Abstract
This is the first prospective study of the effects of human gut microbiota and metabolites on immune checkpoint inhibitor (ICT) response in metastatic melanoma patients. Whereas many melanoma patients exhibit profound response to ICT, there are fewer options for patients failing ICT—particularly with BRAF-wild-type disease. In preclinical studies, specific gut microbiota promotes regression of melanoma in mice. We therefore conducted a study of the effects of pretreatment gut microbiota and metabolites on ICT Response Evaluation Criteria in Solid Tumors response in 39 metastatic melanoma patients treated with ipilimumab, nivolumab, ipilimumab plus nivolumab (IN), or pembrolizumab (P). IN yielded 67% responses and 8% stable disease; P achieved 23% responses and 23% stable disease. ICT responders for all types of therapies were enriched for Bacteroides caccae. Among IN responders, the gut microbiome was enriched for Faecalibacterium prausnitzii, Bacteroides thetaiotamicron, and Holdemania filiformis. Among P responders, the microbiome was enriched for Dorea formicogenerans. Unbiased shotgun metabolomics revealed high levels of anacardic acid in ICT responders. Based on these pilot studies, both additional confirmatory clinical studies and preclinical testing of these bacterial species and metabolites are warranted to confirm their ICT enhancing activity.
Introduction
Immune checkpoint inhibitor therapy (ICT) achieves durable remissions in up to half of patients with metastatic melanoma [1]. However, a substantial number of patients fail to benefit from ICT, and others experience severe autoimmune adverse events including dermatitis, colitis, hepatitis, and hypophysitis [2]. In this setting, investigators have sought to identify host or tumor characteristics that impact ICT response. Tumor mutations, gene expression and protein expression, tumor-associated dendritic cells (DCs) and T-cell infiltration, and levels of circulating lymphocytes/monocytes/eosinophils are associated with ICT resistance and response [3], [4], [5], [6]. To date, no method reproducibly modulates these factors and increases ICT response.
There is mounting evidence that gastrointestinal tract bacteria, collectively known as the gut microbiota, can influence and modulate host immune responses [7], [8]. In preclinical mouse models, the composition of the host gut microbiota is a major factor determining ICT response [8], [9], [10]. Germ-free or antibiotic-treated tumor-bearing mice do not respond to immune therapy [8]. B16 melanoma–bearing mice treated with Bifidobacterium spp. show increased tumor DC antitumor immune gene expression and enhanced anti–PD-L1 immunotherapy response [9]. Furthermore, gut commensals Bacteroides thetaiotamicron or B. fragilis are necessary for anti-CTLA4 antibody anti-B16 melanoma in vivo efficacy [10]. DCs and T cells mixed with either of these Bacteroides species in vitro increased T-cell interferon γ production and in vivo tumor growth inhibition. In all the above studies, the gut bacteria induced maturation of anti-melanoma DCs and T cells.
Based on these observations, we initiated a study of metastatic melanoma patients initiating ICT. Patients were stratified for type of immunotherapy and response to ICT [Response Evaluation Criteria in Solid Tumors (RECIST) criteria]. Fecal samples were collected and analyzed for gut microbiota and metabolite composition. We report the ICT efficacy in 39 metastatic melanoma patients and correlate clinical responses with gut microbiota taxonomic profiles, gut metabolite levels, and patient dietary and antibiotic histories.
Materials and Methods
Patients and Samples
The study design was a single-site, correlative study of the effects of gut microbiota and metabolites on ICT efficacy in 39 adult melanoma patients. The study was approved by the University of Texas Southwestern Medical Center Institutional Review Board (STU 012016-056). The study was conducted in accordance with the Declaration of Helsinki. We enrolled patients with a histologic diagnosis of unresectable or metastatic melanoma that were scheduled to begin ICT and willing to collect stool specimens, store them in a freezer, and deliver them to our facility. In addition, patients had to have measurable disease by RECIST v1.1. Tumor sizes were evaluated within 4 weeks prior to beginning therapy by exams, CT scans, and/or MRIs. Therapy consisted of one of four ICT regimens: 1) outpatient ipilimumab 3 mg/kg IV every 3 weeks for four doses (I); 2) nivolumab 1 mg/kg IV with ipilimumab 3 mg/kg IV every 3 weeks for four doses followed by nivolumab alone at 240 mg IV every 2 weeks (IN); 3) nivolumab alone at 240 mg IV every 2 weeks (N); or 4) pembrolizumab alone at 2 mg/kg IV every 3 weeks (P). Repeat exams and scans were obtained every 2 to 3 months. Demographics, antibiotic use, and probiotic exposure were recorded for each patient. Patient histories were also interrogated for consumption of foods enriched for identified plant xenobiotic in excess in ICT nonprogressors based on metabolomic profiling. Patient response, stable disease, and progression were evaluated by RECIST v1.1 criteria as reported [11].
Fecal specimens were collected at patient's homes and immediately frozen, transferred to our clinic on ice, and immediately stored at −80°C until sample processing. Fecal gDNA was extracted as previously described [12], [13]. Briefly, ~ 200-mg fecal aliquots were suspended in 0.7 ml extraction buffer (200 mM NaCl, 200 mM Tris, 20 mM EDTA, 6% SDS) and 0.5 ml phenol-chloroform-isoamyl alcohol, pH 7.9 (Ambion). Cells were lysed by bead-beating with 0.1-mm diameter zirconia/silica beads (Biospec) subjected to additional phenol-chloroform-isoamyl alcohol extractions. Crude DNA extracts were treated with RNAseA (Qiagen) and column-purified (PCR Purification Kit, Qiagen). DNA was assayed for purity by spectroscopy, and DNA concentrations were quantified by a fluorescence-based assay (Quan-iT PicoGreen dsDNA, Life Technologies).
Metagenomic Shotgun Sequencing (MSS) and Analysis
MSS data (mean of 85,339,022 reads per sample; range 39,703,594 to 121,707,762 reads) were generated from sequencing fecal gDNA fragments from adult melanoma patients (n = 44; 39 samples prior to ICT therapy and 5 repeat samples within 1 month of starting ICT) on an Illumina HiSeq 2000 (100-bp pair-end reads) at the University of Texas Southwestern Medical Center Genomics Core Facility.
Taxonomic and functional analysis of MSS data was performed as previously described [13], [14]. Briefly, raw MSS data were quality controlled using NGS-QC (http://www.nipgr.res.in/ngsqctoolkit.html), and human sequences were removed with the NCBI BMTagger Human Contamination Screening Tool (ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/). Taxonomic composition was performed by using the computational tool MetaPhlAn [15]. Functional pathway abundance was calculated using HUMAnN [16] and FMAP [17]. The open-source software package QIIME [18] was used to measure the diversity indexes using the species-level MetaPhlAn profiles as input. Linear discriminate analysis coupled with effect size measurements (LEfSe) was used to quantitate differential taxonomic and functional pathway abundance between groups (responders versus nonresponders) as previously reported by our laboratory [13], [14].
Unbiased Gut Metabolomic Profiling with Ultrahigh Performance Liquid Chromatography–Tandem Mass Spectroscopy (UPLC-MS/MS)
Unbiased gut metabolomic profiling by UPLC-MS/MS was performed by Metabolon, Inc. (Durham, NC) as previously described [19]. Briefly, aliquots of the fecal samples used for MSS were sent to Metabolon; lyophilized; subjected to methanol extraction; and split into aliquots for analysis by UPLC-MS/MS in the positive (two methods), negative, and polar ion mode, followed by normalization to account for differential volume extracted. Compounds were identified by automated comparison to reference chemical library entries with subsequent visual inspection for quality control as previously described [20]. Peaks were quantified using area under the curve. For studies spanning multiple days, a data normalization step was performed to correct variation resulting from instrument interday tuning differences. For statistical analyses and data display, any missing values were assumed to be below the limits of detection; these values were imputed with the compound minimum (minimum value imputation). Standard statistical analyses (e.g., Welch's two sample t test) were performed in ArrayStudio (Omicsoft) on log-transformed data; P < .05 was considered significant. An estimate of the false discovery rate (q-value) was also calculated to take into account the multiple comparison that normally occur in metabolomics-based studies, with q < 0.05 used as an indication of high confidence in a result. The current analysis was restricted to the 1901 compounds of known identity (named biochemicals).
Data and Materials Availability
MSS data for this study have been deposited in the NCBI Sequence Read Archive: http://www.ncbi.nlm.nih.gov/sra/SRP115355.
Statistical Analyses
Comparison of alpha diversity metrics was analyzed by Mann-Whitney tests, and when multiple comparisons or more than two groups were analyzed, Bonferroni's correction to the significance level α was invoked. Hierarchical clustering was performed using R (version 2.1.2, package hclust). Wilcoxan signed-rank test was calculated in R (version 2.1.2). Statistical analyses were carried out using the GraphPad Prism Software (San Diego, CA) unless otherwise indicated.
Results
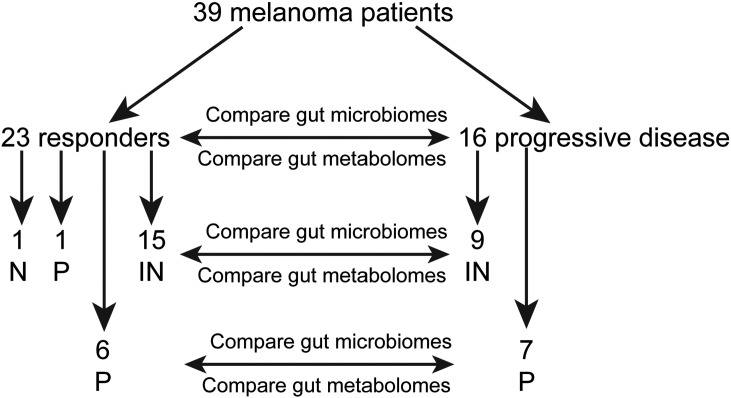
Thirty-nine metastatic melanoma patients were enrolled, consented, provided pretreatment fecal samples, underwent ICT, and had follow-up exams and scans (Table 1). Briefly, patients were predominantly male (n = 30, 77%). Median age was 68 years with a range of 37 to 92. Twenty-two patients had a single site of metastases: 11 lung, 4 nodes, 4 liver, 2 SQ, and 1 bone. Fourteen patients had two sites of metastases, including three lung/liver; three lung/nodes; three nodes/SQ; two nodes/bone; and one each with lung/SQ, SQ/adrenal, and lung/adrenal. And three patients had three sites of metastases: lung/liver/nodes, lung/nodes/SQ, and lung/liver/bone, respectively (Table 1). Twenty-four patients showed RECIST response (19, 49%) or stable (5, 13%) disease to ICT, what we classify as responders in this study, and 15 showed progression. Of those patients treated with IN alone, 16 (67%) were responders (14 RECIST response, 2 stable) and 8 (33%) showed progression. P treatment alone resulted in 6 (46%) responders (3 RECIST response, 3 stable). One patient each was treated with I alone and N alone; each was a responder (Table 2). The disease control rate (response plus stable disease) for both IN and P is consistent with prior studies: a 59% response rate and 13% stable disease rate in a Phase 2 study of IN [21] and a 44% response rate and 8% stable disease rate in a Phase 1 study of IN [22].
Table 1.
Summary of Clinical Characteristics of Melanoma Patients Who Underwent Immune Checkpoint Inhibitor Therapy at the University of Texas Southwestern Medical Center (n = 39)
| Dates of Therapy | 2016-2017 |
|---|---|
| Age (years) | 37-92; median 68 |
| Gender | Female, 9 (23%); Male, 30 (77%) |
| Ethnicity | 37 Caucasian (94%), 1 Hispanic (2%), 1 African-American (2%) |
| Number of metastatic sites | One, 23 (59%); Two, 13 (33%); Three, 3 (8%) |
| Metastases site | Adrenal, 3 (8%); Bone, 3 (8%); Liver, 7 (18%); lung, 21 (54%); Lymph nodes, 15 (38%); SQ, 8 (21%) |
| Antibiotic usage prior to and/or during ICT therapy | 3 (8%) |
| Probiotic therapy | 1 (3%) |
Table 2.
Individual Clinical Characteristics of Melanoma Patients Who Underwent Immune Checkpoint Inhibitor Therapy at the University of Texas Southwestern Medical Center (n = 39)
| Patient Identifier | Sex | Age | Site of Metastases | ICT Therapy | Change in Tumor Size (%) | RECIST Category |
|---|---|---|---|---|---|---|
| P7 | M | 63 | Lung | IN | −4 | Stable |
| P8 | M | 70 | Lung | IN | −55 | Response |
| P10 | M | 75 | Lung | IN | −83 | Response |
| P14 | F | 69 | Lung | IN | −60 | Response |
| P16 | M | 80 | Lung, Nodes | P | 53 | Progression |
| P17 | M | 68 | Nodes | N | −70 | Response |
| P22 | M | 64 | Lung, Liver | P | 81 | Progression |
| P23 | M | 76 | Lung | IN | −55 | Response |
| P24 | M | 44 | Nodes | IN | 136 | Progression |
| P25 | F | 60 | SQ | IN | −100 | Response |
| P28 | F | 68 | Lung, Liver | IN | 85 | Progression |
| P30 | M | 54 | Lung, Liver | IN | 100 | Progression |
| P32 | F | 57 | Nodes, Bone | IN | 100 | Progression |
| P33 | F | 74 | Nodes | P | −36 | Response |
| P34 | M | 57 | Liver | IN | −66 | Response |
| P35 | M | 63 | Nodes | IN | −30 | Response |
| P39 | M | 48 | Nodes, SQ | P | −68 | Response |
| P42 | M | 67 | SQ | P | 100 | Progression |
| P44 | F | 63 | Nodes | P | −27 | Stable |
| P45 | M | 43 | Lung, Nodes, SQ | P | −14 | Stable |
| P46 | M | 68 | SQ, Adrenal | IN | 116 | Progression |
| P48 | M | 86 | Lung | P | 0 | Stable |
| P49 | M | 84 | Lung, Liver, Nodes | P | 125 | Progression |
| P52 | M | 41 | Bone | IN | −100 | Response |
| P53 | M | 74 | Lung, Adrenal | IN | 100 | Progression |
| P54 | M | 79 | Nodes, SQ | IN | 100 | Progression |
| P55 | F | 37 | SQ, Adrenal | IN | −100 | Response |
| P56 | M | 66 | Lung, Nodes | P | 46 | Progression |
| P57 | M | 70 | Liver | I | −100 | Response |
| P58 | M | 52 | Nodes | P | −48 | Response |
| P59 | M | 78 | Lung | IN | −10 | Stable |
| P61 | M | 58 | Lung | IN | −40 | Response |
| P63 | M | 63 | Nodes, SQ | IN | −100 | Response |
| P64 | F | 77 | Lung, Liver, Bone | P | 100 | Progression |
| P65 | M | 69 | Liver | P | 131 | Progression |
| P66 | M | 80 | Lung, Nodes | IN | 61 | Progression |
| P67 | M | 83 | Lung | IN | −34 | Response |
| P68 | M | 92 | Lung | IN | −34 | Response |
| P69 | F | 55 | Lung | IN | −87 | Response |
I, ipilumumab; N, nivolmab; P, pembrolizumab.
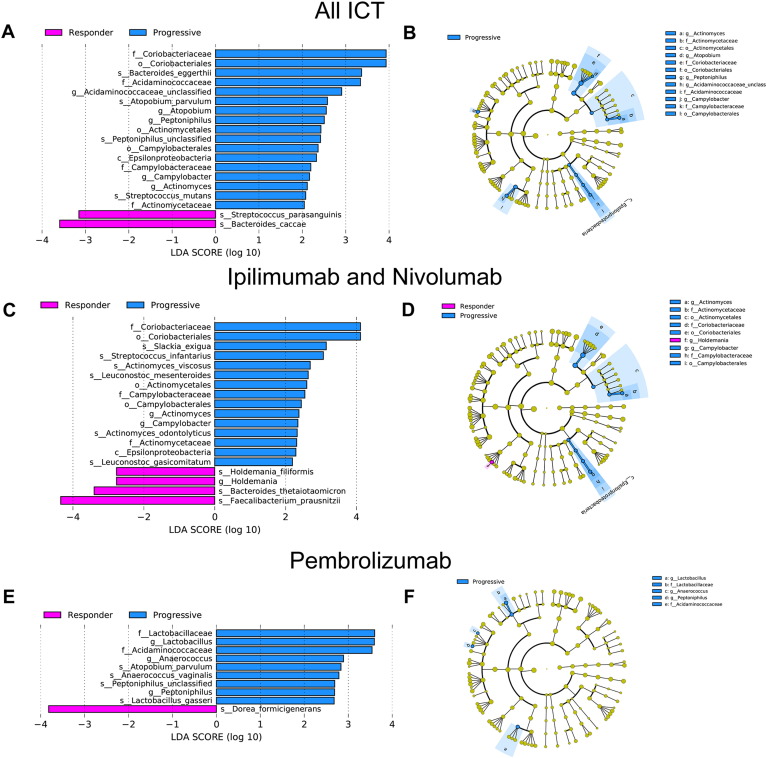
We performed MSS on patient fecal specimens collected prior to ICT to determine if we could detect significant differences in gut microbiota populations between responders and those with progressive disease (Figure 1). MSS has several advantages over the more commonly used 16S rRNA sequencing: 1) elimination of the PCR bias seen with 16S rRNA sequencing, where specific gut microbiota taxa can be either over- or underrepresented depending on the choice of primers and 16S rRNA variable region used for amplification [23], [24], [25]; 2) higher degree of gut microbiome taxonomic resolution, particularly at the species level, which is important since bacteria belonging to the same genus can exhibit significantly different phenotypes or effects on the host [12], [26]; and 3) insight into functional pathways, such as the metabolic potential of the microbiome. From a taxonomic standpoint, among all treated patients, ICT responder microbiomes were significantly enriched with B. caccae (P = .032; linear discriminant analysis coupled with effect size measurements, LEfSe; Kruskal-Wallis test) and Streptococcus parasanguinis (P = .048) compared to those with progression (Figure 2). Among those patients treated only with IN, responder microbiomes were enriched with the Firmicute phylum members Faecalibacterium prausnitzii (P = .032) and Holdemania filiformis (P = .043) and the Bacteroidetes phylum member Bacteroides thetaiotamicron (P = .046) (Figure 2). Among those patients treated only with P, responder microbiomes were enriched with Dorea formicigenerans (P = .045). (Figure 2). Interestingly, despite distinct gut microbiota signatures, overall gut microbiome diversity was not significantly different between responders and those with progressive disease (Supplemental Figure 1).
Figure 1.
Study schema.
Figure 2.
MSS identifies specific bacterial species that are enriched in the gut microbiomes of melanoma patients who are responding to ICT therapy. Relative abundance of gut bacterial taxa as determined by MetaPhlAn analysis of MSS data generated from fecal specimens collected from melanoma patients prior to receiving ipilimumab/nivolumab, pembrolizumab, ipilimumab alone, or nivolumab alone. Differential taxonomic abundance was analyzed by linear discriminate analysis coupled with effect size measurements (LEfSe) projected as a histogram (A, C and E) or cladrogram (B, D and F). All listed bacterial groups were significantly (P < .05, Kruskal-Wallis test) enriched for their respective groups (responder versus progressive).
We then reviewed two specific clinical characteristics that could have a significant effect on the gut microbiomes of our patients: antibiotic exposure and probiotic use. Only three patients on the study received systemic antibiotics immediately before or during the treatment course. Patient 7 (P7, responder) received a 2-week course of ceftriaxone prior to therapy. P22 (progression) received 2 weeks of ciprofloxacin, vancomycin, and metronidazole after two ICT cycles. P44 (responder) received a course of nitrofurantoin after four ICT cycles. Of these various antibiotic treatments, only metronidazole would have activity against (killed) the anaerobic commensals microbiota that were significantly enriched in responder microbiomes (Figure 2) [12], [27]. Unfortunately, we did not collect longitudinal fecal samples for P22 and could not ascertain whether the antibiotic therapy significantly depleted these bacterial species. In terms of probiotics, only one patient (P23) took daily doses of the probiotic Lactobacillus rhamnosus, including prior to ICT therapy. Of note, P23’s microbiome profile did not show any detectable L. rhamnosus. No specific clinical response or toxicity was significantly associated with antibiotic or probiotic use.
Since changes in an individual patient's gut microbiome over time can be associated with physiologic changes, as we have previously shown in other patient populations [13], [14], we performed repeat gut microbiota profiling on five patients (P7, responder; P14, responder; P23, responder; P33, responder; P54, progressive). Repeat samples were obtained within 1 month of starting ICT therapy. In general, the repeat sample clustered (in terms of gut microbiota abundance) with the initial sample, but specific gut microbiota abundances did change during this time (Supplemental Figure 2). Given the limited sample size, additional longitudinal studies will need to be done to determine whether changes in specific gut microbiota populations over the duration of ICT therapy are associated with clinical response.
MSS analysis also revealed differences in microbiome gene content (by identifying the presence/absence and calculating abundance of microbial functional pathways) between responder and progressive microbiomes. Among all ICT recipients, responder microbiomes were significantly enriched with bacterial enzymes involved in fatty acid synthesis (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway ko00061; P = .046; linear discriminant analysis coupled with effect size measurements, LEfSe; Kruskal-Wallis test). Among IN recipients, responder microbiomes were enriched with bacterial enzymes involved in inositol phosphate metabolism (KEGG pathway ko00562; P < .05; LEfSe; Kruskal-Wallis test).
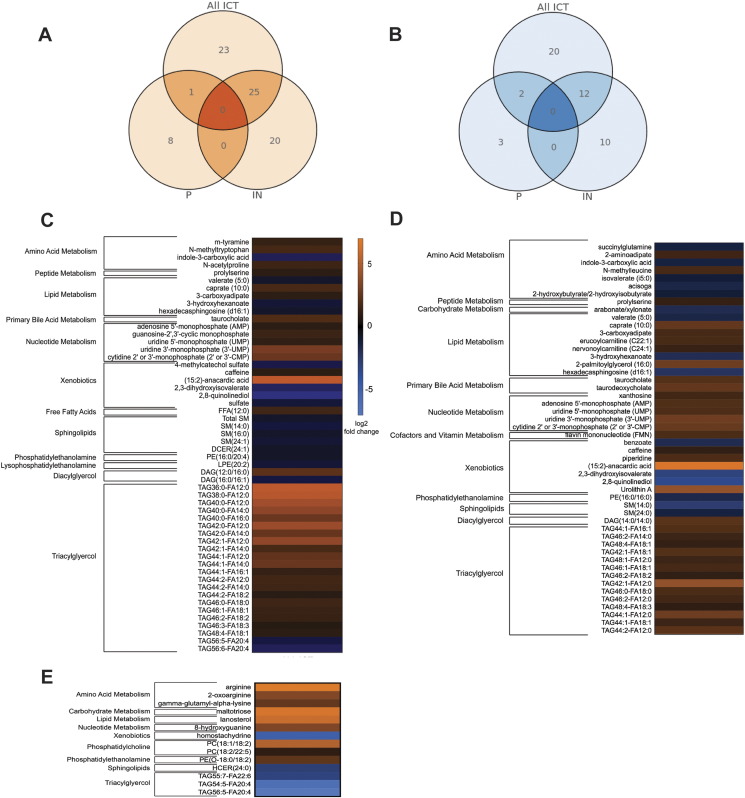
Gut microbiota–derived products, such as short-chain fatty acids, and inositol phosphates, can induce host physiologic changes (e.g., attenuation of colitis and anti-tumor effects) in the absence of bacteria [28], [29]. Hence, we performed unbiased shotgun metabolomic profiling on the same patient fecal specimens used for MSS to determine if we could detect significant differences in gut metabolites between responders and those with progressive disease. Of the 1901 evaluable compounds (known and named biochemicals), among all ICT recipients, 83 metabolites were significantly different when comparing the responder group to the progressive group (49 increased; 34 decreased; P < .05, Welch's two-sample t test). Among those patients treated only with IN, responder metabolomes were significantly enriched in 45 and depleted in 22 metabolites (P < .05, Welch's two-sample t test). Finally, among those patients treated only with P, responder metabolomes were significantly enriched in nine and depleted in five metabolites (P < .05, Welch's two-sample t test) (Figure 3, A and B).
Figure 3.
Unbiased metabolomics analysis of stool metabolites from adult melanoma patients prior to treatment with ICT. UPLC-MS–based global profiling of metabolites in feces of adult melanoma patients receiving immune checkpoint inhibitor therapy (n = 39). Data were log transformed and mean centered. Venn diagrams of metabolites (A) significantly increased or (B) decreased when comparing the ICT responder group versus the progressive group for all ICTs, IN only, and P only. The heat maps show the normalized relative abundances of stool metabolites comparing responders to those with progressive disease for (C) all ICTs, (D) IN only, and (E) P only (q < 0.05, unpaired t test with Welch's correction followed by false discovery rate correction). Orange colors indicate relative abundance increase, and blue indicates relative abundances decrease (responders:progressive, log2 transformed).
Significantly enriched and depleted metabolites in responder metabolomes were involved in numerous metabolic pathways (Figure 3, C and D). Inositol metabolomes were not significantly increased in IN responder metabolomes as our MMS functional analysis would have suggested. Inositol is known to be difficult to detect in unbiased metabolic screening assays, but targeted inositol mass spectrometry approaches can accurately measure inositol concentrations [30]. Thus, additional studies will be needed to determine if this metabolite is increased in IN responder metabolomes. Strikingly, 15:2 anacardic acid levels were markedly increased in all ICT (62-fold, P = .0077, Welch's two-sample t test) and IN (94-fold, P = .0288) responders. Anacardic acid is an alkyl derivative of salicylic acid and produced in the nutshell of cashews [31] and also in mangos. Interestingly, anacardic acids stimulate phagocytes and can augment bactericidal activity [32], [33]. Since anacardic acid is considered a xenobiotic and not known to be a bacterial byproduct or metabolite, we expanded our patient histories to include queries regarding regular consumption of plant-related products with high levels of anacardic acid including cashews. Of note, five of six patients with the highest 15:2 anacardic acid levels reported consuming cashews at least weekly.
Discussion
This is the first detailed report of human gut microbiome metagenomic and metabolomic profiling in melanoma patients treated with combination anti-PD1 and anti-CTLA4 immunotherapy (IN) as well as anti-PD1 therapy alone (P). Using a combination of MSS and unbiased metabolomics profiling, we were able to identify specific gut microbiota species and numerous gut metabolites that were associated with response to ICT therapy in all patients treated with ICT and also subsets of patients treated with the same regimen (IN and P).
Some data suggest that gut microbiota–induced immune effects are dependent on the specific type of cancer therapy administered [9], [10]. On the other hand, the gut commensal Eubacterium limosum is associated with protection against relapse in adult SCT patients [34] regardless of conditioning regimens and GVHD prophylaxis, suggesting that gut microbiota may have anticancer immune-augmenting effects that are therapy independent. Our findings of distinct gut microbes associated with human ICT response for IN and P are unique and consistent with results in preclinical models [9], [10]. We described four specific bacterial species involved in modulating ICT response. Bacteroides species have been previously reported to enhance anti-CTLA4 immune checkpoint efficacy in mice [10] and are presumed to directly contact and stimulate host DCs and T cells via pathogen-associated molecular patterns. We conjecture that the B. thetaiotamicron found to be enriched in anti-CTLA4 plus anti-PD1 responders in this study may work by a similar immune mechanism. Furthermore, we found a number of bacteria in the Firmicutes phylum (members of the Clostridiales order — Faecalibacterium prauznitzii, Holdemania filiformis, and D. formicigenerans) associated with anti-CTLA4 plus anti-PD1 clinical efficacy. Interestingly, a recent study also reported gut Faecalibacterium prausnitzii synergy with anti-PD1 clinically [35]. Interestingly, patients with increased Faecalibacterium and response also showed greater tumor CD8+ T-cell densities. Carbonnel and colleagues performed 16S rRNA gene sequencing of stool from melanoma patients receiving anti-CTLA4 antibody pretreatment and before each antibody infusion [36]. The bacterial diversity and species abundance were not altered by anti-CTLA4 therapy. Firmicutes including F. prausnitzii L2-6, butyrate-producing bacterium L2–21, and Gemmiger formicilis were enriched in responders.
Our snapshots of patient gut microbiomes pretreatment may not accurately reflect potential variations in taxome distribution over days to months of therapy. In our limited sampling of repeat testing on five patients, while there was evidence for relative stability of the microbiome based on matched hierarchical clustering of species abundances among the patient samples, there was also evidence that changes in composition (trending towards significant) were already ensuing. Thus, while an individual gut microbiome can remain stable for long periods of time, antibiotics [37] and diet changes [38] can rapidly alter the gut microbiome. Future studies of distal gut microbiota in ICT should include sequential monitoring to detect effects of ICT and the malignant disease.
The detailed molecular mechanism for immune enhancement by any of the bacteria in humans remains unknown. However, in vitro studies with DCs have implicated some of these bacterial species with immune modulation. B. thetaiotamicron releases 10- to 80-nm outer membrane vesicles (OMVs) that contain mucin-degrading glycosidase hydrolases and sulfatases [39]. These enzymes degrade the gut mucin and permit the OMVs to reach and be phagocytosed by DCs. The OMVs also contain toxins, adhesins, and enzymes that trigger DC activation. B. caccae and B. thetaiotamicron are anaerobic gram-negative organisms with surface lipopolysaccharide that stimulate DCs in a TLR4-dependent manner [40]. The five response-related bacteria merit in vitro testing with human peripheral blood mononuclear cells in mixed lymphocyte reactions. When bacterial species are combined with ipilimumab and nivolumab in mixed lymphocyte reactions, interleukin-2 and interferon-γ can be measured in the supernatant, providing an indirect assay of DC activation [41]. Furthermore, fulfilling Koch's postulates will require testing the proposed bacteria in antibiotic gut microbiome–depleted C57/BL6 mice with B16/F10 melanoma receiving nivolumab and ipilimumab, as has been successfully applied for testing cGAMP analogs [42].
There was no discernible effect of patient exposure to systemic antibiotics or probiotics, albeit the number of patients receiving antibiotics (3) and probiotics (1) was low. The lack of effect of the probiotic on P3 is consistent with a baseline stable commensal community that inhibits overgrowth of the probiotic organism [43], [44]. No particular response or toxicity was linked to antibiotic or probiotic exposure in this limited study.
In attempt to gain functional insight into changes in the gut microbiome, investigators have used metagenomic functional pathway analysis, but increased or decreased abundance of genes within a given microbiome is difficult to interpret. Gut metatranscriptomics are challenging given the lability of mRNA and difficulty in standardizing sample collection. By KEGG analysis, we observed increased inositol metabolism enzymes among IN responders. Greater patient numbers and accurate measurement of gut inositol levels may clarify the role and significance of inositol-related molecules, but there are multiple prior reports of inositol phosphates in innate immunity and anti-cancer activity [29], [45]. We performed unbiased gut metabolomics profiling in an attempt to gain greater functional insight. Surprisingly, among the 1901 evaluable metabolites, the most dramatic correlation with response was seen with a plant xenobiotic 15:2 anacardic acid. As noted previously, anacardic acids stimulate neutrophils and macrophages [32], [33]. Similar to the effects of particular bacterial species, the activation of macrophages/DCs may enhance T-cell recruitment to tumor metastases and, consequently, enhance ICT. In fact, anacardic acid has been shown to have antitumor effect in several preclinical models [46]. Furthermore, preclinical and clinical studies are warranted on this potentially simple therapeutic intervention.
While these preliminary observations do not establish a causal connection between gut microbiota/gut metabolite and ICT efficacy, we plan to pursue larger follow-up clinical studies and more detailed laboratory investigations. These studies may lay the foundation for optimizing the host response to ICT.
Acknowledgments
Acknowledgements
The authors thank the patients that both gave informed consent and were actively involved in sample collection.
Footnotes
Funding: This work was supported by the Roberta I. and Normal L. Pollock Fund (A.Y.K), the Melanoma Research Fund (A.E.F), Cancer Prevention and Research Institute of Texas (RP150596), T. Boone Pickens Cancer Research Fund (A.E.F.), Cancer Prevention and Research Institute of Texas(RP150596)US National Institute of Health (NIH) grant P30CA142543 (Y.X.), NIH grant 5R01CA152301 (Y.X.), and NIH grant R01CA172211 (Y.X.)
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2017.08.004.
Appendix A. Supplementary data
Supplementary Figures.
References
- 1.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvat TZ, Adel NG, Dang TO, Momtaz P, Postow MA, Callahan MK, Carvajal RD, Dickson MA, D'Angelo SP, Woo KM. Immune-related adverse events, need for systemic immunosuppression, and effects on survival and time to treatment failure in patients with melanoma treated with ipilimumab at Memorial Sloan Kettering Cancer Center. J Clin Oncol Off J Am Soc Clin Oncol. 2015;33:3193–3198. doi: 10.1200/JCO.2015.60.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankel AE, Frankel EP. Melanoma metamorphoses: advances in biology and therapy. J Cancer Sci Ther. 2017;9:325–335. [Google Scholar]
- 4.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, Kaisho T, Bogunovic D, Bhardwaj N, Krummel MF. Critical role for CD103(+)/CD141(+) dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell. 2016;30:324–336. doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salerno EP, Bedognetti D, Mauldin IS, Deacon DH, Shea SM, Pinczewski J, Obeid JM, Coukos G, Wang E, Gajewski TF. Human melanomas and ovarian cancers overexpressing mechanical barrier molecule genes lack immune signatures and have increased patient mortality risk. Oncoimmunology. 2016;5:e1240857. doi: 10.1080/2162402X.2016.1240857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21:808–814. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simms-Waldrip TR, Sunkersett G, Coughlin LA, Savani MR, Arana C, Kim J, Kim M, Zhan X, Greenberg DE, Xie Y. Antibiotic-induced depletion of anti-inflammatory Clostridia is associated with the development of graft-versus-host disease in pediatric stem cell transplantation patients. Biol Blood Marrow Transplant. 2017;23:820–829. doi: 10.1016/j.bbmt.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Piper HG, Fan D, Coughlin LA, Ho EX, McDaniel MM, Channabasappa N, Kim J, Kim M, Zhan X, Xie Y. Severe gut microbiota dysbiosis is associated with poor growth in patients with short bowel syndrome. JPEN J Parenter Enteral Nutr. 2016 doi: 10.1177/0148607116658762. [DOI] [PubMed] [Google Scholar]
- 15.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Kim MS, Koh AY, Xie Y, Zhan X. FMAP: functional mapping and analysis pipeline for metagenomics and metatranscriptomics studies. BMC Bioinformatics. 2016;17:420. doi: 10.1186/s12859-016-1278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS One. 2016;11:e0152126. doi: 10.1371/journal.pone.0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Chem. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodi FS, Chesney J, Pavlick AC, Robert C, Grossmann KF, McDermott DF, Linette GP, Meyer N, Giguere JK, Agarwala SS. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol. 2016;17:1558–1568. doi: 10.1016/S1470-2045(16)30366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy K, Hall MW, Lynch MD, Moreno-Hagelsieb G, Neufeld JD. Evaluating bias of illumina-based bacterial 16S rRNA gene profiles. Appl Environ Microbiol. 2014;80:5717–5722. doi: 10.1128/AEM.01451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay J, Singh K, Fern A, Kirton ES, He S, Woyke T, Lee J, Chen F, Dangl JL, Tringe SG. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771. doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3:463–467. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vucenik I, Shamsuddin AM. Protection against cancer by dietary IP6 and inositol. Nutr Cancer. 2006;55:109–125. doi: 10.1207/s15327914nc5502_1. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Villalta P.W., Sturla S.J. Simultaneous determination of inositol and inositol phosphates in complex biological matrices: Quantitative ion-exchange chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:705–712. doi: 10.1002/rcm.3923. [DOI] [PubMed] [Google Scholar]
- 31.Morais S.M., Silva K.A., Araujo H., Vieira I.G., Alves D.R., Fontenelle R.O., Silva A.M. Anacardic acid constituents from cashew nut shell liquid: NMR characterization and the effect of unsaturation on its biological activities. Pharmaceuticals. 2017;10(1):31. doi: 10.3390/ph10010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gnanaprakasam J.N., Estrada-Muniz E., Vega L. The anacardic 6-pentadecyl salicylic acid induces macrophage activation via the phosphorylation of ERK1/2, JNK, P38 kinases and NF-kappaB. Int Immunopharmacol. 2015;29:808–817. doi: 10.1016/j.intimp.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 33.Hollands A, Corriden R, Gysler G, Dahesh S, Olson J, Raza Ali S, Kunkel MT, Lin AE, Forli S, Newton AC. Natural product anacardic acid from cashew nut shells stimulates neutrophil extracellular trap production and bactericidal activity. J Biol Chem. 2016;291:13964–13973. doi: 10.1074/jbc.M115.695866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, Ling L, Kosuri S, Maloy M, Slingerland JB. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35(15):1650–1659. doi: 10.1200/JCO.2016.70.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopalakrishnan VS, Christine, Reuben Alexandre, Karpinets Tatiana, Hutchinson Diane, Prieto Peter A, Tetzlaff Michael T, Lazar Alexander, Davies Michael A. Association of diversity and composition of the gut microbiome with differential responses to PD-1 based therapy in patients with metastatic melanoma. 2017 ASCO-SITC Clinical Immuno-Oncology Symposium; Orlando, FL, USA. J Clin Oncol. 2017;35 [Google Scholar]
- 36.Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 37.Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezow AB, Ernst RK, Coats SR, Braham PH, Karimi-Naser LM, Darveau RP. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb Pathog. 2009;47:68–77. doi: 10.1016/j.micpath.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selby MJ, Engelhardt JJ, Johnston RJ, Lu LS, Han M, Thudium K, Yao D, Quigley M, Valle J, Wang C. Preclinical development of ipilimumab and nivolumab combination immunotherapy: mouse tumor models, in vitro functional studies, and cynomolgus macaque toxicology. PLoS One. 2016;11:e0161779. doi: 10.1371/journal.pone.0161779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, Chen ZJ. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci U S A. 2017;114:1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 44.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lou HR, Mondal S. Molecular control of PtdIns(3,4,5)P3 signaling in neutrophils. EMBO Reports. 2015;16(2):149–163. doi: 10.15252/embr.201439466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemshekhar M. M Sebastin Santhosh, K Kemparaju and KS Girish, Emerging roles of anacardic acid and its derivatives: a pharmacological overview. Basic Clin Pharmacol Toxicol. 2012;110:122–132. doi: 10.1111/j.1742-7843.2011.00833.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures.