Abstract
Nuclear transfection of DNA into mammalian cells is challenging yet critical for many biological and medical studies. Here, by combining cell squeezing and electric-field-driven transport in a device that integrates microfluidic channels with constrictions and microelectrodes, we demonstrate nuclear delivery of plasmid DNA within 1 hour after treatment, the most rapid DNA expression in a high-throughput setting (up to millions of cells per minute per device). Passing cells at high speed through microfluidic constrictions smaller than the cell diameter mechanically disrupts the cell membrane, allowing a subsequent electric field to further disrupt the nuclear envelope and drive DNA molecules into the cytoplasm and nucleus. By tracking the localization of the ESCRT-III (endosomal sorting complexes required for transport) protein CHMP4B, we show that the integrity of the nuclear envelope is recovered within 15 minutes of treatment. We also provide insight into subcellular delivery by comparing the performance of the disruption-and-field-enhanced method with those of conventional chemical, electroporation, and manual-injection systems.
Intracellular delivery, the introduction of exogenous materials into cells, is essential to many studies in basic biology and biomedical research (1, 2). A multitude of techniques exists for cell transfection, including biological, chemical, and physical methods (3). Biological/chemical methods usually rely on carriers such as viruses, vesicles, peptides or nanoparticles (4–7). Physical methods primarily use membrane-disruption techniques such as microinjection, electroporation, laser optoporation, and particle bombardment for gene delivery (8–13). Although electroporation has been widely used for DNA transfection since the early 1980s, the underlying mechanism of delivery is not completely understood in nucleated mammalian cells (14–19). In the electroporation process, DNA molecules accumulate and interact with the electropermeabilized plasma membrane during the electric pulse to form aggregates. Afterwards, those DNA aggregates are internalized into the cytoplasm and subsequently expressed (20–26). It is unlikely that DNA plasmids navigate through the viscous and crowded cytoplasm to reach the nucleus simply by diffusion (27, 28). Microtubule and actin networks have been proposed to play an important role in DNA transportation within the cytoplasm, and the time-scale of such processes can be hours long depending on the cell type (22). The lack of detailed mechanistic understanding and the complex nature of DNA transfer between the plasma membrane and nucleus limit our ability to enhance electroporation performance. Moreover, the strong fields used in current electroporation techniques can lead to significant damage or death (25, 26). Nano structure based methods have demonstrated potential for effective gene transfection by penetrating DNA-loaded nanoneedles into the cell, or by diffusion/electrophoresis through a nanostraw (29–31). However, such methods typically have relatively low throughput. Moreover, the nuclear envelope rupture is not well investigated. Hence, substantial interest remains in creating techniques that can quickly and directly deliver DNA to the nucleus in a large number of cells with controllable nuclear envelope damage.
One example of a feasible strategy would involve the transient disruption of the cell plasma membrane and nuclear envelope followed by entry of the target material before resealing. Cell squeezing is a representative technique that enables delivery of a diversity of materials to numerous cell types by mechanically disrupting the plasma membrane and allowing diffusion to transport materials of interest into the cell cytosol (32–35). DNA delivery is, however, more complicated because DNA must enter the nucleus to perform its function and the cytosolic delivery results in the degradation of DNA before it can reach the nucleus, as reported for microinjection (11). Thus, passive diffusion of DNA is likely insufficient and active transport of the DNA to the nucleus is necessary to initiate gene expression. To address this challenge, we combine disruption of both plasma membrane and nuclear envelope with electric fields to enhance delivery – Disruption and Field Enhanced (DFE) delivery. Recent studies show that moderate nuclear envelope rupture could be rapidly repaired in an ESCRT (endosomal sorting complexes required for transport)-dependent manner, indicating a potential of reversible nuclear envelope rupture (36–39). Here, we use a microfluidic device to create rapid mechanical deformation by cell squeezing to disrupt the plasma membrane, followed by exposing the cell to an electric field that generates reversible nuclear envelope rupture and drives the negatively charged DNA into the nucleus and cytoplasm. With this device, we show a significant increase in efficiency and speed of DNA expression, as well as rapid nuclear localization akin to microinjection. Moreover, DNA plasmids are successfully delivered to both nucleus and cytoplasm at throughputs up to millions of cells per minute per device in continuous flow. We further investigate the disruption and repair of both plasma membrane and nuclear envelope, and their relation to intracellular and nuclear delivery. The DFE system also proves useful in co-delivery of DNA, RNA, and proteins.
DFE Design and Characterization
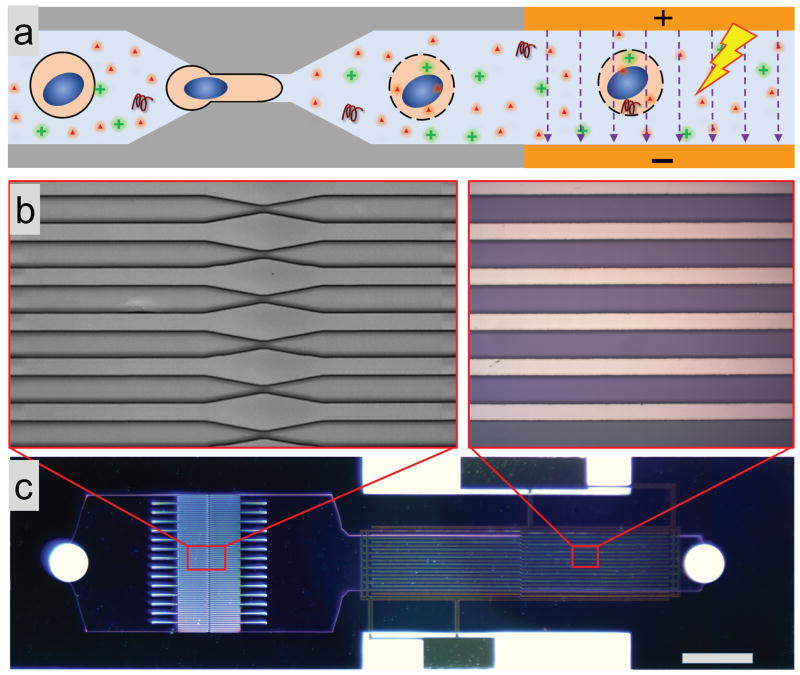
In order to explore whether addition of an electric field would be able to promote delivery of DNA after squeezing, we constructed a microfluidic device with an electric pulse zone downstream of the constriction zone (Figure 1). Each device consisted of a set of parallel, identical constriction channels and a set of electrodes. Consistent with the existing squeeze design (32), 75 parallel channels with constriction zones were etched into a silicon wafer using deep reactive ion etching (DRIE). Subsequently, electrodes were incorporated into the DFE device section by anodic bonding of Pyrex patterned with electrodes (see figure S1 for more details of fabrication). Based on our previous observation for the best efficiency in intracellular delivery (32, 33) with cell squeezing, the width and length of the constriction were in the ranges 4 – 10 μm and 10 – 30 μm, respectively. The length, width, and gap space between each electrode were 8 mm, 60 μm, and 40 μm, respectively, allowing the generation of sufficiently high electric field (~2 kv/cm) with low applied voltage. The duration and duty cycle of the electrical pulse applied to the device ranged from 50 – 500 μs and 1% – 10%, commensurate with values commonly used for electroporation (26).
Fig. 1.
Device structure and working mechanism. (a) Schematic drawing illustrating the working principle: (i) mechanical disruption of the cell membrane as the cell passes through the constriction, and (ii) he subsequent electric pulses driving DNA into the cytoplasm and nucleus through the disrupted membrane. (b) Magnification of a set of identical parallel microfluidic constriction etched into a silicon wafer (left), and a set of electrodes deposited on a Pyrex wafer (right). (c) An optical image of a finished device realized by bonding silicon and Pyrex wafers. The scale bar is 1 mm. Details of device fabrication are in Figure S1.
We typically operated the DFE device at a throughput of 100 000 – 500 000 cells/s per chip per run (each run took 5–20 seconds). Each data point was the mean value of three collections. Each device could be reused 5–10 times before cell debris clogged the channels. At that point, devices were cleaned for reuse by flushing the device with 10% bleach buffer. A 50 – 100 μL mixture of cells and materials to be delivered was placed into the inlet reservoir and driven through the device with nitrogen pressure controlled by a regulator (32). The exposure time of the cells in the electrical field typically ranged from 10 – 100 ms, depending on the flow rate.
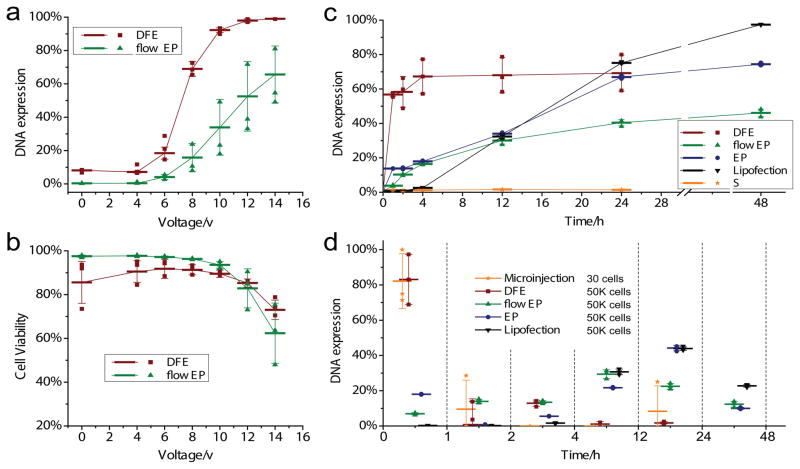
Many parameters governed the performance of mechanical disruption and electrical delivery, including cell speed, constriction dimension, electrical pulse profile, strength, and number of pulses. We first performed a number of experiments to characterize DNA transfection with the DFE technique by treating a mixture of Hela cells and GFP plasmid DNA at different pulse amplitudes. After treatment, cells were incubated at 37 °C for 24 hours. GFP fluorescence measurements by flow cytometry characterized DNA expression. These experiments used a device with a constriction length of 10 μm and a constriction width of 6 μm, denoted as a 10-6 DFE device. Our experimental results show that cell transfection reached above 60% and 90% when the applied amplitude increased to 8V and 10V respectively (Figure 2a red columns). As a control group, we treated cells using a device with the same electrical field and cell speed but no constriction structure (squeezing), corresponding to electroporation in microfluidic devices (flow EP). With cells experiencing only an electric field, the DNA transfection efficacy reached 60% after the applied amplitude increased to 14V (Figure 2a green columns). Both cases had similar cell viability (Figure 2b), suggesting that mechanical disruption dramatically enhanced the DNA delivery at lower field intensities while causing negligible additional damage to the cells.
Fig. 2.
DNA transfection performance and expression dynamics depend on the applied electric field and methods. DNA transfection efficiency (a) and cell viability (b) at 24 h after treatment as a function of applied electric amplitude (voltage). The introduction of mechanical disruption prior electrical delivery significantly enhances the DNA transfection, while bringing negligible damage to cell viability. GFP plasmid DNA transfection efficiency and cell viability were measured by flow cytometry 24 hours after delivery treatment using propidium iodide staining. (c) GFP expression efficiency as a function of time post treatment. Efficiency is defined as the GFP expressing cells over total live cells after treatment. A 10-7 chip is used for mechanical disruption (S: cell squeeze) and DFE. A pulse of 0.1ms/10V at a frequency of 200Hz is used for flow EP (Microfluidic based flow electroporation), and a single pulse of 15ms/15000V is used in EP. (d) The dynamics of DNA expression is analyzed by measuring differential GFP expressing at different time points after treatment. More than 80% of transfected cells expressed GFP within 1 h after treatment in microinjection and DFE. In contrast, most of transfected cells in flow EP (60%), EP (70%) and LP2000 (95%) express GFP 4 – 48 h after treatment. The number of Hela cells in every treatment for each method is shown as well, indicating the throughput of each technique. Each data point is the mean value of triplicate and error bars represent ± s.d.
We also investigated the influence of cell speed on the transfection. Under an applied pulse of 10V, the DNA expression decreased with increasing cell speed due to the reduced number of pulses the cell received as it traveled through the electric field (Figure S2). At cell speeds ~300 mm/s, cell viability and DNA expression seem to best balance severe electric damage at low speed and mechanical damage at high speed. The delivery efficiency of membrane-impermeable Cascade Blue labeled 3-kDa dextran molecules to live Hela cells first dropped and then grew with increasing cell speed, indicating the dominant mechanism of delivery for this molecule switches from electrical field at low speed to mechanical disruption at high speed. The difference in delivery behavior between the 3kDa dextran and DNA molecules further highlight the significance of electrical field effect for DNA transfection.
DFE versus Microinjection, Flow EP, EP, and Cell Squeezing
In order to gain further insight into the mechanism of DFE delivery, we carried out a comparative study with four widely used DNA transfection techniques: microinjection (Eppendorf microinjector 5242), Lipofection (lipofectormine 2000 from ThermoFisher), flow EP (microfluidics-based flow electroporation) and EP (conventional electroporation, using the NEON electroporation system from ThermoFisher, a common commercial electroporation tool). DNA expression was analyzed using flow cytometry after treatment with each technique. EP and flow EP showed similar expression kinetics as GFP gradually expressed throughout 24 hours after treatment (Figure 2c and d). 70% of GFP expressing cells (cells that express GFP fluorescence after 48 hours) expressed GFP between 4–48 hours. In contrast, with microinjection and DFE, more than 80% of GFP expressing cells had measureable expression within the first hour post treatment, indicating that DNA transcription/translation occurred soon after treatment. The remaining 20% of ultimately GFP expressing cells had detectable expression 1 to 4 hours post treatment. Microinjection is broadly accepted as a means of facilitating direct injection of materials into the nucleus. The similar DNA expression kinetics for microinjection and DFE suggest that DNA delivered by DFE becomes accessible for transcription in the nucleus.
For the Lipofection case, we found minimal GFP fluorescence in the first 4 hours post treatment, and more than 95% of transfected cells expressed GFP between 4 – 48 h after treatment. Fluorescence images of GFP expressed cells by DFE, EP, and Lipofection are shown in Figure S3. We further compared the fluorescence intensity of GFP in Hela cells as measured by flow cytometry (Figure S4). The DFE data display a peak in the fluorescence intensity that keeps increasing in intensity within the first 4 hours after treatment indicating that the DNA transcription/translation in most of cells occurred from a similar starting time point soon after treatment. In contrast, for EP and Lipofection, the fluorescence intensity has a flat distribution in the first 12 hours, suggesting that the time required for the DNA migration to nucleus and release from carrier varies from 4 to 24 hours or even longer. DFE has lower RSD (relative standard deviation) and higher NMI (normalized mean intensity) than EP, indicating DFE has better transfection uniformity and level (more transfection) than EP. Lipofection has the highest NMI, which could be due to the fact the DNA molecules are confined in lipid nano particles while in DFE and EP DNA molecules are distributed throughout the solution.
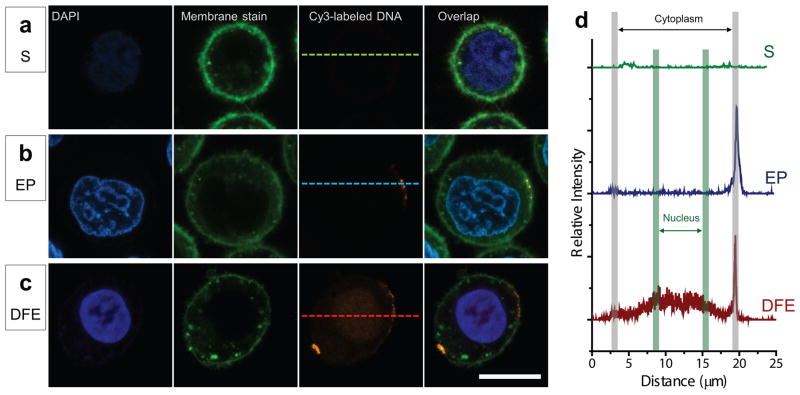
To further explore the working mechanism of DFE, we directly visualized the distribution of DNA at single cell level using CY3 labeled plasmid DNA (Figure 3). Cells were first incubated with DAPI (ThermoFisher) and Cell Mask green plasma membrane stain (ThermoFisher) for nuclear and membrane staining, and then mixed with labeled DNA right before treatment with DFE, EP, and Squeezing. After treatment, cells were incubated in culture medium for 3–5 minutes and then fixed. Optical measurements were carried out using a Nikon A1R confocal microscope. When an electric pulse of 15ms/1200V, known to permeabilize cells, was applied in NEON electroporation system, a sharp CY3 fluorescence spots appeared at the plasma membrane level, indicating the absorption and accumulation of DNA on the membrane (Figure 3b, see Figure S5 for additional results). This result is consistent with previous studies that demonstrate embedding of DNA into the plasma membrane. In Squeezing, no fluorescence of labeled DNA was detected in the cytoplasm with the confocal microscope. In DFE, we found labeled DNA fluorescence distributed in the cytoplasm, nucleus, and plasma membrane (Figure 3c, see Figure S6 for additional results). The bright spots on the plasma membrane are DNA complexes formed as in conventional electroporation. Importantly, the direct visualization of DNA in the cytoplasm and nucleus further supports the hypothesis that DFE is capable of more effective delivery of DNA and indicates that the mechanism of action of the DFE system is likely distinct from electroporation or squeezing alone. These observations are further characterized by the relative fluorescence intensity profiles (Figure 3d) showing the DNA distribution along the dashed line across the single cells in Figures 3a–c treated by squeezing, EP, and DFE. The higher fluorescence intensity in the nucleus than in cytoplasm after DFE could be attributed to (1) degradation of DNA in the cytoplasm by the surrounding DNase and subsequent outward diffusion of Cy3 dye detached from the degraded DNA; or (2) more DNA trapping in the dense nucleus than surrounding cytosol as DNA transits through the cell in the electric field. Combined with our observations of rapid DNA expression, either case demonstrates that disruption and field enhanced delivery facilitates efficient delivery of DNA directly to the nucleus.
Fig. 3.
Visualization of the delivery of fluorescence labeled plasmid DNA (LDNA) to Hela cells. After nucleus and plasma membrane staining, Cells were mixed with Cy3 labeled plasmid DNA before transfection. After treatment, cells were washed with OPTI MEM, fixed with cell fixation kit, and ready for confocal imaging. (a) by only cell squeezing (S), no LDNA signal was detected in the cell, A 10-7 chip was used at cell speed of 500mm/s. (b) in electroporation (EP), an electric pulse of 15ms/1200V was applied using NEON electroporation system. DNA accumulation was found on the plasma membrane. (c) in DFE, a significant Cy3 fluorescence was observed filling cytoplasm and nucleus.10-7 DFE chip was used at cell speed of 500mm/s with applied electric pulse of 0.1ms (200Hz) at 10 V. Labeled DNA was detected in plasma membrane, cytoplasm and nucleus. (d), Relative fluorescence intensity profile over the dashed line across single cells in a, b, and c, shows the labeled DNA molecule distribution after treatments of squeezing (S), EP, and DFE. Scale bar is 10 μm. Original confocal fluorescent images are shown.
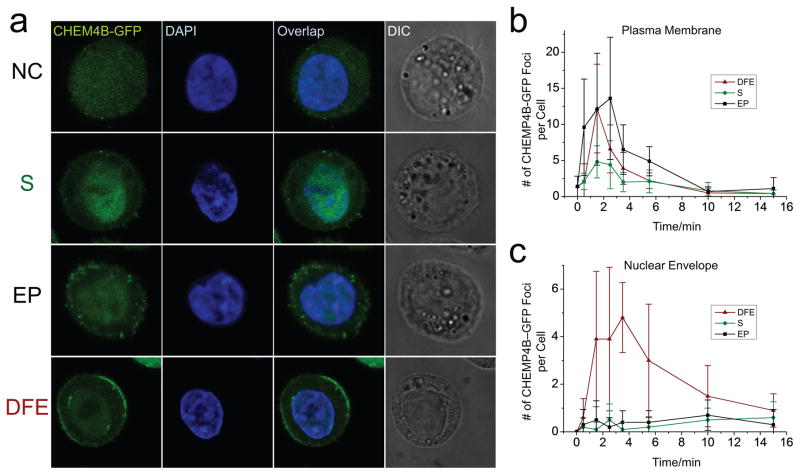
The ESCRT III complex is involved in repair of both plasma membrane and nuclear envelope rupture (36–39). We used HeLa cells expressing GFP-tagged CHMP4B, an important ESCRT III complex subunit, to study the nuclear envelope rupture and repair in DFE. We can induce recruitment of CHMP4B-GFP at the wounding site near the nuclear envelope using microinjection (Figure S7). In DFE, CHMP4B-GFP formed transient foci at the site of both nuclear envelope and plasma membrane (Figure 4a). In EP and Squeezing, CHMP4B-GFP was only recruited to plasma membrane. CHMP4B-GFP localized to the site of both plasma and nuclear membrane at 1–2 minutes right after membrane disruption to repair damage, and decreased after resealing (Figure 4b and c, figure S8). This suggests that DFE could generate reversible disruption on both plasma membrane and nuclear envelope, a critical step for nuclear delivery.
Fig. 4.
ESCRT-III recruitment for plasma membrane and nuclear envelope repair. CHMP4B-GFP expressing Hela cells were stained with DAPI to visualize the nucleus before treatment. After treatments of squeezing (S), EP, or DFE, cells were incubated in culture media, and fixed with a cell fixation kit at 0.5, 1.5, 2.5, 3.5, 5.5, 10, and 15 minutes after treatment, and processed for confocal imaging. (a) Confocal images of representative cells at 1.5 minutes after no treatment (NC, Negative Control), squeezing (S), electroporation (EP), and DFE. CHMP4B-GFP foci are visible at both plasma membrane and nuclear envelope in DFE, but only at the plasma membrane in S and EP. Number of CHMP4B-GFP foci at plasma membrane (b) and nuclear envelope (c) after treatment of S, EP and DFE (10 cells at each data point). Original confocal fluorescent images are shown. Each data point represents the mean value of 10 cells and error bars represent ± s.d..
Discussion
Intracellular delivery of nucleic acids is a challenging first step for an abundance of biological studies and applications. However, the current leading methods for DNA transfection, such as electroporation and lipofection, rely on a delayed trafficking of DNA to the nucleus. On the other hand, mechanical and other permeabilization techniques that deliver DNA directly to the cytoplasm often fail to achieve nuclear penetration and subsequent expression. Here, the proposed DFE concept combines the efficacy of mechanical membrane disruption with the driving force of a field – thus potentially maintaining the robust and rapid delivery capabilities of mechanical disruption while enhancing nuclear delivery of plasmids. DFE performance is likely a nonlinear combination of mechanical disruption and electrical delivery, each of which are influenced by complex sets of parameters. The choice of the buffer solution medium is a challenge. Commercial available buffers (including electroporation buffer from NEON and Eppendorf) works well for electric delivery, but not for cell squeezing based membrane disruption. Phosphate buffered saline (PBS) works well for cell squeezing, but not for electric delivery owing to its high conductivity, which produces electrolytic effects including changes in temperature, pH, and the chemical composition of the solution in proportion to the applied voltage. The hypo-osmolar buffer we used here is compatible with both mechanical disruption and electric delivery process. As expected, we found that hypo-osmolar buffer enlarged cell size, made the plasma membrane more susceptible to disruption (40), and subsequently lowered the cell velocity required for mechanical disruption. The use of hypo-osmolar buffer also facilitated effective electric delivery (41). Lower electric field strength for electric delivery could be used at low cell speeds to protect electrodes and avoid electrolysis.
DNA transport from plasma membrane to nucleus and subsequent transcription is a complex, most likely active, process that can take hours and may vary dramatically among different cell types. This process is essential in electroporation and carrier-based methods such as Lipofection. We have demonstrated that DFE is able to deliver DNA directly into the nucleus by providing a driving force to move DNA across a mechanically disrupted plasma membrane. The mechanical disruption decreases the plasma membrane barrier function to achieve an enhanced electrical delivery. The post-squeezing electric field might also alter distribution of openings in the membrane (42), to facilitate intracellular delivery. To our knowledge, the DFE experiments represent the most rapid expression of plasmid DNA in a high throughput setting (up to a few million cells per device per minute). The throughput could be further improved by adding more parallel microchannels on the chip or operating multiple devices in parallel. As shown in Fig. S2, the increase of diffusion-based delivery coincided with decreasing DNA transfection when increasing cell speed, this is due to the inverse dependence between cell squeezing speed and electric field exposure. High cell speed enhances strong mechanical disruption and thus more diffusive delivery, it also leads to a shorter exposure time in the electric field, causing lower electrical field driven delivery. Such interdependence is a limitation of the current version of the device.
The DNA expression dynamics of Lipofection in Figures 2c and d reveal that DNA transfer to the nucleus and subsequent transcription can require over 4 hours in Hela cells. DNA expression with conventional electroporation was slightly faster. There is ongoing debate regarding how DNA migrates into the nucleus during the electroporation process (15). Some hypothesize that electric pulse permeabilizes the cell membrane and electrophoresis drives DNA directly into the nucleus, while others observe that DNA first form aggregates at electropermeabilized areas of the plasma membrane during electric pulse and then migrates toward the nucleus through a biologically active process (17, 18, 25, 26). In our EP results, 20% of transfected cells expressed GFP within the first hour and 80% expressed throughout the next 20 hours. This could be an indication that both of the aforementioned mechanisms occurred in EP. The small portion of cells that express GFP immediately after treatment may involve direct electrophoresis of DNA into the nucleus while the majority of cells that expressed GFP after 4 hours must first transport the DNA to the nucleus for expression, as in the case of Lipofection. Future studies could integrate a cell trap structure in the electric field downstream of the mechanical disruption to visualize DNA migration through the mechanically induced disruption in the plasma membrane. Recently ESCRT-III proteins have been shown to be involved in both plasma and nuclear membrane repair (36–39). We used HeLa cells expressing GFP-tagged CHMP4B, an important subunit of ESCRT-III complex, to monitor and measure the membrane dynamics in DFE. The recruitment of CHMP4B-GFP foci at both plasma and nuclear membrane after DFE treatment reveals that both membrane systems were disrupted and resealed, which is a critical step for nuclear delivery. In Squeezing and EP, however, CHMP4B-GFP only localized at the plasma membrane, indicating that nuclear envelope rupture did not occur at detectable levels. ESCRT proteins, including CHMP4B, are involved in the repair of small membrane wounds less than 100 nm (36). This may explain why fewer CHMP4B-GFP foci were observed after squeezing compared to EP and DFE. It is known that electric fields usually generate pores in the range of a few nm or tens of nm, while mechanical disruption in Squeezing at high flow rates could generate larger wounds or prompt membrane plasma repair via pathways other than ESCRT-mediated processes (32). The recruitment of CHMP4B-GFP to both plasma and nuclear membrane wounds occurred from1–2 minutes after the disruption, and nuclear membrane repair was completed by about 10–15 minutes post disruption, about 5 minutes slower than plasma membrane resealing. Such repair dynamics are consistent with recently published reports (36–38).
DFE delivery combines microfluidic-based membrane disruption and field effects to achieve potentially greater efficacy than either individual approach. The mechanical disruption techniques, such as squeezing, have shown significant success in delivery of a variety of materials, including proteins and nanomaterials, to a diversity of cell types with minimal toxicity, but have had limited success with DNA, presumably due to ineffective nuclear delivery. In DFE, we have demonstrated a successful co-delivery of DNA plasmid, mRNA, and protein (APC mouse IgG1 k Isotype Ctrl antibody, Figure S9). The results show that DNA and mRNA are significantly dependent on the electric field, while the protein delivery is more dependent on mechanical disruption. In mouse Embryonic Stem Cells (mESC), we achieved DNA transfection of 19% and 36% at 10V and 14V respectively in DFE (Figure S10 a–c). By combining mechanical disruption and electric field effects, our DFE concept has demonstrated reversible nuclear and plasma membrane disruption for both nuclear and cytosolic delivery, and is capable of co-delivering proteins and nucleic acids, characteristics that are difficult to accomplish with any of the aforementioned methods individually.
Outlook
Our DFE system is able to deliver DNA molecules into the nucleus and cytoplasm at high throughput with minimal cell damage. We anticipate that such direct and rapid nuclear delivery will find utility in studies of fundamental biology and biomedical applications such as the implementation of more robust DNA transfection for cell-based therapies. Future work will focus on developing a deeper understanding of the interplay between the membrane disruptions, electric field enhanced transportation, and the cell response, thus facilitate effective implementation in more challenging cell types and applications. One would expect the parameter space of DFE systems to be more complex than squeezing or electroporation alone as its efficacy relies on the synergistic interplay of relevant parameters from both techniques. Nuclear envelope disruptions in DFE can be explored further. Nevertheless, the DFE concept shows potential to enable more rapid delivery and expression compared to existing transfection technologies, and paves the way toward more targeted delivery strategies at the subcellular level.
METHODS
Device Fabrication and Experimental Setup
A silicon wafer was bonded to a Pyrex wafer to form the DFE microfluidic device. Two major steps were involved in the fabrication: (1) the fabrication of microfluidic channels on silicon wafer, and (2) the fabrication of microfluidic electrodes on Pyrex wafer (see Supporting Information for more details). The device was mounted onto a holder with inlet and outlet reservoirs (more details in ref. 29). Electric pulses were generated from a function generator (Agilent 33220A) and gained through an amplifier to drive the device through the wire bonded to the electrode pads using conductive epoxy. Cells were suspended to a density of 1–5 × 106 cells/mL in a modified buffer (25mM KCl, 0.85 mM K2HPO4, 0.3mM KH2PO4, 36mM myo-inositol, PH 7.2, conductivity 3.5mS/cm at 25 °C, ref. 37) for experiment. Solutions of cells, mixed with desired delivery material, were placed in the inlet reservoir. This reservoir was then connected to a compressed air line controlled by a regulator. A pressure (0–20psi) was used to drive the fluid through the device, and at the same time, electric pulses were applied to the device as cells flowed through. Cells were collected from outlet reservoir and subjected to further treatment.
Cell culture
HeLa cell line (Hela-Kyoto) was provided by Hyman Lab, Max Planck Institute, Dresden, Germany. Authentication Test was performed by Multiplex. Cells were routinely cultured in 75 T flasks containing 20mL of DMEM culture medium supplemented with 10% fetal bovine serum (FBS, Invitrogen 16000), at 37 °C in a humidified atmosphere containing 5% CO2. hESCs (H13, WiCell, Wisconsin) were grown on an inactivated mouse embryonic fibroblast (MEF) feeder layer, provided by Jeff Karp Lab in Brigham and Women’s Hospital, Harvard Medical School. mESCs (129P2/OlaHsd, provided by Richard Sherwood lab from Brigham and Women’s Hospital, Harvard Medical School) were maintained on gelatin-coated plates in mESC medium composed of Knockout DMEM (Life Technologies) supplemented with 15% defined FBS (HyClone), 0.1 mM nonessential amino acids (Life Technologies), 1% Glutamax (Life Technologies), 0.55 mM 2-mercaptoethanol (Sigma), 1× ESGRO LIF (Millipore), 5 nM GSK-3 inhibitor XV and 500 nM UO126. Cells were regularly tested for mycoplasma. HeLa cells expressing CHMP4B-GFP were a generous gift of the Tony Hyman lab (43). They were maintained in geneticin-containing media and FACS sorted prior to experiments.
Delivery Materials
Fluorescently labeled molecules, including dextran and plasmid DNA, were mixed with cell solution at a concentration of 0.1 mg/mL. GFP DNA plasmid was used to measure the DNA transfection.
Flow Cytometry
For quantitative analysis of cells after DNA delivery, cells were treated with trypsin 24 hours after delivery experiment and washed twice with PBS (200 uL per well in V-bottom 96-well plate). They were then resuspended in PBS solution supplemented with 3% FBS, 1% F-68 Pluronics, and 1 ug/mL propidium iodide (Sigma) for the analysis using LSR Fortessa (BD Biosceinces).
Lipofection
Lipofectamine 2000 DNA transfection kit was used to represent Lipofection technique. The DNA-lipid complex was prepared by combining 2 uL of Lipofection 2000 reagent in 100 uL of Opti-MEM medium with 2 ug of DNA plasmid in 100 uL of Opti-MEM medium, followed by 5 minutes of incubation in room temperature. The DNA-lipid complex solution was added to cell sample at the ratio of 1:30. More details can be found in the product protocol (Lipofection 2000, Life Technologies).
Microinjection
The microinjection of DNA plasmid into Hela cells was operated by experienced staff. 30 cells were injected for each condition. The DNA concentration in the buffer for injection is 0.1 ug/mL. A pressure of 60hPa and a duration of 0.2 s were used for each injection.
Supplementary Material
Acknowledgments
We thank Peimin Qi from Division of Comparative Medicine at MIT for performing the microinjection, thank Dr. Qing Chen from Richard Sherwood lab at Brigham and Women’s Hospital for providing mouse embryonic stem cells, and thank Dr. Xiaolei Yin from Jeff Karp Lab at David Koch Institute for Integrative Cancer Research at MIT for providing human embryonic stem cells. The assistance and expertise of Glenn Paradis and personnel in the flow cytometry core at the Koch Institute and the Microsystem Technology Laboratory at MIT are highly acknowledged. This research was supported by National Institutes of Health (R01GM101420-01A1), and device fabrication was performed at the Microsystem Technology Laboratory at MIT.
Footnotes
AS, RL, and KFJ have a financial interest in SQZ Biotechnologies.
Author Contributions: X.D., M.S., A.S., R.L, and K.J. designed the research, X.D. and M.S. performed the experiments, X.D. fabricated the devices. X.D. M.S., A.S., J.W., R.L., and K.J. analyzed the data. X. D., M. S., A. S., J.W., R. L., and K. J. wrote the article.
Data availability. The authors declare that all data supporting the findings of this study are available within the paper and the Supplementary Information.
References
- 1.Yin H, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 2.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. The New England journal of medicine. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart MP, Sharei A, Ding X, Sahay G, Langer R, Jensen KF. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 4.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu SC, Huang GYL, Liu JH. Production of retrovirus and adenovirus vectors for gene therapy: a comparative study using microcarrier and stationary cell culture. Biotechnol Progr. 2002;18:617–622. doi: 10.1021/bp020026p. [DOI] [PubMed] [Google Scholar]
- 6.Schmid RM, et al. Liposome mediated gene transfer into the rat oesophagus. Gut. 1997;41:549–556. doi: 10.1136/gut.41.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien JA, Lummis SCR. Biolistic transfection of neuronal cultures using a hand-held gene gun. Nature Protoc. 2006;1:977–981. doi: 10.1038/nprot.2006.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells DJ. Gene therapy progress and prospects: electroporation and other physical methods. Gene Ther. 2004;11:1363–1369. doi: 10.1038/sj.gt.3302337. [DOI] [PubMed] [Google Scholar]
- 10.Meacham JM, Durvasula K, Degertekin FL, Fedorov AG. Physical methods for intracellular delivery: practical aspects from laboratory use to industrial-scale processing. J Lab Autom. 2014;19:1–18. doi: 10.1177/2211068213494388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 12.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Laboratory; 2003. [Google Scholar]
- 13.Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- 14.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1:841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escoffre JM, et al. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol Biotechnol. 2009;41:286–95. doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- 16.Vasilkoski Z, Esser AT, Gowrishankar TR, Weaver JC. Membrane electroporation: The absolute rate equation and nanosecond time scale pore creation. Phys Rev E. 2006;74:021904. doi: 10.1103/PhysRevE.74.021904. [DOI] [PubMed] [Google Scholar]
- 17.Klenchin VA, Sukharev S, Serov SM, Chernomordik LV, Chizmadzhev YA. Electrically induced DNA uptake by cells is a fast process involving DNA electrophoresis. Biophys J. 1991:60. doi: 10.1016/S0006-3495(91)82115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver JC, Smith KC, Esser AT, Son RS, Gowrishankar TR. A brief overview of electroporation pulse strength-duration space: a region where additional intracellular effects are expected. Bioelectrochemistry. 2012;87:236–43. doi: 10.1016/j.bioelechem.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan CA, Neumann E, Sowers AE, editors. Electroporation and electrofusion in cell biology. Springer Science & Business Media; 2013. [Google Scholar]
- 20.Golzio M, Teissie J, Rols MP. Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA. 2002;99:1292–1297. doi: 10.1073/pnas.022646499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paganin-Gioanni A, et al. Direct visualization at the single-cell level of siRNA electrotransfer into cancer cells. Proc Natl Acad Sci USA. 2011;108:10443–7. doi: 10.1073/pnas.1103519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosazza C, et al. Intracellular Tracking of Single-plasmid DNA Particles After Delivery by Electroporation. Mol Ther. 2013;21:2217–2226. doi: 10.1038/mt.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boukany PE, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–54. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 24.Teissie J, Golzio M, Rols MP. Mechanisms of cell membrane electropermeabilization: a minireview of our present (lack of ?) knowledge. Biochim Biophys Acta. 2005;1724:270–80. doi: 10.1016/j.bbagen.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Yarmush ML, Golberg A, Serša G, Kotnik T, Miklavčič D. Electroporation-based technologies for medicine: principles, applications, and challenges. Annu Rev Biomed Eng. 2014;16:295–320. doi: 10.1146/annurev-bioeng-071813-104622. [DOI] [PubMed] [Google Scholar]
- 26.Geng T, Lu C. Microfluidic electroporation for cellular analysis and delivery. Lab Chip. 2013;13:3803–21. doi: 10.1039/c3lc50566a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lechardeur D, Verkman a, Lukacs G. Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Deliv Rev. 2005;57:755–767. doi: 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Dowty ME, Williams P, Zhang G, Hangstrom JE, Wolff JA. Plasmid DNA entry into post-mitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalek AK, et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci USA. 2010;107:1870–5. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X, et al. Nanostraw–Electroporation System for Highly Efficient Intracellular Delivery and Transfection. ACS Nano. 2013:4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, et al. Poking cells for efficient vector-free intracellular delivery. Nat Commun. 2014;5:4466. doi: 10.1038/ncomms5466. [DOI] [PubMed] [Google Scholar]
- 32.Sharei a, et al. A vector-free microfluidic platform for intracellular delivery. Proc Natl Acad Sci USA. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Sharei A, Sim WY, Adamo A, Langer R, Jensen KF, Bawendi MG. Nonendocytic delivery of functional engineered nanoparticles into the cytoplasm of live cells using a novel, high-throughput microfluidic device. Nano letters. 2012;12:6322–6327. doi: 10.1021/nl303421h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kollmannsperger A, et al. Live-cell protein labelling with nanometre precision by cell squeezing. Nat Commun. 2016;7:10372. doi: 10.1038/ncomms10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szeto GL, et al. Microfluidic squeezing for intracellular antigen loading in polyclonal B-cells as cellular vaccines. Sci Rep. 2015;5:10276. doi: 10.1038/srep10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 37.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–62. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 38.Denais CM, Gilbert RM, Isermann P, McGregor AL, Lindert M, te Weigelin B, Davidson PM, Friedl P, Wolf K. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groulx N, Boudreault F, Orlov SN, Grygorczyk R. Membrane reserves and hypotonic cell swelling. The Journal of membrane biology. 2006;214:43–56. doi: 10.1007/s00232-006-0080-8. [DOI] [PubMed] [Google Scholar]
- 41.Wei Z, et al. A laminar flow electroporation system for efficient DNA and siRNA delivery. Anal Chem. 2011;83:5881–5887. doi: 10.1021/ac200625b. [DOI] [PubMed] [Google Scholar]
- 42.Son RS, Gowrishankar TR, Smith KC, Weaver JC. Modeling a conventional electroporation pulse train: decreased pore number, cumulative calcium transport and an example of electrosensitization. Transactions in Biomedical Engineering. 2015 doi: 10.1109/TBME.2015.2466234. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 43.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nature methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.