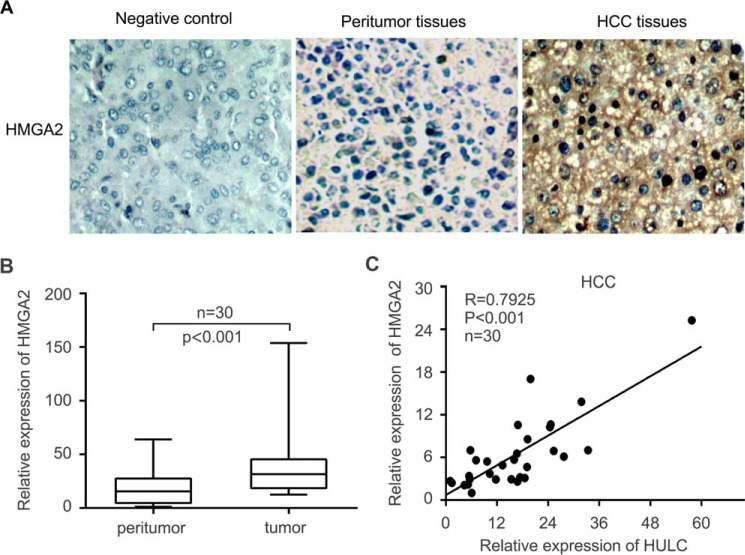
HULC is positively correlated with HMGA2 in clinical HCC tissues and up-regulates HMGA2 in HCC cells
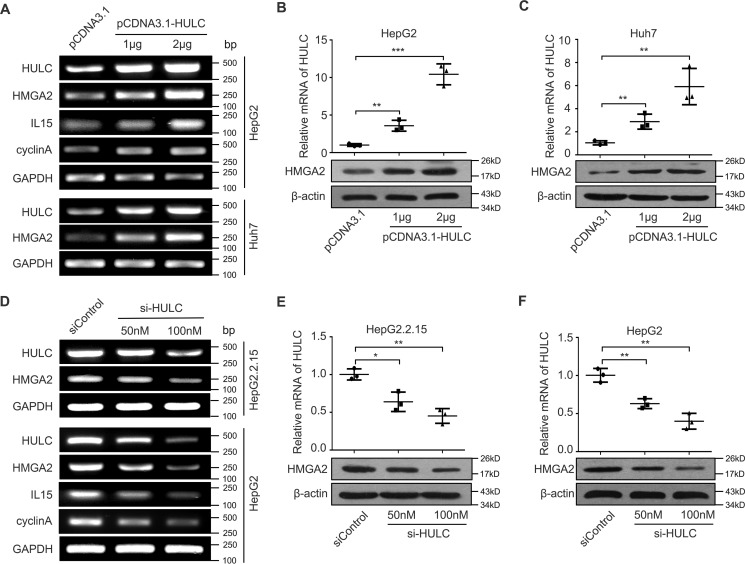
It has been reported that HULC plays important roles in the development of HCC (12). Given that HMGA2 is a crucial oncogene (25, 29), we supposed that HULC might modulate the expression of HMGA2 to promote hepatocarcinogenesis. To demonstrate the significance of HMGA2 in liver cancer, we examined the expression of HMGA2 by immunohistochemistry (IHC) staining using microarrays of clinical HCC tissues. Our data showed that the positive rate of HMGA2 was 71.2% (141 of 198) in HCC tissues (Fig. 1A and supplemental Table S1). Interestingly, the mRNA levels of HMGA2 were significantly increased in 30 clinical HCC tissues relative to their corresponding peritumor tissues (p < 0.001, Wilcoxon's signed-rank test; Fig. 1B). Moreover, quantitative real-time PCR analysis revealed that the expression levels of HULC were positively associated with those of HMGA2 in 30 cases of clinical HCC tissues (r = 0.7925, p < 0.001, Pearson's correlation; Fig. 1C), suggesting that HULC may up-regulate HMGA2 in HCC cells. Next, we evaluated the effect of HULC on HMGA2 in HCC cells. Interestingly, our data indicated that HULC could markedly up-regulate the expression of HMGA2 in HepG2 and Huh7 cells at the levels of mRNA and protein in a dose-dependent manner (Fig. 2, A–C). The transfection efficiency of HULC was validated in these cells. Furthermore, we observed that HULC increased the expression of interleukin-15 (IL15) and cyclin A, two downstream genes of HMGA2, in HepG2 cells in a dose- dependent manner (Fig. 2A), indicating that HULC is able to up-regulate HMGA2. Conversely, inhibition of HULC by siRNA led to the reduction of the expression of HMGA2 in HepG2 and HepG2.2.15 cells (highly expressing endogenous HULC) in a dose-dependent manner at mRNA and protein levels. Meanwhile, the interference efficiency of si-HULC was validated in these cells (Fig. 2, D–F). In addition, si-HULC lowered the expression of IL15 and cyclin A in HepG2 cells in a dose-dependent manner as well (Fig. 2D). Therefore, we conclude that HULC is positively correlated with HMGA2 in clinical HCC tissues and up-regulates HMGA2 in HCC cells.
Figure 1.
HULC is positively correlated with HMGA2 in clinical HCC tissues. A, the expression of HMGA2 was examined by IHC staining in clinical HCC tissues. The negative control means that HMGA2-positive HCC tissue is negative for HMGA2 when the anti-HMGA2 antibody was not incubated. B, the mRNA levels of HMGA2 were evaluated by quantitative real-time PCR analysis in 30 pairs of clinical HCC tissues and their corresponding peritumor tissues (p < 0.001, Wilcoxon's signed-rank test). C, the correlation between HULC and HMGA2 mRNA levels was measured by quantitative real-time PCR analysis in 30 cases of clinical HCC tissues (p < 0.001, r = 0.7925, Pearson's correlation coefficient). Error bars, S.D.
Figure 2.
HULC up-regulates the expression of HMGA2 in HCC cells. A, HepG2 and Huh7 cells were transfected with HULC or vector using Lipofectamine 2000 reagent. RNA was extracted 24 h after transfection. The mRNA levels of HMGA2 and the downstream genes of HMGA2 (such as IL15 and cyclin A) were measured by RT-PCR analysis (n = 3). B and C, HepG2 and Huh7 cells were transfected with HULC or vector using Lipofectamine 2000 reagent. Protein was extracted 48 h after transfection. The protein levels of HMGA2 were examined by Western blot analysis (n = 3). D, HepG2.2.15 and HepG2 cells were transfected with si-HULC or si-control using Lipofectamine 2000 reagent. RNA was extracted 24 h after transfection. The mRNA levels of HMGA2 and the downstream genes of HMGA2 (such as IL15 and cyclin A) were tested by RT-PCR analysis (n = 3). E and F, HepG2.2.15 and HepG2 cells were transfected with si-HULC or control using Lipofectamine 2000 reagent. Protein was extracted 48 h after transfection. The protein levels of HMGA2 was evaluated by Western blot analysis (n = 3). Statistically significant differences are indicated: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***), Student's t test. Error bars, S.D.
miR-186 inhibits the expression of HMGA2 by directly targeting the 3′-UTR of HMGA2 mRNA
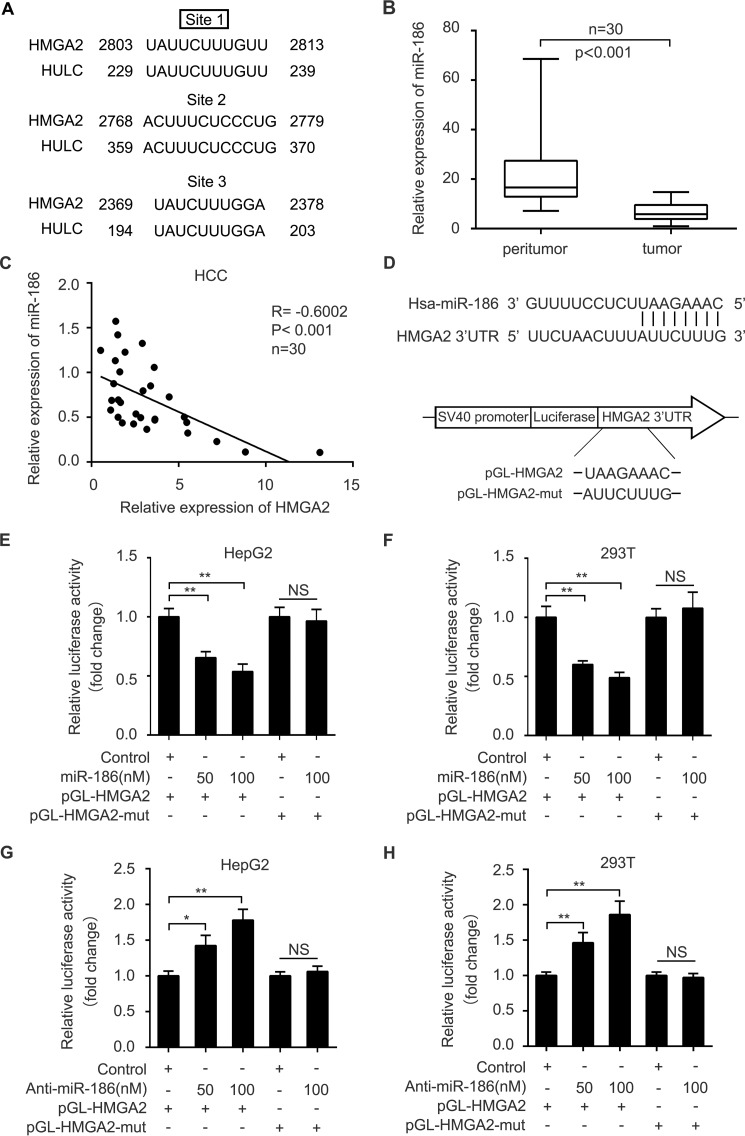
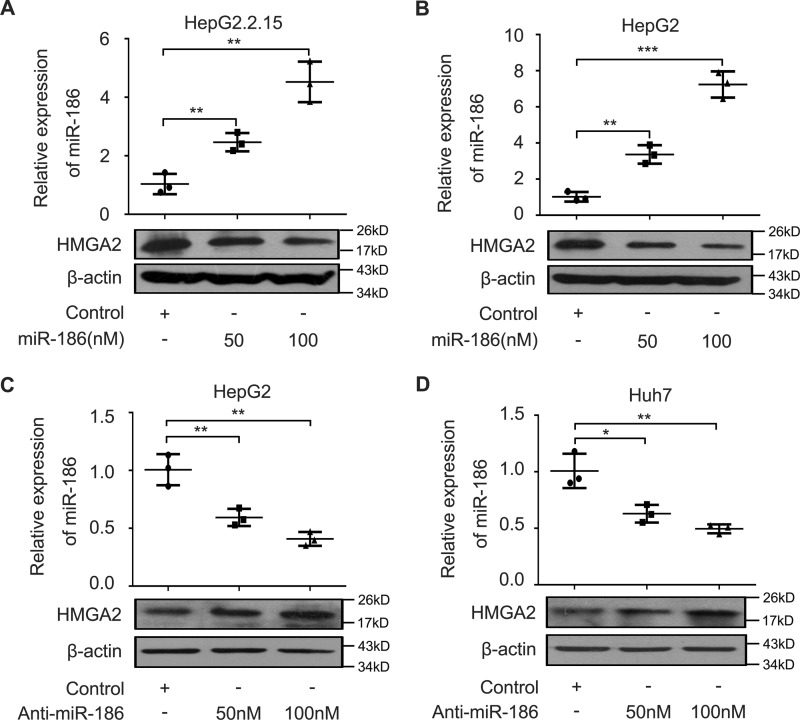
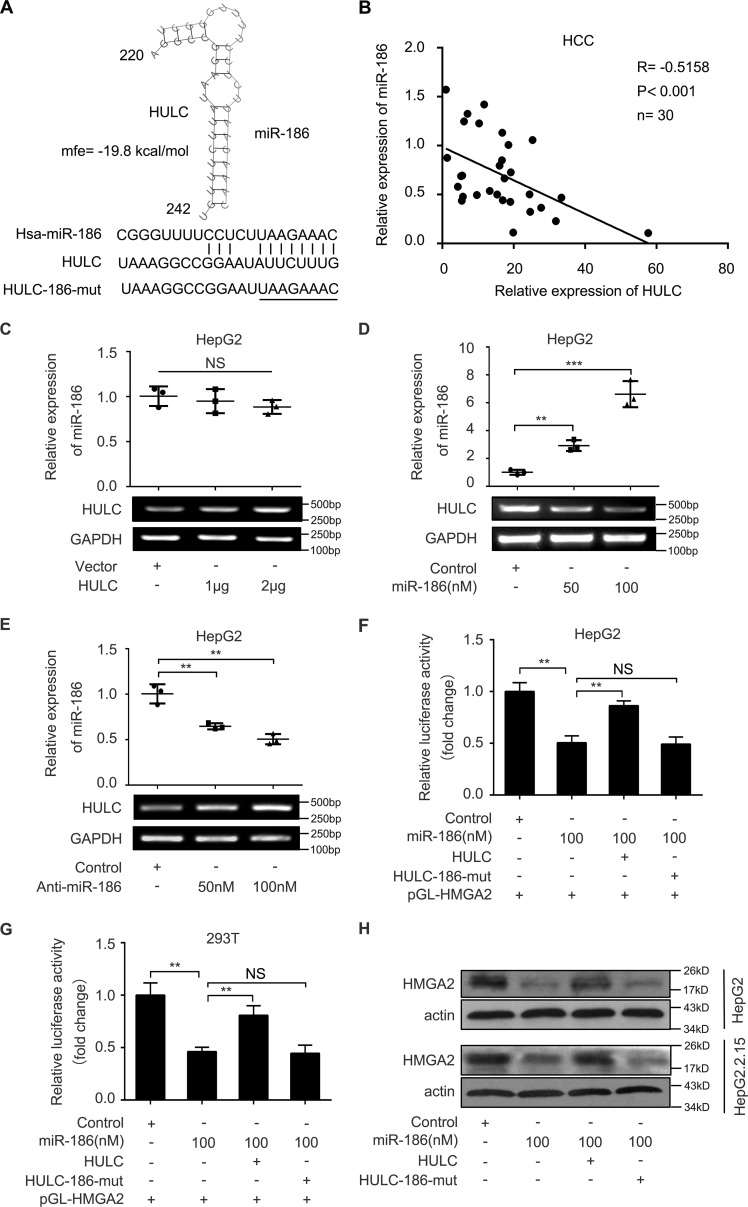
Given that lncRNAs can act as a molecular sponge to sequester miRNAs (30), we speculated that HULC might modulate HMGA2 through miRNAs. We first compared the sequences of HMGA2 mRNA 3′-UTR with those of HULC using the BLAST tool of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi#45219875). Interestingly, bioinformatics analysis identified that three sites of the HULC sequence were the same as those of the HMGA2 mRNA 3′-UTR (Fig. 3A). Furthermore, we identified that miR-186 could potentially bind to site 1 of the HMGA2 mRNA 3′-UTR using miRanda (http://www.microrna.org/microrna/home.do)4 and TargetScan (http://www.targetscan.org/).4 We analyzed the expression levels of miR-186 in 30 paired clinical liver tissues. We found that the expression levels of miR-186 were lower in 30 HCC tissues relative to those in their corresponding peritumor liver tissues by quantitative real-time PCR analysis (Fig. 3B). Then we evaluated the correlation between miR-186 and HMGA2 in clinical HCC tissues. Quantitative real-time PCR analysis demonstrated that the expression levels of HMGA2 were negatively associated with those of miR-186 in 30 clinical HCC tissues (r = −0.6002, p < 0.001, Pearson's correlation; Fig. 3C), suggesting that miR-186 may inhibit HMGA2 in HCC cells. Based on the bioinformatics prediction, we cloned the miR-186 binding site in the 3′-UTR of HMGA2 mRNA and its mutant into luciferase reporter gene vector pGL3-control, termed pGL-HMGA2 and pGL-HMGA2-mut (Fig. 3D). Luciferase reporter gene assays revealed that miR-186 could decrease the luciferase activities of pGL-HMGA2 in HepG2 and 293T cells in a dose-dependent manner, rather than pGL-HMGA2-mut (Fig. 3, E and F). Conversely, the inhibition of miR-186 increased the luciferase activities of pGL-HMGA2 in HepG2 and 293T cells in a dose-dependent manner, but the pGL-HMGA2-mut did not (Fig. 3, G and H), suggesting that miR-186 is able to directly bind to the 3′-UTR of HMGA2 mRNA. Furthermore, Western blot analysis showed that miR-186 was capable of suppressing the expression of HMGA2 in HepG2.2.15 and HepG2 cells in a dose-dependent manner (Fig. 4, A and B), whereas the reverse outcome was obtained when the HepG2 and Huh7 cells were treated with the inhibitor of miR-186 (Fig. 4, C and D). Taken together, we conclude that miR-186 inhibits the expression of HMGA2 by directly targeting the 3′-UTR of HMGA2 mRNA.
Figure 3.
MiR-186 is able to target the 3′UTR of HMGA2 mRNA in HCC cells. A, the same sequences between HULC and the 3′-UTR of HMGA2 mRNA were shown. B, the expression levels of miR-186 were evaluated by quantitative real-time PCR analysis in 30 pairs of clinical HCC tissues and their corresponding peritumor tissues (p < 0.001, Wilcoxon's signed-rank test). C, the correlation between the mRNA levels of HMGA2 and those of miR-186 was examined by quantitative real-time PCR analysis in 30 cases of clinical HCC tissues (p < 0.001, r = −0.6002, Pearson's correlation coefficient). D, a model demonstrates the predicted conserved miR-186 binding site in the 3′-UTR of HMGA2 mRNA. Schematic diagram shows the miR-186 binding site-directed mutant of HMGA2 mRNA 3′-UTR. The HMGA2 mRNA 3′-UTR fragment containing the wild-type or mutant miR-186-binding site was cloned downstream of the luciferase reporter gene in pGL3-control vector. E and F, HepG2 and 293T cells were co-transfected with pGL-HMGA2 and miR-186 or control. The cells were lysed 36 h after transfection. The luciferase activities of pGL-HMGA2 and pGL-HMGA2-mut were assessed by luciferase reporter gene assays (n = 3). G and H, HepG2 and 293T cells were co-transfected with pGL-HMGA2 and miR-186 inhibitor or control. The cells were lysed 36 h after transfection. The luciferase activities of pGL-HMGA2 and pGL-HMGA2-mut were measured by luciferase reporter gene assays (n = 3). Statistically significant differences are indicated: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), no significance (NS), Student's t test. Error bars, S.D.
Figure 4.
MiR-186 inhibits the expression of HMGA2 in HCC cells. A and B, HepG2.2.15 and HepG2 cells were transfected with miR-186 or control using Lipofectamine 2000 reagent. Protein was extracted 48 h after transfection. The protein levels of HMGA2 were detected by Western blot analysis. The transfection efficiency of miR-186 was validated by quantitative real-time PCR analysis (n = 3). C and D, HepG2 and Huh7 cells were transfected with miR-186 inhibitor or control using Lipofectamine 2000 reagent. Protein was extracted 48 h after transfection. The protein levels of HMGA2 were examined by Western blot analysis. The interference efficiency of miR-186 inhibitor was validated by quantitative real-time PCR analysis (n = 3). Statistically significant differences are indicated: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), Student's t test. Error bars, S.D.
HULC is able to increase the expression of HMGA2 by sequestering miR-186
Next, we evaluated the sponge effect of HULC on miR-186 in HCC cells. We predicted the interaction between HULC and miR-186 by using the Bielefeld Bioinformatics Service (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html).4 Bioinformatics analysis showed that HULC could potentially interact with miR-186 through complementary base-pairing reactions (Fig. 5A). Moreover, quantitative real-time PCR analysis demonstrated that the expression levels of HULC were negatively associated with those of miR-186 in 30 clinical HCC tissues (r = −0.5158, p < 0.001, Pearson's correlation; Fig. 5B), suggesting that HULC may interact with miR-186 in HCC cells. We observed that the overexpression of HULC could not inhibit the levels of miR-186 in HepG2 cells (Fig. 5C). However, quantitative real-time PCR analysis revealed that the overexpression of miR-186 could down-regulate the expression of HULC in HepG2 cells, whereas the interference with miR-186 by anti-miR-186 had the opposite effect (Fig. 5, D and E), suggesting that the interaction of miR-186 with HULC lead to the down-regulation of HULC. To confirm whether HULC could enhance the expression of HMGA2 by sequestering miR-186, we constructed the mutant of HULC (HULC-186-mut), which carried a substitution of eight nucleotides within the binding sites of miR-186. Luciferase reporter gene assays showed that HULC could rescue the decrease of pGL-HMGA2 luciferase activities mediated by miR-186 in HepG2 and 293T cells, whereas HULC-186-mut failed to work (Fig. 5, F and G). Furthermore, the similar effect of HULC on the expression of HMGA2 at the protein level was obtained in HepG2 and HepG2.2.15 cells (Fig. 5H). Thus, we conclude that HULC is able to increase the expression of HMGA2 by sequestering miR-186 at the post-transcriptional level.
Figure 5.
HULC is able to increase the expression of HMGA2 by sequestering miR-186. A, bioinformatics prediction of the interaction between HULC and miR-186 through complementary base pairs was shown. The mutant sequence of HULC (residues 220–242) is indicated. B, the correlation between the expression levels of HULC and those of miR-186 was measured by quantitative real-time PCR analysis in 30 cases of clinical HCC tissues (p < 0.001, r = −0.5158, Pearson's correlation coefficient). C, HepG2 cells were transfected with HULC or vector using Lipofectamine 2000 reagent. After 24 h, RNA was extracted and polyadenylated by poly(A) polymerase. The levels of miR-186 were examined by quantitative real-time PCR analysis (n = 3). D and E, HepG2 cells were transfected with miR-186 or miR-186 inhibitor using Lipofectamine 2000 reagent. RNA was extracted 24 h after transfection. The levels of HULC were detected by quantitative real-time PCR analysis. The transfection (or interference) efficiency of miR-186 (or miR-186 inhibitor) was validated by quantitative real-time PCR analysis (n = 3). F and G, HepG2 and 293T cells were co-transfected with pGL-HMGA2 and miR-186 or miR-186 + HULC. The cells were lysed 36 h after transfection. The luciferase activities of pGL-HMGA2 were assessed by luciferase reporter gene assays (n = 3). H, HepG2 and HepG2.2.15 cells were transfected with miR-186 or co-transfected with HULC. Protein was extracted 48 h after transfection. The protein levels of HMGA2 were examined by Western blot analysis (n = 3). Statistically significant differences are indicated: p < 0.01 (**), p < 0.001 (***), no significance (NS), Student's t test. Error bars, S.D.
HULC contributes to the growth of HCC cells in vitro and in vivo through HMGA2
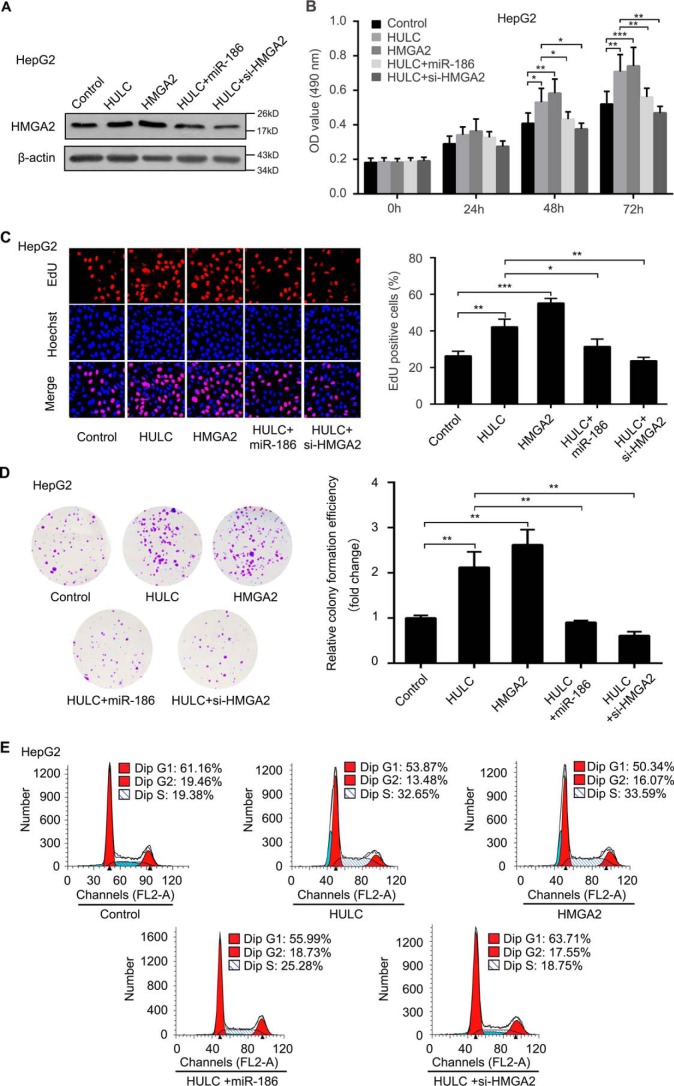
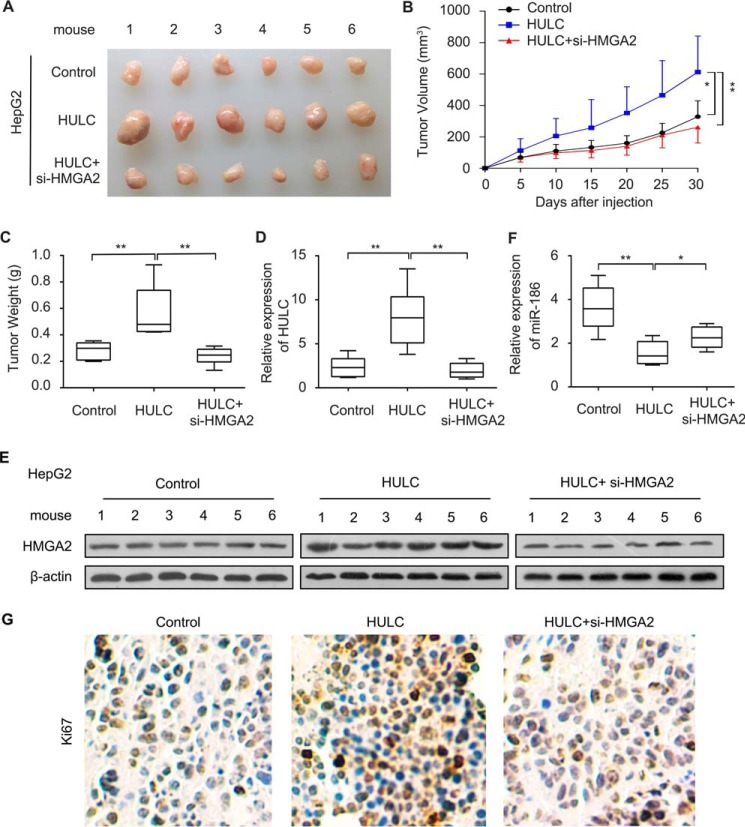
To test whether HMGA2 is involved in the HULC-mediated HCC cells proliferation, we performed 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT), 5-ethynyl-2′-deoxyuridine (EdU), colony formation, and flow cytometry assays in HepG2 cells. The expression of HMGA2 was evaluated by Western blot analysis in the cells after the treatments (Fig. 6A). Our data showed that HULC enhanced the proliferation ability of HCC HepG2 cells, whereas overexpression of miR-186 or inhibition of HMGA2 could suppress the proliferation (Fig. 6, B–E). Based on our data obtained in vitro, we verified our conclusion in vivo using a tumor xenograft model. As expected, we found that overexpression of HULC remarkably increased the volume and weight of the tumors, but the interference of HMGA2 mRNA could block the HULC-enhanced tumor growth in mice (Fig. 7, A–C). Meanwhile, we validated the expression levels of HULC, HMGA2, and miR-186 in the tumor tissues with the pretreatment, respectively. Consistently, the expression levels of HULC were increased in the HULC-overexpressed group, but not for HULC overexpression/si-HMGA2 group (Fig. 7D), which is the outcome of the decreased tumor size and tumorigenesis. The expression levels of HMGA2 were similar to those of HULC (Fig. 7E). The expression levels of miR-186 were opposite those of HULC and HMGA2 (Fig. 7F), which is the outcome of the decreased tumor size and tumorigenesis, suggesting that HULC enhances the growth of hepatoma through HMGA2, in which miR-186 is involved in the event. Accordingly, we examined the expression levels of Ki67, a cell proliferation marker, in the tumor tissues from mice. IHC assays indicated that inhibition of HMGA2 could decrease the expression of Ki67 (Fig. 7G), indicating that HMGA2 is involved in the HULC-mediated growth of HCC cells. Collectively, we conclude that HULC contributes to the growth of HCC cells in vitro and in vivo through HMGA2.
Figure 6.
HULC contributes to the proliferation of HCC cells through HMGA2 in vitro. A, HepG2 cells were transfected with control, HULC, or HMGA2 or co-transfected with HULC and miR-186 or si-HMGA2. Protein was extracted 48 h after transfection. The protein levels of HMGA2 were measured by Western blot analysis (n = 3). B, the proliferation of HepG2 cells transfected with control, HULC, or HMGA2 or co-transfected with HULC and miR-186 or si-HMGA2 was detected by MTT assays (n = 3). C, the proliferation of HepG2 cells transfected with control, HULC, or HMGA2 or co-transfected with HULC and miR-186 or si-HMGA2 was measured by EdU incorporation assays (n = 3). D, the proliferation of HepG2 cells transfected with control, HULC, or HMGA2 or co-transfected with HULC and miR-186 or si-HMGA2 was examined by colony formation assays (n = 3). E, the proliferation of HepG2 cells transfected with control, HULC, or HMGA2 or co-transfected with HULC and miR-186 or si-HMGA2 was assessed by flow cytometry analysis. Error bars, S.D.
Figure 7.
HULC promotes the growth of HCC cells through HMGA2 in vivo. A, dissected tumors from nude mice. B, growth curves of tumors from nude mice. C, average tumor weight of each group. D, the expression levels of HULC were measured by quantitative real-time PCR analysis in tumor tissues from nude mice. E, the expression levels of HMGA2 were examined by Western blot analysis in tumor tissues from nude mice. F, the expression levels of miR-186 were evaluated by quantitative real-time PCR analysis in tumor tissues from nude mice. G, the expression of Ki67 in tumor tissues from nude mice was detected by IHC assays. Error bars, S.D.
Discussion
lncRNAs play pivotal roles in a host of pathologic processes, especially in tumorigenesis (31–33). Our group has reported that lncRNA HULC contributes to the development of HCC by modulating HCC angiogenesis and abnormal lipid metabolism (10, 11). As an oncogene, HMGA2 is highly expressed in many tumor tissues (34, 35). However, the effect of HULC on HMGA2 in hepatocarcinogenesis is not well known. In this study, we are interested in whether HULC is involved in the regulation of HMGA2 in HCC.
To examine the effect of HULC on HMGA2 in the development of HCC, we first evaluated the correlation between expression levels of HULC and HMGA2 in clinical HCC tissues. Our data revealed that the mRNA levels of HMGA2 were much higher in the HCC samples than their corresponding peritumor liver tissues. In addition, the expression levels of HULC were positively associated with those of HMGA2 in clinical HCC samples. This suggests that HULC may increase HMGA2 expression in HCC. As expected, we observed that HULC was able to up-regulate the expression of HMGA2 at mRNA and protein levels in HCC cells. Therefore, this suggests that HULC is involved in the regulation of HMGA2 in liver cancer. Next, we tried to identify the underlying mechanism by which HULC up-regulates HMGA2. Given that lncRNAs can act as an miRNA sponge to promote the development of cancers (30, 36), we presumed that HULC might contribute to the elevation of HMGA2 by sequestering miRNAs. If the miRNA can bind to HMGA2 mRNA and it simultaneously interacts with HULC, then HMGA2 mRNA and HULC will have partially the same sequences. Based on this hypothesis, we compared the sequences of HULC and those of HMGA2 mRNA 3′-UTR. Interestingly, bioinformatics analysis showed that three sites in HULC RNA were completely the same as those in the HMGA2 mRNA 3′-UTR. Furthermore, we predicted that miR-186 could potentially bind with one of those sites, suggesting that miR-186 may be involved in the regulation of HMGA2 mediated by HULC. Increasing evidence has highlighted the significance and molecular mechanisms of miR-186 in many cancers (37). It has been reported that miR-186 suppresses the migration and invasion of human non-small cell lung cancer cells by targeting pituitary tumor transforming gene (PTTG1) mRNA 3′-UTR (38). Overexpression of miR-186 also can decrease the proliferation, invasion, and migration of human gastric cancer through the inhibition of Twist1 expression (39). In our study, we found that the expression levels of miR-186 were negatively associated with those of HMGA2 in clinical HCC tissues, which is consistent with the suppressive function of miR-186 in other cancers. In addition, miR-186 was capable of directly binding to the 3′-UTR of HMGA2 mRNA in HCC cells, resulting in the inhibition of HMGA2 expression. It suggests that miR-186 is able to suppress the expression of HMGA2 in HCC cells. Next, we examined whether HULC could block the decrease of HMGA2 mediated by miR-186. We observed that HULC rescued the inhibition of HMGA2 mediated by miR-186. This suggests that HULC can elevate the expression of HMGA2 through the sponge of miR-186 at the post-transcriptional level. It has been reported that miR-186 can down-regulate some important oncogenes, such as YAP, Twist1, PTTG1, and HIF-1α (15, 17, 38, 40). This suggests that HULC may widely modulate oncogenes through sequestering miR-186 to promote hepatocarcinogenesis. Our group has reported that HULC can sequester miR-107 through a different site from miR-186, leading to an increase in the expression of E2F1. In addition, we have also reported that miR-107 inhibits the expression of HMGA2 in HCC cells (10, 22), implying that HULC may also up-regulate HMGA2 expression by sequestering miR-107. However, our data failed to show that HULC could increase HMGA2 expression through the sponge of miR-107 (data not shown). Functionally, we found that HULC was able to facilitate the growth of HCC cells in vitro and in vivo through HMGA2, in which miR-186 is involved in the event. Therapeutically, HULC and HMGA2 may serve as targets in liver cancer.
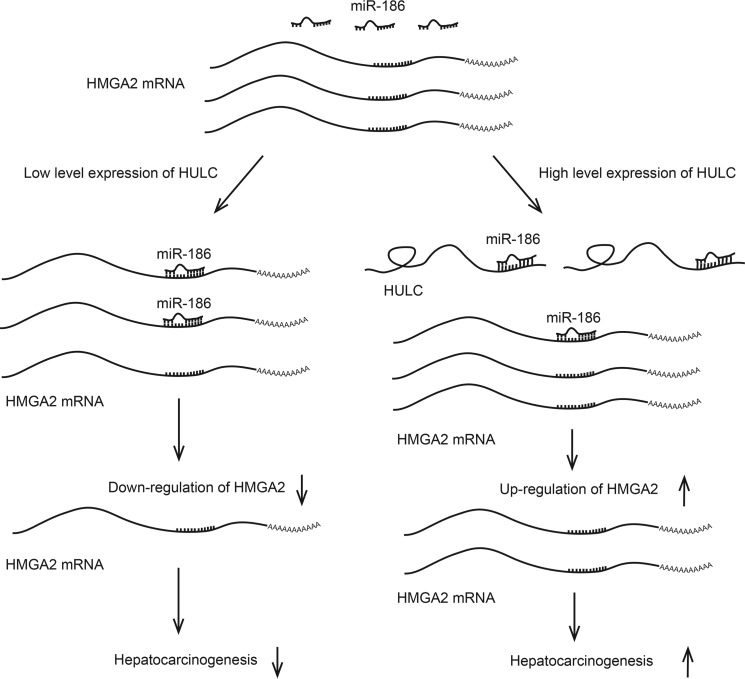
Taken together, we illustrate a novel mechanism by which HULC modulates HMGA2 in hepatocarcinogenesis in this study (Fig. 8). Our findings indicate that HULC is able to up-regulate HMGA2 in HCC cells. Mechanistically, miR-186 is capable of decreasing the expression of HMGA2 through targeting 3′-UTR of HMGA2 mRNA. Strikingly, HULC can sponge miR-186, leading to the increase of HMGA2 expression in hepatocarcinogenesis. Thus, we conclude that HULC increases HMGA2 expression by sequestering miR-186 post-transcriptionally and thereby promotes liver cancer growth. Our finding provides new insights into the mechanism of hepatocarcinogenesis mediated by HULC.
Figure 8.
A model shows that HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the miR-186. miR-186 is capable of decreasing the expression of HMGA2 through targeting the 3′-UTR of HMGA2 mRNA; HULC can sponge miR-186, leading to the increase of HMGA2 expression and resulting in the promotion of hepatocarcinogenesis.
Experimental procedures
Patient samples
HCC tissue samples and their corresponding peritumor liver tissues (n = 30) were obtained during surgery at Tianjin First Center Hospital and Tianjin Tumor Hospital (Tianjin, China). The detailed information on the patients with HCC, obtained from patient records, is presented in supplemental Table S2. Informed consent approving the use of tissue samples for study purposes was obtained from each patient. The study was approved by the Institute Research Ethics Committee at Nankai University and was performed in accordance with relevant guidelines and regulations.
Cell culture
HCC cell lines HepG2, Huh7, and HepG2.2.15 (highly expressing endogenous HULC) and human embryonic kidney cell line 293T were maintained in DMEM (Gibco). All cell lines were supplemented with heat-inactivated 10% FBS (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37 °C.
Plasmid construction
The full-length HULC and HMGA2 were amplified from the cDNA of HepG2 cells and then cloned into pcDNA3.1 vector. A mutant of HULC with a substitution of 8 nucleotides of the miR-186 binding site was constructed into pcDNA3.1, termed HULC-186-mut. Primers for plasmid construction were as follows: HMGA2 CDS, 5′-GGGGTACCGGAGGCAGGATGAGCGCA-3′ (forward primer) and 5′-CCCTCGAGCTAGTCCTCTTCGGCAGACTC-3′ (reverse primer); HULC, 5′-GGGGTACCATGGGGGTGGAACTCATGATGG-3′ (forward primer) and 5′-CCCTCGAGAAGAATGGACATCATTTTATTTCA-3′ (reverse primer); HULC-186-mut, 5′-TGAAGTAAAGGCCGGAATTAAGAAACTTTAAAACATTAAAAACA-3′ (forward primer) and 5′-TGTTTTTAATGTTTTAAAGTTTCTTAATTCCGGCCTTTACTTCA-3′ (reverse primer). The fragment (from 2564 to 3038 bp) containing the miR-186 target site in the 3′-UTR of HMGA2 mRNA was cloned into pGL3-control vector to generate pGL-HMGA2. A mutant of the HMGA2 3′-UTR carrying a substitution of 8 nucleotides within the core seed sequence of miR-186 was constructed, termed pGL-HMGA2-mut. Primers for luciferase reporter gene assays were as follows: pGL-HMGA2, 5′-TGCTCTAGATGACTATGTATTTTTCTATGTT-3′ (forward primer) and 5′-GGGGGCCGGCCCTTGTTGCATCTCACCTA-3′ (reverse primer); pGL-HMGA2-mut, 5′-CAAGTCTGTACCTCAAATCTTAAGAAAAAGGAGATGGACTAATTGAC-3′ (forward primer) and 5′-GTCAATTAGTCCATCTCCTTTTTCTTAAGATTTGAGGTACAGACTTG-3′ (reverse primer). All of the constructs were verified by sequence analysis.
Cell transfection
Cells were cultured in 6-well or 24-well plates for 12 h and then were transfected with plasmids or siRNAs. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The siRNAs of HULC and HMGA2 were used according to the reports (10, 41). The siRNA duplexes targeting HULC and HMGA2, miR-186 mimics, miR-186 inhibitor, and their respective negative controls were synthesized and purified by RiboBio (Guangzhou, China). All siRNA sequences used were as follows: si-control, 5′-AAUGGUCAUGGUCUUAUUCC-3′; si-HULC, 5′-CCUCCAGAACUGUGAUCCA-3′; si-HMGA2, 5′-AGAGGCAGACCUAGGAAAU-3′; mimics NC, 5′-UUCUUCGAAGGUGUGACGU-3′; hsa-miR-186, 5′-CAAAGAAUUCUCCUUUUGGGCU-3′; hsa-miR-186 inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′; hsa-miR-186 inhibitor, 5′-GUUUCUUAAGAGGAAAACCCGA-3′.
RNA extraction, RT-PCR, and quantitative real-time PCR
24 h after transfection, total RNA was extracted from cells (or tumor tissues from mice and patient tissues) using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using the Transcriptor first strand cDNA synthesis kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. For the detection of miR-186 expression, total RNA was polyadenylated by poly(A) polymerase (Ambion, Austin, TX) as described previously (42). Quantitative real-time PCR was applied to detect the expression of HULC, HMGA2, and miR-186 using double-stranded DNA-specific Fast Start Universal SYBR Green Master (Roche Applied Science) following the manufacturer's instructions. Double-stranded DNA specific expression was tested by the comparative Ct method using 2−ΔΔCt (43). All primers used were as follows: GAPDH, 5′-AACGGATTTGGTCGTATTG-3′ (forward primer) and 5′-GGAAGATGGTGATGGGATT-3′ (reverse primer); HMGA2, 5′-TGGGAGGAGCGAAATCTAA-3′ (forward primer) and 5′-GGTGAACTCAAGCCGAAG-3′ (reverse primer); HULC, 5′-ATGGGGGTGGAACTCATGATGG-3′ (forward primer) and 5′-AAGAATGGACATCATTTATTTCA-3′ (reverse primer); IL15, 5′-CAAAGAATGTGAGGAACTGGAG-3′ (forward primer) and 5′-TGTCTAAGCAGCAGAGTGATG-3′ (reverse primer); cyclin A, 5′-GCCATTAGTTTACCTGGACCCAGA-3′ (forward primer) and 5′-CACTGACATGGAAGACAGGAACCT-3′ (reverse primer); U6, 5′-AGAGCCTGTGGTGTCCG-3′ (forward primer) and 5′-CATCTTCAAAGCACTTCCCT-3′ (reverse primer); miR-186, 5′-CAAAGAATTCTCCTTTTGGGCT-3′ (forward primer) and 5′-GCGAGCACAGAATTAATACGAC-3′ (reverse primer).
Western blot analysis
HepG2 (or Huh7) cells were seeded into a 6-well plate and incubated in a humidified 5% CO2 atmosphere at 37 °C in antibiotic-free DMEM supplemented with 10% FBS for 12 h to about 50% confluence. Then cells were transfected with pcDNA3.1-HULC, pcDNA3.1-HMGA2, miR-186, or their corresponding controls using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. 48 h after transfection, Western blot analysis was carried out as described previously (44). Primary antibodies used were rabbit anti-HMGA2 (Genetex, Irvine, CA), mouse anti-β-actin (Sigma). All experiments were repeated three times.
Luciferase reporter gene assays
Cells were transferred into 24-well plates at 3 × 104 cells/well. After 12 h, the cells were co-transfected with the pRL-TK plasmid containing the Renilla luciferase gene (Promega, Madison, WI) used for internal normalization and various constructs containing HMGA2 3′-UTR, pCDNA3.1-HULC, or their corresponding mutants. 36 h after transfection, the luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's protocol. All experiments were performed at least three times.
Analysis of cell proliferation
HepG2 or Huh7 cells were seeded into 96-well plate at 1000 cells/well for 12 h before transfection. The MTT assay was performed as described previously (44). Briefly, cells were cultured after different days (day 0, 1, 2, and 3), and 15 μl of MTT was added to each well, followed by incubation for 4 h. The supernatant was discarded, and 100 μl of dimethyl sulfoxide was added to stop the reaction. Absorbance at 490 nm was assessed using an ELISA reader system (Multiskan Ascent, LabSystems). The EdU incorporation assay was applied by using the Cell-LightTM EdU imaging detecting kit following the manufacturer's instructions (RiboBio). For the colony formation assay, 12 h after transfection, 500 transfected cells were placed into a 6-well plate and maintained in complete medium for 2 weeks. Colonies were fixed with methanol and stained with crystal violet (45). Flow cytometry analysis was processed as described earlier (46).
IHC staining assays
The microarrays including HCC tissues were obtained from Xi'an Aomei Biotechnology Co., Ltd. (Xi'an, China). These microarrays were composed of 198 HCC tissues, which included duplicate core biopsies (1 mm in diameter) from fixed, paraffin-embedded tumors. The medical records of the patients are listed in supplemental Table S1. The IHC staining assay was performed as described previously (47), and the primary antibodies of rabbit anti-HMGA2 (Genetex) and rabbit anti-Ki67 (Thermo Fisher Scientific) were used. The results of IHC were evaluated as described earlier (48). In brief, the staining level of HMGA2 was classified into three groups using a modified scoring method based on the intensity of staining (0, negative; 1, low; 2, high) and the percentage of stained cells (0, 0% stained; 1, 1–49% stained; 2, 50–100% stained). A multiplied score (intensity score × percentage score) of 0 was considered to be negative staining (−), 1 was considered to be low staining (+), 2 was considered to be moderate staining (++), and 4 was considered to be high staining (+++). Categorization of immunostaining intensity was performed by three independent observers.
In vivo tumorigenicity assays
Nude mice were housed and treated in accordance with the guidelines established by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We conducted animal transplantation according to the Declaration of Helsinki principles. All experiments were approved by the Institute Research Ethics Committee at Nankai University. Tumor transplantation in nude mice was performed as described previously (49–51). Briefly, HepG2 cells were pretreated with control, pCDNA3.1-HULC, pCDNA-3.1-HULC, and si-HMGA2 for 24 h. Then the cells were harvested and resuspended at 2 × 107 cells/ml with sterile PBS. Groups of 4-week-old male BALB/c athymic nude mice (Experimental Animal Center of Peking, China; each group, n = 6) were subcutaneously injected at the shoulder with 0.2 ml of the cell suspensions. Tumor growth was measured after 5 days from injection and then every 5 days. Tumor volume was monitored by measuring the length (L) and width (W) with calipers and calculated with the formula, (L × W2)/2. After 30 days, the mice were sacrificed, and the tumors were excised and measured.
Statistical analysis
Each experiment was repeated at least three times. Statistical significance was assessed by comparing mean values ± S.D. using Student's t test for independent groups and was assumed for p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and not significant (NS). Association between expression levels of HULC and HMGA2 in tumor tissues was assessed using Pearson's correlation coefficient. HMGA2 expression in tumor tissues and their corresponding peritumor tissues was compared using the Wilcoxon signed-rank test.
Author contributions
X. Z. and L. Y. conceived the project, designed the experiments, and drafted the manuscript. X. Z. and Y. W. designed the experiments, drafted the manuscript, and performed the experiments. F. C., M. Z., Z. Y., J. L., S. Z., and W. Z. performed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Basic Research Program of China 973 Program Grants 2015CB553703 and 2015CB553905; National Natural Science Foundation of China Grants 81272218, 81372186, 31670769, and 31670771; Project of Prevention and Treatment of Key Infectious Diseases Grant 2014ZX0002002-005; and Project of Prevention and Control of Key Chronic Non-infectious Diseases Grant 2016YFC130340. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ncRNA
- noncoding RNA
- lncRNA
- long ncRNA
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide
- EdU
- 5-ethynyl-2′-deoxyuridine
- HULC
- highly up-regulated in liver cancer
- HCC
- hepatocellular carcinoma
- IHC
- immunohistochemistry.
References
- 1. Ponting C. P., Oliver P. L., and Reik W. (2009) Evolution and functions of long noncoding RNAs. Cell 136, 629–641 [DOI] [PubMed] [Google Scholar]
- 2. Khanduja J. S., Calvo I. A., Joh R. I., Hill I. T., and Motamedi M. (2016) Nuclear noncoding RNAs and genome stability. Mol. Cell 63, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wapinski O., and Chang H. Y. (2011) Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361 [DOI] [PubMed] [Google Scholar]
- 4. Schmitt A. M., and Chang H. Y. (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rinn J. L., and Chang H. Y. (2012) Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panzitt K., Tschernatsch M. M., Guelly C., Moustafa T., Stradner M., Strohmaier H. M., Buck C. R., Denk H., Schroeder R., Trauner M., and Zatloukal K. (2007) Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132, 330–342 [DOI] [PubMed] [Google Scholar]
- 7. Du Y., Kong G., You X., Zhang S., Zhang T., Gao Y., Ye L., and Zhang X. (2012) Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J. Biol. Chem. 287, 26302–26311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui M., Zheng M., Sun B., Wang Y., Ye L., and Zhang X. (2015) A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia 17, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., and Fan Q. (2010) CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38, 5366–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Z., Xiao Z., Liu F., Cui M., Li W., Yang Z., Li J., Ye L., and Zhang X. (2016) Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget 7, 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui M., Xiao Z., Wang Y., Zheng M., Song T., Cai X., Sun B., Ye L., and Zhang X. (2015) Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857 [DOI] [PubMed] [Google Scholar]
- 12. Yu X., Zheng H., Chan M. T., and Wu W. K. (2017) HULC: an oncogenic long non-coding RNA in human cancer. J. Cell Mol. Med. 21, 410–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li C., Chen J., Zhang K., Feng B., Wang R., and Chen L. (2015) Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol. Biochem. 36, 423–434 [DOI] [PubMed] [Google Scholar]
- 14. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu X., Shen H., Yin X., Long L., Xie C., Liu Y., Hui L., Lin X., Fang Y., Cao Y., Xu Y., Li M., Xu W., and Li Y. (2016) miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene 35, 323–332 [DOI] [PubMed] [Google Scholar]
- 16. Liu L., Wang Y., Bai R., Yang K., and Tian Z. (2016) MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis 5, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ruan T., He X., Yu J., and Hang Z. (2016) MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol. Lett. 11, 2941–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morishita A., Zaidi M. R., Mitoro A., Sankarasharma D., Szabolcs M., Okada Y., D'Armiento J., and Chada K. (2013) HMGA2 is a driver of tumor metastasis. Cancer Res. 73, 4289–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fusco A., and Fedele M. (2007) Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 [DOI] [PubMed] [Google Scholar]
- 20. Wu J., Wang Y., Xu X., Cao H., Sahengbieke S., Sheng H., Huang Q., and Lai M. (2016) Transcriptional activation of FN1 and IL11 by HMGA2 promotes the malignant behavior of colorectal cancer. Carcinogenesis 37, 511–521 [DOI] [PubMed] [Google Scholar]
- 21. Young A. R., and Narita M. (2007) Oncogenic HMGA2: short or small? Genes Dev. 21, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Chen F., Zhao M., Yang Z., Zhang S., Ye L., Gao H., and Zhang X. (2016) MiR-107 suppresses proliferation of hepatoma cells through targeting HMGA2 mRNA 3′UTR. Biochem. Biophys. Res. Commun. 480, 455–460 [DOI] [PubMed] [Google Scholar]
- 23. Wu J., Liu Z., Shao C., Gong Y., Hernando E., Lee P., Narita M., Muller W., Liu J., and Wei J. J. (2011) HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 71, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Liu X., Li A. Y., Chen L., Lai L., Lin H. H., Hu S., Yao L., Peng J., Loera S., Xue L., Zhou B., Zhou L., Zheng S., Chu P., Zhang S., Ann D. K., and Yen Y. (2011) Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 17, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu J., Zhang S., Shan J., Hu Z., Liu X., Chen L., Ren X., Yao L., Sheng H., Li L., Ann D., Yen Y., Wang J., and Wang X. (2016) Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett. 376, 284–292 [DOI] [PubMed] [Google Scholar]
- 26. Wu J., and Wei J. J. (2013) HMGA2 and high-grade serous ovarian carcinoma. J. Mol. Med. 91, 1155–1165 [DOI] [PubMed] [Google Scholar]
- 27. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., and Song E. (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 28. Wend P., Runke S., Wend K., Anchondo B., Yesayan M., Jardon M., Hardie N., Loddenkemper C., Ulasov I., Lesniak M. S., Wolsky R., Bentolila L. A., Grant S. G., Elashoff D., Lehr S., et al. (2013) WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol. Med. 5, 264–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaur H., Ali S. Z., Huey L., Hütt-Cabezas M., Taylor I., Mao X.-G. Weingart M., Chu Q., Rodriguez F. J., Eberhart C. G., and Raabe E. H. (2016) The transcriptional modulator HMGA2 promotes stemness and tumorigenicity in glioblastoma. Cancer Letters 377, 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parasramka M. A., Maji S., Matsuda A., Yan I. K., and Patel T. (2016) Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 161, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prensner J. R., and Chinnaiyan A. M. (2011) The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi X., Sun M., Liu H., Yao Y., and Song Y. (2013) Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 339, 159–166 [DOI] [PubMed] [Google Scholar]
- 33. Yang X., Xie X., Xiao Y. F., Xie R., Hu C. J., Tang B., Li B. S., and Yang S. M. (2015) The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 360, 119–124 [DOI] [PubMed] [Google Scholar]
- 34. Agostini A., Brunetti M., Davidson B., Trope C. G., Heim S., Panagopoulos I., and Micci F. (2016) Expressions of miR-30c and let-7a are inversely correlated with HMGA2 expression in squamous cell carcinoma of the vulva. Oncotarget 7, 85058–85062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palumbo A. Jr., Da Costa N. M., Esposito F., De Martino M., D'Angelo D., de Sousa V. P., Martins I., Nasciutti L. E., Fusco A., and Ribeiro Pinto L. F. (2016) HMGA2 overexpression plays a critical role in the progression of esophageal squamous carcinoma. Oncotarget 7, 25872–25884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan J. H., Yang F., Wang F., Ma J. Z., Guo Y. J., Tao Q. F., Liu F., Pan W., Wang T. T., Zhou C. C., Wang S. B., Wang Y. Z., Yang Y., Yang N., Zhou W. P., Yang G. S., and Sun S. H. (2014) A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 [DOI] [PubMed] [Google Scholar]
- 37. Hua X., Xiao Y., Pan W., Li M., Huang X., Liao Z., Xian Q., and Yu L. (2016) miR-186 inhibits cell proliferation of prostate cancer by targeting GOLPH3. Am. J. Cancer Res. 6, 1650–1660 [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Yin C., Zhang B., Sun Y., Shi L., Liu N., Liang S., Lu S., Liu Y., Zhang J., Li F., Li W., Liu F., Sun L., and Qi Y. (2013) PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis 34, 2145–2155 [DOI] [PubMed] [Google Scholar]
- 39. Cao C., Sun D., Zhang L., and Song L. (2016) miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget 7, 79956–79963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L., Wang Y., Bai R., Yang K., and Tian Z. (2016) MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis 5, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao X. P., Zhang H., Jiao J. Y., Tang D. X., Wu Y. L., and Pan C. B. (2016) Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J. Transl. Med. 14, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shan C., Zhang S., Cui W., You X., Kong G., Du Y., Qiu L., Ye L., and Zhang X. (2011) Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis 32, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 43. Schmittgen T. D., and Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 44. Shan C., Xu F., Zhang S., You J., You X., Qiu L., Zheng J., Ye L., and Zhang X. (2010) Hepatitis B virus X protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-ERK1/2. Cell Res. 20, 563–575 [DOI] [PubMed] [Google Scholar]
- 45. Zhang T., Zhang J., You X., Liu Q., Du Y., Gao Y., Shan C., Kong G., Wang Y., Yang X., Ye L., and Zhang X. (2012) Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology 56, 2051–2059 [DOI] [PubMed] [Google Scholar]
- 46. Zhang W., Kong G., Zhang J., Wang T., Ye L., and Zhang X. (2012) MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One 7, e31450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. You X., Liu F., Zhang T., Lv N., Liu Q., Shan C., Du Y., Kong G., Wang T., Ye L., and Zhang X. (2014) Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene 33, 449–460 [DOI] [PubMed] [Google Scholar]
- 48. Shi H., Fang R., Li Y., Li L., Zhang W., Wang H., Chen F., Zhang S., Zhang X., and Ye L. (2016) The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 382, 147–156 [DOI] [PubMed] [Google Scholar]
- 49. You X., Liu F., Zhang T., Li Y., Ye L., and Zhang X. (2013) Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis 34, 1644–1652 [DOI] [PubMed] [Google Scholar]
- 50. Liu Y. W., Xia R., Lu K., Xie M., Yang F., Sun M., De W., Wang C., and Ji G. (2017) LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-mediated H3K4me2 demethylation. Mol. Cancer 16, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu T., Zhu Y., Xiong Y., Ge Y. Y., Yun J. P., and Zhuang S. M. (2009) MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50, 113–121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.