SUMMARY
Selective autophagy eliminates protein aggregates, damaged organelles, and other targets that otherwise accumulate and cause disease. Autophagy receptors mediate selectivity by connecting targets to the autophagosome membrane. It has remained unknown whether receptors perform additional functions. Here, we show that in yeast certain receptor-bound targets activate Atg1, the kinase that controls autophagosome formation. Specifically, we found that in nutrient-rich conditions, Atg1 is active only in a multi-subunit complex comprising constitutive protein aggregates, their autophagy receptor, and a scaffold protein Atg11. Development of a cell-free assay for Atg1-mediated phosphorylation enabled us to activate Atg1 with purified receptor-bound aggregates and Atg11. Another target, damaged peroxisomes, also activated Atg1 using Atg11 with a distinct receptor. Our work reveals that receptor-target complexes activate Atg1 to drive formation of selective autophagosomes. This regulatory logic is a key similarity between selective autophagy and bulk autophagy, which is initiated by a distinct Atg1 activation mechanism during starvation.
INTRODUCTION
Selective autophagy promotes the survival of non-dividing cells, such as neurons, by preventing the accumulation of cytotoxic structures (Hara et al., 2006; Komatsu et al., 2006). Targets of selective autophagy are first detected in the cytosol by receptor proteins (Behrends and Fulda, 2012). Once bound by receptors, targets are sequestered in autophagosomes, double-membrane vesicles that subsequently fuse with lysosomes, resulting in target destruction. The vesicle membrane is the product of a cup-shaped precursor structure that grows around receptor-bound targets until it fuses with itself. Each round of target elimination turns over an autophagosome precursor, but no mechanistic link connecting target detection with precursor biogenesis has been identified.
Though the primary described role of autophagy receptors is to bridge targets to autophagosome-tethered Atg8-family proteins (Ichimura et al., 2000; Rogov et al., 2014; Schreiber and Peter, 2014; Wild et al., 2011), autophagy receptors also bind Atg11-like proteins in both yeast (Yorimitsu and Klionsky, 2005) and higher organisms (Lin et al., 2013; Rui et al., 2015). The function of the interactions between autophagy receptors and Atg11 family proteins is unknown. Intriguingly, Atg11-like proteins also interact with the autophagy kinase (Atg1 in yeast (Yorimitsu and Klionsky, 2005) and ULK1/2 in metazoans (Hara et al., 2008; Rui et al., 2015)), which is required for both selective and non-selective forms of autophagy. Studies of non-selective autophagy in yeast have shown that nutrient depletion causes Atg1 autoactivation by a mechanism dependent on inhibition of Tor kinase signaling (Kamada et al., 2000; Stephan et al., 2009). Once Atg1 phosphorylates its activation loop it becomes catalytically active (Yeh et al., 2010) and able to drive formation of non-selective autophagosome membranes by mechanisms dependent on substrate phosphorylation (Papinski et al., 2014). This regulatory logic enables coordination of nutrient deprivation with nutrient replenishment by cytoplasmic turnover. By contrast, the possibility that Atg1 kinase activity is also regulated in nourished cells to couple target detection with selective autophagosome formation has not been explored. We hypothesized that receptor-bound targets can activate Atg1 using Atg11 as a scaffold for signal transduction (Figure 1A).
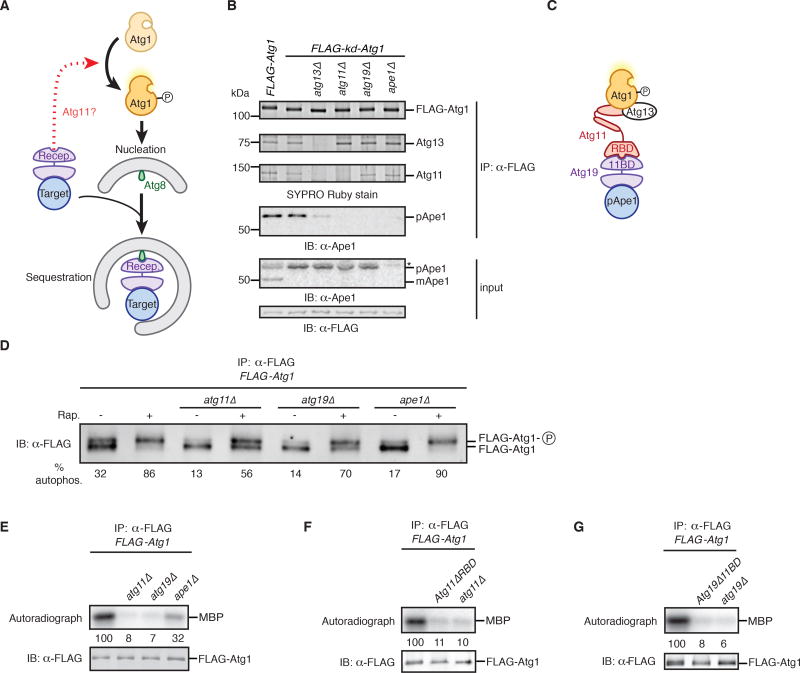
Figure 1. Identification of Ape1, Atg19, and Atg11 as activators of Atg1 kinase activity.
(A) Schematic depicting possible role (dotted red arrow) of receptor-bound targets in inducing autophagosome nucleation by converting Atg1 from an inactive state to an active, autophosphorylated state in an Atg11-dependent manner. Also indicated is the ability of targets to associate with autophagosome precursors via receptor-mediated interactions with Atg8 proteins. Recep., receptor.
(B) Extracts derived from logarithmically growing cells with indicated genotypes were immunoprecipitated (IP) with anti-FLAG magnetic beads. Eluates and extract (input) samples were resolved by SDS-PAGE followed by either SYPRO Ruby staining or immunoblotting (IB) with indicated antibodies. kd, kinase-dead allele; pApe1 and mApe1, precursor and mature (vacuolar) forms of Ape1; *, non-specific band.
(C) Model of the subunit associations within the Atg1 complex based on data in (B) and Figure S1. Also indicated are the Atg11-Binding Domain (11BD) on Atg19 and the Receptor-Binding Domain (RBD) on Atg11.
(D) Logarithmically growing cells with indicated genotypes were treated with rapamycin (Rap.) or mock treated prior to immunoprecipitation (IP) analysis as in part (B) with one notable exception: SDS-PAGE was done for a longer time to resolve Atg1 from its autophosphorylated form (Atg1-P), as visualized by immunoblotting (IB). Autophosphorylation (% autophos.) was calculated as percent of total signal contributed by the upper band. See Figure S1E for statistical analysis.
(E–G) Extracts derived from logarithmically growing cells with indicated genotypes were immunoprecipitated (IP) as in part (B) and then incubated with myelin basic protein (MBP) and ATPγ32P. Kinase reactions were performed in triplicate (one of which is shown) and terminated with loading buffer prior to sample analysis by autoradiography and immunoblotting (IB). Signal from autoradiographs was quantified by densitometry and reported as the average of three reactions, in arbitrary units relative to the wild-type reaction set at 100. See Figure S1 for statistical analysis.
See also Figure S1.
RESULTS
Atg11 Bridges the Atg1–Atg13 Subcomplex with Atg19-Bound Targets
The first key prediction of our hypothesis is that Atg11 scaffolds interactions between receptor-bound targets and Atg1. Budding yeast cells deliver newly-synthesized aminopeptidase precursor pApe1 to the vacuole (yeast lysosome) by a selective autophagy process that depends on the autophagy receptor Atg19, Atg11, Atg1, and Atg13 (an Atg1-binding protein required for Atg1 kinase activity) (Kamada et al., 2000; Kim et al., 2001; Scott et al., 2001). To detect any pApe1 interactions with Atg1, we affinity-purified FLAG-Atg1 from crude, detergent-solubilized extracts using anti-FLAG magnetic beads. Following elution with FLAG peptide, we analyzed the purified material by protein staining (Figure 1B), immunoblotting (Figures 1B and Figure S1A), and quantitative mass spectrometry (Figure S1B). pApe1 co-immunoprecipitated with wild-type Atg1, as well as with kinase-dead Atg1 (kd-Atg1; D211A mutant) and autoactivation-dead Atg1 (ad-Atg1; T226A) (Figure S1A), which is missing a critical phosphorylation site in the activation loop of the kinase domain (Lazarus et al., 2015; Yeh et al., 2010). In addition to pApe1, three other proteins were abundantly associated with Atg1: Atg13, Atg11, and Atg19. By purifying kd-Atg1 from cell extracts lacking individual Atg1-associated proteins, we made three observations arguing that Atg1 is part of a multi-subunit complex in which Atg11 bridges the core kinase subcomplex (comprising Atg1 and Atg13) with Atg19-bound pApe1 (Figure 1B; Figure S1B). First, in the absence of Atg11, Atg1 and Atg13 formed a subcomplex devoid of the other components. Second, in the absence of Atg19, Atg1–Atg13 and Atg11 formed a ternary complex devoid of pApe1. Third, in the absence of Atg13, Atg1 interacted only weakly with Atg11 (Figure S1C) and, hence, Atg19-pApe1. The existence of the multi-subunit Atg1 kinase complex containing pApe1 described here (Figure 1C) has apparently escaped notice in past biochemical studies of immunoprecipitated Atg1, possibly because the Ape1 aggregate is large enough to be sterically excluded from sepharose beads (Inada et al., 2002), and, in the presence of physiological salts, is cleared from cell extracts by centrifugation (>5000 × g) (Scott et al., 2001).
Atg1 Activity is Controlled by Atg19-Bound Targets
The second key prediction of our hypothesis is that Atg1 kinase is turned on in nourished cells by binding to pApe1 targets. In vivo, Atg1 kinase activation causes autophosphorylation resulting in the appearance of a slower-migrating form of Atg1 on SDS polyacrylamide gels (Figure S1D) (Yeh et al., 2010). In wild-type cells a fraction of Atg1 was autophosphorylated (Figure 1D; Figure S1D) and, as predicted, cells lacking either pApe1, Atg19, or Atg11 had a significantly reduced level of autophosphorylated Atg1 (Figure 1D; Figure S1E). Notably, Atg1 autophosphorylation could be induced in ape1Δ, atg19Δ, and atg11Δ cells following treatment with rapamycin, an inhibitor of Tor kinase signaling that mimics starvation (Kamada et al., 2000; Kim et al., 2001; Scott et al., 2001). These data provide strong support for our hypothesis that pApe1–Atg19 complexes control Atg1 activity in nourished cells and confirm that these factors are dispensable for the canonical pathway of Atg1 activation that occurs via repression of Tor kinase signaling.
To verify that that Atg19-bound targets increase the catalytic activity of Atg1, we incubated affinity-purified Atg1 with myelin basic protein (MBP) and ATPγ32P (Kamada et al., 2000). MBP was robustly phosphorylated upon incubation with wild-type Atg1, but not kd-Atg1 or ad-Atg1 (Figure S1F). Consistent with our hypothesis, Atg1 catalytic activity was dependent on Atg11 and Atg19 (Figure 1E; Figure S1G). More precisely, we disrupted the Atg11–Atg19 interaction using either Atg11 lacking the Receptor-Binding Domain (RBD) (Figure S1H) (Yorimitsu and Klionsky, 2005), which is dispensable for the Atg1–Atg11 interaction (Figure S1I), or a mutant version of Atg19 that is unable to bind Atg11 (Atg19Δ11BD) (Figure S1J) (Shintani et al., 2002) and observed a strong decrease in Atg1 catalytic activity (Figures 1F and 1G; Figures S1K and S1L). We also noted that Atg1 activity was only partially dependent on pApe1 (Figure 1E; Figure S1G) and this might reflect Atg19’s ability to recognize other protein aggregates in the absence of pApe1 (Lynch-Day and Klionsky, 2010). In sum, our data demonstrate that “basal” Atg1 activity in nourished cells is in fact dictated by a chain of interactions between the Atg1–Atg13 subcomplex, Atg11, and Atg19-bound targets.
Atg1 Activation by Atg19-Bound Targets Drives Autophagosome Membrane Elongation During Nutrient-Rich Conditions
The mechanistic role of Atg1 kinase activity during autophagy has primarily been studied in the context of the formation of the large (600–900nm) autophagosomes induced by starvation, where it has been shown that Atg1 kinase activity and phosphorylation of Atg9 are required for expansion of the autophagosome membrane (Papinski et al., 2014; Suzuki et al., 2013). Though the kinase activity of Atg1 is also essential for the trafficking of pApe1 to the vacuole (Kijanska et al., 2010; Papinski et al., 2014), the precise step at which Atg1 kinase activity is required during selective autophagy has not been clarified. In particular, it has not been determined whether Atg1 kinase activity is similarly necessary to promote membrane elongation during formation of the much smaller (~150nm) autophagosomes that enwrap pApe1 aggregates, or if it is strictly required for a later step in autophagy progression (Kraft et al., 2012).
To test the hypothesis that Atg1 kinase activation by pApe1 aggregates is required for elongation of selective autophagosome membranes, we first studied the intracellular localization of Atg2, a protein that localizes to the growing rim of cup-shaped autophagosome membranes (Suzuki et al., 2013). In starved cells, localization of Atg2 to the site of autophagosome formation requires Atg1 kinase activity (Papinski et al., 2014). In nourished cells, Atg2-GFP targeting to pApe1 aggregates results in a punctum of fluorescence (Shintani and Klionsky, 2004) and we found that the proportion of mCherry-Ape1 puncta that colocalized with Atg2-GFP puncta was significantly decreased in cells lacking Atg1 or expressing kd-Atg1 or ad-Atg1 (Figure 2A and 2B), whereas mCherry-Ape1 puncta formation was unchanged (Figure S2A). These data argue that autoactivated Atg1 molecules are required to initiate membrane expansion during selective autophagy.
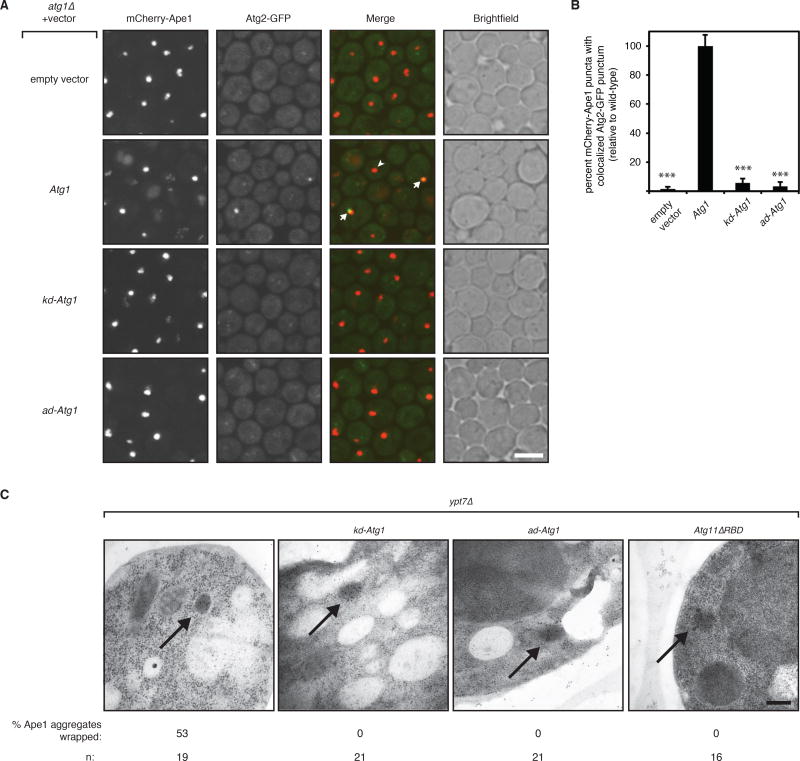
Figure 2. Atg1 kinase activation in nourished cells is required for autophagosome membrane elongation around Ape1 aggregates.
(A) Representative images (maximum intensity projections) of logarithmically growing atg17Δ atg1Δ cells expressing mCherry-Ape1 and Atg2-GFP from their endogenous loci and carrying an empty vector or vector expressing the indicated Atg1 allele. White arrow indicates colocalized mCherry-Ape1 and Atg2-GFP puncta; white arrowhead indicates a lone mCherry-Ape1 punctum. Note that only wild-type Atg1 cells have vacuolar mCherry fluorescence because Atg1 kinase activity is required for pApe1 vacuolar delivery. Scale bar, 5 µm. kd, kinase-dead allele; ad, autoactivation-dead allele.
(B) Image analysis of strains shown in part (A) from three independent experiments (>1000 mCherry-Ape1 puncta per strain per experiment analyzed). Bar graphs report percent mCherry-Ape1 puncta with colocalized Atg2-GFP puncta as the mean and standard deviation (error bars) relative to cells expressing wild-type Atg1 allele. Raw mean percentage of mCherry-Ape1 puncta with colocalized Atg2-GFP puncta in wild-type cells was 3.6%. p values derived from Tukey’s post-test are reported only for comparisons between each mutant and wild-type. *** p < 0.0001.
(C) Representative transmission electron micrographs of logarithmically growing ypt7Δ cells expressing the indicated mutant alleles from their endogenous genomic loci. Black arrows indicate immunogold-labeled Ape1 aggregates. Percentage of Ape1 aggregates (out of n number analyzed) surrounded by autophagosome membranes is indicated. Scale bar, 200 nm.
See also Figure S2.
As an orthogonal means of examining the role of Atg1 activation on autophagosome membrane expansion in cells, we used transmission electron microscopy (TEM) and immunogold labeling of pApe1 aggregates. To visualize fully-extended autophagosome membranes, which have a short half-life in wild-type cells (Geng et al., 2008), we used the ypt7Δ background, in which autophagosomes cannot fuse with the vacuole (Sawa-Makarska et al., 2014). We found that cells expressing wild-type Atg1 exhibited Ape1 clusters that were frequently surrounded by electron-lucent structures that are indicative of double-membrane-bound autophagosomes (Figure 2C; Figure S2B). These structures were never observed in cells expressing Atg11ΔRBD, confirming that they are indeed representative of selective autophagosomes (Figure 2C; Figure S2B). In cells expressing kd-Atg1 or ad-Atg1, we never observed autophagosome membranes around pApe1 aggregates (Figure 2C; Figure S2B). Collectively, these data show that Atg1 kinase activation is required in nourished cells for the membrane expansion step during selective autophagosome formation.
A Chemical Genetics Approach for Studying Atg1 Kinase Regulation in Vitro
The most rigorous test of our hypothesis necessitates biochemical reconstitution of Atg1 kinase activation by autophagy targets. Biochemical reconstitution of the minimal Atg1–Atg13 subcomplex, however, has been elusive. Therefore, we developed a chemical genetics approach that detects Atg1 activity in cell extracts and is amenable to mechanistic dissection by genetic analysis, as well as biochemical reconstitution with pure components. Specifically, we replaced the bulky ‘gatekeeper residue’ in the ATP-binding pocket of Atg1 with a glycine residue (Blethrow et al., 2004) (Figure S3A) to create a functional ATP-analog-sensitive allele of Atg1 (as-Atg1) (Figure S3B). To selectively monitor as-Atg1 activity in cell extracts, we utilized an unnatural, bulky ATP derivative (N6-PhEt-ATPγS) that is inaccessible to wild-type kinases (Figure 3A) but allows for thiophosphorylation of as-kinase substrates (Allen et al., 2007). As Atg1 is expressed at a low level (Geng et al., 2008), we improved detection of its kinase activity in two ways. First, as the endogenous nucleoside diphosphate kinase Ynk1 rapidly consumes ATP analogs (Figure S3C), we used a ynk1Δ genetic background to allow for longer labeling times. Second, we expressed from its endogenous locus a mutant version of Atg13 (8SA) that stabilizes Atg13’s interaction with Atg1 (Kamada et al., 2010), which led to more robust substrate labeling (Figure S3D).
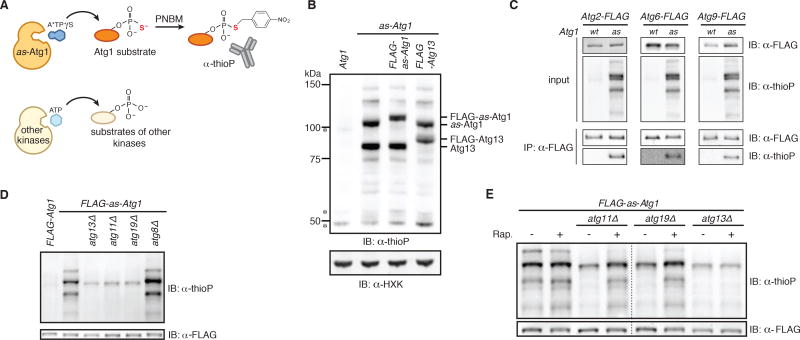
Figure 3. A chemical genetic assay for Atg1 kinase activity.
(A) Schematic of the chemical genetic strategy for monitoring Atg1-dependent thiophosphorylation. Analog-sensitive (as) Atg1 can accept A*TPγS, an N6-substituted ATPγS analog, to thiophosphorylate its substrates while other kinases reject the analog. Following alkylation with para-nitrobenzyl mesylate (PNBM), labeled substrates are immunodetected with anti-thiophosphate ester (thioP) antibody.
(B) Extracts derived from logarithmically growing cells with indicated genotypes were treated as diagrammed in part (A) and analyzed by SDS-PAGE (gel system 1; see Experimental Procedures for details) and immunoblotting (IB) with indicated antibodies. HXK, hexokinase; *, non-specific band.
(C) The indicated extracts were treated as diagrammed in part (A) and subjected to immunoprecipitation (IP) with anti-FLAG agarose resin. Eluates and extract (input) samples were resolved by SDS-PAGE (gel system 2; see Experimental Procedures for details) followed by immunoblotting (IB) with indicated antibodies. wt, wild-type.
(D) Extracts derived from logarithmically growing cells with indicated genotypes were treated as diagrammed in part (A) in triplicate (one of which is shown) and analyzed by SDS-PAGE (gel system 2) and immunoblotting (IB). For quantitation and statistics, see Figure S3F.
(E) Logarithmically growing cells with indicated genotypes were treated with rapamycin (Rap.) or mock treated prior to analysis as in part (D). Dotted line indicates splicing of gel-image data from the same gel. For quantitation and statistics, see Figure S3G.
See also Figure S3.
We grew cells expressing as-Atg1 from its endogenous locus in rich medium and prepared extracts to which we added N6-PhEt-ATPγS. Following incubation, extracts were alkylated and analyzed by immunoblotting with an anti-thiophosphate ester antibody (Figure 3A). The extract containing as-Atg1 yielded a stereotyped banding pattern that was absent from the control wild-type extract (Figure 3B). The two most prominent bands had the expected molecular weights of Atg1 and Atg13, which are known Atg1 substrates (Joo et al., 2011; Yeh et al., 2010). To verify these assignments, we epitope-tagged each protein and observed the expected size increases (Figure 3B). We also verified that as-Atg1 thiophosphorylated the two remaining known Atg1 substrates: Atg2 and Atg9 (Papinski et al., 2014) (Figure 3C). By analogy to mammalian ULK1, which phosphorylates Beclin1 (Atg6 homolog) (Russell et al., 2013), we confirmed that as-Atg1 phosphorylated Atg6 (Figure 3C). Lastly, we confirmed that thiophosphorylation of Atg6 and Atg9 was dependent on their recruitment to the site of autophagosome formation in vivo (Figure S3E) (Backues et al., 2015; Obara et al., 2006). Taken together, these data demonstrate that our cell-free system recapitulates the known substrate specificities of Atg1.
Our extracts were prepared from nourished cells in which Atg1 activity should come specifically from kinase molecules that are part of a multi-subunit complex containing Atg13, Atg11, and Atg19-bound aggregates. Consistent with this notion, Atg1 activity in vitro was severely diminished in atg13Δ, atg11Δ, and atg19Δ extracts. Both Atg1 and Atg19 have Atg8-binding sites but Atg8 was not essential for maintaining Atg1 kinase activity in extracts (Figure 3D; Figure S3F). Critically, pre-treatment of cells with rapamycin restored Atg1-mediated phosphorylation to atg11Δ and atg19Δ extracts (Figure 3E; Figure S3G). By contrast, rapamycin didn’t restore Atg1 activity in atg13Δ extracts reflecting the essential role of Atg13 in all forms of autophagy (Figure 3E; Figure S3G). Collectively, these data show that our cell-free assay recapitulates the known genetic and pharmacological requirements for studying Atg1 kinase activation by both Atg19-bound targets and starvation signals.
Biochemical Reconstitution of Atg1 Activation with Purified Atg11 and Atg19-Bound Targets
To biochemically dissect the mechanism of Atg1 activation by Atg19-bound targets, we first successfully restored Atg1 kinase activity to atg11Δ extracts by supplementing them with purified Atg11 (Figure 4A; Figure S4A). Two lines of evidence argue that the activity of purified Atg11 depends on its interactions with Atg19 endogenous to the extract. First, Atg11 failed to restore robust Atg1 kinase activity to atg11Δ atg19Δ extracts (Figure 4A). Second, purified Atg11ΔRBD did not activate Atg1 in atg11Δ extracts (Figure 4B) but exerted a dominant negative effect on Atg1 kinase activation by purified Atg11 (Figure S4B), consistent with its ability to bind Atg1 independent of Atg19-bound targets. Activation of Atg1 by purified Atg11 required autoactivation of Atg1, as Atg11 failed to stimulate the activity of ad-Atg1 (Figure S4C).
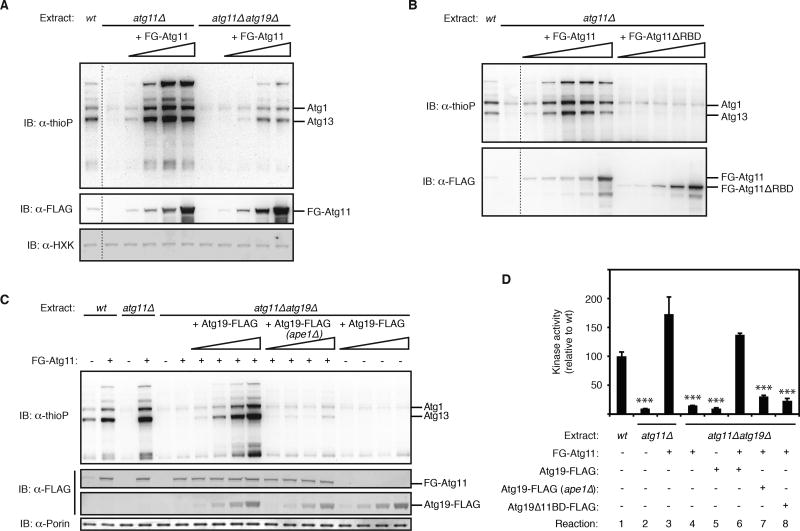
Figure 4. Activation of Atg1 kinase by purified Atg19-target complexes and Atg11.
(A) The indicated as-Atg1 deletion extracts were either pre-incubated with increasing amounts (indicated by wedge) of FLAG-GFP-Atg11 (FG-Atg11) or mock pre-incubated prior to analysis of Atg1 kinase activity (see Experimental Procedures for details). Immunoblotting (IB) with indicated antibodies was used to control for protein add-back and any gel loading differences. wt, wild-type extract derived from as-Atg1 FLAG-GFP-Atg11 (expressed from endogenous locus); HXK, hexokinase. Dotted line indicates splicing of gel-image data from the same gel.
(B) Similar to part (A) except as-Atg1 atg11Δ extract was pre-incubated with either full-length FLAG-GFP-Atg11 or FLAG-GFP-Atg11ΔRBD or mock pre-incubated. wt, wild-type extract derived from as-Atg1 FLAG-GFP-Atg11 (expressed from endogenous locus). Dotted line indicates splicing of gel-image data from the same gel.
(C) Similar to part (A) except FLAG-GFP-Atg11 was added at the optimal concentration along with increasing amounts of affinity-purified Atg19, as indicated. (ape1Δ) indicates Atg19-FLAG was purified from ape1Δ cells (n.b. this purified material did not restore Atg1 activation suggesting that functional Atg19-pApe1 complexes were not reconstituted in situ.). wt, wild-type extract derived from as-Atg1 FLAG-GFP-Atg11 (expressed from endogenous locus). Immunoblotting (IB) against porin (a mitochondrial protein) was used to control for any gel loading differences.
(D) Similar to part (C) except done in triplicate with all proteins added at their optimal concentrations. Bar graphs show the mean kinase activity and standard deviation (error bars), relative to wild-type reaction set to 100. p values derived from Tukey’s post-test for the comparisons between indicated reactions and reaction 1 are shown. ***p < 0.0001.
See also Figure S4.
Next, we purified Atg19 from yeast extracts and used it to restore Atg1 kinase activity to an atg11Δatg19Δ extract provided that we also added purified Atg11 (Figure 4C, 4D; Figure S4D, S4E). Mass spectrometric analysis revealed that purified Atg19 was in fact a complex containing pApe1 and several other target proteins known to be delivered to the vacuole via their association with Atg19-pApe1 (Kageyama et al., 2009; Suzuki et al., 2011; Yuga et al., 2011) (Figures S4F–I). The ability of the purified Atg19 complex to activate Atg1 was dependent on the presence of pApe1 in the complex, consistent with the primacy of the pApe1 aggregate in directing Atg19-mediated selective autophagy under nutrient-rich conditions (Shintani and Klionsky, 2004) (Figure 4C, 4D; Figure S4E). Similarly, a mutant version of Atg19 lacking a coiled coil domain required for the Atg19-pApe1 interaction (Shintani et al., 2002) (Figure S4J) was also unable to support Atg1 kinase activity in extracts (Figure S4K). Lastly, Atg19Δ11BD, which still bound the same cohort of target proteins (Figure S4G), was unable to activate Atg1 (Figure 4D; Figure S4E). In sum, these data demonstrate that Atg19-bound aggregates use Atg11 as a scaffold protein to turn on the Atg1 kinase switch.
Damaged Peroxisomes Activate Atg1 Dependent on Atg11 and the Pexophagy Receptor Atg36
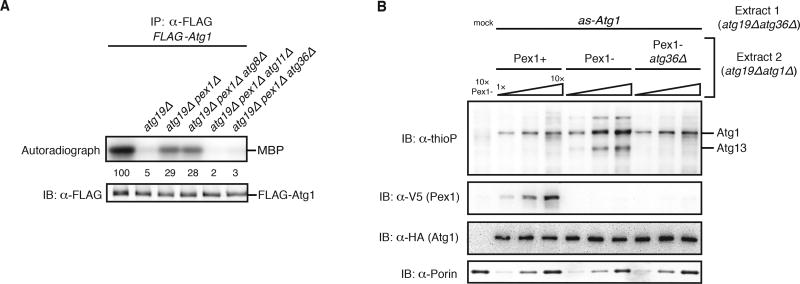
Protein aggregates represent only one class of selective autophagy targets. To test if organelles targeted for destruction by selective autophagy also activate Atg1, we examined the effect of peroxisome damage on Atg1 activation. Wild-type cells growing in rich media have low autophagic turnover of peroxisomes, but cells lacking Pex1, a AAA+ protein required for protein import into the peroxisome matrix, induce selective autophagy of peroxisomes (pexophagy) mediated by Atg11 and the autophagy receptor Atg36 (Nuttall et al., 2014). To facilitate detection of any Atg1 activation due to peroxisome damage, we induced pexophagy, either by PEX1 gene deletion or by engineered degradation of Pex1 using an auxin-inducible degron (Nishimura et al., 2009; Nuttall et al., 2014), in the atg19Δ genetic background, which abolished Atg1 activation by Atg19-bound targets (Figure 5A; Figure S5A). Analysis of MBP phosphorylation by affinity-purified Atg1 revealed that the presence of damaged peroxisomes increased Atg1 kinase activity in a manner wholly dependent on Atg11 and Atg36, but independent of Atg8 (Figure 5A; Figure S5A). As the final test of our starting hypothesis, we biochemically reconstituted Atg1 activation by damaged peroxisomes using an adaptation of our cell-free kinase assay. Mixing of a cell extract containing inactive as-Atg1 with an extract from cells that accumulate damaged peroxisomes (Figure S5B) resulted in kinase activation dependent on Atg36 (Figure 5B). Taken together, these data argue that Atg36-bound damaged peroxisomes use Atg11 to signal activation of the Atg1 kinase.
Figure 5. Activation of Atg1 by damaged peroxisomes.
(A) Myelin basic protein (MBP) phosphorylation by FLAG-Atg1 immunoprecipitated (IP) from the indicated extracts (all in the atg18Δ genetic background to prevent potential destruction of target-bound Atg1 complexes by pexophagy) was carried out as in Figures 1E–G. See Figure S5A for statistical analysis.
(B) Increasing amounts of the indicated extracts derived from the atg19Δatg1Δ genetic background were pre-incubated with HA-as-Atg1 atg19Δatg36Δ extract, or mock buffer, before analysis of kinase activity as in Figure 4A. Immunoblotting (IB) with indicated antibodies was used to control for auxin-induced degradation of Pex1-V5 (see Figure S5B) and any gel loading differences.
See also Figure S5.
DISCUSSION
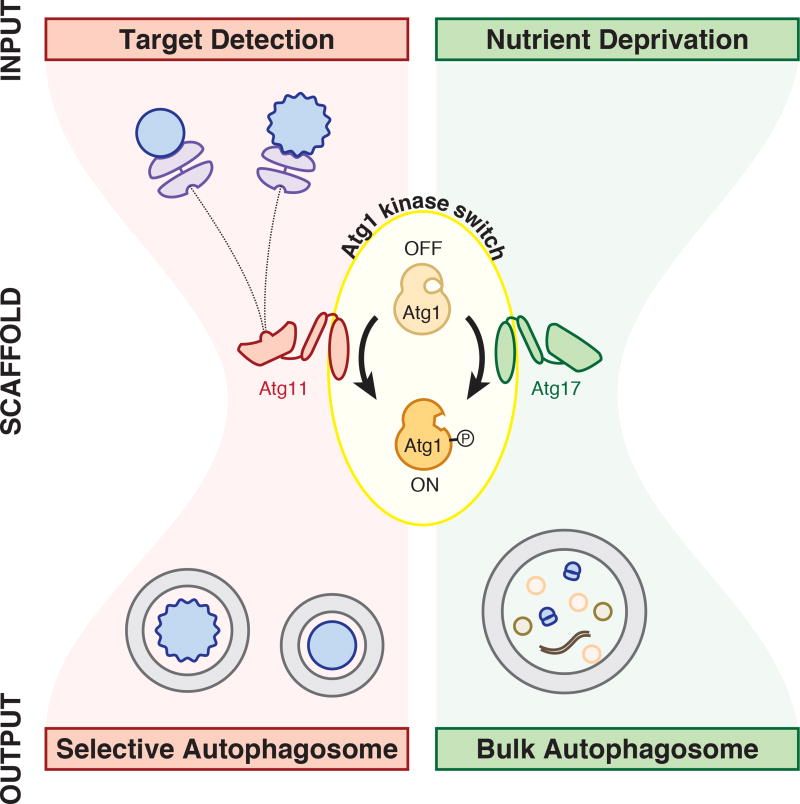
Atg1 kinase activity is essential for selective autophagy of pApe1 aggregates and damaged peroxisomes in nourished cells and, yet, nutrient-sensing pathways repress Atg1 kinase activity under these conditions in order to block non-selective autophagy. How Atg1 overcomes this restriction to promote basal autophagy under nutrient-rich conditions has been a long-standing question. One possibility is that less-catalytically-active Atg1 molecules suffice for the formation of selective autophagosomes. An alternative model is that pApe1 aggregates and damaged peroxisomes activate Atg1 molecules that are target-bound to locally override global Atg1 kinase repression by nutrient-sensing pathways. Here we have presented in vivo and biochemical reconstitution evidence that strongly support the latter model. Our work reveals that disparate autophagic cues (absence of nutrients versus target presence) achieve their objectives (non-selective recycling of cytoplasm versus selective cytoplasmic targeting) using the same signaling currency (Atg1 kinase activation) (Figure 6).
Figure 6. Unified model for Atg1 activation by signals for selective and bulk autophagy.
Autophagy induction is regulated by a bowtie signaling topology with Atg1 kinase activation at its center. Atg11 and Atg17 serve as scaffold proteins required for Atg1 kinase activation by targets and nutrient depletion, respectively. Note that it remains unclear whether Atg11 and Atg17 form mutually exclusive complexes with Atg1. Atg13 and its regulation by upstream kinases are excluded for simplicity. See text for more details.
The definitional role of autophagy receptors is to connect their targets to Atg8 family proteins on the autophagosome membrane (Schreiber and Peter, 2014). Our work provides an explanation for the conservation of receptor interactions with Atg11 family proteins: target-bound receptors interact with Atg11 to cause Atg1 kinase activation, a sine qua non for selective autophagy in nourished cells. Interestingly, Atg11 interaction with Atg19 and Atg36 is dependent on receptor phosphorylation by Hrr25 (a homolog of mammalian casein kinase 1δ) (Pfaffenwimmer et al., 2014; Tanaka et al., 2014). This raises the intriguing possibility that selective autophagy can be programmed by physiological or developmental stimuli using the signaling logic of a kinase cascade (activation of a receptor-specific kinase leading to Atg1 kinase activation). Lastly, we note that not all autophagy receptors interact with Atg11 family proteins. As a case in point, a recent yeast study showed that Cue5 is a receptor for ubiquitinated protein aggregates that does not interact with Atg11 (Lu et al., 2014). Autophagic clearance of Cue5-bound targets was dependent on starvation and we rationalize this as a dependence on starvation cues for Atg1 kinase activation. To conclude, the minimal function of autophagy receptors is to tether their targets to the autophagosome membrane but some receptors additionally interact with scaffold proteins to turn on the autophagy kinase.
How might a scaffolding mechanism for Atg1 kinase activation by selective autophagy targets work? One potential clue is that Atg1 activation by Atg19-bound targets is apparently dependent on autophosphorylation of Thr 226 in the activation loop. Since pApe1 aggregates have many Atg19 binding sites, they have the potential to induce clustering of Atg1 and, hence, facilitate kinase activation by autophosphorylation in trans. Beyond simple clustering, allosteric changes in Atg19 caused by pApe1 binding may also be necessary for Atg1 kinase activation. A recent study showed that pApe1 propeptide binding near the N-terminus of Atg19 increased the binding affinity of Atg8 to the C-terminus of Atg19, which is proximal to the Atg11-binding site (Sawa-Makarska et al., 2014). Regardless of these mechanistic possibilities, our finding that Atg11 is at the core of a versatile molecular switch that receives signal inputs from multiple autophagy receptors is an impetus to obtain structural information about this scaffold protein.
The autophagy kinase activation mechanism we uncovered in budding yeast may be relevant to how selective autophagy in metazoans prevents accumulation of neuronal protein aggregates (Hara et al., 2006; Komatsu et al., 2006), eliminates intracellular bacteria (Ogawa et al., 2005), and clears the C. elegans embryo of paternal mitochondria (Al Rawi et al., 2011; Sato and Sato, 2011). Intriguingly, two metazoan protein families (represented by FIP200 and Huntingtin in humans) with sequence homology to Atg11 (Lin et al., 2013; Ochaba et al., 2014) were shown to bind both autophagy receptors and ULK1 (Hara et al., 2008; Lin et al., 2013; Nagy et al., 2014; Ochaba et al., 2014; Rui et al., 2015). Creation of an analog-sensitive ULK1 allele by mutation of the conserved gatekeeper residue will enable development of a cell-free system for defining the role of candidate scaffold proteins and determining whether receptor-bound target activation of the autophagy kinase is conserved in humans.
EXPERIMENTAL PROCEDURES
Strain construction and PCRs
Yeast strains and plasmids are listed in Tables S1 and S2. Deletion strains were constructed in the BY4741 background (mating type a) by standard PCR-mediated gene knockout. 3 × FLAG, 6 × FLAG-GFP, 13 × MYC, mCHERRY and 3 × HA cassettes were used to modify gene loci using standard PCR-mediated tagging. The ATG11 and PHO8 promoters were replaced with the TDH3 promoter using standard PCR-mediated promoter replacement. For truncation of the ATG11 RBD, a stop codon was introduced after codon 881. For truncation of the ATG19 11BD, epitope tags were introduced after codon 387. A 3 × V5-AID cassette provided by A. Amon was used to modify the PEX1 gene locus. A 3 × GFP cassette provided by J. Nunnari was used to modify the ATG2 locus. To introduce OsTIR1 into the genome, a plasmid containing OsTIR1 provided by A. Amon was digested with PmeI and transformed into yeast for integration at the leu2 locus.
Mutagenesis was performed using QuikChange (Stratagene) mutagenesis. For mutation of the ATG19 ABD, codons 153–191 were deleted. Genomic allelic exchanges were performed using standard URA3 replacement and 5-FOA counter-selection. Primer sequences for all strain constructions are available upon request.
Cell-free Atg1 kinase assay
Frozen lysate powder was mixed with cold 1 × kinase buffer (150 mM KOAc, 10 mM MgOAc, 0.5 mM EGTA, 5 mM NaCl, 20 mM HEPES-KOH [pH 7.3], 5% glycerol) in equal volume (wt/vol), thawed on ice, and resuspended by pipetting. Extracts were cleared twice at 1,000 × g for 5 min at 4 centigrade. Equal volumes of clarified extract and 2 × kinase mix (1 × kinase buffer, 2 × energy mix (90 mM creatine phosphate, 2.2 mM ATP, 0.45 mg/ml creatine kinase), 0.2 mM N6-phenylethyl-ATPγS (N6-PhEt-ATPγS)) were combined and incubated for 1 hr at room temperature. Reactions were quenched with 1/20th volume 0.5 M EDTA. Thiophosphorylated extracts were alkylated with 1/20th volume 50 mM para-nitrobenzyl mesylate (PNBM, Abcam) for 1 hr at room temperature, heated in loading buffer and analyzed by one of two SDS-PAGE systems. Gel system 1 was Novex NuPAGE 4–12% Bis-Tris SDS-PAGE (Life Technologies). This lower percentage gel system was ideal for detecting the small shift in Atg1 mobility due to FLAG-tagging. Gel system 2 was Novex 4–20% Tris-Glycine SDS-PAGE (Life Technologies). Thiophosphorylated substrates were identified by immunoblotting with a rabbit anti-thiophosphate ester primary antibody [51-8] (Abcam) and an anti-rabbit HRP-conjugated secondary antibody (Bio-Rad). Blot imaging was done using an AlphaImager Gel Imaging System (Alpha Innotech).
Purified proteins were pre-incubated with extracts for 30 min at room temperature. FLAG-GFP-Atg11 was incubated at final concentrations of 1.6, 8, 40, and 200 nM in Figure 4A and, alongside FLAG-GFP-Atg11ΔRBD, at 2, 6.3, 20, 63 and 200 nM in Figure 4B. In Figure S4B, FLAG-GFP-Atg11 was used at 8 nM and FLAG-GFP-Atg11ΔRBD was used at 8 nM (1 ×) or 80 nM (10 ×). In Figure S4C, FLAG-GFP-Atg11 was used at 40 nM. In Figure 4C, 0.1, 0.3, 1, and 3 µl of affinity purified Atg19-target complexes were incubated with 10 µl extracts in 15 µl reactions containing 20 nM FLAG-GFP-Atg11. For Figure 4D, 1.5 µl of affinity-purified Atg19-target complexes were incubated with 2.5 µl extracts in 5 µl reactions containing 20 nM FLAG-GFP-Atg11. After pre-incubation, reactions were pelleted at 20,000 × g for 20 min, and pellets were resuspended in kinase buffer containing 1 × energy mix and 0.1 mM N6-PhEt-ATPγS before analysis of kinase activity as above. The pelleting step improved the signal-to-noise ratio but we obtained qualitatively similar results by analyzing the effect of add-backs on kinase activity in total extracts (data not shown).
Peroxisome-containing extracts were prepared from lysate powder by clearing twice for 10 min at 1,000 × g, as above. One, 3 or 10 µl of extracts were pre-incubated for 30 min at room temperature with 10 µl atg19Δatg36Δ HA-as-ATG1 extract, which had been pre-cleared at 20,000 × g for 20 min. Reactions were then pelleted for 20 min at 20,000 × g before resuspension and analysis of kinase activity as above.
Immunoprecipitation of Atg1 substrates after kinase reaction
Following the 1 hr room temperature incubation in the kinase assay, each sample was combined with two volumes of 1 × IP buffer (50 mM HEPES-KOH (pH 6.8), 150 mM KOAc, 2 mM MgOAc, 1 mM CaCl2, 15% glycerol) with 1% NP-40. The resulting solution was incubated with 20 µl washed anti-FLAG M2 affinity gel (Sigma) for three hr at 4 centigrade. The resin was washed three times with 500 µl 1 × IP buffer with 1% NP-40 and incubated for 30 min on ice with 30 µl 1 mg/ml 3 × FLAG peptide (Sigma) in 1 × IP buffer containing 1% NP-40. When appropriate, the eluted material was alkylated with 2.5 mM PNBM prior to SDS-PAGE and immunoblotting to detect thiophosphorylation.
For further experimental procedures, see Supplemental Experimental Procedures in Supplemental Information.
Supplementary Material
Acknowledgments
We thank A. Sanchez for her technical work in support of this project; D. Klionsky, A. Amon, and J. Nunnari for reagents; R. Kunz and B. Budnik for mass spectrometry analysis; A. Gutu and N. Weir for fluorescence microscopy assistance; T. Giddings, M. Morphew and University of Colorado Boulder EM Service for transmission electron microscopy; J. Weissman, A. Murray, E. O’Shea, D. Kahne, and B. Stern for critical reading of the manuscript; and members of the Denic laboratory for scientific advice. This work was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1144152 (to R.A.K.), a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation under Grant No. DRG2130-12 (to C.J.S.), and Harvard University.
Footnotes
AUTHOR CONTRIBUTIONS
R.A.K. and C.J.S. constructed yeast strains and performed the experiments: C.J.S. developed the in vitro kinase assay, R.A.K. performed the in vitro reconstitutions. R.A.K. and C.J.S. constructed figure panels. R.A.K., C.J.S. and V.D. analyzed the data and wrote the manuscript. V.D. directed the project.
References
- Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hübner A, Chou W-H, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nat. Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Orban DP, Bernard A, Singh K, Cao Y, Klionsky DJ. Atg23 and Atg27 Act at the Early Stages of Atg9 Trafficking in S. cerevisiae. Traffic. 2015;16:172–190. doi: 10.1111/tra.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Fulda S. Receptor proteins in selective autophagy. Int. J. Cell Biol. 2012 doi: 10.1155/2012/673290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow J, Zhang C, Shokat KM, Weiss EL. Design and use of analog-sensitive protein kinases. Curr. Protoc. Mol. Biol. 2004;(Unit 18.11) doi: 10.1002/0471142727.mb1811s66. Chapter 18. [DOI] [PubMed] [Google Scholar]
- Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura SI, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Yates JR, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al. Hsp90-Cdc37 Chaperone Complex Regulates Ulk1- and Atg13-Mediated Mitophagy. Mol. Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T, Suzuki K, Ohsumi Y. Lap3 is a selective target of autophagy in yeast, Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2009;378:551–557. doi: 10.1016/j.bbrc.2008.11.084. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6:1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MB, Novotny CJ, Shokat KM. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem. Biol. 2015;10:257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Yang P, Huang X, Zhang H, Lu Q, Zhang H. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J. Cell Biol. 2013;201:113–129. doi: 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. Autophagic clearance of PolyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Kárpáti M, Varga Á, Pircs K, Venkei Z, Takáts S, Varga K, Érdi B, Hegedus K, Juhász G. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10:453–467. doi: 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 2014;10:835–845. doi: 10.4161/auto.28259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, Mao K, Lau AL, Yeung SY, Humbert S, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014;15:862–870. doi: 10.15252/embr.201438932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Rui Y-N, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang Y-Y, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat. Cell Biol. 2014;16:425–433. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta - Mol. Cell Res. 2014;1843:163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Yeh Y-Y, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective Autophagy Regulates Insertional Mutagenesis by the Ty1 Retrotransposon in Saccharomyces cerevisiae. Dev. Cell. 2011;21:358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Tanaka C, Tan L-J, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh-Nakatogawa M, Ohsumi Y, Nakatogawa H. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 2014;207:91–105. doi: 10.1083/jcb.201402128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.