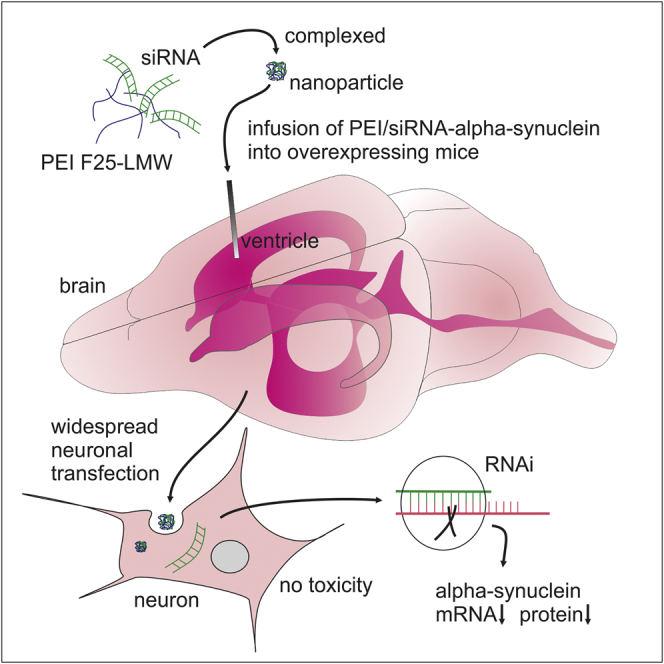
Abstract
RNA interference (RNAi)-based strategies that mediate the specific knockdown of target genes by administration of small interfering RNAs (siRNAs) could be applied for treatment of presently incurable neurodegenerative diseases such as Parkinson’s disease. However, inefficient delivery of siRNA into neurons hampers in vivo application of RNAi. We have previously established the 4–12 kDa branched polyethylenimine (PEI) F25-LMW with superior transfection efficacy for delivery of siRNA in vivo. Here, we present that siRNA complexed with this PEI extensively distributes across the CNS down to the lumbar spinal cord after a single intracerebroventricular infusion. siRNA against α-synuclein (SNCA), a pre-synaptic protein that aggregates in Parkinson’s disease, was complexed with PEI F25-LMW and injected into the lateral ventricle of mice overexpressing human wild-type SNCA (Thy1-aSyn mice). Five days after the single injection of 0.75 μg PEI/siRNA, SNCA mRNA expression in the striatum was reduced by 65%, accompanied by reduction of SNCA protein by ∼50%. Mice did not show signs of toxicity or adverse effects. Moreover, ependymocytes and brain parenchyma were completely preserved and free of immune cell invasion, astrogliosis, or microglial activation. Our results support the efficacy and safety of PEI nanoparticle-mediated delivery of siRNA to the brain for therapeutic intervention.
Keywords: RNA interference, nanoparticles, intracerebroventricular, α-synuclein, mouse model, Parkinson’s disease, transfection
Graphical Abstract
Introduction
Parkinson’s disease (PD), the most common movement disorder, is characterized by a progressive loss of dopaminergic neurons in the substantia nigra1 that coincides with α-synuclein (SNCA) pathology in the midbrain.2 PD is a severely debilitating, eventually fatal, and presently incurable disease. Available symptomatic treatments with dopamine precursors do not change disease progression and fail to improve many motor and non-motor symptoms.3, 4 Consequently, novel disease-modifying treatments for PD are highly warranted.
While the cause of neurodegeneration in PD remains elusive, neuroprotective strategies require the exploration of therapeutic targets that are present early in disease pathogenesis. An intriguing target is SNCA, a pre-synaptic protein which self-associates into toxic oligomers. It aggregates in intraneuronal inclusion bodies in PD and in several other incurable neurodegenerative diseases termed synucleinopathies.5, 6, 7 Our previous studies in a well-established animal model of PD, based on the overexpression of human wild-type SNCA under the Thy-1 promotor (Thy1-aSyn),8 revealed increased transcript levels of genes involved in pro-apoptotic pathways in nigral dopaminergic neurons and their target region prior to dopamine loss.9, 10 Thus, reduction of expression of SNCA or its pathological downstream effectors represents a promising therapeutic strategy.4 Specific reduction of gene expression can be achieved by using synthetic non-coding small interfering RNAs (siRNAs) against the target mRNA, thereby taking advantage of triggering endogenous RNA interference (RNAi). However, efficient delivery of siRNA into neurons in vivo remains challenging due to biological barriers, degradation, low transfection efficacy, and insufficient distribution.11, 12 Virus-mediated expression of siRNA is a potential option, but disadvantages include the local restriction of the effect to the injection site, difficulties in titrating dose response of RNAi, issues with regard to drug standardization and safety, and potential toxicity.12, 13, 14 RNAi-based therapeutic approaches in the brain by siRNA delivery have not been extensively explored so far, also due to poor transfectability of neuronal cells. Given the potentially high relevance of SNCA, we aimed at knocking down human SNCA, which to the best of our knowledge has not been explored so far in a transgenic model of PD using PEI as a non-viral delivery system.
The knockdown of the expression of a given target gene using unmodified, unformulated “naked” siRNAs is not feasible because of their instability, quick degradation, and low penetration of cellular membranes.15, 16 The polyethylenimine (PEI) complexation of siRNA into PEI/siRNA nanoparticles provides a non-viral nucleic acid delivery platform that has been shown efficacious in other pathological models, especially in tumors.17, 18 PEIs are positively charged polymers that form non-covalent complexes with nucleic acids, thus protecting siRNAs from degradation, mediating cellular uptake, and efficiently promoting lysosomal protection and escape into the cytoplasm.19, 20 We have previously established the 4–12 kDa branched PEI F25-LMW, a low-molecular weight PEI with superior transfection efficacy and low toxicity, for delivery of small RNA molecules in vivo and in vitro.21, 22, 23, 24 While very effective and safe for targeting diverse tissues in vivo, we have now extended their application toward neuronal targeting in a therapeutically relevant model of neurodegeneration.
Thus, the aim of this study was to develop a novel therapeutic strategy to silence the neuronal expression of a relevant candidate gene in the brain in vivo by using PEI nanoparticle-mediated delivery of siRNA into the cerebrospinal fluid. In the context of a proof-of-concept study with therapeutic implication, we silenced SNCA in the Thy1-aSyn transgenic mouse model of PD.8, 25, 26 Our approach includes analyses of knockdown efficacy and biocompatibility/absence of toxicity of the PEI/siRNA complexes (1) in vitro in the neuroblastoma cell line SH-SY5Y and (2) in vivo after intracerebroventricular (ICV) injection in Thy1-aSyn mice. Efficacy was determined by specific mRNA knockdown of human SNCA and the verification at protein level.
Results
Confirmation of Cellular Uptake of PEI/siRNA Complexes In Vitro
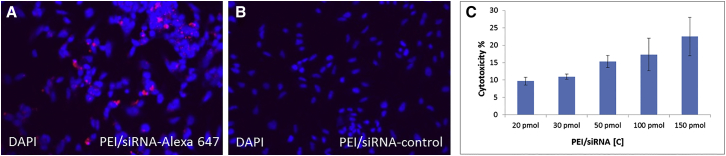
In order to test in vitro transfection efficiency, PEI F25-LMW/siRNA complexes were analyzed in human neuroblastoma cell line SH-SY5Y (Figures 1A–1C). SH-SY5Y cells seeded in 24-well plates on glass were transfected using 50 pmol Alexa-647-coupled control PEI/siRNA (QIAGEN) as a marker for cellular uptake. Punctuate staining patterns indicate transfection (Figure 1A), whereas negative controls incubated with PEI-complexed unlabeled siRNA show no fluorescent signal and no background (Figure 1B). Cells were treated with PEI/siRNA against SNCA or an unrelated siRNA (without a target in vertebrate genome) as negative control (against luciferase). Cytotoxicity of PEI/siRNA complexes was examined in SH-SY5Y cells by lactate dehydrogenase (LDH) assay (Figure 1C). SH-SY5Y cells seeded in 12- or 24-well plates were transfected by the addition of SNCA or control PEI F25-LMW/siRNA complexes (20, 30, 50, 100, and 150 pmol of siRNA/well). Cytotoxicity was 9.7%, 11.0%, 15.3%, 17.3%, and 22.5%, respectively, after 24 hr of incubation in the presence of 10% fetal calf serum in the medium. Morphological changes in cell body size and shape, staining of the stroma, as well as formation of dendrites, were absent (data not shown).
Figure 1.
Confirmation of Neuronal Uptake of PEI/siRNA Complex In Vitro
(A and B) Fluorescence microscopy with LSM microscope of (A) uptake of 50 pmol PEI/siRNA-Alexa 647 (red) and (B) uptake of 50 pmol PEI/siRNA-control in SH-SY5Y (undifferentiated). DAPI, 4′,6-diamidino-2phenylindole (blue nuclei, 20× magnification). (C) Concentration-dependent cytotoxicity of PEI/siRNA-control in SH-SY5Y determined by lactate dehydrogenase assay (Promega; mean ± SD, n = 2; 3 replicates each).
Dose Dependence of Toxicity of PEI/siRNA Application In Vivo
As DNA transfection and siRNA-mediated RNAi efficacies do not necessarily correlate,27 PEI F25-LMW/siRNA complex application in vivo is essential to analyze gene silencing potency of the delivery system. Therefore, we used male Thy1-aSyn mice, which overexpress the human wild-type SNCA in neurons.
First, by unilateral ICV injection (Figures 2A and 2B) of 0.125–2.5 μg PEI/Luc-siRNA in transgenic mice (n = 1–2 mice/dose), we determined the highest dosage of siRNA complexed with PEI F25/LMW that did not exert acute toxic effects. At doses of 0.75 μg Luc-siRNA/2.5 μL or lower, no adverse effects were observed up to 5 days post-injection in any of the injected mice. All mice showed normal weight gain after surgery and no decline in general health. Dosages above 0.75 μg resulted in dose-dependent delayed recovery after surgery and lower weight gain. At the highest dose of 2.5 μg, mice showed moderate ataxia, circling, and increased breathing rate during the first hour after surgery. Gross pathological examinations did not reveal any overt lesions or bleeding in brain tissue, lungs, or other organs, which otherwise would have supported a specific toxic effect of the infused siRNA at higher dosages. Subsequently, PEI F25-LMW/SNCA-siRNA complexes were applied ICV at several dosages ranging from 0.125 to 0.75 μg in 2.5 μL (n = 1–2 mice/dose). PEI-complexed SNCA-siRNA at these dosages was well tolerated by the mice and did not affect general health or weight gain.
Figure 2.
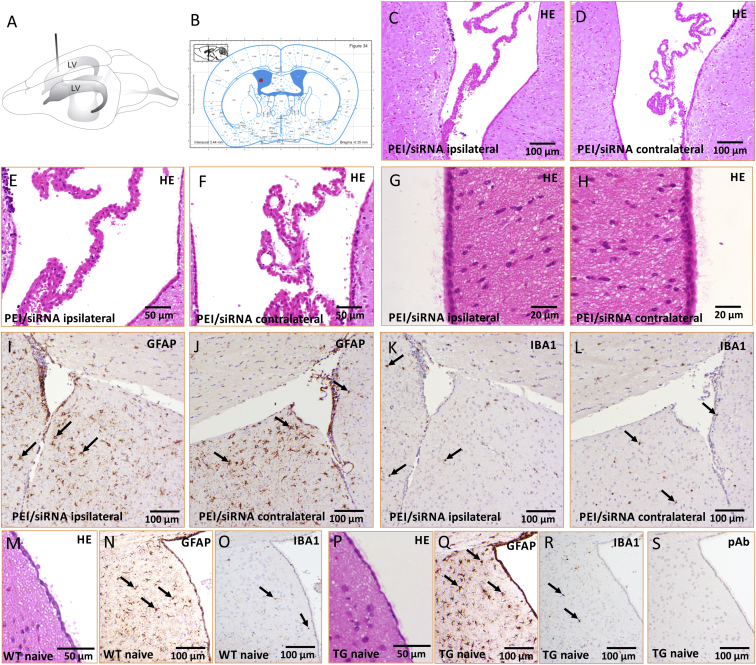
ICV Infusion of PEI/siRNA Does Not Affect the Integrity of Ependymal Cells and Brain Parenchyma
(A) Drawing of a mouse brain with localization of lateral ventricles (LV) and the infusion cannula. (B) Schematic of coronal mouse brain section (Paxinos and Franklin64) at distance to bregma −0.35 mm and localization of infusion cannula. (C–H) Light microscopic images (HE staining) after injection of 0.75 μg PEI/siRNA of a representative coronal brain section ipsilateral (C, E, and G) and contralateral (D, F, and H) to the infusion site (distance to bregma –0.4 mm). The ventricles (C and D), the choroid plexus (E and F), and the ventricular ependymocytes with cilia and microvilli (G and H) were intact without alterations. There was no infiltration of immune cells/inflammatory cells within the ventricle or the adjacent brain parenchyma. The adjacent parenchyma of ipsilateral and contralateral ventricles were devoid of overt astrogliosis (GFAP; I and J) and microglial proliferation or activation (IBA1; K and L). (M–R) Representative corresponding images of naive wild-type controls (M–O) and naive Thy1-aSyn transgenics (TG; P–R). Arrows indicate positive cells. (S) Negative control for immunostaining, primary antibodies were replaced by a rabbit non-immune serum (polyclonal antibodies, pAb) diluted in the same concentration as the primary antibodies.
In order to confirm absence of toxicity, we next performed pathomorphological investigations on brains derived from two transgenic mice injected with the two higher of the well-tolerated dosages (0.5 μg and 0.75 μg, respectively) of PEI/SNCA-siRNA complexes. Brain tissue from a naive untreated transgenic mouse and a corresponding wild-type littermate at same age and gender was used as control. As shown in Figures 2C–2L, there were no signs of degeneration or inflammation after unilateral ICV injection of PEI F25-LMW/siRNA. In particular, the ventricular ependymocytes as well as the choroid plexus were intact without alterations (Figures 2C–2F), and the brain parenchyma showed no evidence of tissue degeneration or inflammation compared to the control. Specifically, the integrity of ependymocytes, including their microvilli and cilia, was preserved after nanoparticle injection (Figures 2G and 2H). We observed no migration of immune cells into brain parenchyma of PEI/siRNA-injected mice and no overt signs of astrogliosis or microglial activation (Figures 2I–2L) in comparison to naive transgenic and wild-type control mice (Figures 2M–2R). Negative control for immunostaining is shown in Figure 2S.
PEI/siRNA Complexes Distribute across the CNS
Penetration of therapeutically active siRNA into target cells is essential for efficient induction of RNAi. To assess in vivo distribution of PEI/siRNA complexes across brain and spinal cord, tissues were sectioned 5 days after ICV injection of fluorescence-labeled PEI/siRNA complexes at different dosages (0.125 μg [n = 1], 0.75 μg [n = 2], 2.5 μg [n = 1]) and qualitatively evaluated for the presence of fluorescence signals. The highest fluorescence intensity was found around the ventricles, confirming successful ICV injection and penetration of PEI/siRNA complexes through ependymocytes (Figure 3A). A single injection of all evaluated dosages resulted in a consistently robust fluorescent staining around neurons (NeuN-positive cells) across the CNS as shown in Figure 3B for the olfactory bulb, the striatum, the cerebral cortex, the brain stem, and the lumbar spinal cord at the dose of 0.75 μg. Importantly, siRNA was also present in dopaminergic neurons (tyrosine hydroxylase [TH] positive) of the substantia nigra pars compacta (Figure 3C). Striatal sections from mice infused with unlabeled PEI/siRNA did not exhibit fluorescence signal in the far-red spectrum (negative control, Figure 3D).
Figure 3.
Distribution of PEI/siRNA Complexes across the CNS
PEI-complexed negative control siRNA labeled with fluorescence dye Alexa 647 (red) 5 days after a single ICV administration of 0.75 μg. DAPI for nuclei staining (blue), NeuN as neuronal marker, and TH as marker for dopaminergic neurons (green) were used to visualize cellular uptake. Confocal microscopy images from coronal brain regions reveal the respective fluorescence dye in and around both lateral ventricles (A; scale bar 40 μm), close to nuclei of neurons (B; scale bars 20 μm) across the CNS from the olfactory bulb to the lumbar spinal cord, and in TH-positive neurons of the substantia nigra (C; scale bar 10 μm). White arrows point at PEI-siRNA granules. (D) Negative control, striatal section of mouse infused with unlabeled PEI/siRNA does not exhibit signal in the far-red spectrum, and omission of the primary antibody resulted in signal loss in the Alexa 488 channel (scale bar 20 μm).
Dose-Dependent Knockdown of SNCA mRNA and Protein by PEI/siRNA Complexes in Brain
In order to determine if non-toxic dosages of specific siRNA, complexed with PEI F25-LMW, are sufficient to decrease SNCA mRNA, Thy1-aSyn mice received unilateral ICV injections of PEI/siRNA complexes against SNCA or unrelated siRNA against luciferase (negative controls). Control siRNA was chosen based on the absence of sequence homologies with the target gene or with any other vertebrate gene. SNCA mRNA levels in the dissected striatum were determined by qRT-PCR (Figures 4A and 4B). Single ICV injection of PEI F25-LMW/siRNA (0.75 μg) complex against SNCA resulted in a 67% knockdown of SNCA mRNA (p = 0.003, Mann-Whitney U rank-sum test, n = 8) compared to negative controls (n = 5). In contrast, the low dose of 0.125 μg PEI/siRNA complexes did not show significant effects on mRNA expression compared to the control group (Figure 4B).
Figure 4.
Knockdown of SNCA mRNA and Protein in Thy1-aSyn Mice by a Single ICV Injection of PEI/SNCA-siRNA Nanoparticles
(A and B) SNCA mRNA expression in the striatum, 5 days after ICV administration of PEI/siRNA (against SNCA or luciferase (Luc) as control). (A) Schematic of coronal mouse brain section (bregma 0.01 mm; Paxinos and Franklin64) with location of dissected striatal tissue highlighted in red (about 400 μm rostral to infusion site). (B) Relative quantification of SNCA mRNA in tissue of the whole striatum; values normalized to respective PEI/Luc control. PEI complexed SNCA-siRNA (gray bars) at 0.75 μg (n = 8; right) but not at 0.125 μg (n = 8; left) reduced SNCA mRNA expression by 67% compared to respective control (white bars, Luc-siRNA at 0.75 μg, n = 5 and 0.125 μg, n = 9). Mean % + SEM, **p = 0.003 Mann-Whitney U rank-sum test. (C) Schematic of coronal mouse brain section (bregma 0.37 mm; Paxinos and Franklin64). Red squares depict locations of microscopic image acquisition for SNCA protein expression quantification (about 800 μm rostral to infusion site, same animals as used for mRNA quantification). (D–J) Quantification of SNCA IHC mean pixel fluorescence intensity in medial (med) and lateral (lat) striatum (STR), cortex (CX), medial septum (MS), and corpus callosum (CC). (D) PEI complexed SNCA-siRNA at 0.75 μg (gray bars) significantly reduced SNCA protein expression compared to control (white bars) in all examined brain regions except corpus callosum, which only shows a little SNCA overexpression. Mean % + SEM, n = 4/group, **p < 0.01 compared to respective control, #p < 0.05 compared to other regions in controls, two-way RM ANOVA, Holm-Sidak. (E–J) Fluorescence microscopy images of sections stained for SNCA of one mouse infused with control PEI/Luc-siRNA 0.75 μg, which shows high expression of SNCA (E and H), one mouse infused with PEI/SNCA-siRNA 0.75 μg with overt knockdown of SNCA (F and I), and IHC negative control where the primary antibody was omitted (G and J). Scale bar is 40 μm. Quantification was performed on the complete region as covered by the presented images. Confirmation of SNCA protein knockdown in western blot analysis of samples from the motor cortex contralateral to the infusion site of the same mice used for immunohistochemical analysis (K). On the right side, a blot with bands corresponding to SNCA or actin protein is shown for respective treatment groups. Bar graph on the left side shows densitometric quantification (OD, optical density) normalized to actin. Mean + SEM, n = 4/group, *p < 0.05, Student’s t test, compared to control.
Knockdown of SNCA mRNA upon treatment of mice with 0.75 μg PEI-complexed SNCA-siRNA also resulted in less SNCA protein, as determined by immunohistochemistry (IHC) analysis of SNCA fluorescence staining intensity in several brain regions (Figures 4C–4J). The primary antibody detects both mouse and human SNCA, but due to several-fold overexpression of human SNCA in transgenic mice, quantification largely reflects the transgenic protein.8 About 1 mm rostral to the ICV infusion site, knockdown of SNCA protein was up to 50% in both hemispheres in the striatum, cortex, and medial septum, further supporting that PEI-complexed SNCA-siRNA effectively distributed from the lateral ventricle across the brain. Specifically, we first used paired t tests in order to compare ipsilateral and contralateral regions. As expected, no differences between hemispheres were found in PEI/Luc-siRNA controls. In PEI/SNCA-siRNA-infused mice, significantly less SNCA protein expression was determined in the medial (p = 0.018, 31%) and lateral (p = 0.047, 13%) striatum ipsilateral to the ICV injection site compared to contralateral (Figure S1A), while cortex and corpus callosum were not different between hemispheres (paired Student’s t test). ANOVA across treatment groups (PEI/SNCA-siRNA versus PEI/Luc-siRNA) and brain regions (medial striatum, lateral striatum, cortex, septum, corpus callosum) was significant for both factors: treatment F(1,24) = 23.9, p = 0.003 and regions F(4,24) = 9.1, p < 0.001. Holm-Sidak post hoc analysis revealed significant knockdown of SNCA protein for all regions (p ≤ 0.001, ∼50%) except the corpus callosum (p = 0.104) (Figure 4D). In PEI/Luc-siRNA-injected transgenic mice, the corpus callosum showed significantly less SNCA overexpression compared to all other regions (p < 0.05), as expected. After knockdown of SNCA with PEI/SNCA-siRNA, staining intensities were no longer different across brain regions (p > 0.05), indicating successful silencing of SNCA overexpression (Figures 4D–4J). Brain tissues of the same mice were used for mRNA and protein expression analysis. Levels of mRNA and protein were positively correlated between mice across treatment groups (spearman correlation, correlation coefficient 0.75, p = 0.038; Figure S1B). Protein knockdown (p < 0.05, 42%) was further confirmed using western blot analysis on the motor cortex contralateral to the infusion site (Figure 4K).
Discussion
Neuroprotective therapy using non-coding RNAs can extend the therapeutic portfolio for so-far-incurable neurodegenerative diseases toward highly specific gene-targeting techniques. The major challenge to overcome is the efficient delivery of RNAi-inducing agents such as siRNA to the target site.11, 12 Here, we present proof-of-concept evidence that the PEI nanoparticle-mediated delivery of siRNA into the brain substantially and extensively reduced the expression of the target gene without any signs of toxicity.
While brain disorders could greatly profit from RNAi-based therapies, transfer to the clinic is so far hampered by insufficient nucleic acid delivery and concerns regarding toxicity12 requiring further in vivo studies of novel delivery systems. Non-viral strategies for knockdown of target genes in neurodegenerative diseases mainly include local application of single-stranded antisense oligonucleotides (ASOs) or siRNA into the cerebrospinal fluid or brain parenchyma.15 While these approaches showed some efficacy in animal models of neurodegenerative diseases,15, 28 hurdles to translating this approach into the clinic may include the need for chronic infusions of large amounts of ASO into the CNS, and the requirement to adequately distribute siRNA to target regions in a large brain.15 Notably, while infusion of naked siRNA against SNCA into the substantia nigra of primates had no adverse consequences,29 viral vectors for SNCA silencing in rodents exerted neurodegeneration and inflammation.30, 31, 32 Polymeric nanoparticles such as PEI complexes are non-metallic, non-pathogenic, and avoid various issues related to viral vectors. PEIs are a delivery platform for siRNA in vitro and in vivo and, besides lipid-based nanoparticles, are among the most promising delivery systems with therapeutic potential in vivo.33, 34, 35 PEIs may be modified for targeting specific tissues and cells, thus increasing therapeutic selectivity.36 The present study extends the application of non-targeted PEIs toward neuronal cells and brain tissue and thus provides a proof of concept for further development of non-viral RNAi-based therapeutic strategies. This is particularly relevant since neuronal cells are notoriously hard to transfect, and hence their efficient in vitro transfection without overt toxicity, as seen in our experiments (Figure 1), could not be taken for granted.
Further studies on dose-dependent toxicity were performed in vivo, since toxicity of nanoparticles is often higher in vitro than in vivo (nanomaterial paradox).37 We first injected different dosages of siRNA against luciferase, complexed with PEI F25-LMW, to investigate toxic nanoparticle effects unrelated to specific target RNAi. We determined 0.75 μg Luc-siRNA ICV to be the highest dosage that did not exert adverse effects in any of the mice injected in our study. This was confirmed by fully preserved integrity of ventricular ependymal cells and absence of immune cell activation or invasion in these mice. At this dose, we do not expect induction of molecular inflammatory pathways, since long-term intrathecal siRNA infusion of 4 μg/day has been shown not to trigger molecular events of interferone response.38 Furthermore, by using fluorescently labeled siRNA complexed with PEI, we could show that the siRNA indeed penetrates into the brain tissue after ICV infusion (Figure 3). Tagged siRNA was visible around the ventricle and across the brain down to the lumbar spinal cord, where fluorescence was also concentrated in large neurons with morphology and location of motoneurons. Furthermore, we demonstrated transfection of dopamine neurons of the substantia nigra. Thus, a single injection of PEI/siRNA was sufficient to deliver siRNA across the CNS. In previous studies, the distribution of drugs or particles after intracerebral or ICV injection was described to be limited to the region surrounding the injection site.12 In contrast, PEI F25-LMW complexation likely facilitated entry of siRNA into brain tissue. This finding is also in line with recent studies in ex vivo tissue slice models demonstrating the penetration of PEI/siRNA complexes into solid tissues like tumors39, 40 and suggests that significant siRNA distribution could be achieved in the human brain, thus supporting further development of our approach. Most neurodegenerative diseases affect large brain regions or spread across the entire CNS. For example, in PD, the specific loss of dopamine neurons in the substantia nigra leads to the cardinal motor symptoms; however, SNCA pathology spreads throughout the nervous system with disease progression and contributes to motor and non-motor symptoms.2 Thus, therapeutic strategies that can distribute effectively are required.
Importantly, siRNA delivered in PEI nanoparticles resulted in efficient target knockdown already after a single ICV injection. Human SNCA as target for confirmation of in vivo efficacy was chosen for two main reasons. First, accumulation and aggregation of SNCA is a pathological hallmark of synucleinopathies such as PD and likely contributes to progressive neurodegeneration, making the reduction of its expression a major and promising therapeutic strategy.4, 5 Mutations or multiplications in SNCA cause familial PD,41 and genetic variants of SNCA that likely increase its expression are major risk factors for sporadic PD.42, 43, 44, 45 Second, SNCA pathology is widespread in brains of patients, thus requiring the extensive distribution as achieved by our approach.46, 47 In order to provide first evidence of nanoparticle efficacy and functionality, we chose a well-established mouse model of PD with overexpression of human SNCA under the Thy-1 promotor (Thy1-aSyn).8, 25, 26 We demonstrate that a single ICV infusion of 0.75 μg PEI/SNCA-siRNA resulted in 67% reduction of SNCA mRNA in the striatum and ∼50% reduction in SNCA protein in all regions investigated (striatum, medial septum, and cortex). Thy1-aSyn mice overexpress human SNCA between 3- and 10-fold, depending on the brain region. Therefore, it can be expected that the efficient dosage used here is more than sufficient to knock down SNCA in human tissue of sporadic PD patients, which express endogenous levels of SNCA, thus providing a sufficiently wide therapeutic window. As accumulation of monomeric SNCA protein results in oligomerization and fibrillation of the protein in Thy1-aSyn mice, the extensive knockdown of our approach will likely reduce the buildup of toxic species and thus slow progression. To this purpose, early intervention is likely more beneficial; however, future studies will explore if reduction in protein load can also reverse already-formed oligomers and fibrils, e.g., by freeing capacity for protein degradation. Based on our experience in pre-clinical studies in Thy1-aSyn mice, the reduction of SNCA expression by 50% is sufficient for improvement of motor deficits in this model.8, 48, 49, 50 Thus, we abstained from behavioral tests in this initial study in order to minimize effects of handling and stress on toxicity and efficacy endpoints measured. There are few studies attempting widespread RNAi induction in brain in vivo without using a viral approach, and to our knowledge there are no other studies using siRNA to silence SNCA in a transgenic mouse model, but efficacy of PEI-mediated transfection and knockdown in brain appears high compared to similar studies. For example, 5 μg siRNA complexed with branched PEIs achieved similar knockdown of N-methyl-D-aspartate (NMDA) receptor NR2B subunit in spinal cord after intrathecal injection,51 while higher dosages of 10 μg siRNA in lipid nanoparticles were infused ICV bilaterally in rats in order to knock down an endogenous target gene at a distance over 4 mm from the injection site.16 Without a delivery system, siRNA requires modification for enhanced stability even after local application. For example, 4 μg/day intrathecal infusion of such modified siRNA over several weeks were required for modest ∼15% knockdown of the target protein, while higher dosages triggered interferon response.38 Importantly, our results show that a single bout of siRNA stabilized in PEI nanoparticles is efficient several days after injection, which eliminates the invasive requirement of continuous infusion. In non-dividing cells, such as neurons, siRNA concentration is not diluted by cell division, resulting in effective RNAi for several weeks after a single injection.52 It is conceivable that use of modified siRNA with increased half-life complexed with PEIs will further reduce the frequency of drug application. The mechanism of PEI clearance is not yet completely clarified, but no cumulative toxicity has been observed in any in vivo study, and the 4 to 12 kDa PEI used here is among the lowest molecular weight PEIs that can be used for siRNA delivery.53 The extensive in vivo experience and absence of immunogenicity of PEIs is advantageous to other shuttle systems, such as the large complexes with viral vectors or loading of siRNA onto exosomes purified from engineered dendritic cells.54 Notably, a recent study introduced a novel relatively small peptide vector targeted to neurons, which achieved substantial knockdown of mouse SNCA in a toxin-induced model of PD after repeated systemic injections at a relatively high dose of 50 μg siRNA.55
Our results support the usability and safety of PEI nanoparticle-mediated delivery of siRNA for extensive reduction of target-gene expression in brain. The established method is the basic pre-requisite for the next step of applying small non-coding RNA to the CNS of disease models for investigation of effects on phenotypes as pre-clinical evidence of efficacy. Notably, regulation of gene expression by endogenous non-coding RNAs such as micro RNAs likely contributes to disease pathogenesis, as their levels were altered in plasma and post-mortem brain samples of patients with neurodegenerative diseases,56, 57, 58 adding further potential therapeutic targets for our approach. We have previously used PEI complexes successfully for miRNA replacement therapy, but application to neurons remains to be investigated.59 While cerebrospinal-fluid-based delivery using ICV or cisternal or lumbar intrathecal administration is feasible in human patients, future strategies will examine less-invasive routes of delivery, such as intranasal application. Furthermore, PEI polyplexes can be targeted by modifications to cross the blood-brain barrier after systemic application.60, 61 For this purpose, our recently developed liposome-PEI complexes may have advantageous properties.53
Materials and Methods
Complexation of siRNA in PEIs
SiRNA against human SNCA (silencer Pre-Designed siRNA 217011; Ambion, Life Technologies), siRNA against luciferase serving as negative control (siRNA Luciferase GL3; Promega, synthesized by BioSpring; sense, 5′-CUUACGCUGAGUACUUCGAdTdT-3′, antisense, 5′-UCGAAGUACUCAGCGUAAGdTdT-3′) and AllStars negative control siRNA (QIAGEN) for experiments requiring fluorescence coupling were used. Complexation of siRNAs was performed as described previously.24 In short, siRNA and PEI F25-LMW were each dissolved in 0.015 M NaCl (pH 7.4). After incubation for 5 min at room temperature (RT), the PEI solution was added to the siRNA-containing vial and mixed by vortexing. Complex formation is based on electrostatic interactions between the positively charged PEI and the negatively charged siRNAs. For complete complexation, the complexes were incubated at RT for 30 min and vortexed again prior to use. Alternatively, complexes were stored at −80°C. Prior to use they were thawed, incubated at RT for 30 min, and vortexed again.
Confirmation of Neuronal Uptake of PEI/siRNA Complexes In Vitro
Uptake of siRNA into SH-SY5Y cells was monitored using Alexa-647-labeled control siRNA (QIAGEN). SH-SY5Y cells were maintained in a 1:1 mixture of Ham’s F12 and DMEM (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (FCS) (GIBCO, Thermo Fisher Scientific, Germany), 365.3 mg/l L-glutamine and 1% penicillin/streptomycin (1000 U/mL; Biochrom, Berlin, Germany). SiRNA (50 pmol) was incubated with PEI at a 1:50 molar ratio of siRNA to PEI. The PEI/siRNA complexes were added to SH-SY5Y cells (1.2 × 105 cells per well in 24-well plates). After incubation for 24 hr at 37°C, cells were fixed with 4% paraformaldehyde and examined via confocal microscopy (laser scanning microscope [LSM] 510 META; Carl Zeiss, Oberkochen, Germany). Toxicity was examined by measuring LDH released into the medium according to the supplier’s protocol (LDH assay; Promega, Madison, WI, USA). Determination of nucleic acid uptake in vitro was done by visualization of fluorescence-labeled cells at 20×.
Animals
Animal care was provided in accordance with the guidelines of the EU Directive 2012/63/EU and the German Animal Welfare Agency. All experiments were approved by the Ethics Committee of the University of Leipzig under protocol number TVV06/14.
Mice overexpressing human wild-type SNCA under the murine Thy1-promoter (Thy1-aSyn mice) maintained on a hybrid C57BL6/DBA2 background8, 62 were used for in vivo delivery experiments of PEI/siRNA complexes. The genotypes of all Thy1-aSyn mice and wild-type littermates were determined at 3 weeks of age and confirmed at the end of the experiment by PCR amplification analysis of ear DNA. Male transgenic littermates from multiple litters were used in this study after performing a power analysis to determine the sample size (Sigma Plot 12.5). Experiments were designed to minimize the number of animals required and to minimize animal suffering. Female Thy1-aSyn transgenics are not suitable for this study due to their low and variable transgene expression.26
Mice used in this study were bred and housed in the institute’s facility. Animals were grouped with not more than six housed in each cage on standard bedding (shredded wood) and maintained on a reverse 12-hr light/12-hr dark cycle. RT in the mouse holding room was 22°C ± 2°C and relative humidity was about 60%; values were recorded during the daily animal check. Food (Altromin standard diet) and water were available ad libitum, and material for nest building (paper rolls, red plastic houses) was provided.
Stereotactic Injection and PEI Delivery In Vivo
Stereotaxic surgery was performed as described previously63 with modifications for ICV infusion. Male Thy1-aSyn transgenic mice (10–16 weeks old, n of 5–9 per group) were deeply anesthetized with 4.5% isoflurane (CP-Pharma, Burgdorf, Germany) and maintained at 1.6%–2.0% isoflurane at a flow rate of 160–200 mL/min with 21% O2 (Univentor 1200 Anaesthesia Unit; Univentor, Malta) inside a stereotaxic frame (Stoelting, Wood Dale, IL, USA). Ophthalmic ointment was applied for eye protection and 0.1% bupivacaine (Jenapharm, Jena, Germany) injected subcutaneously at the surgery site for peri- and post-operative analgesia of skin and periosteum. A scalp incision was made midline, and a hole was drilled in the skull at coordinates for ICV infusion according to Paxinos and Franklin64 (Figure 2B), i.e., with distance to bregma anterioposterior −0.4 mm, mediolateral −1.3 mm, and dorsoventral 2.5 mm. PEI/siRNA complexes were injected ICV in 2.5 μL volume into the right ventricle. Injections were made manually over 5 min with a gauge needle (Type 1701 RN Neuros) attached to a Hamilton syringe (7000, Hamilton Company, Reno, NV, USA). The needle was left in place for 5 min and gradually withdrawn over an additional 5 min to prevent leakage from the needle track. To prevent hypothermia, mice were placed on isothermal pads (Braintree Scientific, MA, USA), and body temperature was recorded with a rectal probe (ThermoWorks, UT, USA). The skin was closed with intracutaneous sutures, which allowed social housing after surgery.
For analysis of CNS distribution of PEI/siRNA complexes, fluorescent dyes (Alexa 488 [green] or Alexa 647 [far-red]) were coupled to negative control siRNA (QIAGEN), complexed with PEI F25-LMW, and ICV injected at 0.125 μg (n = 1), 0.75 μg (n = 2), or 2.5 μg (n = 1) siRNA, respectively, in mice. For toxicity and efficacy experiments, siRNA against SNCA or luciferase (negative control) was complexed with PEI F25-LMW and injected at different concentrations (for detailed description see Results section).
Immunohistochemistry for Analysis of CNS Distribution of PEI/siRNA Complexes
Mice infused with PEI/siRNA complexes coupled to fluorescent dyes were euthanized 5 days post-injection by cervical dislocation under isoflurane anesthesia. Brains and spinal cords were removed immediately, embedded in Tissue Tek (Sakura Finetek Europe B.V., the Netherlands), frozen in 2-methylbutane at −30°C and stored at −80°C. The entire brain and the spinal cord were cut in 16 μm serial coronal sections at −20°C using a cryostat (Zeiss, Histoserve Hypa X C50), mounted on charged glass slides, and immunostained quickly for NeuN or TH as described previously9 with modifications. Sections were post-fixed in 4% paraformaldehyde in PBS (for NeuN) or in 80% ethanol (for TH) for 5 min followed by washing in TRIS-buffered saline (TBS). Sections were incubated for 20 min in mouse immunoglobulin G (IgG) blocking reagent (M.O.M. kit, Vector Laboratories, CA, USA) for NeuN or in TBS/5% normal donkey serum for TH and then incubated with mouse anti-NeuN or rabbit anti-TH primary antibody (Millipore/Merck, Darmstadt, Germany), diluted 1:100 in TBS/2% normal donkey serum at RT for 30 min. Negative control sections were incubated in the same solution without primary antibody. Sections were washed in TBS and then incubated with the secondary antibody, Alexa-488-labeled anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Europe, Suffolk, UK) at 1:600 dilution in TBS/2% normal donkey serum at RT for 10 min protected from light. Sections were washed in TBS and coverslipped with mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vectashield, Vector Laboratories, CA, USA). A confocal microscope (Olympus Fluoview FV 1200, Software Olympus Fluoview 4.1, Olympus, Germany) was used for visualization and image acquisition.
Histopathological Investigation
The histopathological investigation was performed on coronal brain slices (bregma −0.4 mm) of two mice that were euthanized 5 days after unilateral ICV injection of PEI/siRNA complexes (0.5 and 0.75 μg siRNA, respectively) in comparison with an age-matched untreated naive Thy1-aSyn transgenic mouse and a wild-type littermate. After fixation of the brain slices in 4% paraformaldehyde for 24 hr, these were embedded in paraffin, processed routinely, sectioned at 3 μm with a sledge microtome (MicroTec 4055, Walldorf, Germany), and stained with hematoxylin-eosin (HE) or processed for immunohistochemistry. For this, the peroxidase anti-peroxidase complex method and 3,3′-diaminobenzidine-tetrahydrochloride (DAB; Fluka Feinchemikalien, Neu Ulm, Germany) as chromogen were used. As primary antibodies, polyclonal antisera against glial fibrillary acidic protein (marker for astrocytes, 1: 250, no antigen retrieval; Dako, Hamburg, Germany) and IBA1 (marker for microglial cells and macrophages; 1:100, citric acid pre-treatment; Wako Chemicals, Neuss, Germany) were applied. In brief, after deparaffinization and rehydration of the sections, the endogenous peroxidase was blocked. For this, sections were incubated in methanol with 3% hydrogen peroxide for 30 min at RT. Subsequently, sections were rinsed and incubated with the primary antibodies overnight at 4°C. Then, sections were washed and treated with the secondary antibody swine anti-rabbit IgG (1:100; 30 min, RT; Dianova, Hamburg, Germany). This was followed by a further washing step and the application of rabbit-PAP-complex (1:500; 30 min, RT; Dianova, Hamburg, Germany). Afterward, sections were rinsed again, incubated with the chromogen (10 min, RT), and counterstained with Papanicolaou’s solution (Merck, Darmstadt, Germany). All washing steps were performed with TBS containing 1% BSA. For the dilution of the antibodies and the PAP complex, TBS with 1% BSA was used. In the negative controls, the primary antibodies were replaced by the solvent or a rabbit non-immune serum diluted in the same concentration as the primary antibodies. Internal positive controls were the astrocytes (glial fibrillary acidic protein [GFAP]) and microglia (IBA1) of the brain. Tissue sections were investigated by light microscopy (BH2 microscope, Olympus, Munich, Germany) to evaluate the presence of a tissue reaction due to ICV injection of PEI/siRNA. In treated animals, histomorphology of the ipsilateral (injected) ventricle and periventricular parenchyma were compared to those of the contralateral (not injected) ventricle and adjacent parenchyma. In particular, the morphology of the ventricular ependyma as well as the presence of degenerative or inflammatory changes was evaluated in comparison to naive wild-type and transgenic controls.
Quantification of SNCA mRNA Expression
For tissue preparation, mice (n = 5–9/group) were euthanized 5 days post-injection by cervical dislocation under isoflurane anesthesia. Brains and spinal cords were removed immediately, embedded in Tissue Tek (Sakura Finetek Europe B.V., the Netherlands), frozen in 2-methylbutane at −30°C and stored at −80°C. The striatum was dissected from 100 μm tissue slices at −20°C using a cryostat (Zeiss, Histoserve Hypa X C50) and stored until further analysis at −80°C. Motor cortex was dissected from the same slices and stored at −80°C for western blot analysis. The remaining tissue was used for immunohistochemistry.
Expression of human wild-type SNCA mRNA was determined in striatal tissue of male Thy1-aSyn mice by qRT-PCR following the manufacturer’s protocol (QIAGEN, Venlo, the Netherlands) as described previously.9, 26 In brief, RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN). First-strand cDNA synthesis was performed using PrimeScript RT Master Mix Perfect Real Time kit (Takara Bio Clontech Laboratories, CA, USA) and random primers according to the manufacturer’s protocol. Specific TaqMan Gene expression assays were used with probes that span exon-exon junctions (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). Efficacy of the respective assay was evaluated by the manufacturer, but specific sequences are proprietary. One microliter of cDNA was added to the Premix ExTaq kit (Takara Bio, Clontech Laboratories, CA, USA) for a total reaction volume of 5 μL, and amplification was performed on a PikoReal 96 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocols. Data were analyzed using the PikoReal Software 2.1. GeNorm (https://genorm.cmgg.be/) was used to analyze candidate reference genes Atp5b, Eif4a2, Gapdh, and Hprt according to average expression stability as described previously and the normalization factor derived from the most stable genes was chosen for normalization.9, 26
Immunohistochemistry and Quantification of SNCA Protein
Coronal brain sections (16 μm) were cut on a cryostat (Zeiss, Histoserve Hypa X C50), mounted on glass slides, and processed for immunohistochemical analyses. For quantification of SNCA protein, sections of n = 4 mice per group were processed for immunohistochemistry as described previously48 with modifications for snap-frozen mounted sections. Sections were post-fixed in 4% paraformaldehyde in PBS for 10 min followed by washing in TBS. Sections were incubated for 1 hr in mouse IgG blocking reagent (M.O.M. kit, Vector Laboratories, CA, USA) and then incubated with monoclonal mouse anti-SNCA primary antibody (anti-Syn1, #610787; BD Biosciences, Heidelberg, Germany), diluted 1:100 in TBS/2% normal donkey serum/0.5% Triton-X, at RT for 1 hr. Negative control sections were incubated in the same solution without primary antibody. Sections were washed in TBS and then incubated with the secondary antibody, Alexa-488-labeled anti-mouse IgG (Jackson ImmunoResearch Europe, Suffolk, UK) at 1:400 dilution in TBS/2% normal donkey serum/0.5% Triton-X at RT for 1 hr protected from light. Sections were washed in TBS and coverslipped with mounting medium containing DAPI (Vectashield, Vector Laboratories, CA, USA). A Zeiss Axioskop microscope coupled with a stereo investigator software (MBF Bioscience, VT, USA), a Retiga 2000R CLR.12 color digital camera (QImaging, Surrey, Canada), and an LED light source (CoolLED) were used for visualization and image acquisition. Quantification was performed as described previously using fluorescence intensity measurement of ImageJ (NIH) on representative images for each region of interest.48, 65
Western Blot Analysis for SNCA Protein
Analysis was performed following previously published protocols50 on the motor cortex contralateral to the ICV infusion site of the same mice used for immunohistochemical analyses. In brief, protein was extracted in 20 mM Tris-HCl, 150 mM NaCl (pH 7.4), and protein concentration was determined by the BCA protein assay (Thermo Fisher Scientific, Germany).
Western blotting was performed by SDS-PAGE and proteins fractionated on 4%–15% SDS-polyacrylamid gel (Mini-PROTEAN TGX, Bio-Rad) and transferred to PVDF membrane with a semi-dry transfer unit (Bio-Rad). The membrane was blocked (StartingBlock T20, Thermo Scientific; #37539) for 1 hr and incubated with mouse anti-SNCA monoclonal antibody (anti-Syn1, #610787, BD Biosciences, Heidelberg, Germany) at a 1:1,000 dilution or mouse anti-actin (MAB1501; dilution 1:2,000; Merck Chemicals) in blocking buffer for 1 hr at RT. Then, the membranes were washed with PBST (0.2% Tween 20 in 0.1 M PBS) and incubated with secondary, horseradish peroxidase-conjugated antibody (1:5,000; Jackson Immunoresearch, Suffolk, UK) for 1 hr at RT, washed with PBST, and developed with a chemiluminescent substrate (SuperSignal West Pico; Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions. A chemiluminescence imager was used to visualize the blots (FUSION Advance, Peqlab, Germany) and to exclude saturation of the signal. The resulting bands were quantified using ImageJ analysis software (NIH). The optical density values (OD) were normalized to loading control actin. Data are expressed as mean + SEM.
Statistical Analysis
Shapiro-Wilk test was used to test for normality. Non-parametric gene expression data were analyzed with a Mann-Whitney U rank-sum test. Parametric fluorescence intensity values were compared using a two-way repeated-measures ANOVA (treatment × region) followed by multiple comparison correcting post hoc test (Holm-Sidak). For comparison of ipsilateral and contralateral hemisphere, paired Student’s t test was used. Values are significantly different at p < 0.05 (Sigma Plot 12.5).
Author Contributions
Conceptualization, A.A., A.R., F.R.; Methodology, C.H., S.H., A.A., F.R.; Investigation, Formal Analysis, and Visualization, C.H., S.H., A.B., S.S., J.B., M.G., S.A.F., F.R.; Writing – Original Draft, C.H., F.R.; Writing –Review and Editing, C.H., A.A., A.R., F.R.; Funding Acquisition, Resources, and Supervision, A.A, A.R., F.R.
Acknowledgments
We thank Eliezer Masliah for the Thy1-aSyn mice and Steffi Fuchs and Ina Hochheim for excellent assistance with animal husbandry and genotyping. The authors are grateful to the staff members of the histology laboratory of the Institute of Veterinary Pathology for excellent technical support. We thank Iddo Magen for advice during the planning phase of the project.
Footnotes
Supplemental Information includes one figure and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.013.
Supplemental Information
References
- 1.Kordower J.H., Olanow C.W., Dodiya H.B., Chu Y., Beach T.G., Adler C.H., Halliday G.M., Bartus R.T. Disease duration and the integrity of the nigrostriatal system in Parkinson’s disease. Brain. 2013;136:2419–2431. doi: 10.1093/brain/awt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braak H., Del Tredici K., Rüb U., de Vos R.A., Jansen Steur E.N., Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 3.Meissner W.G., Frasier M., Gasser T., Goetz C.G., Lozano A., Piccini P., Obeso J.A., Rascol O., Schapira A., Voon V. Priorities in Parkinson’s disease research. Nat. Rev. Drug Discov. 2011;10:377–393. doi: 10.1038/nrd3430. [DOI] [PubMed] [Google Scholar]
- 4.Schapira A.H., Olanow C.W., Greenamyre J.T., Bezard E. Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: future therapeutic perspectives. Lancet. 2014;384:545–555. doi: 10.1016/S0140-6736(14)61010-2. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini M.G., Goedert M. The alpha-synucleinopathies: Parkinson’s disease, dementia with Lewy bodies, and multiple system atrophy. Ann. N Y Acad. Sci. 2000;920:16–27. doi: 10.1111/j.1749-6632.2000.tb06900.x. [DOI] [PubMed] [Google Scholar]
- 6.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 7.Winner B., Jappelli R., Maji S.K., Desplats P.A., Boyer L., Aigner S., Hetzer C., Loher T., Vilar M., Campioni S. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. USA. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesselet M.F., Richter F., Zhu C., Magen I., Watson M.B., Subramaniam S.R. A progressive mouse model of Parkinson’s disease: the Thy1-aSyn (“Line 61”) mice. Neurotherapeutics. 2012;9:297–314. doi: 10.1007/s13311-012-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter F., Gao F., Medvedeva V., Lee P., Bove N., Fleming S.M., Michaud M., Lemesre V., Patassini S., De La Rosa K. Chronic administration of cholesterol oximes in mice increases transcription of cytoprotective genes and improves transcriptome alterations induced by alpha-synuclein overexpression in nigrostriatal dopaminergic neurons. Neurobiol. Dis. 2014;69:263–275. doi: 10.1016/j.nbd.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabeza-Arvelaiz Y., Fleming S.M., Richter F., Masliah E., Chesselet M.F., Schiestl R.H. Analysis of striatal transcriptome in mice overexpressing human wild-type alpha-synuclein supports synaptic dysfunction and suggests mechanisms of neuroprotection for striatal neurons. Mol. Neurodegener. 2011;6:83. doi: 10.1186/1750-1326-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi C.R., Labhasetwar V., Ghorpade A. Destination brain: the past, present, and future of therapeutic gene delivery. J Neuroimmune Pharmacol. 2017;12:51–83. doi: 10.1007/s11481-016-9724-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franich N.R., Fitzsimons H.L., Fong D.M., Klugmann M., During M.J., Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington’s disease. Mol. Ther. 2008;16:947–956. doi: 10.1038/mt.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y.C., Miller A., Lins L.C., Han S.W., Keiser M.S., Boudreau R.L., Davidson B.L., Narayanan N.S. RNA interference of human α-synuclein in mouse. Front. Neurol. 2017;8:13. doi: 10.3389/fneur.2017.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magen I., Hornstein E. Oligonucleotide-based therapy for neurodegenerative diseases. Brain Res. 2014;1584:116–128. doi: 10.1016/j.brainres.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Rungta R.L., Choi H.B., Lin P.J., Ko R.W., Ashby D., Nair J., Manoharan M., Cullis P.R., Macvicar B.A. Lipid nanoparticle delivery of siRNA to silence neuronal gene expression in the brain. Mol. Ther. Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussif O., Lezoualc’h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B., Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neu M., Fischer D., Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J. Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 19.Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs. Curr. Opin. Mol. Ther. 2007;9:345–352. [PubMed] [Google Scholar]
- 20.Akinc A., Thomas M., Klibanov A.M., Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 21.Höbel S., Koburger I., John M., Czubayko F., Hadwiger P., Vornlocher H.P., Aigner A. Polyethylenimine/small interfering RNA-mediated knockdown of vascular endothelial growth factor in vivo exerts anti-tumor effects synergistically with Bevacizumab. J. Gene Med. 2010;12:287–300. doi: 10.1002/jgm.1431. [DOI] [PubMed] [Google Scholar]
- 22.Ibrahim A.F., Weirauch U., Thomas M., Grünweller A., Hartmann R.K., Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 23.Weirauch U., Gutsch D., Höbel S., Aigner A. Polymer-based delivery of RNA-based therapeutics in ovarian cancer. Methods Mol. Biol. 2013;1049:443–465. doi: 10.1007/978-1-62703-547-7_34. [DOI] [PubMed] [Google Scholar]
- 24.Werth S., Urban-Klein B., Dai L., Höbel S., Grzelinski M., Bakowsky U., Czubayko F., Aigner A. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112:257–270. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Chesselet M.F., Richter F. Modelling of Parkinson’s disease in mice. Lancet Neurol. 2011;10:1108–1118. doi: 10.1016/S1474-4422(11)70227-7. [DOI] [PubMed] [Google Scholar]
- 26.Gerstenberger J., Bauer A., Helmschrodt C., Richter A., Richter F. The novel adaptive rotating beam test unmasks sensorimotor impairments in a transgenic mouse model of Parkinson’s disease. Behav. Brain Res. 2016;304:102–110. doi: 10.1016/j.bbr.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Malek A., Czubayko F., Aigner A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. J. Drug Target. 2008;16:124–139. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]
- 28.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormack A.L., Mak S.K., Henderson J.M., Bumcrot D., Farrer M.J., Di Monte D.A. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS ONE. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorbatyuk O.S., Li S., Nash K., Gorbatyuk M., Lewin A.S., Sullivan L.F., Mandel R.J., Chen W., Meyers C., Manfredsson F.P., Muzyczka N. In vivo RNAi-mediated alpha-synuclein silencing induces nigrostriatal degeneration. Mol. Ther. 2010;18:1450–1457. doi: 10.1038/mt.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodr C.E., Becerra A., Han Y., Bohn M.C. Targeting alpha-synuclein with a microRNA-embedded silencing vector in the rat substantia nigra: positive and negative effects. Brain Res. 2014;1550:47–60. doi: 10.1016/j.brainres.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khodr C.E., Sapru M.K., Pedapati J., Han Y., West N.C., Kells A.P., Bankiewicz K.S., Bohn M.C. An α-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson’s disease, but displays toxicity in dopamine neurons. Brain Res. 2011;1395:94–107. doi: 10.1016/j.brainres.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schäfer J., Höbel S., Bakowsky U., Aigner A. Liposome-polyethylenimine complexes for enhanced DNA and siRNA delivery. Biomaterials. 2010;31:6892–6900. doi: 10.1016/j.biomaterials.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 34.de Fougerolles A., Vornlocher H.-P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park T.G., Jeong J.H., Kim S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Kwon E.J., Lasiene J., Jacobson B.E., Park I.K., Horner P.J., Pun S.H. Targeted nonviral delivery vehicles to neural progenitor cells in the mouse subventricular zone. Biomaterials. 2010;31:2417–2424. doi: 10.1016/j.biomaterials.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joris F., Manshian B.B., Peynshaert K., De Smedt S.C., Braeckmans K., Soenen S.J. Assessing nanoparticle toxicity in cell-based assays: influence of cell culture parameters and optimized models for bridging the in vitro-in vivo gap. Chem. Soc. Rev. 2013;42:8339–8359. doi: 10.1039/c3cs60145e. [DOI] [PubMed] [Google Scholar]
- 38.Wang H., Ghosh A., Baigude H., Yang C.S., Qiu L., Xia X., Zhou H., Rana T.M., Xu Z. Therapeutic gene silencing delivered by a chemically modified small interfering RNA against mutant SOD1 slows amyotrophic lateral sclerosis progression. J. Biol. Chem. 2008;283:15845–15852. doi: 10.1074/jbc.M800834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ewe A., Höbel S., Heine C., Merz L., Kallendrusch S., Bechmann I., Merz F., Franke H., Aigner A. Optimized polyethylenimine (PEI)-based nanoparticles for siRNA delivery, analyzed in vitro and in an ex vivo tumor tissue slice culture model. Drug Deliv. Transl. Res. 2017;7:206–216. doi: 10.1007/s13346-016-0306-y. [DOI] [PubMed] [Google Scholar]
- 40.Merz L., Höbel S., Kallendrusch S., Ewe A., Bechmann I., Franke H., Merz F., Ainger A. Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur. J. Pharm. Biopharm. 2017;112:45–50. doi: 10.1016/j.ejpb.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Devine M.J., Gwinn K., Singleton A., Hardy J. Parkinson’s disease and α-synuclein expression. Mov. Disord. 2011;26:2160–2168. doi: 10.1002/mds.23948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simón-Sánchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 44.Edwards T.L., Scott W.K., Almonte C., Burt A., Powell E.H., Beecham G.W., Wang L., Züchner S., Konidari I., Wang G. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Tredici K., Braak H. Lewy pathology and neurodegeneration in premotor Parkinson’s disease. Mov. Disord. 2012;27:597–607. doi: 10.1002/mds.24921. [DOI] [PubMed] [Google Scholar]
- 47.Braak H., Rüb U., Del Tredici K. Cognitive decline correlates with neuropathological stage in Parkinson’s disease. J. Neurol. Sci. 2006;248:255–258. doi: 10.1016/j.jns.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Richter F., Fleming S.M., Watson M., Lemesre V., Pellegrino L., Ranes B., Zhu C., Mortazavi F., Mulligan C.K., Sioshansi P.C. A GCase chaperone improves motor function in a mouse model of synucleinopathy. Neurotherapeutics. 2014;11:840–856. doi: 10.1007/s13311-014-0294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming S.M., Mulligan C.K., Richter F., Mortazavi F., Lemesre V., Frias C., Zhu C., Stewart A., Gozes I., Morimoto B., Chesselet M.F. A pilot trial of the microtubule-interacting peptide (NAP) in mice overexpressing alpha-synuclein shows improvement in motor function and reduction of alpha-synuclein inclusions. Mol. Cell. Neurosci. 2011;46:597–606. doi: 10.1016/j.mcn.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter F., Subramaniam S.R., Magen I., Lee P., Hayes J., Attar A., Zhu C., Franich N.R., Bove N., De La Rosa K. A molecular tweezer ameliorates motor deficits in mice overexpressing α-synuclein. Neurotherapeutics. 2017 doi: 10.1007/s13311-017-0544-9. Published online June 5, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan P.H., Yang L.C., Shih H.C., Lan K.C., Cheng J.T. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 2005;12:59–66. doi: 10.1038/sj.gt.3302376. [DOI] [PubMed] [Google Scholar]
- 52.Bartlett D.W., Davis M.E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ewe A., Panchal O., Pinnapireddy S.R., Bakowsky U., Przybylski S., Temme A., Aigner A. Liposome-polyethylenimine complexes (DPPC-PEI lipopolyplexes) for therapeutic siRNA delivery in vivo. Nanomedicine (Lond.) 2017;13:209–218. doi: 10.1016/j.nano.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Cooper J.M., Wiklander P.B., Nordin J.Z., Al-Shawi R., Wood M.J., Vithlani M., Schapira A.H., Simons J.P., El-Andaloussi S., Alvarez-Erviti L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord. 2014;29:1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Javed H., Menon S.A., Al-Mansoori K.M., Al-Wandi A., Majbour N.K., Ardah M.T., Varghese S., Vaikath N.N., Haque M.E., Azzouz M., El-Agnaf O.M. Development of nonviral vectors targeting the brain as a therapeutic approach for Parkinson’s disease and other brain disorders. Mol. Ther. 2016;24:746–758. doi: 10.1038/mt.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Cardo L.F., Coto E., de Mena L., Ribacoba R., Moris G., Menéndez M., Alvarez V. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J. Neurol. 2013;260:1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 57.Wong G., Nass R. miRNAs and their putative roles in the development and progression of Parkinson’s disease. Front. Genet. 2012;3:315. doi: 10.3389/fgene.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodall E.F., Heath P.R., Bandmann O., Kirby J., Shaw P.J. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front. Cell. Neurosci. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Höbel S., Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013;5:484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 60.Morris V.B., Labhasetwar V. Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials. 2015;60:151–160. doi: 10.1016/j.biomaterials.2015.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko Y.T., Bhattacharya R., Bickel U. Liposome encapsulated polyethylenimine/ODN polyplexes for brain targeting. J Control Release. 2009;133:230–237. doi: 10.1016/j.jconrel.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Rockenstein E., Mallory M., Hashimoto M., Song D., Shults C.W., Lang I., Masliah E. Differential neuropathological alterations in transgenic mice expressing alpha-synuclein from the platelet-derived growth factor and Thy-1 promoters. J. Neurosci. Res. 2002;68:568–578. doi: 10.1002/jnr.10231. [DOI] [PubMed] [Google Scholar]
- 63.Richter F., Hamann M., Richter A. Moderate degeneration of nigral neurons after repeated but not after single intrastriatal injections of low doses of 6-hydroxydopamine in mice. Brain Res. 2008;1188:148–156. doi: 10.1016/j.brainres.2007.09.083. [DOI] [PubMed] [Google Scholar]
- 64.Paxinos G., Franklin K. Fourth Edition. Academic Press; 2012. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- 65.Richter F., Gabby L., McDowell K.A., Mulligan C.K., De La Rosa K., Sioshansi P.C., Mortazavi F., Cely I., Ackerson L.C., Tsan L. Effects of decreased dopamine transporter levels on nigrostriatal neurons and paraquat/maneb toxicity in mice. Neurobiol. Aging. 2017;51:54–66. doi: 10.1016/j.neurobiolaging.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.