Abstract
Production and secretion of pro-metastatic proteins is a feature of many tumor cells. The FAM3C interleukin-like epithelial-to-mesenchymal-transition (EMT) inducer (ILEI) has been shown to be strongly up-regulated in several cancers and to be essential for tumor formation and metastasis in epithelial cells, correlating with a significant decrease in overall survival in colon and breast cancer patients. ILEI has been seen to interact with the γ-secretase presenilin 1 subunit (PS1). However, not much is known about the mechanism-of-action or the detailed ILEI structure. We present here the crystal structures of FAM3C ILEI and show that it exists as monomers but also as covalent dimers. The observed ILEI β-β-α fold confirmed previous indications that the FAM3C proteins do not form classical four-helix-bundle structures as was initially predicted. This provides the first experimental evidence that the interleukin-like EMT inducers are not evolutionarily related to the interleukins. However, more surprisingly, the ILEI dimer structure was found to feature a trans-linked domain swap, converting an intramolecular disulfide to intermolecular. Interestingly, dimeric but not monomeric ILEI was subsequently found to cause a dose-dependent increase in EpRas cell invasiveness comparable with TGF-β, indicating that the dimer might be the active ILEI species. This is in line with a parallel study showing that covalent oligomerization of ILEI is essential for EMT and tumor progression in vivo. The structures and the activity data give some first insight into the relationship between dimerization and ILEI function as well as indicate an intriguing link between ILEI, the PS1-protease, TGF-β, and the TGF-β receptor 1.
Keywords: cancer, crystallography, epithelial-mesenchymal transition (EMT), γ-secretase, presenilin, transforming growth factor β (TGF-β)
Introduction
The FAM3 superfamily was first identified in a search for new cytokines. Based on secondary structure predictions, the FAM3A protein was identified and suggested to exhibit a four-helix bundle structure similar to IL-2, IL-10, and the interferons (1). Three related protein subfamilies were subsequently identified, FAM3B, FAM3C, and FAM3D, with a cross-sequence identity between 30 and 50%. Four cysteines are conserved throughout FAM3A-D and were hypothesized to correspond to the two disulfide bridges between helix 1 and 4 and between helix 2 and 3 of the classical cytokine fold. However, despite the sequence similarity between the different FAM3 subfamilies, no other clear sequence homology could be inferred to previously characterize cytokines or any other proteins.
In 2006, expression profiling of polysome-bound mRNA enabled the identification of FAM3C as a key player in TGF-β-induced EMT3 and late stage progression of carcinoma. Due to the postulated similarity to the interleukins, the human FAM3C protein was thus renamed interleukin-like EMT inducer, ILEI. FAM3C ILEI was subsequently shown to cause EMT onset and tumor progression in a tissue culture model system of non-tumorigenic polarized mammary epithelial (EpH4) cells (2). Endogenous ILEI protein was sufficient to initiate the late steps in metastasis such as extravasation, invasion, and proliferation of tumor cells at remote sites. ILEI was observed to be expressed in normal epithelia of several organs and up-regulated in malignant malformations. More prominent than elevated expression was the detection of altered subcellular distribution of the protein in carcinomas. A shift from granules in close proximity to the nucleus to a higher prevalence of diffuse cytoplasmic localization was shown to be a histological read-out of increased ILEI secretion; that is, ILEI-containing granules representing low secretion levels and an accumulation of ILEI in the Golgi and trans-Golgi network, whereas diffuse ILEI distribution reflecting high secretion levels and a shift of the protein into an extended trans-Golgi network and secretory vesicles (3). Interestingly, the altered subcellular localization of ILEI strongly correlated with a decrease in metastasis-free and overall survival of breast cancer patients (2, 3). The relevance of the shift in subcellular localization of ILEI linked to overall survival was confirmed also in patients with colorectal cancer (4) as well as in patients with hepatocellular carcinoma (5). Additionally, ILEI was found to be a valid prognostic marker in patients with esophageal squamous cell carcinoma (ESCC) (6). The inability of exogenous TGF-β to induce EMT in cells where ILEI had been silenced strengthened the hypothesis of ILEI as an important downstream effector of TGF-β in EMT signaling (2). The importance of ILEI as a pro-metastatic protein was recently further underlined by an analysis of the metastasis promoting secretome (7).
A follow-up study by Howe and co-workers (8) revealed that ILEI as well as the protein disabled-2 (Dab2) are post-transcriptionally regulated by TGF-β. The messenger ribonucleoprotein hnRNP E1 (also called PCBP1) is acting in trans- on cis-regulatory regions in the 3′-UTR of ILEI and Dab2, binding to a stem-loop motif (8). TGF-β activates a kinase cascade leading to the release of hnRNP E1 and ultimately to translation of the transcripts (9). ILEI was also identified as a TGF-β-inducible regulator of EMT in the lung cancer cell line A549, modulated by the same trans-activating pathway involving TGF-β activation and hnRNP E1 release (10).
N-terminal sequence analysis showed that secreted ILEI starts at Ser-42, suggesting that an additional 17 amino acids are removed in a second cleavage after processing of the signal peptide (2). Further investigation of the post-translational processing of ILEI identified plasmin and the urokinase receptor (uPAR) as critical components in the system regulating ILEI secretion, subcellular location, tumor progression, and propensity to metastasize, linked to survival prognosis in breast cancer patients (3).
Somewhat surprisingly, recently published data revealed an unexpected link between ILEI and Alzheimer's disease (AD). High levels of ILEI protein exhibited a strong correlation with a decrease in amyloid-β (Aβ) formation in the brain, indicating ILEI to be a negative regulator of Aβ production (11). Extracellular application of recombinant ILEI was found to decrease Aβ generation, whereas subsequent RNAi-mediated knockdown of endogenous ILEI increased Aβ40 and Aβ42 levels. The first cleavage of the amyloid-β precursor protein (APP) by β-secretase yields an APP C-terminal fragment (CTF-β). This is subsequently cleaved by γ-secretase to form Aβ peptides. Cross-linking experiments revealed that ILEI binds to the presenilin-1 (PS1) subunit of the γ-secretase complex and prevents the second cleavage by destabilizing the interaction between γ-secretase and CTF-β. However, the ILEI-PS1 interaction does not seem to inhibit γ-secretase activity per se, as it maintains the ability to cleave Notch substrates. More recent data from the same laboratory show that neuronal ILEI resides in perinuclear vesicular structures, co-localizing with the trans-Golgi network and that the expression of ILEI in the brain decreases with age (12). This makes ILEI attractive both from its potential as a biomarker but also as a possible therapeutic for AD as interference with Notch signaling is likely to be an issue for therapies targeting all γ-secretase activity (11, 12).
In 2013 we published the first crystal structure of any FAM3 protein, FAM3B PANDER. Interestingly, the PANDER structure was found to exhibit a novel β-β-α fold not shared by any previously determined protein structures and significantly different from the hypothesized four-helix bundle fold. These results suggested that the FAM3 superfamily might represent an unknown class of signaling molecules (13).
We show here that FAM3C ILEI, in contrast to FAM3B PANDER, exists in both monomer and dimer forms. The crystal structures of the ILEI monomers strengthen the view of the FAM3 as a new group of regulatory proteins, distinct from the classical cytokines. The mouse dimer structure combined with mass spectrometry (MS) data shows that ILEI dimerizes via a covalent domain swap. Dimeric ILEI was further shown to be active, increasing the invasiveness of EpRas cells similar to a TGF-β control compared with monomeric protein. This indicates that the dimer might be the relevant species involved in EMT and ILEI-driven tumor progression.
Results
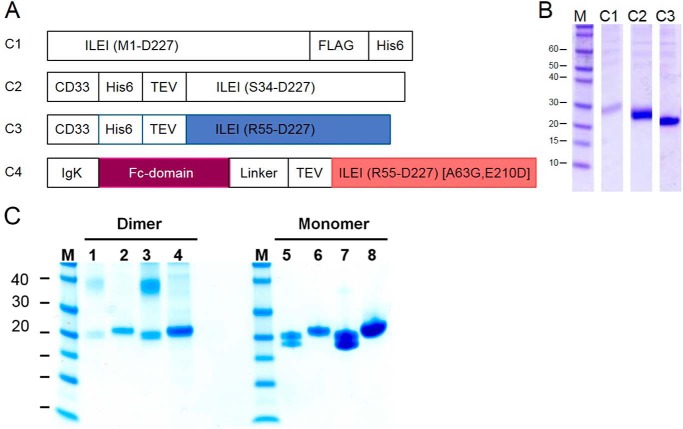
Four FAM3C ILEI protein constructs were designed based on the FAM3B PANDER structure. One full-length construct contained the native signal peptide (C1). Two constructs used a CD33 signal peptide where one encoded a protein starting at Ser-34 after the signal peptide (C2), and one encoded a truncated construct starting at Arg-55 (C3). A Fc fusion construct contained an IgK signal peptide and two point mutations in the ILEI-coding sequence (C4) (Fig. 1A). C1 and C2 were only constructed with the human ILEI sequence, whereas C3 was made in both human and mouse variants and C4 as a mouse variant only. A single letter in front of the construct indicates the species (h or m). Expression tests performed in suspension-adapted HEK cells showed that replacement of the native signal sequence with the CD33 signal sequence increased secretion of ILEI severalfold. This is independent of where the ILEI-coding sequence started (Fig. 1B). Large-scale purification of constructs hC3 and mC3 included an affinity capture step using Ni-NTA followed by TEV cleavage, reverse His-Trap, and size exclusion chromatography (SEC). ILEI expressed as a mix of monomer and dimer, with the monomer the dominant variant (Figs. 1, B and C, supplemental Fig. S6).
Figure 1.
Constructs for recombinant expression of ILEI in suspension-adapted HEK cells. A, construct 1 containing a native signal peptide and the sequence of human ILEI followed by a FLAG tag and a C-terminal His-tag. Constructs 2 and 3 were designed based on expression of FAM3B PANDER (13) with the native signal peptide replaced by CD33 signal peptide. Human and mouse variants of C3 were used for crystallization experiments. C4 was designed with an N-terminal Fc fusion preceded by an IgK signal peptide. In the C4 construct the ILEI coding region also contains two mutations, Ala-63 mutated to Gly and Glu-210 mutated to Asp. Mouse variants of C3 and C4 were coexpressed to produce a chimeric ILEI heterodimer. B, Coomassie-stained SDS-PAGE showing yields of human ILEI protein in the supernatant after small-scale expression and Ni-NTA purification of constructs hC1, hC2, and hC3. C, SDS-PAGE of mouse ILEI dimer and monomer, construct mC3, under non-reducing (lanes 1, 3, 5, and 7) and reducing conditions (lanes 2, 4, 6, and 8). Samples in lanes 3, 4, 7, and 8 are concentrated versions of lanes 1, 2, 5, and 6. Note the presence of the dimer and the split monomer bands under non-reducing conditions compared with the single band monomer under reducing conditions.
With the aim of determining the first three-dimensional structure of the FAM3C proteins, purified human and mouse ILEI (hC3 and mC3) from both monomer and dimer species were used in crystallization trials. After extensive screening, crystals could be obtained for the hC3 ILEI monomer and for both the mC3 ILEI monomer and dimer forms. High resolution diffraction data of these were subsequently obtained to 1.8 Å, 2.0 Å, and 1.9 Å, respectively. The human ILEI monomer was found to crystallize in space group P41 with two molecules per asymmetric unit and the mouse monomer in C2 with two molecules, whereas the mouse dimer construct crystallized in P21 with four molecules. The refinement of the structures converged to an R/Rfree of 0.17/0.21 for the human monomer, 0.22/0.24 for the mouse monomer, and 0.21/0.23 for the mouse dimer with good geometry (Table 1).
Table 1.
Data collection and refinement
| PDB 5LC2 human monomer | PDB 5LC3 mouse monomer | PDB 5LC4 mouse dimer | |
|---|---|---|---|
| Data collection | |||
| Space group | P41 | C2 | P21 |
| Cell dimensions (Å) | 110.7 110.7 30.5 | 163.8 32.0 75.7, 112° | 47.8 118.4 85.4, 99° |
| Resolution (Å) | 39.1-1.59 (1.65) | 44.3-2.0 (2.1) | 48.5-1.84 (1.91) |
| Measured reflections | 240,458 (5580) | 77,440 (5,891) | 237,659 (21,793) |
| Unique reflections | 46,062 (2551) | 23,649 (1729) | 69,589 (6,480) |
| Rmerge | 0.058 (0.60) | 0.042 (0.46) | 0.078 (1.0) |
| 〈I/σI〉 | 14.4 (1.2) | 13.2 (2.1) | 8.2 (0.9) |
| Completeness (%) | 91.7 (53.4) | 98.9 (99.5) | 98.4 (92.7) |
| Redundancy | 5.2 (2.2) | 3.3 (3.4) | 3.4 (3.4) |
| Refinement | |||
| Resolution (Å) | 30.0-1.80 (1.85) | 44.2-2.0 (2.13) | 48.5-1.84 (1.89) |
| Rwork/Rfree | 0.17/0.21 | 0.22/0.24 | 0.21/0.23 |
| No. atoms | |||
| Protein | 2,592 | 2,499 | 5,209 |
| Water | 447 | 105 | 423 |
| Average B-factors | |||
| Protein (Å2) | 25.3 | 61.7 | 39.0 |
| Water (Å2) | 37.2 | 51.1 | 46.3 |
| Ramachandran outliers (%) | 0.6 | 1.3 | 0.8 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.010 | 0.010 | 0.008 |
| Bond angles (°) | 1.06 | 1.20 | 1.00 |
ILEI monomers featured a non-interleukin-like fold
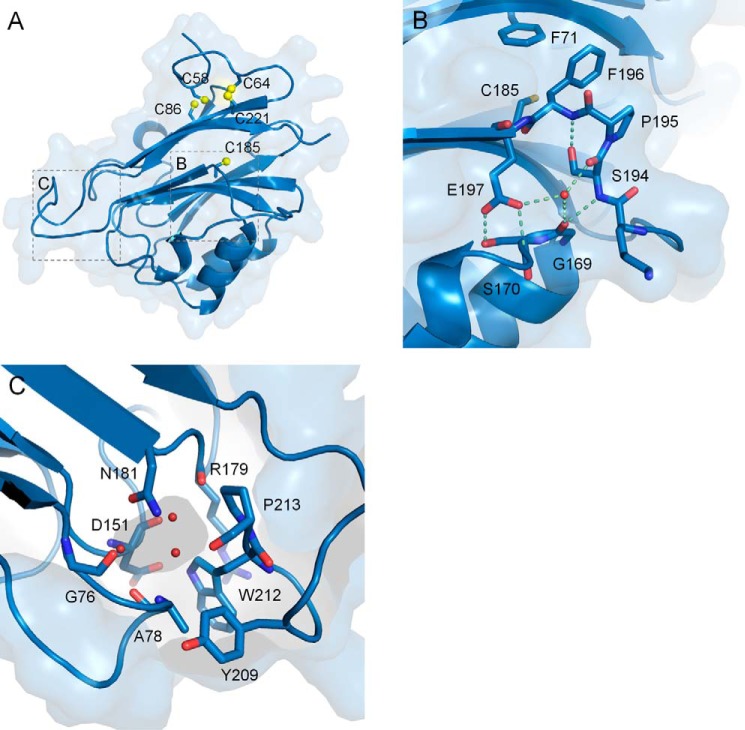
As predicted by modeling based on the PANDER structure (13), the human and mouse ILEI monomers were found to exhibit a globular β-β-α structure. Two anti-parallel β sheets are lined with a distinctly curved N-terminal loop and a short helical loop on one side and a layer of three short helices on the other (Fig. 2). The FAM3C ILEI structure thus shares no obvious relation to any of the previously published interleukin folds. To verify this, a search of the Protein Data Bank (PDB) was performed using DaliLite (14) and PDBeFold (EBI Protein Data Bank; Ref. 15) using the human monomer structure as the query. The top scoring and only relevant hit was the original FAM3B PANDER structure (PDB ID 2YOP and 2YOQ) with a PDBeFold Z-score of 15.1 and a sequence identity of 40%. The remainder of PDB scored low with the Fas apoptotic inhibitory molecule 1 (PDB ID 3MX7) showing some partial similarity with a Z-score of 4.9 and a sequence identity of 4%, well below the significance cut-off of Z > 6.
Figure 2.
The FAM3C ILEI monomer fold. A, the ILEI monomer structure, composed of two layers of β sheets and three short helices. The two cysteine bridges between Cys-58 and Cys-86 as well as between Cys-64 and Cys-221 are indicated in yellow together with a buried cysteine, Cys-185, located on β7. B, a highly conserved SPFE motif breaks the N-terminal end of β8 compared with the FAM3B PANDER structure (supplemental Fig. S1). C, the water-filled ILEI monomer pocket.
The human and mouse ILEI monomers feature a similar disulfide topology as FAM3B PANDER, connecting Cys-58–Cys-86 and Cys-64–Cys-221 (Fig. 3). However, the environment around the latter cysteine bridge is different in the FAM3C compared with the FAM3B proteins, with ILEI His-68 stabilizing the N-terminal loop by a bidentate stacking interaction to Pro-65 and Pro-223. In contrast to PANDER, the ILEI monomers also feature a long kinked loop in between strands β7 and β8, flanking strand β8. The side chains of 194SPFE197 form two distinct kinks, causing a break of strand β8 perpendicular to the central β sheet (Fig. 2B, supplemental Fig. S1). The atypical conformation of this SPFE motif is conserved in both the human and the mouse monomer structures, creating a well-defined circular formation including a central water molecule. In contrast, the rest of the loop from Gly-189–Thr-192 exhibit significant flexibility in some of the chains. To further explore the relationship within the FAM3 superfamily and get some insight into the key areas on the FAM3C ILEI surface, a number of FAM3 sequences were gathered by an iterative procedure and aligned (supplemental Fig. S2A). Similar to FAM3B PANDER, the highest conservation is found on one side of the ILEI molecule around a small enclosed water-filled cavity. This pocket is formed by loops between the end of the two β sheets, lined by the strictly conserved Gly-76, Asp-151, Arg-179, Tyr-209, Trp-212, and Pro-213 and the partially conserved Ala-77 and Ala-78 (Fig. 2C). The ILEI cavity contains three or four enclosed water molecules. This is smaller than the corresponding FAM3B pocket due to the replacement of PANDER Ser-142 by ILEI Asn-181 but is consistent across all four monomer ILEI chains. The conservation in the vicinity of the cavity creates a connected, strictly conserved surface area formed by Asp-151, Phe-178, Arg-179, Asn-207, and Tyr-209 centered around Trp-212 at the tip of the ILEI molecule (supplemental Fig. S2, B and C).
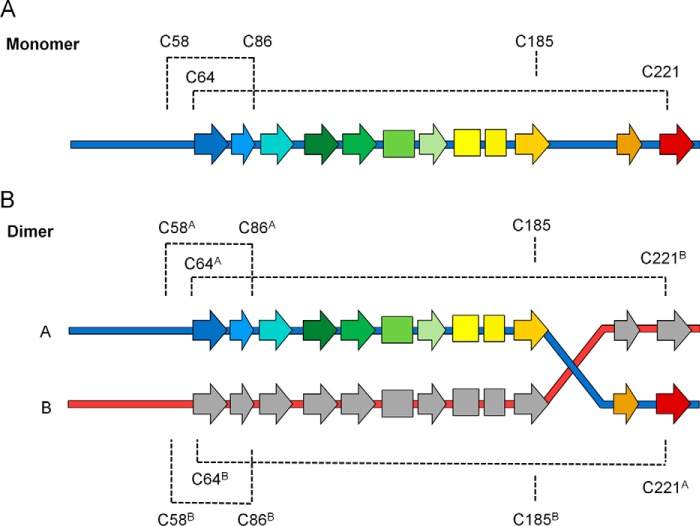
Figure 3.
Disulfide topology diagram of FAM3C ILEI monomers and dimers. Both monomer (A) and dimer (B) ILEI contain two disulfide bridges. The Cys-58–Cys-86 disulfide is conserved in both. However, the Cys-64–Cys-221 bridge is converted into a trans-linked disulfide in the dimer, introducing a covalent bond between the N-terminal strand of one chain with the C-terminal strand of the other.
ILEI dimerized through a covalent domain swap
The mouse dimer crystals were found to contain two ILEI dimers per asymmetric unit, each related by a rough 2-fold symmetry. The overall structures of the individual dimer chains are very similar to the human and mouse monomer structures, with a root mean square deviation of 0.8 Å over the Tyr-56–Lys-188 portion of the mouse monomer. However, the density of strand β7 was found to extend away from the central sheet in all four dimer chains, crossing over to its neighboring molecule. This forms a domain-swapped dimer where strands β8 and β9 from one molecule replace the equivalent two strands in the other via the kinked 194SPFE197 motif (Figs. 3 and 4). Interestingly, strong density was found connecting Cys-64 of each chain to Cys-221 of its dimer partner, covalently linking the N terminus of the first chain to the C-terminal strands of the second (supplemental Fig. S3). The oligomerization thus converts the intramolecular Cys-64–Cys-221 disulfide of the FAM3C ILEI monomer to an equivalent intermolecular cysteine bridge in the dimer.
Figure 4.
FAM3C ILEI dimer fold. A, the domain-swapped FAM3C ILEI dimer structure with the two chains colored in blue and red, respectively. B, surface representation of the dimer. C, a highly conserved SPFE motif located in the dimer interface breaks the N-terminal end of β8. D, the water-filled ILEI pocket split by the two dimer chains.
The swapped β8 and β9 exhibited very similar conformation to the corresponding stretch in the two monomer structures. However, the peptides of the dimer Tyr-209 and Glu-210 are inverted compared with both the two mouse and the two human monomer chains, with the main chain carboxyl of Glu-210 forming a hydrogen bond to the nearby Asn-207. Similarly, the side chain of His-199 is rotated 90 degrees compared with its monomer counterpart, forming an electrostatic interaction to Asp-180. Five residues, Gly-189–Lys-193, bridge the distance between the two dimer partners. Although the domain swap makes the buried area of the dimer interface >2900 Å2, the difference in buried surface area compared with the monomers is small, <400 Å2. The small additional area provided by the Gly-189–Lys-193 bridge indicates that the dimer geometry might be flexible. However, the bridge exhibited very similar geometry in all four dimer chains, and the overall geometry deviated <15 degrees between the two independent dimers in the asymmetric unit. The link region is stabilized by Lys-193, hydrogen bonding to the backbone carbonyl of the conserved Ser-170. Lys-193 directly precedes the 194SPFE197 motif, breaking strand β8 in the two monomer structures. Lys-193, Pro-195, Phe-196, and Glu-197 together with Ser-170 and the neighboring Phe-71 form an extended cluster that is highly conserved not only in FAM3C but also in the FAM3A and FAM3D sequences (supplemental Fig. S2).
The enclosed water pocket of the ILEI monomers is also retained also in the dimeric structure. However, in dimeric ILEI the cavity is split by Gly-76, Ala-77, Ala-78, Asp-151, and Arg-179 from one chain (denoted A) and Tyr-209, Trp-212, Pro-213 from the other (denoted B, Fig. 4D). Interestingly, the ILEI monomers and the four individual molecules of the mouse dimer all show a distinct B-factor distribution, where the area around the closed pocket and the loops around the tip of the β sheets exhibit significantly lower temperature factors than the other side of the molecule, indicating the stability of these areas (supplemental Fig. S4). β8 and β9 have about average B-factors in both the swapped and the un-swapped structures, whereas the loop connecting strand β7 and β8 is significantly more mobile in the human and the mouse monomers compared with the dimer bridge region.
Similar to FAM3B PANDER, the ILEI surface features a large number of charged residues. 21 acids and 23 bases form numerous salt bridges. Several of these electrostatic pairs exist across the ILEI dimer interface; Lys-72–Glu-217–Glu-219 linking β9A to β1B is partially conserved among the FAM3 proteins. Asp-180–Lys-201 connecting the swapped β8 to β7 and the neighboring Lys-208–Glu-214 of β9 are in contrast highly conserved in FAM3A, -C, and -D, whereas the side chain of Lys-156 and the Asp-151–Arg-179 salt bridge are conserved throughout all four FAM3 subfamilies. The latter three residues together with the side chain of Lys-201, form a conserved positively charged patch at the tip of β6 and β7, similar to the ILEI monomers (supplemental Fig. S2, B and C).
ILEI dimer in solution
The amount of dimer seen under non-reducing compared with reducing conditions (Fig. 1C) gave strong support for a cysteine-linked ILEI oligomer. However, to verify that the intermolecular Cys bridge seen in the dimer crystal structure also could be observed in solution, an experiment was designed to produce a chimeric dimer composed of two non-identical chains that could be isolated using two different tags. For this purpose a fourth ILEI construct containing an Fc fusion was produced (mC4 Fc-ILEI) (Fig. 1). This construct included two mutations, A63G and E210D, to distinguish the intramolecular disulfides of the monomers from the intermolecular disulfides of the dimers. The purified chimeric dimer, kept in a non-reducing buffer to maintain the disulfide bridges, was digested with trypsin and analyzed by MS. The MS data showed the presence of covalent trans-linked peptides between Cys-64 from one construct and Cys-221 from the other, verifying the presence of the domain swapped dimer also in solution (supplemental Fig. S7).
Recombinantly expressed ILEI dimer induced invasiveness in EpRas cells
An initial functional evaluation was carried out in vitro using recombinant ILEI mC3 protein. Murine EpRas cells were treated with either purified mC3 monomer or dimer protein (containing a small population of monomer), and the effect on invasive properties toward serum was measured. A significant, dose-dependent increase on invasiveness could be seen on the cells treated with mC3 ILEI dimer, comparable with TGF-β that was used as a positive control. No effect could be detected on cells treated with mC3 monomer (Fig. 5). This is in line with a parallel study by Kral et al.,4 showing that ILEI oligomerization essential for EMT induction in vitro as well as elevated tumor growth, EMT induction, and increased metastasis formation in vivo.
Figure 5.
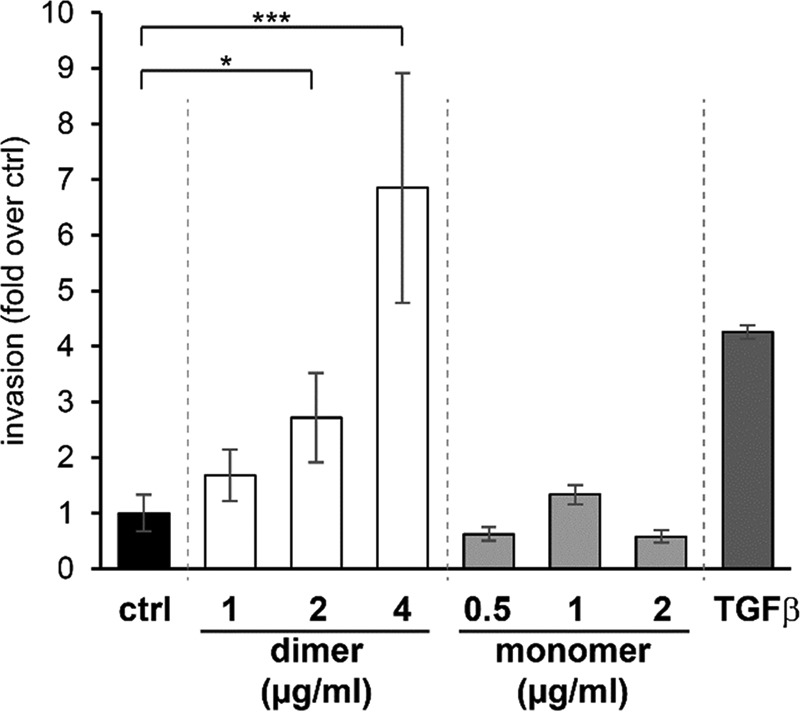
ILEI dimers increase in vitro invasive capability of EpRas cells in a dose-dependent manner. Trans-well invasion assay of EpRas cells treated with increasing concentrations of recombinant mC3 ILEI dimers and monomers toward a 10% FCS attractant. Invasion was performed for 24 h and is shown as -fold change over non-treated control (ctrl). Recombinant human TGF-β1 (hTGFb1) treatment under the same conditions was used as positive control. Data represent the mean ± S.E. of three independent experiments. Statistical significance (*, p ≤ 0.05, *** p ≤ 0.001) as determined by an unpaired two-sided Student's t test.
Discussion
FAM3C interleukin-like EMT inducer ILEI has been shown to be a secreted signaling protein involved in epithelial-mesenchymal transition and tumor progression. ILEI was able to induce tumor growth and metastasis formation in non-tumorigenic epithelial cells, whereas down-regulation of ILEI was found to attenuate TGF-β1-dependent EMT (2). Expression levels and a shift in subcellular localization of ILEI where the ILEI molecules re-localize from the endoplasmic reticulum to endocytic vesicles in the cytoplasm correlated with the tumor growth and overall survival of a population of breast cancer patients, indicating the potential for ILEI as an independent prognostic marker (2–6). In addition, recently published data revealed a strong link between ILEI and Alzheimer's disease, where high levels of ILEI were found to negatively correlate with AD biomarkers (11).
We present here the first crystal structures of human and mouse FAM3C protein in monomeric and dimeric forms. The structures show that the FAM3C interleukin-like EMT inducer ILEI is not related to any of the previously characterized interleukins, instead forming a mixed α/β fold containing two disulfide bridges. Surprisingly, the structure of the mouse dimer showed that ILEI dimerization is achieved by a strand swap where β8 and β9 are replaced in the two chains, creating a covalent trans-linked dimer. This domain swap was subsequently verified by mass spectrometry showing the presence of the intermolecular disulfide also in solution. The two dimer molecules are connected by a short four-amino acid bridge. The Lys-193–Glu-197 stretch at the end of the bridge and the neighboring Ser-170 create a circular structure with unusual geometry, forming a sharp kink away from the central ILEI sheet (Figs. 2B and 4C). 193KSPFE197 are all highly conserved in FAM3ACD but not in FAM3B, whereas Ser-170 is conserved throughout the superfamily. Phe-196 in turn forms a close interaction with the neighboring Phe-71 that is conserved in FAM3ACD (supplemental Fig. S2A). In FAM3B this is replaced by a tyrosine, unable to form a similar edge-face interaction. The geometry of the kinked Lys-193–Glu-197 motif and the observed conservation indicate that this stretch might be important for dimer formation, breaking the strand conformation seen in the FAM3B PANDER structure. This also suggests that the proteins of FAM3A and FAM3D might be able to form similar dimers as FAM3C, whereas the FAM3B proteins will not.
In an initial experiment to evaluate the function of the two ILEI forms, ILEI monomer and dimer were added to EpRas cells to evaluate the effect on cell invasion compared with a TGF-β-positive control. A strong dose-dependent effect on invasion could be seen for dimeric but not for monomeric ILEI, indicating that the dimer is the active ILEI species triggering cell invasion in the EpRas cancer model system (Fig. 5). The effect on invasion was significant even though the dimer population contained a portion of monomer due to the difficulty of separation. The activity of the dimer combined with the significant proportion of monomer observed in the expression raises the question of how the dynamics of oligomerization is controlled. As seen from the structures, β8 and β9 of the ILEI monomer need to open to form the dimer. In a parallel study, Kral et al.4 show that secreted ILEI monomers is found in two forms, a folded native form and a partially unfolded form running at a slightly higher molecular weight. By inhibiting extracellular thiol exchange by DTNB (5,5′-dithiobis(nitrobenzoic acid)) the unfolded monomer and the ILEI dimer could be completely suppressed but not the folded monomer. This indicates that unfolded ILEI monomers might be an intermediate in the formation of extracellular dimers. Interestingly, this is supported by the observation of a double monomer band under non-reducing conditions in our study (Fig. 1C, lanes 1, 3, 5, and 7) in contrast to a merged monomer band under reducing conditions (Fig. 1C, lanes 2, 4, 6, and 8).
In their investigation of the ILEI Cys-bridge topology, Kral et al.4 serendipitously found that modification of a fifth, non-conserved cysteine, Cys-185 inhibits both the opening of the monomer as well as the dimer formation. As seen from the monomer and dimer structures, Cys-185 is not involved in disulfide formation (Figs. 2 and 4). However, Cys-185 is located in the dimer interface, forming short backbone hydrogen bonds to Ser-194 and Phe-196 of the kinked Lys-193 to Glu-197 motif and stacking to the neighboring Phe-71 of β1. It is thus tempting to speculate that the residues lining of the interface and the water-filled cavity introduces some instability into the ILEI monomer, facilitating the opening of the two strands and the domain swap. The structures indicate several interesting positions that could be mutated to probe the stability and the dynamics of ILEI dimerization, not only in the Lys-193 to Glu-197 stretch but also in β1 and β7 of the dimer interface.
In 2014 Hasegawa et al. (11) showed that FAM3C ILEI prevents cleavage of the APP C-terminal fragment by binding the γ-secretase PS1 subunit. Application of recombinant ILEI was found to decrease HEK293 Aβ production, whereas subsequent RNAi-mediated knockdown of endogenous ILEI increased Aβ40 and Aβ42 generation. ILEI overexpression was also seen to reduce Aβ plaque in cerebral cortex and hippocampus in APP/ILEI transgenic mice compared with APP-only control mice, showing that ILEI is able to reduce Aβ overload in an in vivo AD model system. The studies indicated that ILEI destabilizes the β-secretase-cleaved APP C-terminal fragment bound to γ-secretase without inhibiting PS1 protease activity. Analysis of a number of γ-secretase mutants suggested that ILEI interacts with the C terminus of the PS1 domain but could not be rationalized in detail due to the lack of any reliable γ-secretase or FAM3C ILEI models. However, the recently published high resolution cryo-EM structure of the γ-secretase (16) combined with the structures presented here indicate that ILEI might bind in the large extracellular cleft of the γ-secretase (supplemental Fig. S5A). Mutation of presenilin Phe-462, His-463, and Gln-464 reduced PS1-ILEI interactions, whereas mutations of Asp-458, Gln-459, and Leu-460 completely abolished ILEI binding (11). The Asp-458–Leu-460 and Phe-462–Gln-464 stretches are located in the terminal α-helix of PS1, interacting with the extracellular portions of anterior pharynx defective-1 domain (APH-1) and the N-terminal helix of the nicastrin domain (NCT) (supplemental Fig. S5B). The negatively charged PS1 helix and the neighboring positively charged Arg-38 of NCT and Arg-62 of APH-1, could potentially interact with the highly conserved cluster of charged residues outside the water-filled ILEI cavity (supplemental Fig. S2C). This would place ILEI between the extracellular part of the nicastrin and the rim of the transmembrane NCT-APH1-PS1-PEN-2 horseshoe-like structure, potentially preventing nicastrin recognition of the shredded APP (17) without blocking the γ-secretase active site.
No direct link between the observed ILEI inhibition of γ-secretase and the previously described effects on EMT and tumor formation has thus far been shown. However, ILEI is regulated by TGF-β that acts both as a repressor and a promoter of EMT (2). TGF-β has in turn been seen to decrease levels of APP-CTF and Aβ (18) and has been associated with an increased risk of AD (19, 20). Hasegawa et al. (11) showed that this effect could be significantly reduced by ILEI-specific siRNA, indicating that the effect of TGF-β on Aβ production is mediated by ILEI. Interestingly, a recent study shows that the PS1 protease of γ-secretase is ubiquitinylated by the E3 ligase TRAF6. The activated PS1 subsequently mediates intramembrane proteolysis of the TGF-β1 type I receptor (TβRI). The intracellular domain of TβRI then goes into the nucleus and acts as a transcription factor, inducing expression of invasive target genes. Although the details and the direction of regulation are unclear, this presents an intriguing link between TGF-β, FAM3C ILEI, the PS1 protease, and the TGF-β receptor. This could potentially connect the observed AD/γ-secretase ILEI effects to EMT and cancer. However, more studies are needed to investigate whether ILEI also reduces TβRI γ-secretase cleavage as well as to elucidate the ILEI-binding mode on PS1 or on any other potential membrane-bound FAM3C interaction partner.
The human and mouse structures and the initial in vitro characterization confirms previous predictions that FAM3C ILEI are not cytokine-like proteins as well as give an indication on the mechanism of ILEI dimerization and its relation to the observed effect on cell invasion. The recently published link between ILEI and Aβ production highlights the functional diversity of the FAM3 proteins and underlines the need for further investigations of the different subgroups. The presented ILEI structures will be essential tools for future studies of the superfamily as well as for exploring the mode of action of FAM3C.
Experimental procedures
Materials and methods
Columns and column media were from GE Healthcare unless specified. All materials used for the expression of protein were from Thermo Fisher Scientific unless otherwise stated. Empty columns (Poly-Prep) were from Bio-Rad.
Construct design and protein expression
Human ILEI (FAM3C, Uniprot Q92520) starting at Met-1 (C1), Ser-34 (C2), and Arg-55 (C3) and mouse ILEI (FAM3C, Uniprot Q91VU0) starting at Ser-34 (C2), Arg-55 (C3), and Arg-55 with two mutations (A63G, E210D) (C4) were synthesized by GenScript and cloned into neomycin-resistant pDEST12.2 OriP vector (21). C1 contained a C-terminal His6 tag preceded by a FLAG tag (extra sequence: DYKDDDDKGHHHHHH). C2 and C3 contained human CD33 signal peptide (underlined) followed by a His6 tag and TEV recognition site on the N terminus of ILEI: MPLLLLLPLLWAGALAELHHHHHHENLYFQ*[m/hILEI Ser-34–Asp-227/Arg-55–Asp-227], and C4 contained an Ig-κ signal peptide (underlined) followed by an Fc domain, a linker, and a TEV recognition site and an ILEI-coding sequence of Arg-55–Asp-227 (METDTLLLMVLLLWVPGSTGDKTHTCPPCPAPEFEGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPASIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGKGGGGSGGGGSGGGGSGGGGSAENLYFQ*[mILEI Arg-55–Asp-227].
Proteins were transiently expressed in the Expi293TM system (Thermo Fisher Scientific) and used the manufacturer's protocols and transfection reagents (ExpiFectamineTM reagent and enhancers 1 and 2). Cells were harvested 6 days after transfection. For the co-transfection experiments the two plasmids were transfected in a 1:1 ratio at a final concentration of 1.7 μg plasmid/ml cell culture at a cell density of 2 × 106 cells/ml.
Protein purification
Expressed proteins were affinity-purified from the supernatants as described in Johansson et al. (13). In brief: proteins from constructs C1, C2, and C3 were batch-purified on Ni-NTA (Qiagen) by incubating the supernatant (volume 1.6 liters) with the Ni-NTA matrix (15 ml of resin) at 4 °C overnight, and the bound protein was eluted with buffer containing 500 mm imidazole. After buffer exchange to 30 mm HEPES, 150 mm NaCl, 5% glycerol, pH 7.5, the His tag was removed by cleavage with TEV protease (22) at a ratio of 10:1 protein:TEV at 4 °C overnight. TEV protease, cleaved His tag, and uncleaved protein was removed on reverse His-Trap. The flow-through was purified in a final step by size exclusion chromatography using Superdex 75, aliquoted, and flash-frozen in LN2. For the purification of co-expressed mouse C3 and C4, the supernatant (500 ml) was first loaded onto a 1-ml HisTrap FF Crude column equilibrated in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 15 mm Imidazole. After extensive washing with the same buffer, His-tagged proteins were eluted with the buffer containing 300 mm imidazole. The sample was desalted on a desalting column and into a buffer containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl, and the eluted volume was adjusted to 14 ml. The mixture was incubated overnight at 4 °C with 1 ml of MabSelect Sure resin. The resin with bound protein was transferred to an empty column, and after extensive washing by gravity flow with the buffer, bound protein was eluted with 0.1 m glycine, pH 3.5, and 150 mm NaCl, and immediately pH was neutralized by the addition of 1 m Tris-HCl, pH 8, to the eluted fractions (9:1, eluate:neutralizing buffer). Eluted fractions were analyzed by SDS-PAGE, and the relevant fractions were pooled and further purified on a Superdex 75 HiLoad (16/600) column pre-equilibrated with 30 mm HEPES, pH 7.5, 150 mm NaCl, and 5% glycerol. After SDS-PAGE analysis, relevant SEC fractions were pooled and concentrated using an Amicon Ultra centrifugal filter (10-kDa cutoff filter, Millipore). The SEC pools before and after concentration were analyzed by non-reducing and reducing SDS-PAGE (supplemental Fig. S6B), and the concentrated (non-reduced) sample was analyzed by MS.
Liquid chromatography-mass spectrometry
A non-reduced sample (0.4 mg/ml) was diluted in 25 mm ammonium bicarbonate, pH 7.8, and cleaved by trypsin (Promega) overnight at 37 °C. The protein-enzyme ratio was 1:50. A nanoflow HPLC instrument (Easy nLC, Thermo Fisher Scientific) was coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific) with a nanoelectrospray ion source. The peptide mixture (4 μl) was loaded onto a precolumn (Acclaim Pep-Map C18, 3 μm, 100 Å, 75 μm × 2 cm, Thermo Fisher Scientific) with buffer A (0.2% formic acid in water) and separated on an EASY-Spray column (PepMap, C18, 3 μm, 100 Å, 75 μm × 15 cm, Thermo Fisher Scientific) with a linear gradient of 3–45% buffer B (0.2% formic acid in acetonitrile) at a flow rate of 300 nl/min over 30 min. The column was washed with 95% buffer B for 5 min. The Q Exactive instrument was operated in data-dependent mode to switch between full MS and MS/MS acquisition. The scan range was 300–2000 m/z, and the resolution was 70,000 with a target ion intensity of 1 × 106 or 400 ms maximum injection time. The four most intense ions per full scan were targeted for fragmentation (3 m/z isolation window and intensity threshold 8.3 × 103) with a target ion intensity of 1 × 105 or 60-ms maximum injection time at 17,500 resolution. Fragmentation was performed with a normalized collision energy of 28 with a dynamic exclusion of 30 s. The capillary temperature was set to 275 °C with S-Lens RF at 60.
Crystallization
Initial crystal hits for mouse ILEI monomer and dimer as well as human monomer were obtained in several conditions in the JBScreen Classic HTS I and II (Jena Bioscience) using vapor diffusion. Optimized crystals were obtained in the following conditions; mouse ILEI monomer at 30 mg/ml + 17.5% w/v PEG4000, 10% glycerol, and 82 mm (NH4)2SO4; mouse ILEI dimer at 10 mg/ml + 3.3 m (NH4)2SO4, 1% 2-methyl-2,4-pentanediol 100 mm MES, pH 6.5, human ILEI monomer at 30 mg/ml + 25% PEG monomer ether 550, 100 mm MES, pH 6.5, 10 mm ZnSO4. Drops for both screening and optimization were set up using a Mosquito LCP crystallization robot (TTP Labtech) at 0.1 μl of protein + 0.1 μl of reservoir solution. Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop.
Structure determination
X-ray diffraction data for the mouse ILEI monomer was collected at beamline ID23–1, European Synchrotron Research Facility (ESRF), Grenoble, France. The data were indexed and integrated using MOSFLM (23) and subsequently scaled with SCALA (24). An initial model of the mouse ILEI monomer was obtained with molecular replacement using the mouse FAM3B PANDER (PDB id 2YOP) as a search model in PHASER (25). The human ILEI monomer and the mouse ILEI dimer were subsequently determined using the mouse monomer structure as a PHASER search model. MOSFLM, SCALA, and PHASER were used as incorporated in the Collaborative Computational Project 4 (26). The structures were further refined by alternate cycles of model building in Coot (27) and refinement in AutoBuster (28). Complete data collection and refinement statistics can be found in Table 1. A search for structural neighbors was performed using DaliLite version 3 (14) and PDBeFold version 2.59 (15, 29). In both searches the human monomer structure was used as the query.
Cell culture and trans-well invasion assay
Murine EpRas cells (the immortalized murine mammary epithelial cell line EpH4 transformed with oncogenic Ras; Ref. 30) were cultured on plastic dishes in growth medium (a 1:1 mixture of Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 nutrient mixture (F-12) containing 4% FCS (Sigma) and 20 mm HEPES) and subcultured at a 1:3 ratio twice a week. For the invasion assay, cells were seeded into 6-well plates (50,000 cells/well). After 4 h, fresh growth medium was supplemented by mC3 ILEI monomer and dimer purified as described above at concentrations of 0.5, 1, 2, and 4 μg/ml in 30 mm HEPES, pH 7.5, 150 mm NaCl, and 5% glycerol and incubated for 48 h. Control cells were supplemented by recombinant human TGF-β1 (10 ng/ml; Peprotech). After pretreatment, cells were seeded into starvation medium (growth medium without FCS) containing the same factors as for the pretreatment into trans-well invasion chambers with an 8-μm pore size, coated with Matrigel (Corning), and equilibrated in starvation medium (25,000 cells/inlet, each condition in triplicate). The lower chamber had the same medium composition supplemented with 10% FCS in addition as attractant. Cells were allowed to invade for 24 h, and non-invaded cells were removed from the upper side of the chamber. The cells were fixed and stained with DAPI. Total numbers of invaded cells were counted using fluorescent microscopy imaging followed by ImageJ analysis.
ILEI dimer in solution; expression, purification, and mass spectrometry
To investigate whether the covalent trans-linkage between Cys-64 and Cys-221 of the two ILEI dimer chains could be observed also in solution, the mouse constructs C3 and C4 were co-expressed in suspension adapted HEK cells (Fig. 1). A purification protocol was developed to isolate chimeric C3:C4 ILEI dimers based on the His tag of C3 and the Fc domain of C4. The first step involved Ni-NTA to capture all ILEI variants containing a His tag (supplemental Fig. S6C, populations b, c, and d). In the second step Fc-linked ILEI monomer and/or dimer were captured on a MabSelect column. The MabSelect eluate was further purified by size exclusion chromatography. The majority of the fractions were analyzed by SDS-PAGE (supplemental Fig. S6, A and B). The results show that several forms of ILEI could be identified and purified using the His/Fc setup, indicated by arrows a–e in supplemental Fig. S6, A and B. A schematic representation of the different populations is found in supplemental Fig. S6C.
In the first Ni-NTA capture step, pure mC4 Fc-ILEI dimers were found in the Ni-NTA flow-through as expected (supplemental Fig. S6A, lanes 1 and 2, and C, population a). Due to the inherent dimeric nature of the Fc-domain, this band is likely a mC4 Fc-ILEI dimer resulting from dimerization of the two Fc domains. This is supported by the observation that no mC4 Fc-ILEI monomer was detected in the flow-through. mC3 His-ILEI monomer and dimer were captured in the Ni-NTA step (supplemental Fig. S6A, lanes 6 and 7, populations c and d). In addition, a high-molecular weight band was also captured on Ni-NTA, corresponding to a mC3–mC4 ILEI multimer complex (supplemental Fig. S6, A, lanes 6 and 7, and C, population b).
In the following MabSelect step, His-tagged ILEI monomer and dimer are seen in the flow-through (supplemental Fig. S6A, lanes 11 and 12, populations c and d). Elution from the MabSelect resin again suggested the presence of a chimeric mC3–mC4 multimer complex (supplemental Fig. S6A, lanes 16–17). This fraction was further purified on a size exclusion column, and the peak corresponding to the mC3–mC4 multimer was selected (supplemental Fig. S6B, lane 1), concentrated, and analyzed by non-reducing (supplemental Fig. S6B, lane 2) and reducing SDS-PAGE (supplemental Fig. S6B, lane 3). Reduced sample in supplemental Fig. S6B, lane 3 shows that the sample consists of both mC3 and mC4 constructs supporting that the cross-linking cysteine-bridge between the two ILEI molecules in the ILEI dimer is formed also in solution. Intensities of the bands in the reduced sample on the SDS gel suggest that the complex most likely consists of two mC4 Fc-ILEI molecules and one mC3 His-ILEI (supplemental Fig. S6C, population b–I). These results and the results from the mass-spectrometry analysis (below) indicate that only one trans-linked ILEI dimer could be formed using the Fc-dimer scaffold (supplemental Fig. S6C, population b-I). Consequently, an Fc-dimer with two ILEI dimers forming trans-linkage cysteine-bridges could not be isolated.
To further investigate the nature of the covalent ILEI dimer, the purified mC3–mC4 ILEI complex was analyzed by mass spectrometry. The chimeric mC3–mC4 dimer (supplemental Fig. S6B, lane 2) was cleaved into peptides by trypsin while maintaining a non-reducing environment. The peptides were subsequently analyzed by MS. Two modifications were introduced in the ILEI coding sequence in mC4 Fc-ILEI to be able to discriminate between the cis-linked Cys-64–Cys-221 disulfides of the monomer and the trans-linked disulfide seen in the dimer. No mutations were introduced in the mC3 His-ILEI sequence, and it is, therefore, identical to the wild-type (WT) ILEI sequence. The two modifications introduced in mC4 Fc-ILEI, A63G and E210D, were selected to avoid changes in the number of charges and size to minimally impact the structure. This meant that for a peptide with a Cys-64–Cys-221 cysteine bridge, detection of the loss of 14 Da compared with the WT ILEI sequence indicates an intermolecular cysteine bridge between mC4 Fc-ILEI and mC3 His-ILEI, whereas a loss of 28 Da compared with the WT ILEI sequence indicates an intramolecular cysteine bridge within mC4 Fc-ILEI. Both these peptide forms are observed in the MS spectrum of the sample (supplemental Fig. S7C), verifying the existence of the trans-linked cysteine bridge also in solution. As a consequence, these results also confirm the in-solution relevance of the observed β8-β9 domain swap. The presence of a third population is also seen in the mass spectrum, although at much lower intensity (supplemental Fig. S7C). This population has no shift in mass compared with WT ILEI sequence and corresponds to an intermolecular cysteine bridge from mC3 His-ILEI monomer. This is most likely a result from partial separation of the chimeric mC3–mC4 dimer during the experiment. All cysteine-linked peptides were further verified by MS/MS spectra.
Author contributions
A. M. J. and L. H.-S. designed the experiments. A. M. J. expressed, purified, and crystallized the proteins and solved the three structures. A. C. performed the cell studies. J. M. crystallized the protein. A.-C. N. performed the MS studies. E. A. purified the proteins. A. M. J. and P. J. wrote the paper with input from all other authors.
Supplementary Material
Acknowledgment
Gordon Leonard at the European Synchrotron Radiation Facility (ESRF) is acknowledged for assistance during remote data collection.
This work was supported by Discovery Sciences, AstraZeneca. This work was also supported by the Austrian Science Foundation (FWF) Grant P25834-B23) (to A. C.). A. M. J., J. M., A.-C. N., E. A., P. J., and L. H. S. are employees of AstraZeneca, which provided funding.
This article contains supplemental Figs. S1–S6.
The atomic coordinates and structure factors (codes 5LC2, 5LC3, and 5LC4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Kral, M., Klimek, C., Kutay, B., Timelthaler, G., Lendl, T., Neuditschko, B., Gerner, C., Sibilia, M., and Csiszar, A. (2017) Covalent dimerization of interleukin-like epithelial-to-mesenchymal transition (EMT) inducer (ILEI) facilitates EMT, invasion and late aspects of metastasis. FEBS J. 10.1111/febs.14207.
- EMT
- epithelial-to-mesenchymal-transition
- ILEI
- interleukin-like epithelial–mesenchymal transition inducer
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- AD
- Alzheimer's disease
- Aβ
- amyloid-β
- APP
- amyloid-β precursor protein
- CTF
- C-terminal fragment
- PS1
- presenilin-1
- Ni-NTA
- nickel-nitrilotriacetic acid
- SEC
- size exclusion chromatography
- NCT
- nicastrin domain
- APH-1
- anterior pharynx defective-1 domain
- TβRI
- TGF-β1 type I receptor
- TEV
- tobacco etch virus
- h-
- human
- m-
- mouse.
References
- 1. Zhu Y., Xu G., Patel A., McLaughlin M. M., Silverman C., Knecht K., Sweitzer S., Li X., McDonnell P., Mirabile R., Zimmerman D., Boyce R., Tierney L. A., Hu E., Livi G. P., Wolf B., Abdel-Meguid S. S., Rose G. D., et al. (2002) Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 80, 144–150 [DOI] [PubMed] [Google Scholar]
- 2. Waerner T., Alacakaptan M., Tamir I., Oberauer R., Gal A., Brabletz T., Schreiber M., Jechlinger M., and Beug H. (2006) ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239 [DOI] [PubMed] [Google Scholar]
- 3. Csiszar A., Kutay B., Wirth S., Schmidt U., Macho-Maschler S., Schreiber M., Alacakaptan M., Vogel G. F., Aumayr K., Huber L. A., Beug H. (2014) Interleukin-like epithelial-to-mesenchymal transition inducer activity is controlled by proteolytic processing and plasminogen-urokinase plasminogen activator receptor system-regulated secretion during breast cancer progression. Breast Cancer Res. 16, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao Z. H., Lu C., Wang Z. N., Song Y. X., Zhu J. L., Gao P., Sun J. X., Chen X. W., Wang M. X., Dong Y. L., and Xu H. M. (2014) ILEI: a novel target for Epithelial-Mesenchymal Transition and poor prognosis in colorectal cancer. Histopathology 65, 527–538 [DOI] [PubMed] [Google Scholar]
- 5. Lahsnig C., Mikula M., Petz M., Zulehner G., Schneller D., van Zijl F., Huber H., Csiszar A., Beug H., and Mikulits W. (2009) ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene 28, 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu Y.-H., Zhang B., Li M., Huang P., Sun J., Fu J., and Guan X.-Y. (2015) Prognostic significance of FAM3C in esophageal squamous cell carcinoma. Diagn. Pathol. 10, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Halberg N., Sengelaub C. A., Navrazhina K., Molina H., Uryu K., and Tavazoie S. F. (2016) PITPNC1 recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell 29, 339–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaudhury A., Hussey G. S., Ray P. S., Jin G., Fox P. L., and Howe P. H. (2010) TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hussey G. S., Link L. A., Brown A. S., Howley B. V., Chaudhury A., and Howe P. H. (2012) Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PLoS ONE 7, e52624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song Q., Sheng W., Zhang X., Jiao S., and Li F. (2014) ILEI drives epithelial to mesenchymal transition and metastatic progression in the lung cancer cell line A549. Tumour Biol. 35, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 11. Hasegawa H., Liu L., Tooyama I., Murayama S., and Nishimura M. (2014) The FAM3 superfamily member ILEI ameliorates Alzheimer's disease-like pathology by destabilizing the penultimate amyloid-β precursor. Nat. Commun. 5, 3917. [DOI] [PubMed] [Google Scholar]
- 12. Liu L., Watanabe N., Akatsu H., and Nishimura M. (2016) Neuronal expression of ILEI/FAM3C and its reduction in Alzheimer's disease. Neuroscience 330, 236–246 [DOI] [PubMed] [Google Scholar]
- 13. Johansson P., Bernström J., Gorman T., Oster L., Bäckström S., Schweikart F., Xu B., Xue Y., and Schiavone L. H. (2013) FAM3B PANDER and FAM3C ILEI represent a distinct class of signaling molecules with a non-cytokine-like fold. Structure 21, 306–313 [DOI] [PubMed] [Google Scholar]
- 14. Holm L., and Rosenström P. (2010) Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 16. Bai X.-C., Yan C., Yang G., Lu P., Ma D., Sun L., Zhou R., Scheres S. H. W., and Shi Y. (2015) An atomic structure of human γ-secretase. Nature 525, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolduc D. M., Montagna D. R., Gu Y., Selkoe D. J., and Wolfe M. S. (2016) Nicastrin functions to sterically hinder γ-secretase-substrate interactions driven by substrate transmembrane domain. Proc. Natl. Acad. Sci. U.S.A. 113, E509–E518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tesseur I., Zou K., Esposito L., Bard F., Berber E., Can J. V., Lin A. H., Crews L., Tremblay P., Mathews P., Mucke L., Masliah E., and Wyss-Coray T. (2006) Deficiency in neuronal TGF-β signaling promotes neurodegeneration and Alzheimer's pathology. J. Clin. Invest. 116, 3060–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caraci F., Battaglia G., Bruno V., Bosco P., Carbonaro V., Giuffrida M. L., Drago F., Sortino M. A., Nicoletti F., and Copani A. (2011) TGF-β1 pathway as a new target for neuroprotection in Alzheimer's disease. CNS Neurosci. Ther 17, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caraci F., Bosco P., Signorelli M., Spada R. S., Cosentino F. I., Toscano G., Bonforte C., Muratore S., Prestianni G., Panerai S., Giambirtone M. C., Gulotta E., Romano C., Salluzzo M. G., Nicoletti F., et al. (2012) The CC genotype of transforming growth factor-??1 increases the risk of late-onset Alzheimer's disease and is associated with AD-related depression. Eur. Neuropsychopharmacol. 22, 281–289 [DOI] [PubMed] [Google Scholar]
- 21. Stewart R., Thom G., Levens M., Güler-Gane G., Holgate R., Rudd P. M., Webster C., Jermutus L., and Lund J. (2011) A variant human IgG1-Fc mediates improved ADCC. Protein Eng. Des. Sel. 24, 671–678 [DOI] [PubMed] [Google Scholar]
- 22. van den Berg S., Löfdahl P. A., Härd T., Berglund H. (2006) Improved solubility of TEV protease by directed evolution. J. Biotechnol. 121, 291–298 [DOI] [PubMed] [Google Scholar]
- 23. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O., Vonrhein C., and Womack T. (2016) Buster version 2.11.6, United Kingdom Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 29. Krissinel E., and Henrick K. (2005) Computational Life Sciences: First International Symposium, CompLife 2005, September 25–27, 2005 (Berthold R., Glen M., Diederichs R. C., Kohlbacher O., and Fischer I, eds.) Springer Berlin Heidelberg, Berlin, Heidelberg [Google Scholar]
- 30. Oft M., Peli J., Rudaz C., Schwarz H., Beug H., and Reichmann E. (1996) TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10, 2462–2477 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.