Abstract
The intestinal tract contains many commensal bacteria that modulate various physiological host functions. Dysbiosis of commensal bacteria triggers dysfunction of the intestinal epithelial barrier, leading to the induction or aggravation of intestinal inflammation. To elucidate whether microRNA plays a role in commensal microbiome-dependent intestinal epithelial barrier regulation, we compared transcripts in intestinal epithelial cells (IECs) from conventional and germ-free mice and found that commensal bacteria induced the expression of miR-21-5p in IECs. miR-21-5p increased intestinal epithelial permeability and up-regulated ADP ribosylation factor 4 (ARF4), a small GTPase, in the IEC line Caco-2. We also found that ARF4 expression was up-regulated upon suppression of phosphatase and tensin homolog (PTEN) and programmed cell death 4 (PDCD4), which are known miR-21-5p targets, by RNAi. Furthermore, ARF4 expression in epithelial cells of the large intestine was higher in conventional mice than in germ-free mice. ARF4 suppression in the IEC line increased the expression of tight junction proteins and decreased intestinal epithelial permeability. These results indicate that commensal microbiome-dependent miR-21-5p expression in IECs regulates intestinal epithelial permeability via ARF4, which may therefore represent a target for preventing or managing dysfunction of the intestinal epithelial barrier.
Keywords: inflammation, intestinal epithelium, microRNA (miRNA), permeability, tight junction, commensal microbiota
Introduction
Large numbers of commensal bacteria inhabit the intestinal tract and modulate various host physiological functions, as well as help digestion and the absorption of dietary components (1). Dysbiosis of the intestinal microbiota is involved in diverse diseases, such as inflammatory bowel disease (IBD)2 and food allergies (2). These diseases are accompanied by excessive immune activation and disruption of mucosal barrier function, leading to the induction or aggravation of inflammation.
IECs, which play an important role in the absorption of nutrients, separate the intestinal lumen from the intestinal contents. IECs act as a physical barrier to prevent invasion of the intestinal contents, including pathogenic microorganisms. Intestinal epithelial permeability is controlled by tight junctions, which consist of firm contacts between neighboring IECs through cell surface proteins.
Disruption of the epithelial barrier enhances inflammatory responses via activation of the mucosal immune system, increasing the risk of IBD and autoimmune diseases (3). For instance, it has been suggested that the degradation of tight junction proteins on IECs by proteases from commensal bacteria is associated with Crohn's disease (4, 5). Thus, it is required to clarify the mechanism by which commensal bacteria affect and regulate the function of host IECs.
MicroRNAs (miRNAs) are small non-coding RNAs that are first synthesized from non-coding sequences of genomic DNA by RNA polymerase II or III as pri-miRNA. They are then processed by the ribonuclease Drosha into 70-nt-long pre-miRNAs and transported to the cytoplasm. The enzyme Dicer cleaves the pre-miRNAs to produce miRNA/miRNA duplexes. Then, the mature miRNAs are incorporated into RNA-induced silencing complexes that bind to complementary seed sequences in the 3′ untranslated regions of target mRNAs and either facilitate direct degradation of the mRNAs or inhibit their translation into proteins. Posttranscriptional suppression of target genes by RNA interference is involved in various biological processes and diseases. Previous reports have shown that miRNAs modulate the function of IECs. Studies using mice with IEC-specific deletion of Dicer 1 revealed that miRNAs in IECs play a critical role in the induction of the antiparasitic Th2 response (6). Although commensal bacteria have been reported to affect miRNA expression (7, 8), the molecular mechanisms underlying the regulation of IEC function by commensal bacteria through miRNAs have not been fully elucidated. In this study, we evaluated how commensal bacteria affect miRNA expression in IECs and regulate epithelial permeability, which may provide a novel target for the prevention and treatment of diseases related to epithelial barrier dysfunction.
Results
Commensal bacteria regulate miRNA expression in IECs
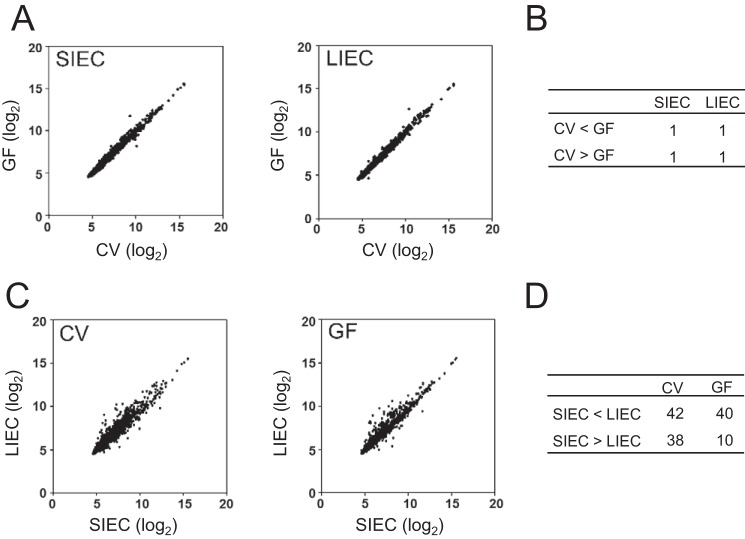
First, we analyzed whether commensal bacteria affect miRNA expression in IECs. We comprehensively analyzed the expression patterns of miRNAs in small and large intestinal epithelial cells (SIECs and LIECs, respectively) from CV and GF mice by microarray. We found differentially expressed miRNAs in both the SIECs and the LIECs of CV versus GF mice (Fig. 1A). The number of miRNAs with a more than 2-fold change in expression in CV versus GF mice is shown in Fig. 1B. These results indicate that commensal bacteria modulate miRNA expression in IECs.
Figure 1.
Commensal bacteria regulate miRNA expression in IECs. A–D, total RNA, including low-molecular-weight RNA, was prepared from the SIECs and LIECs of CV and GF mice. The expression of miRNA was analyzed by microarray testing. The plots compare the expression of miRNAs between CV and GF mice (A) or between SIECs and LIECs (C). The numbers of miRNAs with a more than 2-fold difference in expression are shown in B and D.
In addition, miRNA expression differed in the SIECs versus LIECs of all mice (Fig. 1C). The number of miRNAs with a more than 2-fold change in expression between SIECs and LIECs of the same mice was larger than that between CV and GF mice, indicating that miRNA expression was more dramatically affected by anatomical location than by the presence of commensal bacteria (Fig. 1D).
Commensal bacteria induce miR-21-5p expression in IECs
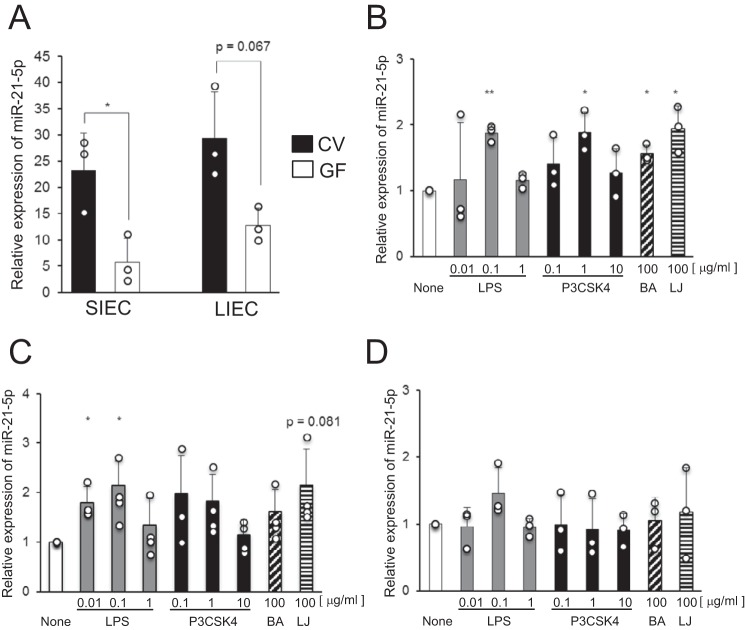
We next investigated the expression of miR-21-5p, which was identified as a potential microbiota-dependent miRNA by microarray analysis, in SIECs and LIECs of CV and GF mice by qRT-PCR. We found that miR-21-5p expression was higher in both SIECs and LIECs of CV mice compared with those of GF mice (Fig. 2A). These results confirmed that miR-21-5p expression in IECs was induced by commensal bacteria.
Figure 2.
miR-21-5p expression in IECs is induced by bacterial components. A, miR-21-5p expression in SIECs and LIECs of CV and GF mice was analyzed by qRT-PCR. Data are expressed as mean ± S.D. of three independent experiments. B–D, IEC lines, HT-29 (B), SW480 (C), and Caco-2 (D), were stimulated with LPS, P3CSK4, and heat-killed BA and LJ for 24 h. miR-21-5p expression was analyzed by qRT-PCR. Relative values compared with cells without bacterial stimulation are shown as mean ± S.D. of three (HT-29 and Caco-2) or four (SW480) independent experiments. *, p < 0.05; **, p < 0.01 (versus cells without stimulation).
Furthermore, we investigated the regulatory mechanism of miR-21-5p expression in the human IEC lines HT-29, SW480, and Caco-2. We stimulated the cells with LPS, Pam3CSK4 (P3CSK4), and the heat-killed intestinal bacteria Bacteroides acidifaciens type A43 (BA) and Lactobacillus johnsonii 129 (LJ). We found that the expression of miR-21-5p in HT-29 and SW480 cells was increased by all bacterial stimuli used in the experiment; neither the LPS nor the P3CSK4 responses were dose-dependent (Fig. 2, B and C). In contrast, miR-21-5p expression in Caco-2 cells, which show low responsiveness to bacterial stimulation because of minimal Toll-like receptor (TLR) expression (9), was unaffected by the bacterial stimuli (Fig. 2D). Treatment with anti-TLR4 antibody inhibited the LPS-induced expression of miR-21-5p (p = 0.130). In addition, the anti-TLR2 antibody partially inhibited the induction of miR-21-5p expression by P3CSK4 (supplemental Fig. S1). The partial effect of the anti-TLR2 antibody may be due to the concentration and/or affinity of the antibody. These results suggest that the induction of miR-21-5p expression by bacterial stimulation is mediated by TLRs.
miR-21-5p affects intestinal epithelial permeability but not IL-8 production from IECs
To clarify the role of miR-21-5p in IECs, we analyzed the effect of the miR-21-5p locked nucleic acid (LNA) inhibitor on intestinal epithelial permeability using filter-grown Caco-2 cells as an intestinal epithelial monolayer model. The transepithelial electrical resistance (TER) of Caco-2 cells treated with miR-21-5p LNA inhibitor was significantly greater than the TER of cells treated with the control LNA (Fig. 3A).
Figure 3.
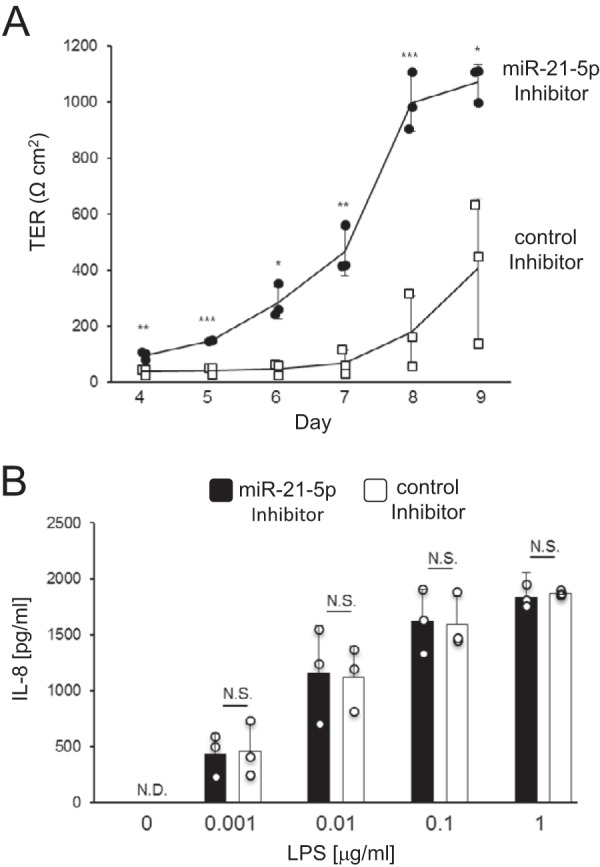
miR-21-5p affects intestinal epithelial permeability but not IL-8 production by IECs. A, Caco-2 cells were seeded on transwell inserts on day 0 and treated with miR-21-5p inhibitor on day 1. TER values were measured on days 4 through 9. The results are expressed as mean ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.005 (versus control). B, SW480 cells were transduced with the miR-21-5p inhibitor. Cells were then stimulated with 0.001–1 μg/ml LPS for 24 h, and the IL-8 concentration in the culture supernatant was measured by ELISA. The results are expressed as mean ± S.D. of three independent experiments. N.D., not detected; N.S., not significant (versus control).
We next evaluated the effect of the miR-21-5p inhibitor on IL-8 production by SW480 cells upon stimulation with LPS. IL-8 production in the culture supernatants of miR-21-5p inhibitor- and control LNA-transduced cells was similar (Fig. 3B). These results indicate that miR-21-5p in IECs is involved in the regulation of intestinal epithelial permeability, but not IL-8 production upon bacterial stimulation.
miR-21-5p increases intestinal epithelial permeability via ARF4
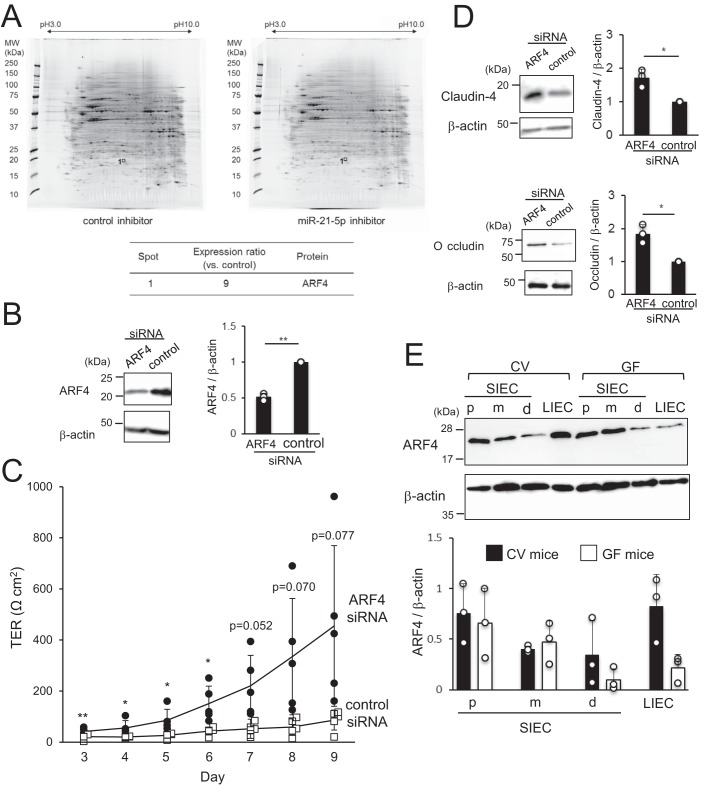
To identify the mechanisms underlying the modulation of intestinal epithelial permeability by miR-21-5p, a proteome analysis was performed on cells treated with miR-21-5p LNA inhibitor or control LNA. Several spots showed markedly different expression in the two groups, indicating that miR-21-5p regulates the expression of specific proteins. One of these spots was identified by MS-MS analysis as ARF4, which was deemed a candidate target molecule of miR-21-5p in IECs as it had 9-fold higher expression in the presence of miR-21-5p inhibitor than in the presence of the control (Fig. 4A). ARF4-specific and control siRNAs were then introduced into Caco-2 cells to analyze the role of ARF4 in controlling intestinal epithelial permeability. We confirmed that the expression of ARF4 was decreased by siRNA (Fig. 4B). Unexpectedly, the TER of Caco-2 monolayers treated with ARF4 siRNA was significantly higher than the TER of cells treated with control siRNA (Fig. 4C). Because the expression of ARF4 and miR-21-5p appears to increase intestinal epithelial permeability, it is possible that ARF4 is under the control of miR-21-5p but not directly down-regulated by this miRNA. In addition, the tight junction–related proteins claudin-4 and occludin were up-regulated in cells treated with ARF4 siRNA compared with those treated with control siRNA (Fig. 4D). These results indicate that ARF4 increases intestinal epithelial permeability via suppression of tight junction–related proteins.
Figure 4.
miR-21-5p increases intestinal epithelial permeability via ARF4. A, protein expression was compared in Caco-2 cells treated with miR-21-5p LNA inhibitor and control LNA by 2D gel electrophoresis. The spots that were differentially expressed in the two groups were subjected to MS-MS analysis. The spots corresponding to ARF4 are indicated. B and D, ARF4 siRNA was introduced into Caco-2 cells. After culture for 72 or 120 h, the expression of ARF4 (72 h), claudin-4 (120 h), occludin (120 h), and β-actin (72 and 120 h) was analyzed by Western blotting. Representative blots (left panel) and the mean ± S.D. of the relative band intensities normalized to β-actin (right panel) of three independent experiments are shown. *, p < 0.05; **, p < 0.005 (versus control). C, Caco-2 cells were seeded on transwell inserts on day 0, and ARF4 siRNA was introduced into the cells on day 1. TER values were measured on days 3 through 9. The results are expressed as mean ± S.D. of five independent experiments. *, p < 0.05; ** p < 0.005 (versus control). E, ARF4 expression in proximal (p), medial (m), and distal (d) SIECs and LIECs from CV and GF mice was analyzed by Western blotting. Representative blots (top panel) and mean ± S.D. of the relative band intensities normalized to those of β-actin (bottom panel) of three independent experiments are shown. *, p < 0.05.
Next, we analyzed the expression of ARF4 in SIECs from the proximal, medial, and distal portions of the small intestine and LIECs of CV and GF mice by Western blotting. ARF4 expression was higher in LIECs of CV mice than in those of GF mice, but it was comparable in SIECs of CV versus GF mice, indicating that ARF4 may be induced in LIECs by the microbiota (Fig. 4E). The finding that both ARF4 and miR-21-5p were induced by the microbiota in LIECs also supports the hypothesis that ARF4 is not a direct target of miR-21-5p. Furthermore, we did not find target sequences in the 3′ region of the ARF4 gene using TargetScan (http://www.targetscan.org/vert_71/)3 (33). Taken together, these data indicate that ARF4 in IECs increases intestinal epithelial permeability in a miR-21-5p-dependent manner, although it is not likely to be a direct target of this miRNA.
ARF4 is regulated by known targets of miR-21-5p: PTEN and PDCD4
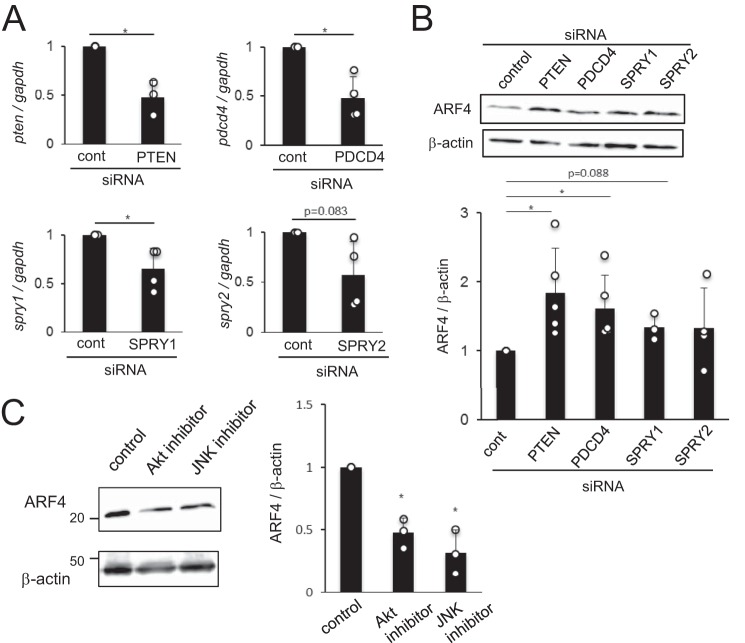
Previous reports have shown that miR-21 targets PTEN, PDCD4, and Sprouty (SPRY) 1 and 2 (10–13), which are known to be negative regulators of the PI3K–Akt, JNK–AP-1, and Erk signaling pathways, respectively (14–16). These pathways regulate intestinal epithelial permeability (17–19). We hypothesized that miR-21-5p targets PTEN, PDCD4, SPRY1, and SPRY2 and activates these pathways to induce ARF4 in IECs, increasing intestinal epithelial permeability. To test this hypothesis, we examined the effects of PTEN, PDCD4, SPRY1, and SPRY2 siRNAs on ARF4 expression in Caco-2 cells. We found that expression of the mRNA for PTEN, PDCD4, SPRY1, and SPRY2 was decreased by the relevant siRNA (Fig. 5A). The expression of ARF4 was significantly increased in cells treated with PTEN and PDCD4 siRNAs compared with those treated with control siRNA (Fig. 5B). ARF4 expression was only slightly increased in cells treated with SPRY1 and SPRY2 siRNAs. This may be due to the reduced efficacy of knockdown by these siRNA constructs compared with the PTEN and PDCD4 siRNA (Fig. 5A). Furthermore, the expression of ARF4 was significantly lower in cells treated with an Akt or JNK inhibitor than in control cells (Fig. 5C). These results suggest that down-regulation of PTEN and PDCD4 by miR-21-5p induces ARF4 expression through the Akt and JNK pathways, which results in increased intestinal epithelial permeability.
Figure 5.
ARF4 is regulated by PTEN and PDCD4, targets of miR-21-5p. A, Caco-2 cells were treated with siRNAs to PTEN, PDCD4, SPRY1, or SPRY2. After culture for 48 h, the expression of PTEN, PDCD4, SPRY1, and SPRY2 mRNA was analyzed by qRT-PCR. Expression relative to GAPDH is expressed as the mean ± S.D. of three (PTEN) and four (PDCD4, SPRY1, and SPRY2) independent experiments. cont, control. B, Caco-2 cells were treated with siRNAs to PTEN, PDCD4, SPRY1, or SPRY2 and cultured for 72 h. ARF4 expression was analyzed by Western blotting. Representative blots (top panel) and mean ± S.D. of the relative band intensities normalized to those of β-actin (bottom panel) of three (SPRY1 siRNA), four (SPRY2 siRNA), or five (PTEN and PDCD4 siRNA) independent experiments are shown. C, Caco-2 cells were treated with Akt or JNK inhibitor. After 48 h of culture, ARF4 expression was analyzed by Western blotting. Representative blots (left panel) and relative band intensities normalized to those of β-actin (right panel) of three independent experiments are shown. *, p < 0.05 (versus control).
Discussion
Intestinal commensal microbiota play an important role in the maintenance of host intestinal epithelial barrier functions. In contrast, dysfunction of the intestinal epithelial barrier is often associated with the development of various diseases. In this study, we demonstrated the molecular mechanisms underlying the regulation of the intestinal epithelial barrier by commensals via miR-21-5p and ARF4.
We found that miRNA expression in IECs was affected by the presence of the commensal microbiota. The effects of commensal bacteria on intestinal miRNA expression have also been shown by Singh et al. (8) using cecal tissue sections from CV and GF mice and by Dalmasso et al. (7) using ilea and colons from GF and microbiota-transferred mice. Singh et al. (8) found 16 miRNAs, whereas Dalmasso et al. (7) found nine miRNAs that were differentially expressed depending on commensal bacteria. Our data clearly showed that the expression of miRNAs in murine IECs is affected by the commensal microbiota, using purified primary IECs. We also found that miRNA expression in IECs was influenced not only by the presence of commensal bacteria but also by location. Lee et al. (20) similarly observed differential miRNA expression in SIECs and LIECs in a mouse colitis model. The physiological significance of this difference is not fully understood, but it may reflect the differences in the structures and functions of the small and large intestinal epithelia.
The expression of miR-21, one of the most highly conserved miRNAs in mammals, was induced by commensal microbiota. Its microbiota-induced expression may be mediated by TLRs, although the effects of the anti-TLR antibodies were not statistically significant. However, a previous report has shown that miR-21 is induced by LPS in a MyD88-dependent manner (21). Recently, it has been reported that miR-21 is highly expressed in inflamed tissues in diseases such as cancer and IBD (22, 23). Because these diseases are known to be associated with dysbiosis of intestinal microbiota, our results suggest that miR-21-5p may become a useful marker for inflammatory diseases associated with intestinal dysbiosis.
Although miR-21-5p is expressed at a physiological level in IECs of CV mice, it does not induce severe inflammation under steady-state conditions. Inflammation is likely to be induced when this miRNA is excessively expressed upon disruption of intestinal symbiosis or infection with pathogenic microbes. In fact, miR-21 was reportedly induced by pathogenic microbes such as Helicobacter pylori, Salmonella Typhimurium, and Mycobacterium species, leading to excessive immune responses (24, 25). Neither LPS nor P3CSK4 acted in a dose-dependent manner, suggesting that cellular mechanisms suppress the excessive induction of miR-21-5p. It is possible that lower concentrations of the ligands induce miR-21-5p in a dose-dependent manner. Microbiota-dependent miRNAs, including miR-21-5p, will become targets for the maintenance of intestinal homeostasis or its restoration following dysbiosis.
We demonstrated that intestinal microbiota-dependent miR-21-5p expression modulates intestinal epithelial permeability. Similarly, miR-21 enhances TNF-α–mediated increases in intestinal epithelial permeability (26). Recently, miR-122 and miR-29 were shown to increase intestinal epithelial permeability by targeting the transcripts for occludin and claudin-1, respectively (27, 28). Our results revealed that a microbiota-induced specific miRNA increases intestinal epithelial permeability. Thus, microbiota-targeted intervention may help to recover or control intestinal epithelial permeability. Probiotic bacteria contribute to the regulation of intestinal barrier function (29), which could be mediated by miRNA.
Our findings demonstrated that ARF4 was controlled by miR-21-5p and modulated intestinal epithelial permeability through the regulation of tight junction proteins such as claudin-4 and occludin. Because proteome analysis revealed that ARF4 expression was increased by the miR-21-5p inhibitor, we hypothesized that ARF4 is a direct target of miR-21-5p, which decreases intestinal epithelial permeability. However, we found that ARF4 was an indirect target; its expression was controlled by the direct targets of miR-21-5p, PTEN, and PDCD4, and it increased intestinal epithelial permeability. We cannot explain this discrepancy at present, but it may involve the side effects of other miR-21-5p targets. We finally concluded that miR-21-5p targets PTEN and PDCD4 to suppress their expression, up-regulating ARF4 via the Akt and JNK pathways, and, as a result, increasing epithelial permeability. Because it was difficult to directly show the ARF4-dependence for miR-21-5p–mediated control of epithelial permeability in our experimental system, we cannot exclude the possibility that there is another mechanism independent of ARF4 by which miR-21-5p increases epithelial permeability in addition to the ARF4-dependent mechanism.
ARF belongs to the Ras superfamily of small GTPases, which play important roles in various biological and cellular processes (30). Some small GTPases are known to control intestinal function and homeostasis. For instance, knockdown of ARF6 suppresses Escherichia coli internalization by the human IEC line T84 (31). On the other hand, PTEN, PDCD4, SPRY1, and SPRY2 are repressors of the Ras signaling pathway (14–16), which is involved in the regulation of intestinal epithelial permeability (17–19). This is the first report to clarify the role of ARF4 in the control of intestinal epithelial permeability as well as the regulation of ARF4 expression by PTEN and PDCD4. Regulation of intestinal epithelial permeability by tight junction proteins is a major mechanism to prevent excessive inflammatory responses and maintain intestinal homeostasis. The growing literature on the mechanisms underlying the regulation of intestinal epithelial permeability may help identify novel targets to treat inflammatory diseases related to intestinal epithelial barrier dysfunction. Further studies in vivo and in vitro will elucidate the interactions between commensal microbiota and their hosts.
Experimental procedures
Mice
BALB/c mice were purchased from CLEA Japan (Tokyo, Japan) and bred under CV conditions. Autogenous GF mice were bred and housed in our GF facilities at the College of Bioresource Sciences, Nihon University. Mice were maintained in a temperature-controlled room with a 12-h light/dark cycle with free access to food and water. Female mice were used at 10–12 weeks of age. All experiments were approved by the Nihon University Animal Care and Use Committee and conducted in accordance with their guidelines.
Preparation of murine IECs
IECs from the small and large intestines of mice were prepared as described previously (32).
Human IEC lines
The human epithelial colonic adenocarcinoma cell lines HT-29 and Caco-2 were purchased from DS Pharma Biomedical (Osaka, Japan). The human epithelial colonic adenocarcinoma cell line SW480 was provided by the Cell Resource Center for Biomedical Research (Institute of Development, Aging, and Cancer, Tohoku University, Miyagi, Japan). Cells were cultured as described in a previous report (9).
Microarray analysis
Total RNA, including low-molecular-weight RNA, was isolated from IECs using the High Pure miRNA isolation kit (Roche). The expression of miRNA was evaluated by miRCURY LNATM microRNA array (Exiqon, Vedbaek, Denmark) after analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Preparation of heat-killed bacteria
BA and LJ were isolated from murine intestinal commensal bacteria and kindly provided by Dr. Itoh (University of Tokyo). BA and LJ were cultured in EG broth supplemented with 0.01% menadione and 0.1% hemin or de Man, Rogosa, and Sharpe (MRS) broth, respectively, for 48 h at 37 °C under anaerobic conditions. The bacteria were collected by centrifugation, washed three times with sterilized water, heated at 80 °C for 50 min, and lyophilized.
Stimulation of IEC lines with bacterial ligands
The human IEC lines Caco-2, HT-29, and SW480 were stimulated with 0.01–1 μg/ml ultra-pure E. coli K12 LPS (Invivogen, San Diego, CA), 0.1–10 μg/ml P3CSK4 (Invivogen), and 100 μg/ml heat-killed BA and LJ. After 24 h of culture, cells were collected to extract total RNA, including low-molecular-weight RNA, to quantify miR-21-5p expression. Alternatively, for the measurement of IL-8 production, SW480 cells were stimulated with 0.001–1 μg/ml LPS for 24 h.
Introduction of miRNA inhibitor and siRNA
The miR-21-5p LNA inhibitor or control LNA (Exiqon) and ARF4, PTEN, PDCD4, SPRY1, SPRY2, or control siRNA (Ambion) (all 50 nm) were introduced into cells using X-tremeGENETM HP DNA transfection reagent (Roche).
Treatment with Akt and JNK inhibitors
The Akt inhibitor LY-294002 (Wako Pure Chemical Industries, Osaka, Japan) and JNK inhibitor SP600125 (Wako Pure Chemical Industries) were added to cells at a concentration of 50 μm for 48 h.
Quantitative RT-PCR (qRT-PCR)
The protocols for reverse transcription and quantitative analysis of mRNA or miRNA expression by qPCR were described in a previous report (32). We normalized mRNA expression to that of GAPDH. Although miRNA expression is usually normalized to U6 or RNU1a1, they were not suitable for this experiment because the miRNA expression of the IECs varied markedly depending on their differentiation status. Therefore, we determined relative expression values using equal amounts of RNA. The nucleotide sequences of the primers used for qPCR are shown in supplemental Table S1.
Measurement of TER
Caco-2 cells were seeded on transwell inserts, and their TER was measured using a Millicell ERS voltohmmeter (Millipore, Darmstadt, Germany). To calculate the TER, we subtracted the resistance of a blank well from the resistance of the sample wells and then multiplied the value by the area of the insert membrane.
Measurement of IL-8 production
The IL-8 concentration in the culture supernatants was measured by human CXCL8/IL-8 immunoassay (R&D Systems Biotech, Minneapolis, MN), according to the instructions of the manufacturer.
Western blotting
Cell lysates were prepared as described previously (32). We used primary antibodies against ARF4 (Proteintech), claudin-4 (Santa Cruz Biotechnology, Dallas, TX), occludin (Abcam, Cambridge, UK), and β-actin (Abcam).
Proteome analysis
Proteome analysis was performed by Takara Bio (Shiga, Japan). Briefly, we compared protein expression in Caco-2 cells treated with the miR-21-5p LNA inhibitor and those treated with control LNA by 2D electrophoresis followed by silver staining. Spots with substantial differences in intensities in inhibitor- and control-treated samples were collected to perform in-gel trypsin digestion and MALDI-TOF/TOF analysis using a 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA).
Statistical analysis
Statistical analyses were performed using two-tailed Student's t test.
Author contributions
K. N. performed the experiments, analyzed the data, and wrote the paper. Y. S. performed the experiments and analyzed and discussed the data. H. N. and T. K. performed the experiments. Y. N., M. T., A. H., S. K., and S. H. analyzed and discussed the data. K. T. designed the study, performed the experiments, analyzed the data, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This study was supported in part by a grant from the Japan Society for the Promotion of Science (KAKENHI 26450165 to K.T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Fig. S1 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- IBD
- inflammatory bowel disease
- IEC
- intestinal epithelial cell
- miRNA
- microRNA
- SIEC
- small intestinal epithelial cell
- LIEC
- large intestinal epithelial cell
- CV
- conventional
- GF
- germ-free
- P3CSK4
- Pam3CSK4
- BA
- Bacteroides acidifaciens type A43
- LJ
- Lactobacillus johnsonii 129
- TLR
- Toll-like receptor
- LNA
- locked nucleic acid
- TER
- transepithelial electrical resistance
- PTEN
- phosphatase and tensin homolog
- SPRY
- Sprouty
- ARF
- ADP ribosylation factor.
References
- 1. Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., and Gordon J. I. (2005) Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 2. Tlaskalová-Hogenová H., Stěpánková R., Kozáková H., Hudcovic T., Vannucci L., Tučková; L., Rossmann P., Hrnčí T., Kverka M., Zákostelská Z., Klimečová K., Pibylová J., Bártová J., Sanchez D., Fundová P., et al. (2011) The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heller F., Florian P., Bojarski C., Richter J., Christ M., Hillenbrand B., Mankertz J., Gitter A. H., Bürgel N., Fromm M., Zeitz M., Fuss I., Strober W., and Schulzke J. D. (2005) Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 129, 550–564 [DOI] [PubMed] [Google Scholar]
- 4. Pruteanu M., Hyland N. P., Clarke D. J., Kiely B., and Shanahan F. (2011) Degradation of the extracellular matrix components by bacterial-derived metalloproteases: implications for inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 5. Pruteanu M., and Shanahan F. (2013) Digestion of epithelial tight junction proteins by the commensal Clostridium perfringens. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G740–G748 [DOI] [PubMed] [Google Scholar]
- 6. Biton M., Levin A., Slyper M., Alkalay I., Horwitz E., Mor H., Kredo-Russo S., Avnit-Sagi T., Cojocaru G., Zreik F., Bentwich Z., Poy M. N., Artis D., Walker M. D., Hornstein E., et al. (2011) Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 12, 239–246 [DOI] [PubMed] [Google Scholar]
- 7. Dalmasso G., Nguyen H. T., Yan Y., Laroui H., Charania M. A., Ayyadurai S., Sitaraman S. V., and Merlin D. (2011) Microbiota modulate host gene expression via microRNAs. PLoS ONE 6, e19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh N., Shirdel E. A., Waldron L., Zhang R. H., Jurisica I., and Comelli E. M. (2012) The murine caecal microRNA signature depends on the presence of the endogenous microbiota. Int. J. Biol. Sci. 8, 171–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takahashi K., Sugi Y., Hosono A., and Kaminogawa S. (2009) Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J. Immunol. 183, 6522–6529 [DOI] [PubMed] [Google Scholar]
- 10. Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., and Allgayer H. (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 [DOI] [PubMed] [Google Scholar]
- 11. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., and Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sayed D., Rane S., Lypowy J., He M., Chen I. Y., Vashistha H., Yan L., Malhotra A., Vatner D., and Abdellatif M. (2008) MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 19, 3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., et al. (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 [DOI] [PubMed] [Google Scholar]
- 14. Bitomsky N., Böhm M., and Klempnauer K. H. (2004) Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene 23, 7484–7493 [DOI] [PubMed] [Google Scholar]
- 15. Hanafusa H., Torii S., Yasunaga T., and Nishida E. (2002) Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 4, 850–858 [DOI] [PubMed] [Google Scholar]
- 16. Zhou X., Ren Y., Moore L., Mei M., You Y., Xu P., Wang B., Wang G., Jia Z., Pu P., Zhang W., and Kang C. (2010) Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab. Invest. 90, 144–155 [DOI] [PubMed] [Google Scholar]
- 17. Aggarwal S., Suzuki T., Taylor W. L., Bhargava A., and Rao R. K. (2011) Contrasting effects of ERK on tight junction integrity in differentiated and under-differentiated Caco-2 cell monolayers. Biochem. J. 433, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langlois M. J., Bergeron S., Bernatchez G., Boudreau F., Saucier C., Perreault N., Carrier J. C., and Rivard N. (2010) The PTEN phosphatase controls intestinal epithelial cell polarity and barrier function: role in colorectal cancer progression. PLoS ONE 5, e15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Wu S., Ma J., Xia Y., Ai X., and Sun J. (2015) Bacterial protein AvrA stabilizes intestinal epithelial tight junctions via blockage of the C-Jun N-terminal kinase pathway. Tissue Barriers 3, e972849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J., Park E. J., Yuki Y., Ahmad S., Mizuguchi K., Ishii K. J., Shimaoka M., and Kiyono H. (2015) Profiles of microRNA networks in intestinal epithelial cells in a mouse model of colitis. Sci. Rep. 5, 18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., and O'Neill L. A. (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 22. Shi C., Yang Y., Xia Y., Okugawa Y., Yang J., Liang Y., Chen H., Zhang P., Wang F., Han H., Wu W., Gao R., Gasche C., Qin H., Ma Y., and Goel A. (2016) Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 65, 1470–1481 [DOI] [PubMed] [Google Scholar]
- 23. Wu F., Zhang S., Dassopoulos T., Harris M. L., Bayless T. M., Meltzer S. J., Brant S. R., and Kwon J. H. (2010) Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm. Bowel Dis. 16, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maudet C., Mano M., and Eulalio A. (2014) MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 588, 4140–4147 [DOI] [PubMed] [Google Scholar]
- 25. Staedel C., and Darfeuille F. (2013) MicroRNAs and bacterial infection. Cell. Microbiol. 15, 1496–1507 [DOI] [PubMed] [Google Scholar]
- 26. Zhang L., Shen J., Cheng J., and Fan X. (2015) MicroRNA-21 regulates intestinal epithelial tight junction permeability. Cell Biochem. Funct. 33, 235–240 [DOI] [PubMed] [Google Scholar]
- 27. Ye D., Guo S., Al-Sadi R., and Ma T. Y. (2011) MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 141, 1323–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Q., Costinean S., Croce C. M., Brasier A. R., Merwat S., Larson S. A., Basra S., and Verne G. N. (2015) MicroRNA 29 targets nuclear factor κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology 148, 158–169.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohland C. L., and Macnaughton W. K. (2010) Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G807–G819 [DOI] [PubMed] [Google Scholar]
- 30. Citalán-Madrid A. F., García-Ponce A., Vargas-Robles H., Betanzos A., and Schnoor M. (2013) Small GTPases of the Ras superfamily regulate intestinal epithelial homeostasis and barrier function via common and unique mechanisms. Tissue Barriers 1, e26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smyth D., McKay C. M., Gulbransen B. D., Phan V. C., Wang A., and McKay D. M. (2012) Interferon-γ signals via an ERK1/2-ARF6 pathway to promote bacterial internalization by gut epithelia. Cell. Microbiol. 14, 1257–1270 [DOI] [PubMed] [Google Scholar]
- 32. Sugi Y., Takahashi K., Kurihara K., Nakata K., Narabayashi H., Hamamoto Y., Suzuki M., Tsuda M., Hanazawa S., Hosono A., and Kaminogawa S. (2016) Post-transcriptional regulation of Toll-interacting protein in the intestinal epithelium. PLoS ONE 11, e0164858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agarwal V., Bell G. W., Nam J. W., and Bartel D. P. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.