Abstract
Yeast frataxin homolog (Yfh1) is the orthologue of human frataxin, a mitochondrial protein whose deficiency causes Friedreich Ataxia. Yfh1 deficiency activates Aft1, a transcription factor governing iron homeostasis in yeast cells. Although the mechanisms causing this activation are not completely understood, it is assumed that it may be caused by iron-sulfur deficiency. However, several evidences indicate that activation of Aft1 occurs in the absence of iron-sulfur deficiency. Besides, Yfh1 deficiency also leads to metabolic remodeling (mainly consisting in a shift from respiratory to fermentative metabolism) and to induction of Yhb1, a nitric oxide (NO) detoxifying enzyme. In this work, we have used conditional Yfh1 mutant yeast strains to investigate the relationship between NO, Aft1 activation and metabolic remodeling. We have observed that NO prevents Aft1 activation caused by Yfh1 deficiency. This phenomenon is not observed when Aft1 is activated by iron scarcity or impaired iron-sulfur biogenesis. In addition, analyzing key metabolic proteins by a targeted proteomics approach, we have observed that NO prevents the metabolic remodeling caused by Yfh1 deficiency. We conclude that Aft1 activation in Yfh1-deficient yeasts is not caused by iron-sulfur deficiency or iron scarcity. Our hypothesis is that Yfh1 deficiency leads to the presence of anomalous iron species that can compromise iron bioavailability and activate a signaling cascade that results in Aft1 activation and metabolic remodeling.
Keywords: Friedreich ataxia, Nitric oxide, Iron-sulfur, Metabolism, Targeted proteomics
Graphical abstract
Highlights
-
•
Frataxin deficiency activates Aft1, promoting iron uptake and metabolic remodeling.
-
•
Yhb1 deficiency and NO donors prevent Aft1 activation by increasing NO levels.
-
•
NO also prevents the metabolic remodeling caused by frataxin deficiency.
-
•
NO does not prevent Aft1 activation caused by iron or Fe/S deficiency.
-
•
Frataxin deficiency activates Aft1 by a pathway non-involving loss of Fe/S clusters.
1. Introduction
Frataxin is a mitochondrial protein highly conserved throughout evolution, with orthologous counterparts in almost all organisms, including mammals, bacteria, fungi and plants [1]. The budding yeast Saccharomyces cerevisiae contains an orthologous protein named Yfh1 (for Yeast Frataxin Homolog 1) and therefore has been widely used to explore frataxin function [13]. It is also well stablished that frataxin deficiency causes Friedreich Ataxia in humans [16], [6].
Several functions have been proposed for frataxins. Most of them are related to iron metabolism, because frataxin deficiency has been found to cause misregulation of this process in several organisms. In this context, the biosynthesis of iron-sulfur clusters is the function that has attracted more support because frataxins have been found interacting with several mitochondrial proteins involved in this process. Among these proteins we can find Nfs1, a cysteine desulfurase which supplies sulfur, and Isu1, a protein that interacts with Nfs1 and acts as an scaffold were nascent iron-sulfur clusters are assembled. The current model of iron-sulfur biogenesis suggests that frataxins would regulate cysteine desulfurase activity and therefore would stimulate iron-sulfur biogenesis [28]. Nevertheless, the analysis of the effects of frataxin deficiency in several models indicates that frataxin is not essential for iron-sulfur biogenesis in vivo and suggests that frataxin may have additional functions [35]. In this regard, frataxin has also been related with heme biosynthesis [32], iron storage and/or detoxification [29], and modulation of iron regulatory protein-1 activation [9]. Frataxin may be also interacting with components of the OXPHOS system [12] and may confer oxidative stress protection [3].
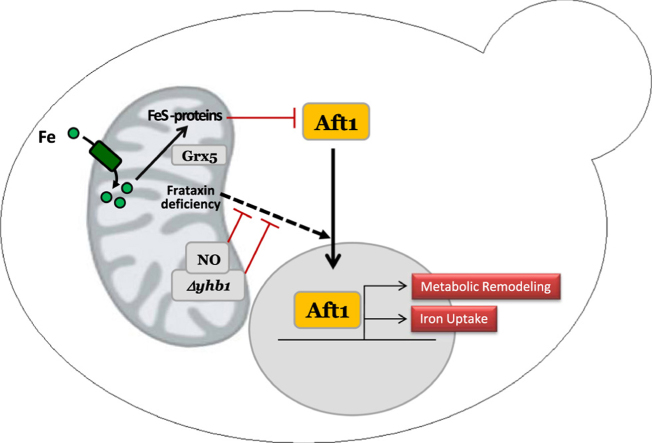
In yeast, frataxin/Yfh1 deficiency induces the expression of several proteins involved in iron uptake. This phenomenon results in iron overload and is caused by activation of the iron regulator Aft1 [14]. Nevertheless, the mechanisms leading to activation of Aft1 in Yfh1 deficient cells have not been investigated in detail. Aft1 is regulated by an iron-sulfur cluster, which under iron-sufficient conditions stabilizes a protein complex that retains Aft1 inactive in the cytosol. Iron scarcity or impaired iron-sulfur biogenesis prevent the formation of such cluster and activate Aft1, resulting in increased iron uptake [23]. In Yfh1 deficient cells, it has been assumed that Aft1 activation would be caused by the loss of such iron-sulfur cluster. However, previous research from our group using conditional Yfh1 mutants provided two observations which challenged this hypothesis: i) activation of Aft1 could be observed earlier than iron-sulfur loss [22]; ii) loss of iron-sulfur containing proteins in Yfh1 deficient yeasts required the presence of Cth2, an Aft1 target that binds to mRNAs from iron-containing proteins and promotes its degradation [21]. Therefore, we hypothesize that in Yfh1 deficient yeast, Aft1 may be activated by a mechanism different than iron-sulfur cluster deficiency.
In this work, we provide further evidences that Aft1 is not activated by iron-sulfur deficiency in Yfh1-deficient yeasts. These evidences have been obtained upon the analysis of the relationship between frataxin and yeast flavohemoglobin (Yhb1). We had previously found this protein induced in a proteomic and transcriptomic analysis of Yfh1-deficient yeast [21]. This protein attracted our attention, as Yhb1 had been found interacting with Yfh1, although the physiological relevance of such interaction was not explored [12]. Yhb1 is a flavoprotein with a globin-like domain as well as NAD and FAD binding sites. It has NO reductase activity and therefore it is believed to act in NO detoxification. Yhb1 is localized in the mitochondria in anoxic conditions, while a dual cytoplasmatic/mitochondrial localization is observed in aerobic conditions [7]. It has also been recently shown to be involved in sulfur detoxification [10]. In mammals, neuroglobin it is structurally related to the globin domain and could conserve some of its functions [2]. After focusing on the analysis of the relationship between frataxin, Yhb1 and NO in yeast, we have found that Aft1 activation in Yfh1-deficient yeast is prevented by NO, further supporting that Aft1 activation upon Yfh1 deficiency occurs through a mechanism not dependent on iron-sulfur deficiency.
2. Results
2.1. Yhb1 is induced upon Yfh1 depletion
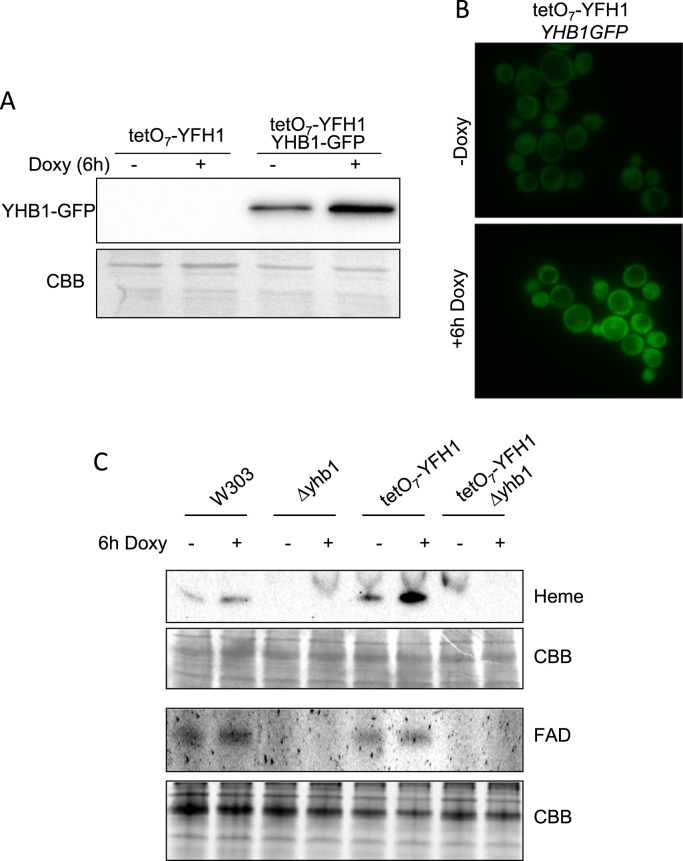
In order to explore in more detail the relationship between Yhb1 and Yfh1, we first confirmed the previously reported induction of Yhb1 in Yfh1-deficient cells [21]. We used a conditional Yfh1 mutant in which YFH1 expression is under the control of a tet promoter (tetO7YFH1 strain). In this strain YFH1 can be repressed by doxycycline addition to the growth media. In addition, this strain is able to grow in non-fermentable carbon sources such as glycerol, avoiding the repressing effect of glucose on several mitochondrial activities [22]. To confirm induction of Yhb1, this protein was GFP-tagged in a tetO7YFH1 strain and its content analyzed by anti-GFP western blot. Fig. 1A indicates that a clear induction of Yhb1-GFP could be observed 6 h after doxycycline addition. We also analyzed cellular localization by fluorescence microscopy. We could observe that Yhb1-GFP was localized both in the cytoplasm and mitochondria, as previously reported by other authors. Microscopic images confirmed the increase in protein content upon Yfh1 depletion, but we did not observe any change in the dual localization of Yhb1-GFP (Fig. 1B). We finally analyzed the presence of the holo form of Yhb1 in both control and frataxin deficient cells. Yhb1 contains a heme group and a FAD binding site. Both groups can be detected using non-denaturing (native) polyacrylamide gels and a Chemidoc XRS imaging system. Flavins can be directly detected in gels due to its fluorescent properties. Heme can be detected after transfer to PVDF membranes due to its peroxidase activity. Thus, cell extracts from control and doxycycline treated tetO7YFH1 cells were separated in native polyacrylamide gels and either imaged to detect the fluorescence from protein-bound flavins or transferred to PVDF membranes to detect heme. Extracts from Δyhb1 mutants were also loaded and were used to identify the flavin and heme bands corresponding to Yhb1. As shown in Fig. 1C, the heme signal corresponding to Yhb1 is strongly increased upon Yfh1 depletion. This result confirms the induction of Yhb1 and indicates that the protein is present in the holo form. Of note, using this analytical approach, the heme from Yhb1 appears as the most intense band, supporting previous data about the abundance of this protein. Yhb1-bound FAD could also be detected in both control and doxycycline treated tetO7YFH1 cells. However, the low signal-to-noise ratio from this flavin signal did not allow reliable quantitative analysis of this band.
Fig. 1.
Yhb1 is induced by Yfh1 deficiency. Yeast cells were grown in YPG and, where indicated, doxycycline was added to the culture media for 6 h in order to repress Yfh1 expression. A, whole cell extracts were analyzed by western blot using anti-GFP antibodies; B, tetO7YFH1 YHB1GFP cells were analyzed by fluorescence microscopy. A punctuate pattern can be appreciated in both images, indicating the dual localization of Yhb1-GFP irrespective of the presence of doxycycline. C, whole cell extracts were loaded on native gels and the presence of the heme group and the flavin from Yhb1 were detected as described under experimental conditions. Protein load was verified by post-western Coomassie Briliant Blue (CBB) staining of the gels or PVDF membranes.
2.2. Increased nitroxidative stress in Δyfh1 cells
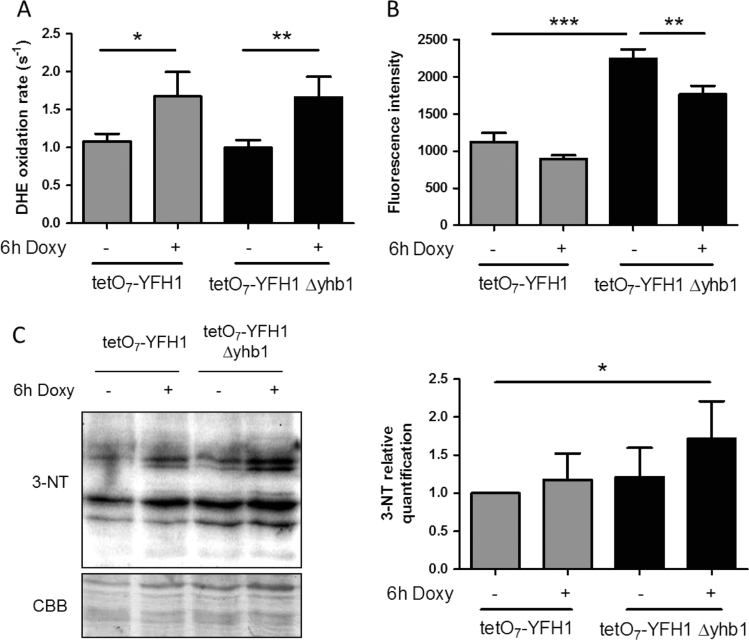
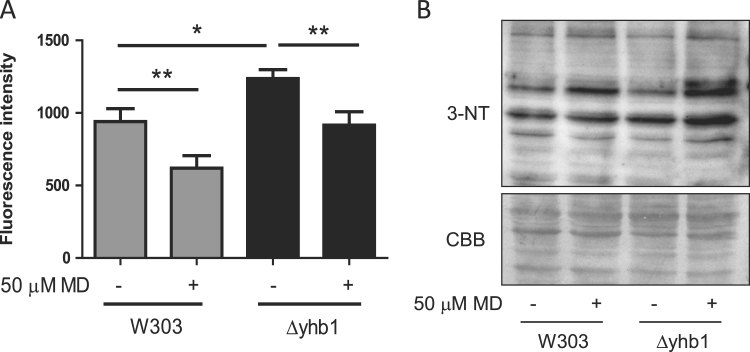
It has been proposed that Yhb1 would have a central role in regulating NO levels in yeast cells, as it has NO oxidase activity and is induced by nitroxidative stress. We hypothesized that induction of Yhb1 in Yfh1-deficient cells would be related to the presence of reactive oxygen or nitrogen species in Yfh1-deficient cells. Indeed, we had previously shown an increased O2- production in Δyfh1 cells [13]. Thus we analyzed O2- and NO levels in tetO7YFH1 and tetO7YFH1Δyhb1 cells treated with doxycycline. Superoxide levels were measured using DHE, while NO levels were measured using DAF-FM DA and flow cytometry. As shown in Fig. 2A, loss of Yfh1 led to increased production of superoxide, while NO levels were increased in Δyhb1 mutants (Fig. 2B), confirming the role of this protein in regulating NO levels. Interestingly, loss of Yfh1 promoted a slight but significant decrease in NO levels in both strains (Fig. 2B). This observation suggested us that O2- could be reacting with NO to form peroxynitrite, a highly reactive compound that can easily react with tyrosines to form nitrotyrosines. To confirm this hypothesis, we analyzed the presence of nitrotyrosines by western blot in tetO7YFH1 and tetO7yfh1Δyhb1 cells and we observed a significant increase in the presence of this postraductional modification when cells where deficient in both Yfh1 and Yhb1 (Fig. 2C–D). These results confirm that Yfh1 loss leads to increased O2- production, which in Yhb1 deficient cells would react with NO to produce peroxynitrite. To further confirm that increased NO levels could be scavenging O2- and lead to nitrotyrosine formation, we exposed wild type and Δyhb1 cells to the superoxide generating agent menadione and analyzed the presence of nitrotyrosines and NO levels in these cells (Fig. 3A–B). Similar to what we had previously observed in doxycycline treated tetO7YFH1 cells, menadione induced a decrease in NO levels and increased presence of nitrotyrosines. These observations confirmed that increased levels of O2- were produced in Yfh1-deficient cells and suggested that NO could be acting as a O2- scavenging agent.
Fig. 2.
Superoxide and NO levels intetO7YFH1andtetO7YFH1Δyhb1strains. Yeast cells were grown in YPG and, where indicated, doxycycline was added to the culture media for 6 h in order to repress Yfh1 expression. A, Superoxide production was measured using DHE; B, NO levels were measured using DAF-FM DA and flow cytometry; C, the presence of nitrotyrosines was evaluated in crude extracts by western blot using antibodies which recognize this prostraductional modification. Histograms represent relative nitrotyrosine content in each condition, which was calculated from the chemiluminescent signal of each lane of the western blot analyzed with Quantity One software (Bio-Rad). Nitrotyrosine levels in tetO7YFH1 control cultures were used as a reference value. From A to C, data are represented as means±SD from 3 independent experiments. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition.
Fig. 3.
NO levels in W303 andΔyhb1strains after menadione treatment. Yeast cells were grown in YPG and, where indicated, menadione was added to the culture media. A, NO levels were measured using DAF-FM DA and flow cytometry. Data are represented as means±SD from 3 independent experiments. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition.; B, the presence of nitrotyrosines was evaluated in crude extracts by western blot using antibodies which recognize this prostraductional modification.
2.3. Yhb1 deficiency and NO prevent Aft1 activation in Yfh1-deficient cells
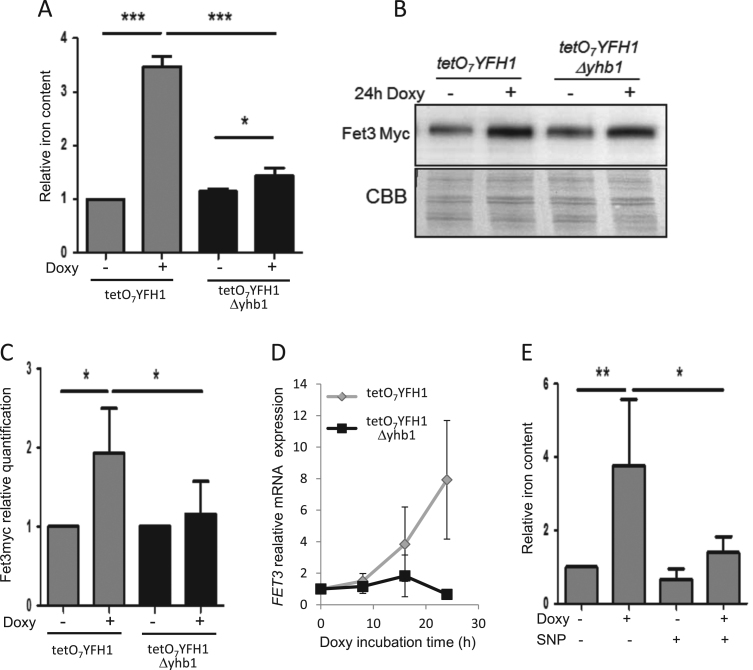
We decided to analyze which was the impact of Yhb1 deficiency on the phenotypes caused by Yfh1 deficiency. First, we analyzed the effect of Yhb1 on iron overload, one of the phenotypes observed in Yfh1 deficient cells. Iron overload in Yfh1-deficient yeast is caused by Aft1 activation, which promotes induction of several proteins involved in iron uptake. One of these proteins is the ferro-oxidase Fet3 [22]. Therefore, iron content was analyzed in tetO7YFH1 and tetO7YFH1Δyhb1 cells treated for 24 h with doxycycline and we observed that Yhb1 loss prevented iron accumulation (Fig. 4A). To confirm that this effect was due to prevention of Aft1 activation, we also analyzed the content of Fet3 by western blot in tetO7YFH1 mutants containing a Fet3-myc tagged version of the protein. As expected, Yfh1 deficiency promoted Fet3 induction. This induction was partially prevented in Yhb1 deficient cells, confirming that Aft1 activation was prevented in Δyhb1 cells (Fig. 4B–C). We also analyzed Fet3 expression by qPCR in both strains at different times after doxycycline treatment (Fig. 4D). This analysis confirmed that activation of Aft1 was prevented in Δyhb1 cells and also confirmed previous results which indicated that activation of the iron regulon after Yfh1 depletion is progressive (expression of the Aft1 targets increases over time) [21]. To further investigate the role of NO in preventing iron overload in Yfh1-deficient cells, we explored the effects of sodium nitroprusside (SNP), an NO donor, on iron accumulation in doxycycline-treated tetO7YFH1 cells. In order to use a non-toxic concentration of this compound, we first analyzed the effects of different concentrations of SNP on generation time of W303 cells. As we observed that SNP had minor effects on generation time at concentrations up to 10 μM (data not shown), we performed the forthcoming experiment at this concentration. Thus, we analyzed the effect of 24 h treatment with 10 μM SNP and doxycycline on iron content of tetO7YFH1 and W303 cells. As indicated in Fig. 4E, SNP prevented iron accumulation in doxycycline treated tetO7YFH1 cells, while had minor effect on non-doxycycline treated cells or wild type cells.
Fig. 4.
Activation of Aft1 by Yfh1-deficiency is prevented inΔyhb1cells. Yeast cells were grown in YPG and, where indicated, doxycycline was added to the culture media for 24 h in order to repress Yfh1 expression. A, whole cell iron content was measured by the BPS method as indicated in experimental procedures. B, Fet3-myc was detected by western blot using anti-myc antibodies. CBB staining was used as loading control. C, Histograms represent relative Fet3-myc content in each condition, which was calculated from the chemiluminescent signal of the western blot analyzed with Quantity One software (Bio-Rad). D, Relative FET3 expression was analyzed by qPCR in YPG-grown tetO7YFH1 (grey) and tetO7YFH1Δyhb1 (black) after addition of doxycycline for the indicated times. Actin expression was used as an internal control to normalize expression levels. E, whole cell iron content measured by the BPS method in SNP-treated cells (10 μM). In A, C, D and E levels in tetO7YFH1 untreated cultures were used as a reference value. Data are represented as means ± SD from 3 independent experiments. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition.
2.4. Activation of the iron regulon by iron scarcity does not require Yhb1
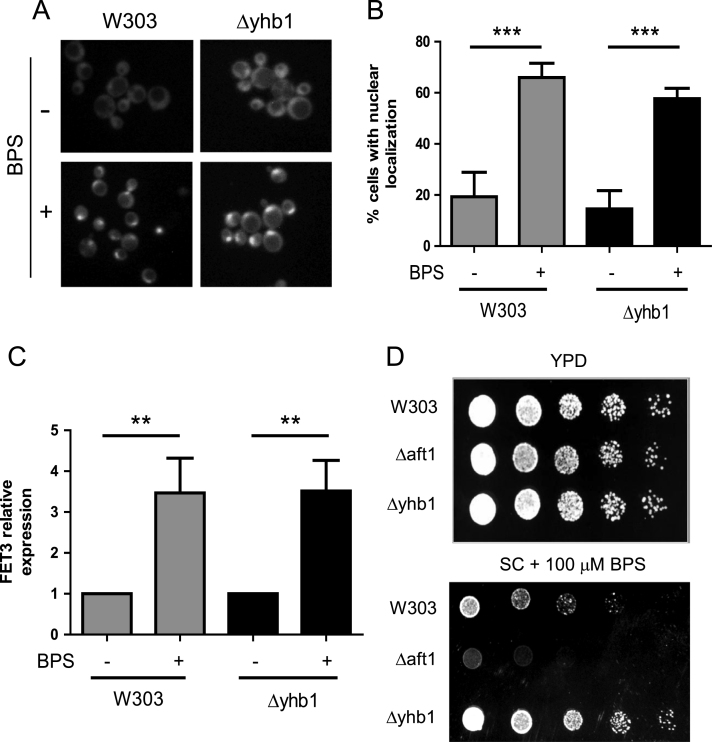
As loss of Yhb1 prevented activation of Aft1 in doxycycline-treated tetO7YFH1 cells we became interested in analyzing which was the effect of Yhb1 deficiency on the ability of the cell to sense iron levels and induce an Aft1-mediated response. Therefore, a wild type and a Δyhb1 strain with Aft1 GFP-tagged were treated with the iron chelator BPS for 4 h and nuclear localization of Aft1 was examined by fluorescence microscopy. Yhb1 deletion had no effect on the nuclear localization of Aft1 (Fig. 5A–B). We also measured the expression levels of FET3 by qPCR. BPS promoted a marked induction of this gene, but in this case induction was not prevented in Δyhb1 cells (Fig. 5C). We finally analyzed the sensitivity of Δyhb1 cells towards BPS in SC agar plates. A Δaft1 mutant was used as a control of mutant with increased sensitivity towards iron-deficiency. As shown in Fig. 5D, Δyhb1 cells did not show increased sensitivity towards iron deficiency. Overall, these results indicate that Yhb1 is not required for the response to iron scarcity, although it modulates Aft1 activation triggered by Yfh1. These observations suggest that Aft1 induction in Yfh1 deficient cells is triggered by a different pathway than Aft1 activation by iron scarcity.
Fig. 5.
Activation of Aft1 by iron scarcity is no prevented inΔyhb1cells. A, the indicated strains were grown in YPD media with or without 100 μM BPS and the localization of Aft1-GFP analyzed by fluorescence microscopy. B, histograms show the percentage of cells showing nuclear localization of Aft1-GFP in each indicated condition. Data are represented as means ± SD from 3 independent experiments in which at least 100 cells were analyzed. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition. C, the indicated strains were grown in YPG media with or without BPS and FET3 mRNA expression was measured by qPCR. Data are represented as means ± SD from 3 independent experiments. D, the indicated strains were grown in YPD to OD600 = 0.5 and serial dilutions (1:5) where plated on YPD plates supplemented or not with BPS.
2.5. Lack of Yhb1 does not prevent iron overload caused by disrupted iron-sulfur biogenesis
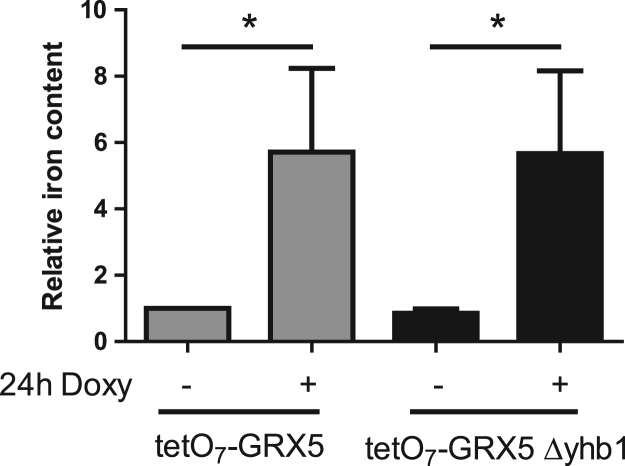
Mutations in proteins involved in iron-sulfur biogenesis are known to cause activation of Aft1 and consequently iron overload. We decided to analyze the effect of a null Yhb1 mutation in the activation of the iron regulon caused by depletion of Grx5, a mitochondrial protein involved in iron-sulfur biogenesis [27]. For that purpose, we used tetO7GRX5 strains, in which Grx5 was under the control of a tetO7 promoter. Addition of doxycycline to this strain promoted an increase in iron accumulation, similar to that observed in tetO7YFH1 cells (Fig. 6). However, contrary to what we had observed in the tetO7YFH1Δyhb1 mutant, iron accumulation was not prevented in tetO7GRX5Δyhb1. These results indicate that NO is not able to prevent Aft1 activation caused by impaired iron-sulfur biogenesis and also confirm that activation of Aft1 in Yfh1 deficient cells is not caused by impaired iron-sulfur biogenesis.
Fig. 6.
Activation of Aft1 by Grx5 deficiency is not prevented by NO. Yeast cells were grown in YPG and, where indicated, doxycycline was added to the culture media for 24 h in order to repress Grx5 expression. Whole cell iron content was measured by the BPS method as indicated in experimental procedures. Data are represented as means±SD from 3 independent experiments. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition.
2.6. Lack of Yhb1 prevents metabolic remodeling in Yfh1-deficient cells
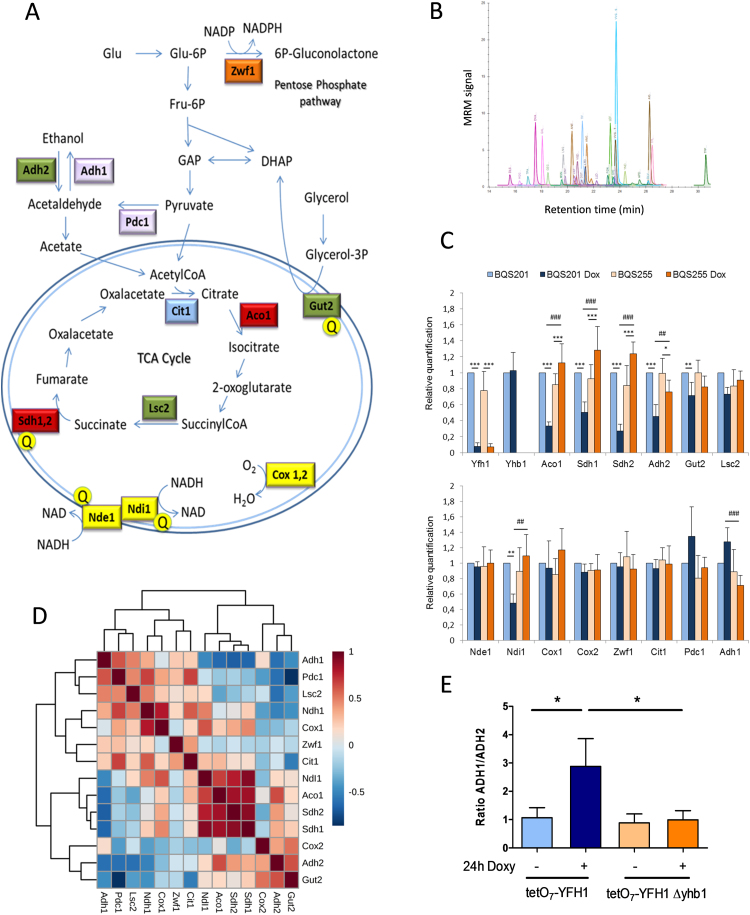
We finally, decided to analyze the consequences of NO on metabolic remodeling caused by Yfh1 deficiency. In a previous work, we reported that Yfh1 deficiency in yeasts triggers a drastic metabolic remodeling governed by the metabolic regulators Adr1 and Cth2. This remodeling finally leads to downregulation of iron-sulfur containing proteins and other proteins involved in respiratory metabolism [21]. We decided to analyze the changes in the content of some key Adr1 and Cth2 targets upon Yfh1 deficiency in tetO7YFH1 and tetO7YFH1Δyhb1 cells. These were three Adr1 targets (Adh2, Gut2 and Lsc2) [33], two iron-sulfur containing enzymes which are Cth2 targets (Aco1 and Sdh2) and a Cth2 target not containing an iron-sulfur (Sdh1) [25]. In order to have a more complete view of the metabolic status of the yeasts, we also analyzed the content of two enzymes involved in alcoholic fermentation (Adh1 and Pdc1), glucose-6-P-dehydrogenase (Zwf1) -a key enzyme of the pentose phosphate pathway-, the Krebs cycle enzyme citrate synthase 1 (Cit1), and the components of the OXPHOS system Nde1, Ndi1, Cox1 and Cox2. Yhb1 and Yfh1 were also measured to confirm its presence or absence. Actin and mitochondrial porin were used as loading controls. Fig. 7 shows a simplified scheme of S. cerevisiae metabolism indicating the pathways in which these proteins are involved. For the detection of these proteins we used an MRM-based targeted approach in which one or two proteotypic peptides from each protein were detected in a LC-Triple quadrupole mass spectrometer. To develop these assays, four proteotypic peptides and five transitions per peptide were selected from SRM atlas (www.srmatlas.com) [24] for each protein of interest. These transitions were analyzed in trypsin-digested crude yeast extracts using an Agilent LC/Triple Quad system. Then, isotope-labeled (heavy) synthetic versions of those peptides showing the best results were synthesized (JPT peptides) and used to confirm the retention times and transitions observed in yeast samples. These heavy peptides were also used as internal standards in subsequent analysis. For the final analysis we selected the two best-responding peptides for each protein, and three transitions were used for detecting each peptide. For some proteins, we could only find a single peptide that fulfilled the quality criteria. Details about the peptides and transitions used can be found in Table 2 and in Supplemental table 1. We next analyzed the relative content of these peptides in trypsin-digested whole cell extracts from tetO7YFH1 and tetO7YFH1Δyhb1 cells exposed or not to doxycicycline for 24 h. Actin and mitochondrial porin were used as loading controls, as previous work had indicated that porin content was not altered after 24 h of doxycycline addition [22]. A summary of the results obtained can be found in Fig. 7 (the complete set of results can be freely browsed through Panaromaweb). It can be observed that peptides from Yfh1 were not detected in doxycycline treated cells, nor Yhb1 peptides in Δyhb1 cells, confirming the quality of the analysis. The content of the Adr1 targets (Adh2, Gut2 and Lsc2) and of the Cth2 targets (Aco1, Sdh1, Sdh2) also decreased in tetO7YFH1 cells after doxycycline treatment, confirming previous observations. Interestingly, loss of Cth2 targets was completely prevented in Δyhb1 cells. Indeed, the content of these proteins was increased in doxycycline-treated tetO7YFH1Δyhb1. In the case of the Adr1 targets, the decrease was more pronounced for Adh2. This effect is not surprising, as Adr1 has a stronger regulatory effect on this protein than on Gut2 or Lsc2 [33]. Recovery in Δyhb1 cells was less pronounced for Adh2 than for Cth2 targets, and therefore did not reach statistically significance. Interestingly, Yfh1 depletion also lead to a slight induction of enzymes involved in alcoholic fermentation (Adh1 and Pdc1), which was also prevented in Yhb1-deficient cells. Such increase may be related to the metabolic remodeling experienced by yeast upon Yfh1 deficiency, which tries to direct its metabolism to fermentation. In this sense, the ratio Adh1/Adh2 is clearly higher in Yfh1 deficient cells, and is restored in Yfh1-deficient Δyhb1 cells (Fig. 7E). Regarding the OXPHOS system, marked changes were observed in complex II, as the Cth2 targets Sdh1 and Sdh2 decreased. Such decrease was prevented in Yhb1-deficient cells. We also noticed a decrease in Ndi1, but neither in Nde1 nor in the complex IV subunits Cox1 and Cox2. The reasons and consequences of NdiI1 decrease are not known, as neither Adr1 nor Cth2 have been described to regulate Ndi1 expression. No major changes were observed in the Krebs cycle enzymes Cit1 nor in the pentose phosphate pathway enzyme Zwf1. We also performed an analysis of covariance of the complete dataset, which included the four samples and three replicates per sample (Fig. 7D). Clustering of these data indicates a strong correlation among the Cth2 targets and that Ndi1 showed a better correlation with these Cth2 targets than with Adr1 targets. A good correlation could also be observed between the fermentative enzymes Adh1 and Pdc1, and also between the Adr1 targets Adh2 and Gut2. Overall, the results obtained from these analyses confirm the previous observed decrease in Adr1 and Cth2 targets. They also show that such decrease is specific, as there is not a general loss of mitochondrial proteins. Finally, the analysis also indicates that NO can prevent metabolic remodeling associated with Yfh1 deficiency.
Fig. 7.
Analysis ofS. crerevisiaemetabolism by targeted proteomics. A, a simplified scheme of S. cerevisiae metabolism showing the proteins analyzed. Note that many intermediate reactions are not indicated. Colors indicate the following functional groups: red, Cth2 targets; green, Adr1 targets; blue, TCA enzymes; yellow, OXPHOS components; purple, enzymes involved in fermentation; orange, pentose phosphate pathway. The yellow Q indicates that these enzymes provide electrons to ubiquinone. Nde1 is known as the external NADH dehydrogenase, as it oxidizes NADH from the cytosol and the intermembrane space. Ndi1 is the internal NADH dehydrogenase and oxidizes NADH from the mitochondrial matrix. Both dehydrogenases transfer electrons to Ubiquinone. GAP, Glyceraldehyde-3P; DHAP, dihydroxyacetone phosphate. B, a representative chromatogram showing the MRM trace of the different peptides analyzed. C, Histograms show the relative content of the measured proteins. Protein levels in tetO7YFH1 control cultures were used as a reference value. Data are represented as means±SD from 3 independent experiments. * and ** indicate significant (p < 0.05) or highly significant (p < 0.01) differences, respectively, when compared to control condition. D, correlation patterns of the indicated proteins among different samples. Dark red in the heatmap indicates strong correlation, while dark blue indicates poor correlation. E, Adh1/Adh2 ratio in each condition analyzed.
Table 2.
Peptides sequences from the proteins analyzed in the targeted proteomics experiments.
| Gene | Uniprot entry | Protein name | Peptide sequence |
|---|---|---|---|
| Adh2 | sp|P00331|ADH2_YEAST | Alcohol dehydrogenase 2 | ANGTVVLVGLPAGAK |
| VVGLSSLPEIYEK | |||
| Gut2 | sp|P32191|GPDM_YEAST | Glycerol-3-phosphate dehydrogenase | MSNYLVQNYGTR |
| TPLDFLLR | |||
| Yfh1 | sp|Q07540|FRDA_YEAST | Yeast Frataxin Homolog | LTDILTEEVEK |
| QIWLASPLSGPNR | |||
| Yhb1 | sp|P39676|FHP_YEAST | Flavohemoprotein | VGAQPNALATTVLAAAK |
| ATVPVLEQQGTVITR | |||
| Sdh2 | sp|P21801|DHSB_YEAST | Succinate dehydrogenase iron-sulfur subunit | DLVPDLTNFYQQYK |
| DGTEVLQSIEDR | |||
| Sdh1 | sp|Q00711|DHSA_YEAST | Succinate dehydrogenase flavoprotein subunit | GEGGFLVNSEGER |
| DVAAPVTLK | |||
| Zwf1 | sp|P11412|G6PD_YEAST | Glucose-6-phosphate 1-dehydrogenase | FGNQFLNASWNR |
| TFPALFGLFR | |||
| Cox2 | sp|P00410|COX2_YEAST | Cytochrome c oxidase subunit 2 | LNQVSALIQR |
| LLDTDTSMVVPVDTHIR | |||
| Cox1 | sp|P00401|COX1_YEAST | Cytochrome c oxidase subunit 1 | APDFVESNTIFNLNTVK |
| Por1 | sp|P04840|VDAC1_YEAST | Mitochondrial outer membrane protein porin 1 | VSDSGIVTLAYK |
| LEFANLTPGLK | |||
| Nde1 | sp|P40215|NDH1_YEAST | External NADH-ubiquinone oxidoreductase 1 | GALAYIGSDK |
| Act1 | sp|P60010|ACT_YEAST | Actin | VAPEEHPVLLTEAPMNPK |
| SYELPDGQVITIGNER | |||
| HQGIMVGMGQK | |||
| Ndi1 | sp|P32340|NDI1_YEAST | Rotenone-insensitive NADH-ubiquinone oxidoreductase | YNDLGALAYLGSER |
| Pdc1 | sp|P06169|PDC1_YEAST | Pyruvate decarboxylase 1 | TPANAAVPASTPLK |
| Lsc2 | sp|P53312|SUCB_YEAST | Beta subunit of succinyl-CoA ligase | DLSQEDPDEVK |
| Aco1 | sp|P19414|ACON_YEAST | Mitochondrial aconitase | TIFTVTPGSEQIR |
| QNVETLDIVR | |||
| Cit1 | sp|P00890|CISY1_YEAST | Citrate synthase | AIGVLPQLIIDR |
| Adh1 | sp|P00330|ADH1_YEAST | Alcohol dehydrogenase 1 | ANELLINVK |
| VVGLSTLPEIYEK |
3. Discussion
Yfh1 deficiency activates Aft1 and promotes iron uptake. It has been assumed that such activation is caused by impaired iron-sulfur biogenesis as occurs under conditions of iron scarcity or when certain proteins involved in the biosynthesis of iron-sulfur clusters are mutated. However, previous results from our group suggested that Aft1 activation in Yfh1-deficient yeasts was not caused by iron-sulfur deficiency, as such activation occurred earlier than iron-sulfur deficiency [22]. In the present work we have shown that increased NO levels can prevent Aft1 activation in Yfh1-deficient yeasts. This has been demonstrated in Δyhb1 cells, where increased NO levels are due to the absence of this NO detoxifying enzyme, and by exogenous exposure to SNP, an NO donor. Remarkably, this phenomenon is only observed when Aft1 activation is triggered by Yfh1 deficiency, while it is not observed when Aft1 is activated by iron scarcity or impaired iron-sulfur biogenesis (Grx5 deficiency). Therefore, this work confirms that the mechanism that leads to Aft1 activation in Yfh1-deficient yeasts differs to the one promoted by that other conditions.
We hypothesize that Aft1 activation in Yfh1-deficient yeast may be caused by the presence of anomalous iron species and that NO would chelate such iron, form iron-nitrosyl complexes and limit its toxicity. The ability of NO to chelate free iron and form these complexes has been observed in several biological systems experiencing increased NO concentrations [36], [17], [37]. Interestingly, iron-nitrosyl complexes may have beneficial effects, such as preventing iron from generating free radicals or increasing iron bioavailability. It has been proposed that these complexes can also contribute to iron mobilization as they could be transported to different organs [4]. Therefore, our hypothesis (summarized in Fig. 8) is that Yfh1 deficiency would lead to the presence of anomalous iron species in mitochondria that could compromise iron bioavailability and activate a signaling cascade resulting in Aft1 activation. This activation would be progressive. An initial mild Aft1 activation would slightly increase iron uptake. Then, a vicious cycle will be initiated in which the increased presence of anomalous iron species would further activate Aft1. NO would be able to chelate these iron species and therefore avoid Aft1 activation. Interestingly, some authors have described that respiratory function in Yfh1-deficient yeasts can be rescued by limiting iron toxicity, either expressing human mitochondrial ferritin [5] or overexpressing the vacuolar iron transporter CCC1 [8]. Therefore, our results further support that iron toxicity plays a central role in frataxin-deficient cells.
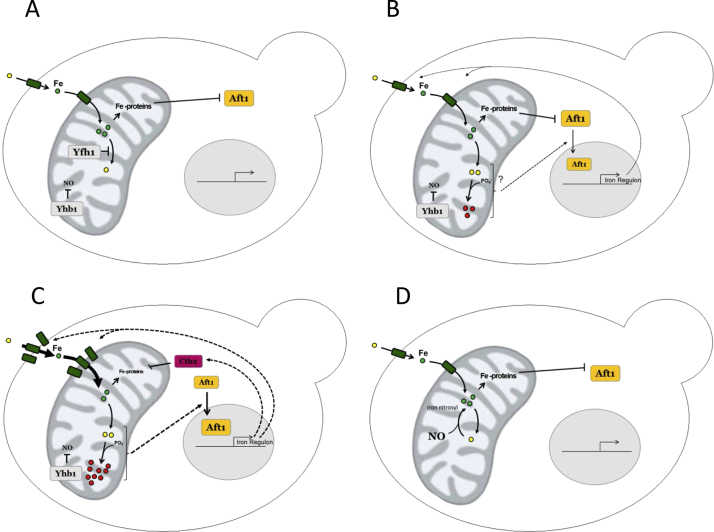
Fig. 8.
Working model to explain the protective effect of NO in Yfh1-deficient cells. A, Yfh1 contributes to maintain mitochondrial iron in a reduced and/or safe form. Fe-containing proteins (notably Fe-S proteins) retain Aft1 in the cytosol. B, loss of Yfh1 leads to increased presence of anomalous iron species which trigger a mild activation of Aft1 trough unknown mechanisms. A moderate increase in iron uptake proteins can be found. C, a vicious cycle takes place: the more iron enters the cell, the more anomalous iron species are formed and Aft1 is activated more strongly. Furthermore, Cth2 is expressed, leads to decreased expression of Fe-containing proteins and the repressing effect of these Fe-proteins on Aft1 is lost. A high increase in iron uptake proteins can be detected. D, In the absence of Yhb1, NO levels increase and NO can form iron-nitrosyl complexes which can prevent the formation of anomalous iron species.
And how can frataxin deficiency be linked to the presence of anomalous iron species? We can envisage at least two different but not mutually exclusive ways. The first one would be related to its role as a regulator of cysteine desulfurase activity [28]. Loss of such regulation could lead to an imbalance between iron delivery and iron-sulfur formation that could trigger the formation of reactive iron species. The second one would be related to its role as an iron chaperone: frataxins would keep iron in a non-toxic, easy-deliverable form. Therefore, its deficiency would increase the presence of free or toxic iron forms [29], [32]. Regarding the nature of these anomalous iron species and its capacity to activate Aft1, Seguin et al. [30], showed that Yfh1-deficient yeast cells accumulated iron in the mitochondria in the form of amorphous nanoparticles of ferric phosphate. In a conditional Yfh1 mutant, these phosphate precipitates appeared early, whereas iron accumulation was more progressive. Moreover, iron accumulation was prevented by phosphate supplementation, suggesting that Aft1 activation in Yfh1-deficient yeasts could be related to decreased phosphate bioavailability. The relevance of this iron-phosphate connection in frataxin deficient cells merits further investigation.
Regarding the relationship between NO and frataxin, our results are reminiscent of that found in frataxin-deficient Arabidopsis thaliana, where Martin and collaborators found that elevated NO levels protected this plant against oxidative stress caused by frataxin deficiency [19]. However, some differences may exist between the mechanisms of NO protection in plants and yeast. In plants, Martin et al. detected an increase in NO production after frataxin deficiency, and induction of the iron regulon was prevented by NO scavengers. In yeast, we have not detected an increase in NO upon Yfh1 depletion and we have shown that NO can prevent activation of the iron regulon. These differences may be related to the fact that NO is a signaling molecule in plants which acts as a mediator in the response to iron scarcity. It is produced upon iron deficiency and is required for the expression of genes involved in iron metabolism [19], [4]. This may not be the case in yeast,
Another interesting point of this work is the analysis of the effect of NO on the metabolic remodeling triggered by Yfh1-deficiency in yeast. The deployed targeted proteomics approach allowed us to test the metabolic status of the yeast cell in a rapid and efficient way. Some of the targeted proteins (or their mRNAs) had been analyzed previously in this same tetO7YFH1 model by western blot (Aco1, Sdh2, Yfh1) or qPCR (Adh2) [21]. We have been able to reproduce these previous results, demonstrating the validity of the approach. In addition, we have monitored several additional proteins, providing a more complete view of the yeast metabolic status. An interesting indicator is the Adh1/Adh2 ratio. Adh1 reduces acetaldehyde to ethanol during glucose fermentation, while Adh2p catalyzes the conversion of ethanol to acetaldehyde. Adh1 accounts for most of the Adh activity in the presence of glucose, while Adh2 is repressed in the presence of glucose and strongly induced in the presence of non-fermentable carbon sources such as glycerol or ethanol [31]. Therefore the Adh1/Adh2 ratio is an indicator of whether the yeasts are focused on fermentation (high Adh1/Adh2 ratio) or respiration (low Adh1/Adh2 ratio). In our model, we can observe that this ratio increases in Yfh1-deficient tetO7YFH1 yeasts due to both decreased Adh2 and increased Adh1. This ratio is restored in Yfh1-deficient tetO7YFH1Δyhb1 yeasts due to partial recovery of both Adh1 and Adh2 control levels. The decrease in Adh2 is caused by inactivation of Adr1, a transcription factor required for Adh2 expression which is inactivated upon Yfh1 deficiency [21]. The partial recovery of Adh2 suggests that NO may partially prevent Adr1 inactivation. As this inactivation can be caused by oxidative stress, the observed effect could be a consequence of the O2- scavenging properties of NO (as shown by the increased presence of nitrotyrosines in double Yfh1 and Yhb1-deficient cells).
The targeted proteomics approach also indicates that the consequences of the other known actor in metabolic remodeling, Cth2, are clearly prevented in Δyhb1 cells. This is not surprising, as Cth2 activation depends on Aft1 activation (which is in turn prevented by NO) and we had previously demonstrated that Aco2 and Sdh2 content (and activity) were preserved in a Yfh1-deficient Δcth2 strain [21]. Therefore, our results confirm that loss of iron-sulfur proteins in Yfh1-deficient cells is a secondary event caused by Cth2 induction.
4. Conclusions
A major conclusion of this work is that Yfh1 deficiency activates the iron regulon by a different pathway than iron-sulfur loss. Therefore, alternative pathways for Aft1 activation may exist in yeast which, as discussed above, could be related to the presence of anomalous iron species. We have also observed that metabolic status can be efficiently monitored using a targeted proteomics strategy focusing on key metabolic enzymes. Finally, the observation that NO can mitigate the defects caused by Yfh1 deficiency supports the hypothesis that Yfh1 loss leads to the increased presence of anomalous iron forms, and that this anomalous iron plays a central role in the events caused by Yfh1 deficiency. It also suggests that NO donors could have a therapeutic effect in FRDA patients. NO donors such as SNP or nitroglycerin have been used for more than a century for controlling congestive heart failure associated with heart attack or lowering blood pressure during surgery [20]. Beyond its vasodilating action, neuroprotective properties have been demonstrated for NO in an iron-induced model of Parkinson's disease [26]. This observation, together with the results presented in this work, opens the possibility to explore the potential therapeutic effect of NO donors in mammalian models of FRDA.
5. Materials and methods
5.1. Yeast strains and culture conditions
The strains used in this work are summarized in Table 1. All the mutants are derived from the strain W303-1A (Mata ura3-1 leu2-3, 112 trp1-1 his3-11,15 ade2-1) considered the wild-type strain in this work. Cells were grown using rich media preferentially containing glycerol (1% yeast extract, 2% peptone and 3% glycerol (YPG)) or glucose when stated (1% yeast extract, 2% peptone and 2% glucose (YPD)). Cells were cultured in a rotary shaker at 30 °C. All the experiments described in this work were performed using exponentially growing cells at optical densities ranging from 0,4 to 0,7 (λ = 600 nm, 1 cm light path).
Table 1.
S. cereviasiae strains used in this work.
| Strain | Name used in text | Relevant genotype | Comments |
|---|---|---|---|
| W303-1A | Wild-type | MATa ura3-1 leu2-3, 112 trp1-1 his3-11,15 ade2-1 | Wild-type |
| BQS201 | tetO7-YFH1 | W303-1A tetO7-YFH1::kanMX4 | Chromosomal YFH1 promoter replaced with tetO7 promoter [22] |
| BQS254 | Δyhb1 | W303-1A yhb1::natMX4 | Deletion of Yhb1 in W303-1A |
| BQS255 | tetO7-YFH1 Δyhb1 | BQS201 yhb1::natMX4 | Deletion of YHB1 in BQS201 |
| BQS280 | tetO7-YFH1 Yhb1-GFP | BQS201 YHB1-GFP::hphNT1 | Chromosomal YHB1 tagged with GFP in BQS201 |
| BQS267 | tetO7-YFH1 Fet3-Myc | BQS201 FET3-MYC::hphNT1 | Chromosomal FET3 tagged with Myc-tag in BQS201 |
| BQS268 | tetO7-YFH1 Δyhb1 Fet3-Myc | BQS255 FET3-MYC::hphNT1 | Chromosomal FET3 tagged with Myc-tag in BQS255 |
| MML348 | Δaft1 | W303-1A aft1::URA3 | Deletion of AFT1 in W303-1A [27] |
| BQS269 | W303 Aft1-GFP | W303-1A AFT1-GFP::hphNT1 | Chromosomal AFT1 tagged with GFP in W303 |
| BQS270 | Δyhb1 Aft1-GFP | BQS254 AFT1-GFP::hphNT1 | Chromosomal AFT1 tagged with GFP in BQS254 |
| MML313 | tetO7-GRX5 | W303-1A tetO7-GRX5::kanMX4 | Chromosomal GRX5 promoter replaced with tetO7 promoter [27] |
| BQS279 | tetO7-GRX5 Δyhb1 | MML313 yhb1::natMX4 | Deletion of YHB1 in MML313 |
5.2. Plasmids
The plasmid pAG25 was used to construct deletion cassettes. This plasmid was a gift from John McCusker (Addgene plasmid # 35121) [11]. Plasmids pYM20 and pYM25 were used for Myc and GFP tagging the c-terminus of selected proteins [15].
5.3. Western Blot analysis
Whole cell protein extracts were prepared in the following way. Cells were resuspended in 50 mM Tris-HCl buffer pH 7,5 containing a mixture of protease inhibitors (Merck-Millipore, ref 539133) and disrupted using glass beads in a BioSpec Mini-Beadbeater. To this lysate, an equal volume of SDS 7,5% and β-mercaptoethanol 7,5% was added. After vortexing, the lysate was centrifuged at 12,000 rpm for 10 min. Protein extracts were separated in SDS-polyacrylamide gels and transferred to PVDF membranes. The following primary antibodies were used: GFP (Clontech, 632381), Nitrotyrosine (Biovision, 5416-50) and Myc (Millipore, 06–549). Anti-Rabbit and anti-mouse antibodies conjugated to peroxidase were used for detection. Images were acquired in a Chemidoc XRS (Bio-Rad) and chemiluminescent data was analyzed using Image lab software (Bio-Rad). Comassie Blue staining of the membrane after detection was used as protein load control.
5.4. Heme and Flavin detection
To detect the holo form of Yhb1, cells were resuspended in PBS containing a mixture of protease inhibitors and disrupted using glass beads in a BioSpec Mini-Beadbeater. These protein extracts were centrifuged (3000 rpm for 5 min) and 20 μg of protein were loaded in polyacrylamide gels and separated in native conditions (without SDS). After electrophoresis, gels were directly imaged in a ChemiDoc XRS to detect Flavin fluorescent signal (Excitation: Blue Epi Illumination; Emission: 530/28 filter) or transferred to a PVDF membrane, washed once with TBS buffer and incubated with HPR substrate to detect heme peroxidase activity. The chemiluminescent signal was acquired in a ChemiDoc XRS.
5.5. RNA isolation and gene expression analysis
Gene expression was analyzed using quantitative Real-Time PCR (RT-qPCR). Total RNA was extracted using RNeasy kit (Qiagen) according to manufacturer's instructions. 1 μg of total RNA was converted into cDNA using SuperScript II system (Invitrogen) and 50 ng of the produced cDNA were used for each qPCR reaction. These reactions were carried out in an iCycler (BioRad) using TaqMan Universal PCR Master Mix kit (Applied Biosystems). Primers and probes for each gene were provided by Applied Biosystems (Taqman Gene Expression Assays) and actin (ACT1) was used as an internal control. Quantification of the acquired data was performed using the Bio-Rad CFX Manager (version 3.1, Bio-Rad). Relative expression values were calculated based on the Ct values with efficiency correction based on multiple samples.
5.6. Targeted proteomics (MRM)
Cells were resuspended in 50 mM Tris-HCl buffer pH 7,5 plus 1 mM EDTA and a mixture of protease inhibitors and were disrupted using glass beads. After, SDS was added to a final concentration of 2% and protein extracts were vortexed, boiled and centrifuged (12,000 rpm 10 min). Proteins were quantified using the BCA assay (Thermo Scientific) and 30 μg were precipitated with cold acetone (9 volumes) and resuspended in 8 M urea, 0.1 M ammonium bicarbonate. Then, proteins were subjected to reduction by 12 mM DTT and alquilation by 40 mM iodoacetamide. Samples were diluted with 0.1 M ammonium bicarbonate to a final concentration of 1.5 M urea and mass spectrometry grade trypsin (Trypsin Gold, Promega) was added to a final enzyme:substrate ratio of 1:100. After digestion, 0,8 ul from a heavy peptide standards mixture was added to the sample. The approximate concentration of each heavy peptide in this mixture was 3,5 μM. Heavy peptides were obtained from JPT (SpikeTidesTM_L). The resulting peptide mix was purified and enriched using C18 columns (Pierce C-18 Spin Columns, Thermo Scientific). Eluted fraction from the C18 column was evaporated using a Concentrator Plus (Eppendorf) and peptides were resuspended in 5% acetonitrile plus 0,1% formic acid. All peptide samples were analyzed on a triple quadrupole spectrometer (Agilent 6420) equipped with an electrospray ion source. Chromatographic separations of peptides were performed on an Agilent 1200 LC system using a Supelco Bioshell A160 Peptide C18 column (1 mm × 15 cm). Peptides (up to 15 micrograms of protein digest) were separated with a linear gradient of acetonitrile/water, containing 0.1% formic acid, at a flow rate of 75 μl/min. A gradient from 5% to 60% acetonitrile in 45 min was used. The mass spectrometer was operated in multiple reaction monitoring mode. Transitions were obtained from SRM atlas and imported into Skyline software [18], which was also used to analyze results. In the SRM assays validation phase, the transitions obtained from SRM atlas were analyzed in several runs. Each MRM acquisition was performed with Q1 and Q3 operated at unit resolution. Once validated and optimized, the SRM assays were used to quantify all the analyzed peptides using scheduled SRM mode in a single run (retention time window, 120 s; cycle time, 1 s). For calculating protein content, the light to heavy ratio of each peptide was normalized to this same value from the housekeeping proteins (actin and porin). Finally, the value obtained in the control sample (tetO7YFH1 minus doxycycline) was used as a reference value.
5.7. Other methods
To analyze subcellular localization of GFP-tagged proteins, exponentially growing cells were washed once and resuspended in PBS. After, they were visualized by fluorescence microscopy (Olympus DP30 BW) using the U-MNUA3 filter. Total cellular iron was quantified as previously described in [34]. Briefly, cells were digested in 3% nitric acid and total iron was quantified using bathophenanthroline sulfonate as a chelator. To determine the rate of superoxide production, exponentially growing cells were centrifuged and washed twice with water and finally resuspended in PBS plus 0,1% glycerol and 5 μg/mL dihydroethidium (DHE, Fluka). The rate of the increase in DHE fluorescence (excitation 520 nm, emission 590 nm) was measured during 20 min using a fluorescence microplate reader (Infinite M200, Tekan) and was used to calculate superoxide production rate. Nitric Oxide levels were measured using the fluorescent dye DAF-FM DA (Life Technologies). 10 μM DAF-FM DA was added to exponentially growing cells during 1 h at 30 °C. After, cells were washed twice and resuspended in PBS. Fluorescence intensity was measured in a flow-cytometer (FACS Canto II, Becton Dickinson).
5.8. Statistical analysis
All experiments were performed in at least 3 completely independent culture preparations. The data obtained from the independent experiments was analyzed using GraphPad Prism v5.01. Data was analyzed using Students t-test or, when more than two samples were compared, using 1 way ANOVA followed with ad hoc Tukey's test. The p-values lower than 0.05(*), 0.01(**) or 0001 (***) were considered significant. Covariance analysis of the targeted proteomics data was performed with MetaboAnalyst using the Pearson correlation coefficient [38].
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants SAF2013-44820-R to J.R. (from Ministerio de Economia y Competitividad (Spain) and SGR2009-00196 from the Generalitat de Catalunya). We thank Roser Pané for technical assistance.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.09.001.
Appendix A. Supplementary material
Supplementary material
References
- 1.Adinolfi S., Trifuoggi M., Politou A.S., Martin S., Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 2002;11:1865–1877. doi: 10.1093/hmg/11.16.1865. [DOI] [PubMed] [Google Scholar]
- 2.Astuti R.I., Watanabe D., Takagi H. Nitric oxide signaling and its role in oxidative stress response in Schizosaccharomyces pombe. Nitric Oxide. 2016;52:29–40. doi: 10.1016/j.niox.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Bayot A., Santos R., Camadro J.-M., Rustin P. Friedreich's ataxia: the vicious circle hypothesis revisited. BMC Med. 2011;9:112. doi: 10.1186/1741-7015-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buet A., Simontacchi M. Nitric oxide and plant iron homeostasis. Ann. N.Y. Acad. Sci. 2015;1340:39–46. doi: 10.1111/nyas.12644. [DOI] [PubMed] [Google Scholar]
- 5.Campanella A., Isaya G., O’Neill H.A., Santambrogio P., Cozzi A., Arosio P., Levi S. The expression of human mitochondrial ferritin rescues respiratory function in frataxin-deficient yeast. Hum. Mol. Genet. 2004;13:2279–2288. doi: 10.1093/hmg/ddh232. [DOI] [PubMed] [Google Scholar]
- 6.Campuzano V., Montermini L., Lutz Y., Cova L., Hindelang C., Jiralerspong S., Trottier Y., Kish S.J., Faucheux B., Trouillas P., Authier F.J., Durr A., Mandel J.-L., Vescovi A., Pandolfo M., Koenig M. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 7.Cassanova N., O’Brien K.M., Stahl B.T., McClure T., Poyton R.O. Yeast flavohemoglobin, a nitric oxide oxidoreductase, is located in both the. J. Biol. Chem. 2005;280:7645–7653. doi: 10.1074/jbc.M411478200. [DOI] [PubMed] [Google Scholar]
- 8.Chen O.S., Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich's ataxia by limiting mitochondrial iron accumulation. J. Biol. Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- 9.Condo I., Malisan F., Guccini I., Serio D., Rufini A., Testi R. Molecular control of the cytosolic aconitase/IRP1 switch by extramitochondrial frataxin. Hum. Mol. Genet. 2010;19:1221–1229. doi: 10.1093/hmg/ddp592. [DOI] [PubMed] [Google Scholar]
- 10.Gardner P.R., Gardner D.P., Gardner A.P. Globins scavenge sulfur trioxide anion radical. J. Biol. Chem. 2015;290:27204–27214. doi: 10.1074/jbc.M115.679621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Cabo P., Vazquez-Manrique R.P., Garcia-Gimeno M.A., Sanz P., Palau F. Frataxin interacts functionally with mitochondrial electron transport chain proteins. Hum. Mol. Genet. 2005;14:2091–2098. doi: 10.1093/hmg/ddi214. [DOI] [PubMed] [Google Scholar]
- 13.Irazusta V., Cabiscol E., Reverter-Branchat G., Ros J., Tamarit J. Manganese is the link between frataxin and iron-sulfur deficiency in the yeast model of Friedreich ataxia. J. Biol. Chem. 2006;281:12227–12232. doi: 10.1074/jbc.M511649200. [DOI] [PubMed] [Google Scholar]
- 14.Irazusta V., Obis E., Moreno-Cermeño A., Cabiscol E., Ros J., Tamarit J. Yeast frataxin mutants display decreased superoxide dismutase activity crucial to promote protein oxidative damage. Free Radic. Biol. Med. 2010;48:411–420. doi: 10.1016/j.freeradbiomed.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 16.Koeppen A.H. Friedreich's ataxia: pathology, pathogenesis, and molecular genetics. J. Neurol. Sci. 2011;303:1–12. doi: 10.1016/j.jns.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster J.R., Hibbs J.B. EPR demonstration of iron-nitrosyl complex formation by cytotoxic activated macrophages. Proc. Natl. Acad. Sci. USA. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M., Colman M.J.R., Gómez-Casati D.F., Lamattina L., Zabaleta E.J. Nitric oxide accumulation is required to protect against iron-mediated oxidative stress in frataxin-deficient Arabidopsis plants. FEBS Lett. 2009;583:542–548. doi: 10.1016/j.febslet.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 20.Miller M.R., Megson I.L. Recent developments in nitric oxide donor drugs. Br. J. Pharmacol. 2007;151:305–321. doi: 10.1038/sj.bjp.0707224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno-Cermeno A., Alsina D., Cabiscol E., Tamarit J., Ros J. Metabolic remodeling in frataxin-deficient yeast is mediated by Cth2 and Adr1. Biochim. Biophys. Acta. 2013;1833:3326–3337. doi: 10.1016/j.bbamcr.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Moreno-Cermeno A., Obis E., Belli G., Cabiscol E., Ros J., Tamarit J. Frataxin depletion in yeast triggers up-regulation of iron transport systems before affecting iron-sulfur enzyme activities. J. Biol. Chem. 2010;285:41653–41664. doi: 10.1074/jbc.M110.149443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Outten C.E., Albetel A.N. Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details. Curr. Opin. Microbiol. 2013;16:662–668. doi: 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picotti P., Clément-Ziza M., Lam H., Campbell D.S., Schmidt A., Deutsch E.W., Röst H., Sun Z., Rinner O., Reiter L., Shen Q., Michaelson J.J., Frei A., Alberti S., Kusebauch U., Wollscheid B., Moritz R.L., Beyer A., Aebersold R. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puig S., Vergara S.V., Thiele D.J. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauhala P., Andoh T., Chiueh C.C. Neuroprotective properties of nitric oxide and S-nitrosoglutathione. Toxicol. Appl. Pharmacol. 2005;207:91–95. doi: 10.1016/j.taap.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Manzaneque M.T., Tamarit J., Belli G. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouault T.A. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 29.Schagerlof U., Elmlund H., Gakh O., Nordlund G., Hebert H., Lindahl M., Isaya G., Al-Karadaghi S. Structural basis of the iron storage function of frataxin from single-particle reconstruction of the iron-loaded oligomer. Biochemistry. 2008;47:4948–4954. doi: 10.1021/bi800052m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seguin A., Santos R., Pain D., Dancis A., Camadro J.-M., Lesuisse E. Co-precipitation of phosphate and iron limits mitochondrial phosphate availability in Saccharomyces cerevisiae lacking the yeast frataxin homologue (YFH1) J. Biol. Chem. 2011;286:6071–6079. doi: 10.1074/jbc.M110.163253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Smidt O., du Preez J.C., Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res. 2008;8:967–978. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 32.Soderberg C.G., Gillam M.E., Ahlgren E.-C., Hunter G.A., Gakh O., Isaya G., Ferreira G.C., Al-Karadaghi S. The structure of the complex Between yeast frataxin and ferrochelatase: characterization and pre-steady state reaction of ferrous iron delivery and heme synthesis. J. Biol. Chem. 2016;291:11887–11898. doi: 10.1074/jbc.M115.701128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tachibana C., Yoo J.Y., Tagne J.-B., Kacherovsky N., Lee T.I., Young E.T. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamarit J., Irazusta V., Moreno-Cermeño A., Ros J. Colorimetric assay for the quantitation of iron in yeast. Anal. Biochem. 2006;351:149–151. doi: 10.1016/j.ab.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Tamarit J., Obis È., Ros J. Oxidative stress and altered lipid metabolism in Friedreich ataxia. Free Radic. Biol. Med. 2016;100:138–146. doi: 10.1016/j.freeradbiomed.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Toledo J.C., Bosworth C.A., Hennon S.W., Mahtani H.A., Bergonia H.A., Lancaster J.R. Nitric oxide-induced conversion of cellular chelatable iron into macromolecule-bound paramagnetic dinitrosyliron complexes. J. Biol. Chem. 2008;283:28926–28933. doi: 10.1074/jbc.M707862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanin A.F., Svistunenko D.A., Mikoyan V.D., Serezhenkov V.A., Fryer M.J., Baker N.R., Cooper C.E. Endogenous superoxide production and the nitrite/nitrate ratio control the concentration of bioavailable free nitric oxide in leaves. J. Biol. Chem. 2004;279:24100–24107. doi: 10.1074/jbc.M312601200. [DOI] [PubMed] [Google Scholar]
- 38.Xia J., Wishart D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016;55 doi: 10.1002/cpbi.11. (14.10.1-14.10.91) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material