Abstract
Human metabolomics has great potential in disease mechanism understanding, early diagnosis, and therapy. Existing metabolomics studies are often based on profiling patient biofluids and tissue samples and are difficult owing to the challenges of sample collection and data processing. Here, we report an alternative approach and developed a computation-based prediction system, MetabolitePredict, for disease metabolomics biomarker prediction. We applied MetabolitePredict to identify metabolite biomarkers and metabolite targeting therapies for rheumatoid arthritis (RA), a last-lasting complex disease with multiple genetic and environmental factors involved.
MetabolitePredict is a de novo prediction system. It first constructs a disease-specific genetic profile using genes and pathways data associated with an input disease. It then constructs genetic profiles for a total of 259,170 chemicals/metabolites using known chemical genetics and human metabolomic data. MetabolitePredict prioritizes metabolites for a given disease based on the genetic profile similarities between disease and metabolites. We evaluated MetabolitePredict using 63 known RA-associated metabolites. MetabolitePredict found 24 of the 63 metabolites (recall: 0.38) and ranked them highly (mean ranking: top 4.13%, median ranking: top 1.10%, P-value: 5.08E–19). MetabolitePredict performed better than an existing metabolite prediction system, PROFANCY, in predicting RA-associated metabolites (PROFANCY: recall: 0.31, mean ranking: 20.91%, median ranking: 16.47%, P-value: 3.78E–7). Short-chain fatty acids (SCFAs), the abundant metabolites of gut microbiota in the fermentation of fiber, ranked highly (butyrate, 0.03%; acetate, 0.05%; propionate, 0.38%). Finally, we established MetabolitePredict’s potential in novel metabolite targeting for disease treatment: MetabolitePredict ranked highly three known metabolite inhibitors for RA treatments (methotrexate:0.25%; leflunomide: 0.56%; sulfasalazine: 0.92%).
MetabolitePredict is a generalizable disease metabolite prediction system. The only required input to the system is a disease name or a set of disease-associated genes. The web-based MetabolitePredict is available at:http://xulab.case.edu/MetabolitePredict.
Keywords: Human metabolomics, Metabolomic biomarker discovery, Human gut microbiome, Metaboloite inhibitor, Rheumatoid arthritis
1. Introduction
Human metabolome is the complete set of small-molecule metabolites found in the human body. Human metabolomics is the study of metabolome using patient biofluids and tissue samples in order to find molecular profiles associated with diseases or health status. Metabolomics has potential for early disease diagnosis, monitoring therapy and understanding disease pathogenesis [1,2].
Profiling human metabolome is challenging. The human metabolomes are affected not only by intrinsic factors such as host genetics, but also by many external factors, including lifestyle, pollutants, diet, medications, exercise, gut microbiota, and age [3]. In addition, metabolites are highly heterogeneous and include lipids, small peptides, amino acids, organic acids, vitamins, carbohydrates, nucleic acids, as well as metabolites derived from drugs, environmental contaminants, food additives, toxins, cosmetics, and other xenobiotics [4]. Since human metabolome is affected by not only intrinsic but also many external factors, sample collection, storage, processing and data analysis is crucial for reproducibility and knowledge generalization.
Here we report a novel disease metabolite prediction system, MetabolitePredict, that performs de novo prediction of disease-associated metabolites and metabolite targeting therapies via simultaneous integrative analysis of vast amounts of human disease genetics, chemical genetics, human metabolomic data, and genetic pathways. MetabolitePredict complements current clinical sample-based metabolomics studies: current human metabolomics characterize clinically significant metabolite profiles from patient samples; MetabolitePredict contextualizes disease metabolite biomarker discovery with vast amounts of existing system-level genetic and molecular data. MetabolitePredict is also different from existing computation-based metabolite prediction systems, including PROFANCY [5] and MetPriCNet [6], which identify additional disease metabolites based on known disease-associated metabolites, therefore cannot perform predictions for diseases without known metabolites. MetabolitePredict is a de novo prediction system that can predict metabolite biomarkers for any diseases without the need of known disease-associated metabolites. We demonstrated that MetabolitePredict performs better than PROFANCY in prioritizing RA-associated metabolites. We recently developed algorithms that prioritize human gut microbial metabolite biomarkers for colorectal cancer (CRC) [7] and Alzheimer’s disease [8] based on genetic relevance between diseases and microbial metabolites (171 microbial metabolites). MetabolitePredict incorporated our previous algorithms and developed new algorithms for large-scale prioritization of metabolites (259,170 chemicals/pathways) based on pathway profile similarity. In addition, MetabolitePredict has the additional capability in identifying metabolic inhibitors for novel disease treatments. To the best of our knowledge, MetabolitePredic represents the first de novo prediction system for both metabolomic biomarker discovery and metabolite targeting-based drug discovery.
We applied MetabolitePredict to rheumatoid arthritis (RA) for both metabolomics biomarker discovery and metabolite targeting for two reasons. First, RA is a common, chronic, systemic, inflammatory disorder. RA affects up to 1% of the population worldwide [9]. The cause of RA remains unknown, with multiple genetic and environmental factors involved [10–12]. Second, the availability of known RA-associated metabolites and metabolite inhibitor-based treatments allows us to robustly evaluate MetabolitePredict’s functionalities. We tested MetabolitePredict using 63 RA-associated metabolites extracted from published metabolomics studies [3,13] and from the Human Metabolome Database (HMDB) [4].
We evaluated MetabolitePredict in identifying human gut microbial metabolites that may be involved in RA pathogenesis. Human gut microbiota (>1014 microbial cells comprising about 1000 different species) are important modifiable environmental factors that we are exposed to continuously [14]. These microbiota exist in symbiotic relationship with a human host by metabolizing compounds that humans are unable to utilize and by controlling the immune balance of the human body [15]. Evidence increasingly suggests that gut microbiota and their metabolites exert profound effects on the host immune system, and are implicated in the initiation and progression of many common complex diseases, including RA [16,17]. We demonstrated that MetabolitePredict has the potential to identify which and how human gut microbial metabolites are associated with RA.
Disease-specific metabolomic profiles are a promising source of drug targets. Considerable efforts have been focused on combining metabolic modulators with conventional therapies for cancer [18] and other diseases. Metabolic inhibitors such as methotrexate, leflunomid and sulfasalazine have been used to treat RA [19–22]. In this study, we established MetabolitePredict’s potential in novel metabolite targeting for diseases.
2. Data and methods
2.1. Data
MetabolitePredict incorporated a large amount of data, including human metabolome, disease genetics, chemical genetics, functional protein interactions and signaling pathways. The system is highly flexible and additional datasets can be easily included.
2.1.1. Disease genetics and genomics data
MetabolitePredict incorporates disease genetics from two complementary data resources: (1) The Catalog of Published Genome-Wide Association Studies (GWAS Catalog), an exhaustive source containing descriptions of disease-/trait-associated single nucleotide polymorphisms (SNPs) from published GWAS data [23]. Currently, the GWAS Catalog contains 22,470 disease/trait-gene pairs, representing 8,689 genes and 881 common complex diseases/traits, including RA and 95 RA-associated genes; and (2) The Online Mendelian Inheritance in Man database (OMIM), the most comprehensive source of disease genetics for Mendelian disorders [24]. Currently, OMIM includes 15,462 disease-gene pairs for 5,983 diseases and 8,831 genes, including RA and 20 RA-associated genes. We used these two complementary resources of disease genetics to demonstrate the robustness of MetabolitePredict.
2.1.2. Chemical genetics data
We used the STITCH (Search Tool for Interactions of Chemicals) database to obtain chemical/metabolite-gene associations. STITCH is a database of known and predicted interactions between chemicals and proteins [25]. STITCH contains data on the interactions between 300,000 small molecules and 2.6 million proteins from 1133 organisms. In this study, we used chemical-gene associations found in human body, which include 1,466,636 chemical-gene pairs, 259,171 chemicals, and 15,620 human genes.
2.1.3. The Human Metabolome Database (HMDB)
HMDB contains detailed information about 41,993 small molecule metabolites found in the human body and is intended for applications in metabolomics, biomarker discovery and other applications [4]. We used HMDB to obtain a list of metabolites found in human body, including human gut microbial metabolites.
2.1.4. Genetic pathway data
We used the rich pathway information from the Molecular Signatures Database (MSigDB) to construct pathway profiles for diseases and metabolites. MSigDB is currently the most comprehensive resource for 10,295 annotated pathways and gene sets [26].
2.2. Methods
2.2.1. Overview of MetabolitePredict
Currently, MetabolitePredict implemented two prioritization algorithms: (1) gMetabolitePredict, which prioritizes metabolites based on gene set profile similarities; and (2) pMetabolitePredict, which prioritizes metabolites based on pathway profile similarities.
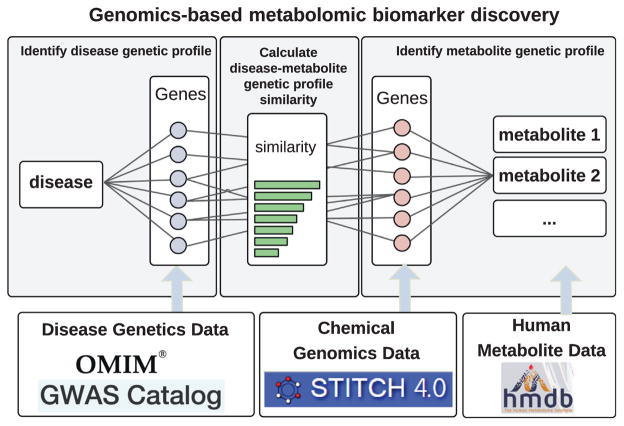
gMetabolitePredict is shown in Fig. 1 and consists of the following components: (1) gMetabolitePredict constructs a genetic profile for an input disease, which is the set of disease-associated genes; (2) gMetabolitePredict then constructs genetic profiles for a total of 259,170 chemicals/metabolites from STITCH. The genetic profile for a metabolite is a set of metabolite-associated genes extracted from STITCH; (3) gMetabolitePredict prioritizes metabolites for an input disease based on the genetic profile similarity between disease and metabolites. Currently, gMetabolitePredict implemented three commonly used set similarity measures: (a) overlap, (b) Jaccard similarity coefficient, and (c) cosine similarity [27].
Fig. 1.
gMetabolitePredict: a genetics-based disease metabolite prediction system.
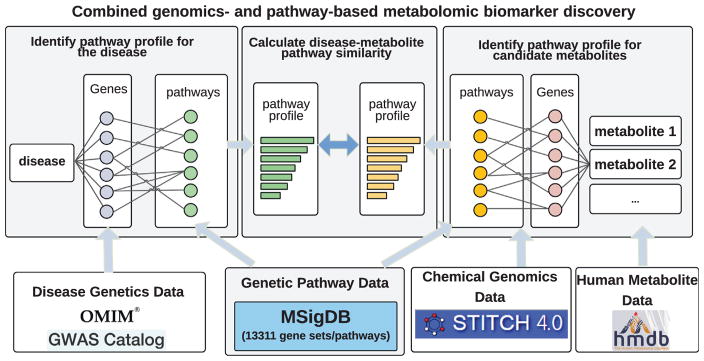
pMetabolitePredict is shown in Fig. 2 and consists of the following components: (1) pMetabolitePredict first constructs a pathway profile for a given disease by performing pathway enrichment analysis for a set of disease-associated gene; (2) pMetabolitePredict constructed pathway profiles for a total of 259,170 chemicals/metabolites. This step was only performed once and the data were stored in MetabolitePredict database; and (3) pMetabolitePredict prioritizes metabolites based on the pathway profile similarity (overlap, Jaccard coefficient, and cosine similarity) between disease and metabolites.
Fig. 2.
pMetabolitePredict: a pathway-based disease metabolite prediction system.
2.2.2. Construct genetic and pathway profile for input disease
For gMetabolitePredict, the genetic profile for a disease is the set of disease-associated genes (Fig. 1). For RA, we used 20 RA-associated genes from the OMIM database and 95 genes from the GWAS Catalog to build two gene profiles for RA. For pMetabolitePredict, pathway enrichment analysis was performed to identify genetic pathways significantly enriched for the set of disease associated genes (Fig. 2): pathways associated with for each gene first were obtained from MSigDB. For each pathway, the probability of this pathway associated with a set of genes (disease or metabolite-associated genes) was assessed by comparing to that for the same number of randomly selected genes. We repeated the random process 1000 times and performed a t-test to assess the enrichment significance. For example, the pathway profile for RA consists of these 266 significantly enriched pathways.
2.2.3. Construct genetic and pathway profiles for chemicals/metabolites
Similarly, MetabolitePredict built gene and pathway profiles for 259,170 chemicals from the STITCH database. For example, butyric acid, a human gut microbial metabolite, is associated with 669 genes, for which a total of 609 pathways are significantly enriched. The genetic profile for butyric acid is the set of 669 genes and the pathway profile is the set of 609 significantly enriched pathways.
2.2.4. Prioritize metabolites for input diseases
MetabolitePredict prioritizes metabolites based on the gene profile (gMetabolitePredict) or pathway profile (pMetabolitePredict) similarity between disease and metabolites. Currently, MetabolitePredict implements three commonly used set similarity measures: overlap, Jaccard coefficient, and cosine similarity. Additional similarity measures can be easily incorporated.
2.2.5. Evaluation using known RA-associated metabolites
We evaluated MetabolitePredict in identifying and prioritizing metabolite biomarkers for RA using 63 RA-associated metabolites extracted from published metabolomics studies [3,13] and from HMDB [4]. Recall, mean ranking, and median rankings were used for performance measures. Significance was calculated by comparing to random expectation (based on random expectation, these metabolites shall have an average ranking of 50%).
A good prioritization algorithm shall enrich true positives among top-ranked entities. We compared enrichments of true positives at 12 different ranking cutoffs (top 1%, 5%, 10%, 20, 30, …, 100%). We used enrichment curves instead of precision-recall curves because the large number of prioritized chemicals/metabolites (259,170) and the relative small number of known RA metabolites (large denominator and small numerator) make precision at each ranking cutoff extremely small. At each ranking cutoff, we calculated the enrichment fold by dividing the precision at the cutoff by the precision at ranking cutoff of 100% (which is the precision of random ranking). For example, the precision at ranking cutoff of top 1% is 0.0028, which is small. However it represents 45-fold enrichment as compared to the precision of 6.21E-05 at ranking cutoff of 100%. We compared gMetabolitePredict to pMetabolitePredict in prioritizing/enriching true positives at 12 ranking cutoffs.
2.2.6. Compare to an existing metabolite prediction system
We compared MetabolitePredict to PROFANCY in prioritizing RA-associated metabolites. From the web-based PROFANCY application, we obtained a list of 6574 prioritized metabolites for RA. We evaluated these predictions using the 63 known RA-associated metabolites. Recall, mean ranking, and median rankings were calculated. Significance of these rankings was calculated by comparing to random expectation.
2.2.7. Understanding how human gut microbial metabolites are involved in RA pathogenesis
Animal studies show that the short-chain fatty acids (SCFAs), the abundant metabolites of gut microbiota in the fermentation of fiber, have a role in the suppression of inflammation in RA [28,29]. We tested MetabolitePredict in prioritizing three known RA-associated SCFAs (butyrate, acetate, and propionate). We then analyzed top-ranked human gut microbial metabolites and identified genetic pathways significantly enriched for these top-ranked metabolites. We first identified genes associated with top ranked microbial metabolites (ranked within top 20%) using chemicalgene associations from STITCH database. Pathway enrichment analysis was then performed to find genetic pathways significantly enriched for this set of genes.
2.2.8. Evaluate MetabolitePredict’s potential in identifying metabolite inhibitors for RA treatment
Currently, there are three FDA-approved metabolite inhibitors for the treatment of RA. We prioritized 259,171 chemicals from STITCH based on their genetic relevance to RA pathogenesis. These chemicals include not only metabolites but also metabolite inhibitors. We evaluated the rankings of three known metabolite inhibitors (methotrexate, leflunomid and sulfasalazine) among 259,171 prioritized chemicals.
3. Results
3.1. pMetabolitePredict ranking RA-associated metabolites highly
We compared gMetabolitePredict and pMetabolitePredict in prioritizing 63 known RA-associated metabolites. As shown in Table 1, pMetabolitePredict performed much better than gMetabolitePredict for Jaccard and overlap similarity measures. In addition, the overlap-based measure has best performance.
Table 1.
Evaluation of pMetabolitePredict, gMetabolitePredict, and PROFANCY in prioritizing 63 RA-associated metabolites. The 20 RA-associated genes from the OMIM data based were used as input for both gMetabolitePredict and pMetabolitePredict.
| Prediction System | Similarity Measure | Mean Ranking (top %) | Median Ranking (top %) | P value |
|---|---|---|---|---|
| gMetabolitePredict | Overlap | 19.05% | 2.80% | 1.74E–5 |
| Jaccard | 19.18% | 2.81% | 1.77E–5 | |
| Cosine | 19.18% | 2.81% | 1.77E–5 | |
| pMetabolitePredict | Overlap | 4.43% | 1.37% | 4.55E–20 |
| Jaccard | 6.83% | 3.85% | 5.71E–20 | |
| Cosine | 47.99% | 56.91% | 0.818 | |
| PROFANCY | 20.9% | 16.5% | 3.78E–7 |
We also compared both systems to PROFANCY [5]. PROFANCY has recall of 0.31, a mean ranking of 20.9%, and a median ranking of 16.5%. These results show that pMetabolitePredict performed better than PROFANCY.
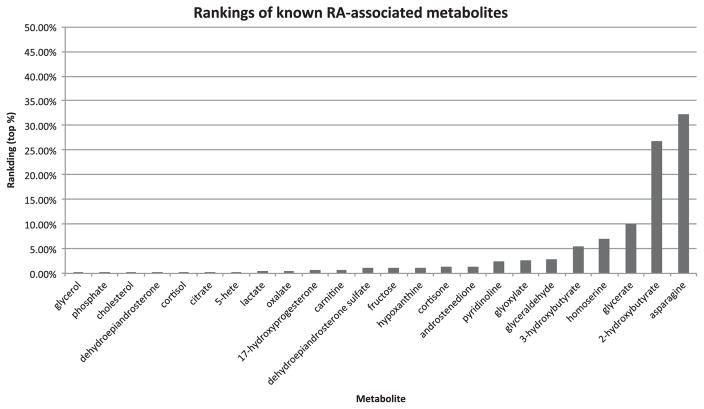
Fig. 3 shows the actual rankings of the 24 identified (out of 63) known metabolites among prioritized chemicals. The other 39 metabolites are not in either STITCH or HMDB database, therefore not identified by the systems. All 24 metabolites were ranked within top 35% and the mean and median rankings are 4.13% and 1.10%, respectively.
Fig. 3.
The rankings of 24 known RA-associated metabolites among 259,170 prioritized chemicals from STITCH.
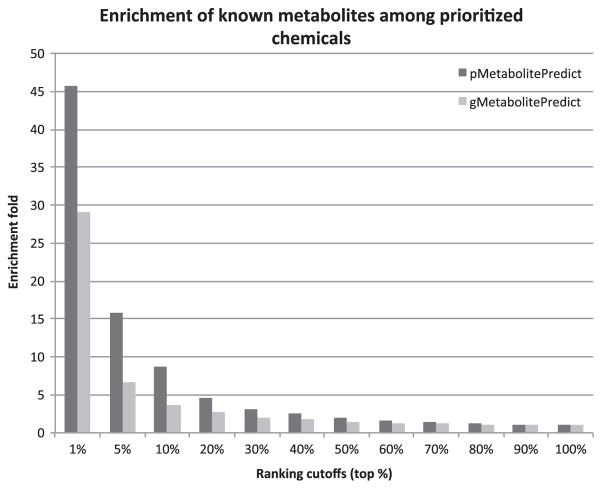
We further compared the prioritization capabilities of gMetabolitePredict and pMetabolitePredict at 12 ranking cutoffs. As shown in Fig. 4, both prioritization systems enriched true positives among top ranked metabolites. For example, pMetabolitePredict has a enrichment fold of 45.8 at the cutoff of top 1%, which is much higher than the enrichment fold of 2 at the cutoff of top 50%. The similar trend was observed for gMetabolitePredict. pMetabolitePredict has better enrichment performance than gMetabolitePredict at all ranking cutoffs. For instance, pMetabolitePredict has an enrichment fold of 45.8 at the cutoff of 1%, which is much higher that the 29.3 for pMetabolitePredict at the same cutoff.
Fig. 4.
Enrichment of true positives among prioritized metabolites at 12 ranking cutoffs for both gMetabolitePredict and pMetabolitePredict. The 95 RA-associated genes from the GWAS Catalog and chemical genetics from STITCH were used.
3.2. MetabolitePredict is robust across different disease genetics data resources
The only required input to MetabolitePredict is a disease name or a set of disease-associated genes. We then investigated how robust pMetabolitePredict is when different disease genetics data were used (the OMIM database and the GWAS Catalog). Table 2 shows that pMetabolitePredict was able to rank known RA-associated metabolites significantly highly across two complementary disease genetics databases.
Table 2.
The performance of pMetabolitePredict across two disease genetics (the OMIM database and the GWAS Catalog).
| Disease Genetics Database | Recall | Mean Ranking (top %) | Median Ranking (top %) | P value |
|---|---|---|---|---|
| OMIM | 38.10% | 4.32% | 1.37% | 4.55E–20 |
| GWAS | 38.10% | 4.13% | 1.10% | 5.08E–19 |
3.3. Systematic analysis of significant human gut microbial metabolites
From the 259,170 chemicals/metabolites prioritized by MetabolitePredict using RA as input, we identified a set of 65 metabolites originated from human gut microbiome (based on HMDB classification). 50 of these 65 microbial metabolites ranked within top 20%, indicating that gut microbial metabolism in general is related to RA. Short-chain fatty acids (SCFAs), the abundant metabolites of gut microbiota in the fermentation, ranked highly: butyrate, top 0.03%; acetate, top 0.05%; propionate, top 0.38%. These results indicate that fiber in food as well as the capability of human gut microbiota in fiber fermentation may be implicated in RA pathogenesis and that alternating these modifiable environmental factors may present a practical disease prevention strategy for RA. Our findings are consistent with recent studies showing that SCFAs have a role in the suppression of inflammation in RA [28,29].
We examined functional commonalities of the 50 top-ranked RA-associated microbial metabolites. We first identified genes associated with these metabolites, and then identified 78 genetic pathways significantly enriched for these genes. The top 20 pathways (Table 3) indicate that human gut microbial metabolites may be mechanistically linked to RA through glycolysis, amino acid metabolism, TCA cycle, and fatty acid bio-oxidation. The identification of microbial metabolites and the understanding of their role as key mediators through which these bacteria promote/protect against RA will provide insight into the basic mechanisms of RA etiology, facilitate our understanding of the complex host genome-microbiome interactions in RA, and enable/activate new possibilities for RA diagnosis, prevention, and treatment.
Table 3.
Top 20 genetic pathways significantly enriched for top-ranked RA-associated human gut microbial metabolites.
| Pathway | Enrichment | Pathway | Enrichment |
|---|---|---|---|
| Ethanol oxidation | 41 | Feeder Pathways for Glycolysis | 33 |
| Digestion of dietary carbohydrate | 29 | Glycine, serine and threonine metabolism | 27 |
| Glycolysis Pathway | 23 | Tyrosine metabolism | 16 |
| Galactose metabolism | 15 | Pyruvate metabolism | 14 |
| Tryptophan metabolism | 14 | The Citric Acid Cycle (Krebs pathway) | 14 |
| Glycolysis/Gluconeogenesis | 14 | Phenylalanine metabolism | 13 |
| Phase 1 – Functionalization of compounds | 12 | Fatty acid metabolism | 12 |
| Mitochondrial Fatty Acid Beta-Oxidation | 12 | Starch and sucrose metabolism | 12 |
| Valine, leucine and isoleucine degradation | 11 | Metabolism of polyamines | 11 |
| Peroxisomal lipid metabolism | 11 | Free Radical Induced Apoptosis | 11 |
3.4. Known metabolite inhibitors for RA treatments ranked highly
Methotrexate, leflunomid and sulfasalazine are three metabolic inhibitors used to treat RA [19]. Methotrexate is a folate inhibitor and currently the most important and most frequently prescribed medication for the treatment of RA. Methotrexate inhibits DNA and RNA synthesis in lymphocytes by preventing de novo purine and pyrimidine synthesis [20]. Leflunomide is an isoxazole derivate that inhibits the mitochondrial enzyme dehydroorotate dehydrogenase and prevents de novo synthesis of pyrimidine in lymphocytes [22]. Sulfasalazine inhibits folate-dependent enzyme and induces apoptosis of neutrophils and macrophages [21].
MetabolitePredict prioritized a total of 259,171 chemicals derived from STITCH based on their genetic relevance to RA pathogenesis. These chemicals include not only metabolite biomarkers but also metabolite inhibitors. Among the prioritized chemicals, the three metabolite inhibitors ranked highly: methotrexate, top 0.25%; leflunomide, top 0.56%; sulfasalazine, top 0.92%. These results demonstrate MetabolitePredict’s potential in not only metabolite biomarker discovery but also identifying novel therapies for metabolic targeting in RA.
4. Discussion
MetabolitePredict is a general approach and can perform de novo predictions of metabolites, microbial metaboites as well as metabolite-targeting therapies for any diseases. The input can be a disease name or a set of disease-associated genes. The web-based MetabolitePredict is publicly available at: http://xulab.case.edu/MetabolitePredict. We evaluated MetabolitePredict for RA metabolomic biomarker discovery, gut microbial metabolite identification, and metabolite inhibitor discovery, because metabolomics, microbiome studies, and metabolite targeting therapy in RA are relatively well studied. However, we did not tailor the system for RA in any way. Therefore, we expect that MetabolitePredict would be equally effective for other diseases.
The de novo prediction system MetabolitePredict is different from existing computation-based metabolite prediction systems [5,6], which identify disease metabolites based on known disease-associated metabolites and cannot perform predictions for diseases without known metabolites. Though we demonstrated that MetabolitePredict performs better than PROFANCY in prioritizing RA-associated metabolites, the de novo prediction system has its inherent limitation since it ignores our existing knowlege of disease-associated metabolites. In the future, we will further improve MetabolitePredict by taking into account of the increasingly available knowlege of disease-associated metabolites. For example, we can further prioritize a metabolite for a disease if the metbolite share genetic or pathway profile with known disease-associated metabolites.
Rapid environmental changes and modern lifestyles are the driving factors to many common complex diseases, including RA. While significant progress has been made in understanding genetic, molecular, and cellular mechanisms of RA, however, little is known about which environmental factors are important in RA. Human gut microbiota are important modifiable environmental factors that are part of the ecosystem of our bodies. We demonstrated that MetabolitePredict has the potential to identify which and how human gut microbial metabolites are associated with RA.
Another advantage of MetabolitePredict is that it can simutaneously predict both disease metaboilte biomarkers and metabolite targeting for disease treatment. We showed that MetabolitePredict identified and ranked highly three known metabolite inhibitors for RA treatments.
MetabolitePredict does not replace existing patient sample-based metabolomics studies, instead, it largely complements existing metabolomics profiling by contextualizing metabolite biomarker discovery with vast amounts of existing knowledge of diseases, genes, pathways, and metabolites. We believe that MetabolitePredict fills an important need by simultaneous identifying and understanding metabolite biomarkers for diseases, understanding how modifiable environment factors such as human gut microbiome are involved in disease mechanisms, and by translating metabolomic data into relevant biological knowledge and drug treatments.
With the vast amounts of knowledge built into MetabolitePredict and it can take a list of genes as input, we expect that MetabolitePredict can be applied to identify metabolite signatures unique for disease subtypes, disease progression, as well as treatment response given the involved genes are available.
Acknowledgments
RX was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under the NIH Director’s New Innovator Award number DP2HD084068, Case Western Reserve University/Cleveland Clinic CTSA Grant (UL1TR000,439), Research Scholar Grant (RSG-16-049-01 - MPC) from the American Cancer Society, 2015 Landon Foundation-AACR INNOVATOR Award for Cancer Prevention Research (Grant No. 15-20-27-XU), and Pfizer Investigator- Initiated Research Grant (WI206753).
Footnotes
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
RX and QW have jointly conceived, designed and implemented the algorithms and wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Nicholson JK, Wilson ID. Understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discovery. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 3.Guma M, Tiziani S, Firestein GS. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat Rev Rheumatol. 2016;12(5):269–281. doi: 10.1038/nrrheum.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2013;41(D1):801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang D, Li C, Yao Q, Yang H, Xu Y, Han J, Li J, Su F, Zhang Y, Zhang C, Li D, Li X. Prioritizing candidate disease metabolites based on global functional relationships between metabolites in the context of metabolic pathways. PLOS ONE. 2014;9(8):104934. doi: 10.1371/journal.pone.0104934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Q, Xu Y, Yang H, Shang D, Zhang C, Zhang Y, Sun Z, Shi X, Feng L, Han J, Su F. Global prioritization of disease candidate metabolites based on a multiomics composite network. Sci Rep. 2015;5(17201) doi: 10.1038/srep17201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu R, Wang Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genom. 2015;16(suppl 7):4. doi: 10.1186/1471-2164-16-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu R, Wang Q. Towards understanding brain-gut-microbiome connections in Alzheimer’s disease. BMC Syst Biol. 2016;10(suppl 3):63. doi: 10.1186/s12918-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. Rheumatoid Arthritis. < http://www.cdc.gov/arthritis/basics/rheumatoid.htm>.
- 10.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. New Engl J Med. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 11.Taneja V. Arthritis susceptibility and the gut microbiome. FEBS Lett. 2014;588(22):4244–4249. doi: 10.1016/j.febslet.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlson EW, Deane K. Environmental and gene-environment interactions and risk of rheumatoid arthritis. Rheum Dis Clin N Am. 2012;38(2):405–426. doi: 10.1016/j.rdc.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Priori R, Scrivo R, Brandt MJ, Valerio, Casadei L, Valesini G, Manetti C. Metabolomics in rheumatic diseases: the potential of an emerging methodology for improved patient diagnosis, prognosis, and treatment efficacy. Autoimmun Rev. 2013;12(10):1022–1030. doi: 10.1016/j.autrev.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 15.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 16.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7(10):569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity. Curr Opin Rheumatol. 2014;26(1):101–107. doi: 10.1097/BOR.0000000000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discovery. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 19.Meier FM, Frerix M, Hermann W, Müller-Ladner U. Current immunotherapy in rheumatoid arthritis. Future Med. 2013;5(9):955–974. doi: 10.2217/imt.13.94. [DOI] [PubMed] [Google Scholar]
- 20.Kremer JM, Alarcón GS, Lightfoot RW, Willkens RF, Furst DE, Williams HJ, Dent PB, Weinblatt M. Methotrexate for rheumatoid arthritis. Arthritis Rheum. 1994;37(3):316–328. doi: 10.1002/art.1780370304. [DOI] [PubMed] [Google Scholar]
- 21.Hirohata S, Ohshima N, Yanagida T, Aramaki K. Regulation of human B cell function by sulfasalazine and its metabolites. Int Immunopharmacol. 2002;2(5):631–640. doi: 10.1016/s1567-5769(01)00186-2. [DOI] [PubMed] [Google Scholar]
- 22.Fox RI, Herrmann ML, Frangou CG, Wahl GM, Morris RE, Strand V, Kirschbaum BJ. Mechanism of action for leflunomide in rheumatoid arthritis. Clin Immunol. 1999;93(3):198–208. doi: 10.1006/clim.1999.4777. [DOI] [PubMed] [Google Scholar]
- 23.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(D1):1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(suppl 1):514–517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn M, Szklarczyk D, Pletscher-Frankild S, Blicher TH, von Mering C, Jensen LJ, Bork P. STITCH4: integration of protein–chemical interactions with user data. Nucleic Acids Res. 2013;42(D1):401–407. doi: 10.1093/nar/gkt1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir PH, Tamayo, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Kamber M, Pei J. Morgan Kaufmann series in data management systems. Vol. 3. Elsevier; Morgan Kaufmann: 2012. Data Mining: Concepts and Techniques. [Google Scholar]
- 28.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discovery. 2009;8(12):969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]