Abstract
A novel fungal laccase produced by the ascomycete Chaetomium sp. isolated from arid soil was purified and characterized and its ability to remove dyes was determined. Extracellular laccase was purified 15-fold from the crude culture to homogeneity with an overall yield of 50% using ultrafiltration and anion-exchange chromatography. The purified enzyme was found to be a monomeric protein with a molecular mass of 68 kDa, estimated by SDS-PAGE, and with an isoelectric point of 5.5. The optimal temperature and pH value for laccase activity toward 2,6-DMP were 60 °C and 3.0, respectively. It was stable at temperatures below 50 °C and at alkaline conditions. Kinetic study showed that this laccase showed higher affinity on ABTS than on 2,6-DMP. Its activity was enhanced by the presence of several metal ions such as Mg2+, Ca2+ and Zn2+, while it was strongly inhibited by Fe2+, Ag+ and Hg2+. The novel laccase also showed high, remarkable sodium chloride tolerance. Its ability to decolorize different dyes, with or without HBT (1-hydroxy-benzotriazole), as redox mediator, suggests that this protein may be useful for different industrial applications and/or bioremediation processes.
Keywords: Chaetomium, Screening, Fungi, Laccase, Oxidoreductase
Introduction
An intense research on extreme laccases has been performed in the recent years due to its resistance to the harsh conditions of several biotechnological and industrial applications (Giardina et al. 2010; Rodgers et al. 2010; Dwivedi et al. 2011). As reported previously, these versatile enzymes play an interesting role in many industrial sectors, including paper biopulping and biobleaching, decolourization of textile dyes and detoxification of contaminated soils (Rodríguez Couto and Toca Herrera 2006), in food industries, and bioremediation and organic synthesis (Rodríguez Couto et al. 2006; Sakurai and Kataoka 2007). Laccases (benzenediol oxygen oxidoreductase, EC 1.10.3.2) are copper-containing enzymes belonging to the blue oxidases’ group. The laccases are mostly extracellular glycoproteins (Kunamneni et al. 2007) which can be found under three forms: monomeric, dimeric or tetrameric. Laccases contain usually three types of copper sites (types 1, 2, and 3) and four copper atoms that catalyze the oxidation of suitable substrates with the concomitant reduction of O2 to water (Frasconi et al. 2010). They have received much attention during the last decades due to their relatively wide substrate specificity. Laccases are widespread in nature and are present in fungi, some bacteria, insects and higher plants (Hakulinen et al. 2002; Riva 2006). However, fungal laccases are the most studied enzymes (Baldrian 2006; Sharma et al. 2007), especially from the Basidiomycetes class (Claus 2004). To our knowledge, only few previous studies are reported on Ascomycetes and illustrated that this group of fungi often presented a multigene family of laccases but without much information regarding the regulation mechanisms of laccase expression (Levasseur et al. 2010; Fan et al. 2014). Laccases from ascomycetes have been little investigated, especially the ones produced from thermophilic and thermotolerant ascomycetes. This class of fungi constitutes a potent natural source of thermostable enzymes (Boonlue et al. 2003) which are expected to be powerful tools for sustainable developments in industrial and biotransformation processes under harsh conditions (Maheshwari et al. 2000). Phylogenetic analyses showed that the known thermophilic ascomycetes fungi, from the orders Sordariales, Eurotiales, and Mucorales (Berka et al. 2011) belong to the Chaetomiaceae family. Chaetomium species are well known as cellulolytic filamentous fungi participating in decomposition of cellulose-rich materials (Phonkerd et al. 2008). This broad genus constitutes the predominant components of the mycoflora in various soils (Rodríguez et al. 2002; Wijeratne et al. 2006). Chaetomiaceae family includes species that play potent roles in biology and biotechnological fields (Soni and Soni 2010), taxonomy (Wang and Zheng 2005), and molecular studies (Aggarwal et al. 2008). For decades, huge number of new compounds (bioactive metabolites) have been isolated and reported from this genus (Park et al. 2005). Due to the increasing demand for fungal extracellular enzymes that are able to work under harsh conditions (Krishnamurthy et al. 2008), some species of Chaetomium have been studied for their ability to produce several enzymes with potential applications (Abd El-Azeem and Salem 2012). Many species of Chaetomium are known for their effective ability to suppress the growth of bacteria and fungi through mycoparasitism, competition and combination of various mechanisms (Park et al. 2005; Zhang and Yang 2007; Marwah et al. 2007). They have also been reported as potential biocontrol species against many pathogenic microorganisms including Fusarium, Helminthosporium and Phytophthora (Soytong et al. 2001) and as interesting producers of cellulases, e.g., Chaetomium murorum (Reanprayoon and Pathomsiriwong 2012). In the present study, we report the purification of laccase from the ascomycete fungal strain, Chaetomium sp., and its biochemical characterization. The purified enzyme was evaluated for its ability for dye decolourisation.
Materials and methods
Chemicals
The compounds used as laccase substrates were: 2,6-dimethoxyphenol (2,6-DMP), 2.2′-azinobis-(3-ethyl benzthiazoline-6-sulphonate) (ABTS), guaiacol and the synthetic dyes including Acid Orange 51, Direct Red 75, Malachite Green, Direct Blue 86 and Remazol Brilliant Blue R. All chemicals were of analytical grade and were from Sigma-Aldrich (St. Louis, MO, USA).
Isolation and screening of laccase-producing fungal strains
The soil samples used in this study were collected from arid regions in southern Tunisia. Strains were isolated by streaking several times until pure fungal colonies were obtained. After purification, fungal strains were maintained on 2% (w/v) malt extract agar (MEA) plates and incubated at 37 °C for 5–7 days. For the detection of laccase activity, all collected strains were grown on MEA plates at 37 °C containing 5 mM of 2,6-DMP and 150 µM copper sulfate. Colonies, with red-brown halos, were picked out as potential positive strains and transferred into flasks of a basal medium with 150 µM CuSO4, as inducer (M7 medium), for further characterization (see below, point 2.4).
Molecular identification of fungal strains
Fungi screened out in our work were isolated in our laboratory from arid soil in southern Tunisia. These collected strains were subjected to molecular identification of their ITS sequences. The genomic DNA was extracted from the fungal cells using DNeasy Plant Mini Kit (Qiagen). To perform the sequence analysis, the 18S rDNA region was amplified by polymerase chain reaction (PCR) using the universal internal transcribed spacers combination ITS1/ITS4 as primers (Damm et al. 2008). PCR amplification was performed in a thermocycler system with an initial denaturation step at 95 °C for 1 min 30 s, followed by 35 cycles consisting of denaturation (94 °C for 45 s), annealing (52 °C for 45 s), elongation (72 °C for 1 min 30 s), final extension (72 °C for 5 min) and ending at 4 °C; 400–600 pb amplicons were obtained and loaded onto a 1% agarose gel. The amplified DNA was purified using QIAquick® Gel Extraction Kit and sequenced using an automated ABI Prism 3730 DNA sequencer (Applied Biosystems,…). The nucleotide sequences were analyzed by comparing the known 18S ribosomal reference sequences in the National Center for Biotechnology Information (NCBI) GenBank database by BLAST analysis (Altschul et al. 1997). Multiple sequence alignment was achieved using ClustalW (Thompson et al. 1994). The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004). Phylogenetic tree was inferred using the neighbor-joining method (Saitou and Nei 1987) in the MEGA7 program (Kumar et al. 2016) with bootstrap values based on 1000 replicates (Felsenstein 1985). All positions with less than 95% site coverage were eliminated.
Fungal strain and culture conditions
The Chaetomium sp. strain was grown on malt extract agar (MEA) plates incubated at 45 °C for 5–7 days. Chaetomium sp. preculture was prepared by placing three mycelial plugs from a 5-day-old culture (45 °C) on malt extract agar plates, in 250 mL Erlenmeyer flasks containing 50 mL of M7 medium. After 4 days, 2% (v/v) of the homogenized mycelial suspension was used to inoculate liquid media. Cultures were incubated at 37 °C in a rotary shaker at 160 rpm. Cultures were carried out on M7 medium of the following composition per liter: glucose, 10 g; yeast extract, 1 g; soya peptone, 5 g; KCl, 0.5 g; MgSO4, 7H2O, 0.5 g; KH2PO4, 1 g; ammonium tartrate, 2 g; trace elements solution, 1 mL. The trace elements solution composition per liter was as follow: B4O7Na2·10H2O, 0.1 g; CuSO4·5H2O, 0.01 g; FeSO4·7H2O, 0.05 g; MnSO4·7H2O; 0.01; ZnSO4·7H2O, 0.07 g; (NH4)6Mo7O24·4H2O, 0.01 g. The pH of the solution was adjusted to 5.5. The basal medium was supplemented with 150 µM CuSO4 as an inducer of laccase activity on the third day (Dittmer et al. 1997).
Enzyme assays and protein determination
Laccase activity was measured spectrophotometrically with 2,6-dimethoxyphenol (2,6-DMP) and 2.2′-azinobis-(3-ethyl benzthiazoline-6-sulphonate) (ABTS) as substrates. The enzymatic reaction mixtures were determined by using 5 mM ABTS in 100 mM sodium acetate buffer, pH 5.0 (ε 436 = 29.300 M−1 cm−1) (Rodríguez et al. 2008), or 5 mM 2,6-DMP in 100 mM citrate buffer, pH 5.0 (ε 469 = 27,500 M−1 cm−1) (Jaouani et al. 2005). The enzymatic assays were carried out at room temperature. Protein concentration was determined using BCA method (Bio-Rad) (Smith et al. 1985).
Laccase purification
Laccase produced from Chaetomium sp. was purified from 8-day-old cultures containing CuSO4. Mycelia were removed from culture liquid by filtration through filter paper. The filtrate was concentrated and dialyzed against 10 mM sodium phosphate (pH 6.5) (Buffer A) by ultrafiltration (Filtron; 10 kDa cutoff membrane). The concentrated enzyme solution was loaded onto a Hitrap QFF cartridge column (16 × 25 mm, GE Healthcare) pre-equilibrated with the same buffer A at a flow rate of 1 mL min−1. Retained enzymes were eluted at the same flow rate with a linear NaCl gradient (0–1 M) in 10 mM sodium phosphate (pH 6.5) described as Buffer B. The fractions containing highest laccase activity were pooled, desalted using PD-10 column and concentrated by 30 kDa molecular weight cut-off ultrafiltration concentrator. 1-mL samples were subjected to a mono-Q-anion exchange column (Q-Sepharose GL 5/50) (GE Healthcare Life Sciences) equilibrated with the same buffer A. Retained proteins were eluted with the same buffer B at the same flow rate. The laccase peak was collected, concentrated and stored at 4 °C for subsequent characterization. An SDS-PAGE was performed to check the purity of the purified laccase. The protein content and laccase activity of each purification step were measured.
Laccase characterization
The apparent molecular weight of the purified laccase was estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and gel filtration. SDS-PAGE was performed on 12% gel as described by Laemmli (1970) using high-molecular-mass protein marker (Bio-Rad). Gel filtration was carried out on Sepharose G-75 column calibrated with conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa) and aprotinin (6.5 kDa) to estimate the molecular weight of the native protein. A volume of 0.1 mL of concentrated protein was loaded onto this column, equilibrated with 10 mM sodium phosphate buffer containing 100 mM NaCl, pH 6.5, and eluted isocratically at a flow rate of 1 mL min−1. The isoelectrofocusing was performed on 5% polyacrylamide gel with pH range from 3.5 to 10 (Chakroun et al. 2010). Protein bands after SDS-PAGE were visualized by staining with Coomassie Brilliant Blue R-250. Zymogram analysis was carried out under non-denaturing conditions and activity bands were obtained using 10 mM 2,6-DMP in 100 mM sodium acetate buffer (pH 5.0) after washing the gel for 10 min with the same buffer (Zouari-Mechichi et al. 2006). The UV–Visible absorption spectrum (300–800 nm) of the purified laccase was recorded in 10 mM sodium phosphate buffer, pH 6.5 using the spectrophotometer (Neifar et al. 2010).
Effect of pH and temperature on laccase activity and stability
Optimum pH for laccase activity was determined towards ABTS and 2,6-DMP at a pH range from 2.0 to 8.0. The pH level was adjusted using citrate–phosphate buffer with a final concentration of 50 mM. pH stability was tested after incubating the laccase at different pH values (pH 2.0–9.0) for 24 h at 4 °C. To determine the optimal temperature, laccase activity was tested at various temperatures (30–90 °C) at optimal pH. Thermal stability was investigated by incubating the purified enzyme at different temperatures (40, 50, 60, 70 and 80 °C) for 6 h. Residual activities were measured using 2,6-DMP as substrate. The estimated activity at starting time was considered as 100% stability.
Effect of sodium chloride on laccase activity
The halotolerance test of the purified laccase was investigated in presence of 250–3000 mM NaCl. Controls were carried out under the same conditions, without NaCl. The relative activities were measured using 2,6-DMP as substrate.
Effect of metal ions and inhibitors on laccase activity
The effect of metal ions, used at three concentrations 10, 100 mM and 200 mM on laccase activity was determined by incubating the enzyme with each metal at 4 °C for 15 min before measuring the residual activity. The tested ions were: Ca2+ (CaCl2), Cu2+ (CuSO4), Mg2+ (MgSO4), Zn2+ (ZnSO4), Hg2+ (HgSO4), Ag+ (Ag2SO4) and Fe2+ (FeSO4). The effect of some inhibitors on laccase activity was tested as described above using 2,6-DMP assay. Inhibitors were used at different concentrations as follows: ethylenediaminetetra-acetic acid (EDTA) (0.5, 10 and 30 mM); dithiothreitol (DTT) (1 and 2.5 mM); β-mercaptoethanol (0.05 mM); l-cysteine (0.01 mM) and sodium azide (NaN3) (0.01 and 0.1 mM). Laccase activity, in the absence of metal ions and inhibitors, was used as the control.
Kinetic parameters of the purified laccase
The substrate specificity of the purified laccase was investigated for ABTS and 2,6-DMP with concentrations ranging from 1.25 µM to 1 mM in 100 mM sodium acetate buffer at optimal pH. K m and k cat values were estimated using the Lineweaver–Burk plots of Michaelis–Menten equation (Lineweaver and Burk 1934; Lu et al. 2013).
Decolourization of synthetic dyes
Dye decolourization capability of the pure laccase obtained from Chaetomium sp. was evaluated using various synthetic dyes such as Remazol Brilliant Blue R (RBBR) (592 nm); Direct Blue 86 (594 nm); Acid Orange 51 (446 nm); Direct Red 75 (600 nm) and Malachite Green (630 nm). The reaction mixture (5 mL) included citrate–phosphate buffer (pH 5.0; 100 mM), laccase (0.3 U mL−1) and dye (50 mg L−1). Reaction was carried out at 28 °C for 4 h and 24 h in the darkness under mild shaking conditions (Ben Younes et al. 2007; Schückel et al. 2011). Decolourization was assayed with and without HBT (1-hydroxy-benzotriazole) as the most common laccase mediator at a final concentration of 0.5 mM. The control samples were carried out in parallel without adding the enzymes. All experiments were performed in triplicate. The decolourization percentage was measured spectrophotometrically as the relative decrease of absorbance at the maximal absorbance–wavelength of each dye (Wang et al. 2010).
Results and discussion
Isolation, screening, and identification of laccase producing fungal strains
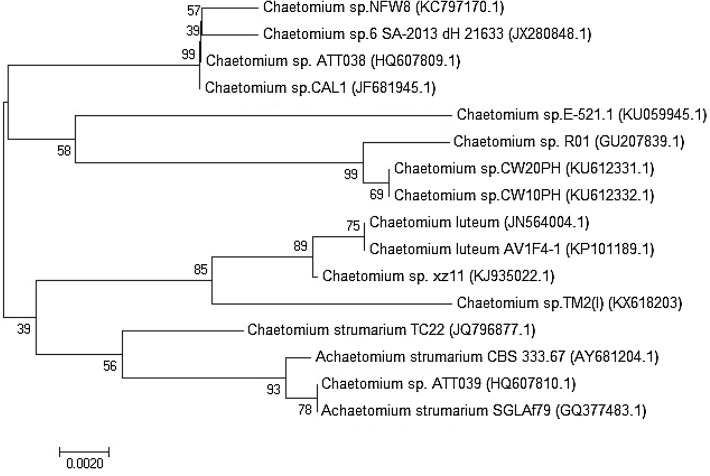
Sixteen fungal strains were isolated from arid Tunisian soils. The most interesting strain was identified based on the analysis of the amplified nucleotide sequences of the nuclear ribosomal ITS1-5.8-ITS4 region (~600 bp). The phylogenetic tree was inferred using neighbor joining plot after alignment of the DNA amplified sequences using ClustalW software (Fig. 1) with their putative homologue sequences from GenBank. The analysis of ITS sequences similarity revealed that the fungus, designated as TM2, is closely related to members of the genus Chaetomium. This strain was selected for further study. The dendogram obtained from the alignment showed that the maximum identity (97%) of TM2 was found with Chaetomium sp. ATT038 (Accession number HQ607809). This new fungus showed a homology with other close strains including, Chaetomium luteum (Accession number JN564004) (97% identity), Chaetomium sp. xz11 (Accession number KJ935022) (98% identity), and Chaetomium sp. R01 (accession number: GU207839) (96% identity). It is clear that the selected strain, Chaetomium sp. TM2, belongs to the cluster that included laccase-producing ascomycetes. Laccase has been reported in some well-known ascomycetes from Sordariomycetes class (Kittl et al. 2012), such as Myceliophtora thermophila (Berka et al. 1997) and Melanocarpus albomyces (Kiiskinen et al. 2002). However, the genus Chaetomium is a representative model of cellulolytic filamentous fungi, while few data have been reported as laccase producers (Chefetz et al. 1998; Qasemian et al. 2012; Manai et al. 2016).
Fig. 1.
Phylogenetic tree constructed by NJ method based on the alignment of ITS sequences of the isolated fungus, identified as a Chaetomium sp. strain, with homologue sequences obtained from the NCBI GenBank
Purification of extracellular laccase
The purification process of laccase obtained from Chaetomium sp. culture is summarized in Table 1. Liquid culture broth was concentrated, dialyzed and purified using two different chromatographic columns. During the first chromatographic step, the purification was carried out using a weak anion exchanger (Q-Cartridge) with a linear gradient of NaCl in the range 0–1 M. Laccase activity was separated from most proteins and brown pigments which showed strong absorbance at 280 nm. Afterward, a high efficiency exchange anion column (Mono-Q) was used to further separate the laccase obtained from the last step after concentration by using Amicon system. Chromatography through Mono-Q column resolved a single protein peak corresponding to a blue fraction with the isolated laccase, which was purified 15.11-fold with a final yield of 50.39%.
Table 1.
Summary of laccase purification from Chaetomium sp
| Volume (mL) | Total activity (U) | Protein (µg) | Specific activity (U mg−1) | Yield (%) | Purification fold | |
|---|---|---|---|---|---|---|
| Culture filtrate | 1500 | 480 | 100,000 | 4.8 | 100 | 1 |
| Concentration | 200 | 435.15 | 83333.33 | 5.22 | 90.65 | 1.08 |
| UF + Dialyse | 40 | 414.67 | 68266.67 | 6.07 | 86.39 | 1.26 |
| HiTrap QFF | 30 | 325.08 | 25,000 | 13 | 67.72 | 2.7 |
| Mono-Q | 1 | 241.9 | 3333.33 | 72.56 | 50.39 | 15.11 |
Laccase characterization
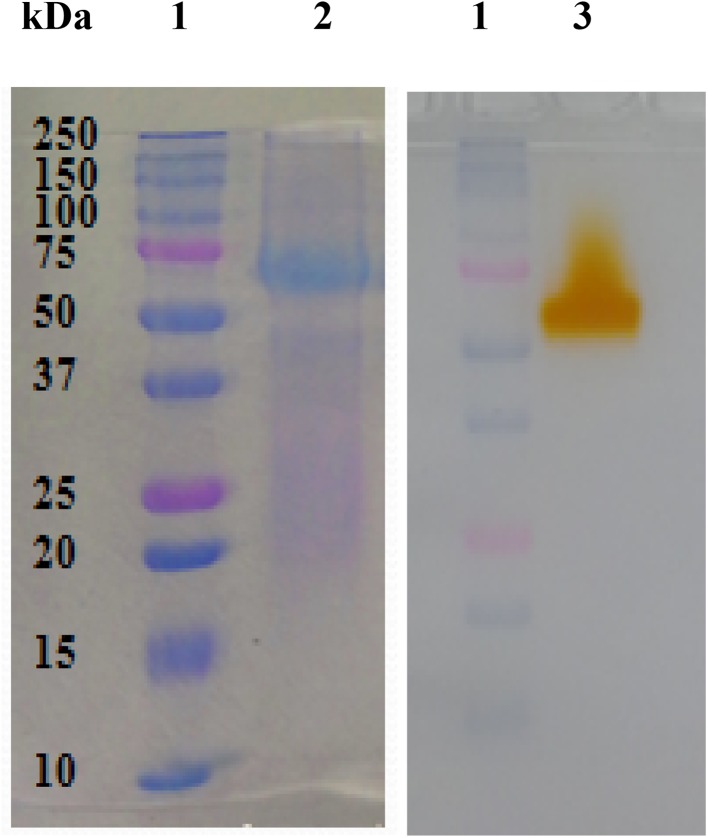
To verify homogeneity of the purified laccase and to determine its molecular mass, it was analyzed by SDS-PAGE, showing an apparent molecular mass of 68 kDa (Fig. 2a). The zymogram of this protein showed that it maintains its activity after the SDS-PAGE (Fig. 2b). The gel filtration chromatography of this protein, using Sepharose G-75 column, showed a single peak which showed a molecular mass around 80 kDa, indicating that this laccase was a monomeric protein (data not shown). These results agree with data reported in many typical fungal laccases, described as monomeric proteins with molecular weight ranging from 50 to 130 kDa (Morozova et al. 2007). The estimated molecular weight of this laccase was very close to that of laccases from Xylaria polymorpha (67 kDa) (Liers et al. 2007), Cerrena unicolor (LaccI, 64 kDa) (Michniewicz et al. 2006), Pycnoporus sanguineus CS43 (66 and 68 kDa) (Ramírez-Cavazos et al. 2014) as well as Pleurotus eryngii (61–65 kDa) (Muñoz et al. 1997). Although it has been reported the production of several laccase isoforms in fungi (Baldrian 2006; D’Souza-Ticlo et al. 2009; Chakroun et al. 2010), the isolated Chaetomium sp. produced a single laccase with a pI value of 5.5 (data not shown). This value is higher than those reported in other fungal laccases as those from the basidiomycetes Coriolopsis sp (pI 3.5–4.3), Coriolus sp (pI 4.0–4.6) (Baldrian 2006), Trametes trogii (pI 3.3–3.6) (Garzillo et al. 1998) or the ascomycetes Melanocarpus albomyces (pI 4.0) (Kiiskinen et al. 2002). Baldrian (2006) showed that typical fungal laccases are proteins with acidic isoelectric point around pH 4.0. Moreover, the pI of the laccase from the new Chaetomium sp. isolated strain was similar to that reported for the laccase produced by Chaetomium thermophilum (pI 5.1) (Chefetz et al. 1998). The purified laccase has a blue color typical for copper containing oxidases. The UV–Vis spectrum of the pure laccase (Fig. 3) showed a peak at 580 nm, corresponding to type 1 copper centre which gives it its characteristic blue color, and a shoulder at 320 nm, indicating the presence of type 3 trinuclear cluster. These results are similar to that for other typical fungal blue laccases (Dwivedi et al. 2011; Yang et al. 2014). The absorbance ratio 280 nm/610 nm was determined to be 20.0, which is consistent with that reported for these enzymes (15–20) (Ademakinwa and Agboola 2016). This laccase has some properties similar to or in the range of most reported ascomycetes and basidiomycetes fungal blue laccases (Chakroun et al. 2010; Ben Younes and Sayadi 2011).
Fig. 2.
Electrophoretic analysis of the purified laccase from Chaetomium sp. strain. a SDS-PAGE analysis. Lane 1: protein marker; Lane 2: purified laccase. b Zymogram of the purified laccase. Lane 1: protein marker; Lane 3: Laccase zymogram against 2,6-DMP used as substrate
Fig. 3.
Absorption spectrum of the purified Chaetomium sp. laccase
Effect of pH and temperature on laccase activity and stability
The effect of pH on laccase activity was tested at different pH values ranging from 2.0 to 8.0 by using 2,6-DMP and ABTS as substrates. As shown in Fig. 4a, the optimal pH for the oxidation of both substrates was 3.0. This result is in agreement with data reported by Baldrian (2006) and Morozova et al. (2007) for both substrates used. A previous study reported that most fungal laccases have optimal pH values in the acidic range using ABTS as substrate (Siroosi et al. 2016). It is in a linear response to pH. This result is consistent with many previous works reported by Schückel et al. (2011); Halaburgi et al. (2011) and Yang et al. (2014). No activity was observed above 7.0, which is in contrast to that of Melanocarpus albomyces laccase (more than 70% of the residual activity remained at pH 8.0) (Kiiskinen et al. 2002). The pH stability of the purified Chaetomium sp. laccase is shown in Fig. 4b. The purified laccase retained 100% of its relative activity over a wide range of pH values from 4.0 to 9.0, which is similar to many reported fungal laccases from ascomycetes and basidiomycetes. Laccases from the thermophilic ascomycete Melanocarpus albomyces (Kiiskinen et al. 2002); Fusarium proliferatum (Fernaud et al. 2006), Trichoderma atroviride (Chakroun et al. 2010) and Cladosporium cladosporioides (Halaburgi et al. 2011) showed tolerance to neutral and slightly alkaline conditions (pH 7.0–9.0). The studies of the effect of pH on the purified laccase from Streptomyces psammoticus showed that it is stable in the alkaline pH range 6.5–9.5 (Niladevi et al. 2008). This remarkable property makes this laccase a promising candidate for industrial applications where alkaline conditions predominate. Temperature dependence of enzyme was studied in a wide temperature range from 30 to 90 °C. The highest activity of the purified laccase using 2,6-DMP as substrate, was observed at 60 °C (Fig. 4c). Similar temperature was reported for ascomycetes laccases from Scytalidium thermophilum (T opt 65 °C) (Ogel et al. 2006), Xylaria polymorpha (T opt 60 °C) (Liers et al. 2007), Trametes sp. (T opt 60 °C) (Daâssi et al. 2013) and Melanocarpus albomyces (T opt 60–70 °C) (Kiiskinen et al. 2002). Generally, the typical range of optimum temperature that characterized the fungal laccases is from 50 to 70 °C (Baldrian 2006; Morozova et al. 2007; Chakroun et al. 2010; Wu et al. 2010; Santhanam et al. 2011). The thermal stability was studied for 6 h at temperatures in the range 40–80 °C. As shown in Fig. 4d, the purified laccase was not a thermostable enzyme similar to that produced from the ascomycete Trichoderma reesei (Lavasseur et al. 2010). At 40 °C, no loss of laccase activity was recorded, which indicated that the purified enzyme was highly stable at this temperature. At 50 °C, the enzyme retained around 85% of its activity after 90 min of incubation. Above 50 °C, the loss of laccase activity was much more pronounced with a half life (IC50) of 90 min at 60 °C, which is close to those observed in other fungal thermostable laccases (Baldrian 2006; Chakroun et al. 2010; Yang et al. 2014). This result is similar to that reported by Daâssi et al. (2013) which showed that laccase from Trametes sp. retained 50% of its activity after 90 min of incubation at 55 °C. Moreover, thermostability studies, reported by Niladevi et al. (2008), showed that laccase from Streptomyces psammoticus is not stable at temperatures above 50 °C during prolonged incubation time. It was found that after 90 min of incubation, the residual activities of the enzyme were reduced to 85 and 69% at 55 and 60 °C, respectively. This activity loss at high temperatures could be associated with thermal inactivation of the protein and could be related with a modification in its three-dimensional structure (by release of copper ions from the active site) (Palonen et al. 2003; Rich et al. 2013).
Fig. 4.
Effect of pH and temperature on the activity and stability of the purified laccase from Chaetomium sp. strain: a optimum pH using 2,6-DMP (filled square) and ABTS (filled diamond) as substrates. b pH stability, after 24 h incubation at 4 °C. (c) Optimum temperature using 2,6-DMP as substrate. (d) Thermostability of the purified enzyme at 40 °C (filled diamond), 50 °C (filled square), 60 °C (filled triangle), 70 °C (small letter x) and 80 °C (cyrillic Zhe)
Effect of chloride on laccase activity and stability
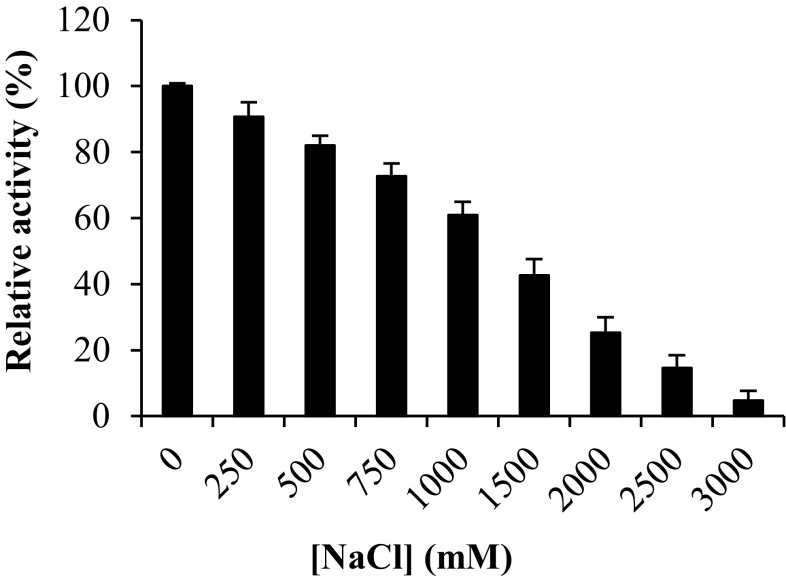
The most important obstacle reducing the application of laccases within different biotechnological and industrial processes is its sensitivity to high chloride ion concentrations (Xu 1996; Kittl et al. 2012). A lot of studies report that most fungal laccases were strongly inhibited at chloride ion concentrations below 100 mM (Farnet et al. 2008) due to its sensitivity towards halides (Fang et al. 2011). It has been proved that an interruption of internal electron transfer from the T1 to the T2/T3 centers caused by the binding of chloride to T2/T3 trinuclear center (Ruijssenaars and Hartmans 2004). So that, the dependence of Chaetomium sp. laccase on NaCl concentration was evaluated using 2,6-DMP as substrate (Fig. 5). The purified Chaetomium sp. laccase showed tolerance towards sodium chloride, it retained 60% of its initial activity at 1 M NaCl which is similar to that reported in Picnoporus sanguineus (Lu et al. 2013). Compared with laccases produced from Chaetomium sp. and Xylogone sphaerospora (two ascomycota), the purified enzyme showed a high tolerance to the presence of sodium chloride ions (Qasemian et al. 2012). Qasemian et al. (2012) showed that laccases from Chaetomium sp. remained 50% of its initial activity at 100 mM. With Xylogone sphaerospora strain, IC50 was obtained for 200 mM NaCl. This remarkable chloride tolerance may give a preference to this laccase in some industrial applications where a high chloride concentration is needed, such as pulp biobleaching and textile dye decolorization (Rodríguez Couto et al. 2006; Vanhulle et al. 2008; Rodríguez et al. 2008).
Fig. 5.
Effect of NaCl on the purified laccase activity. The relative activity of samples was determined using 2,6-DMP as substrate at 4 °C
Effect of metal ions and inhibitors on laccase activity
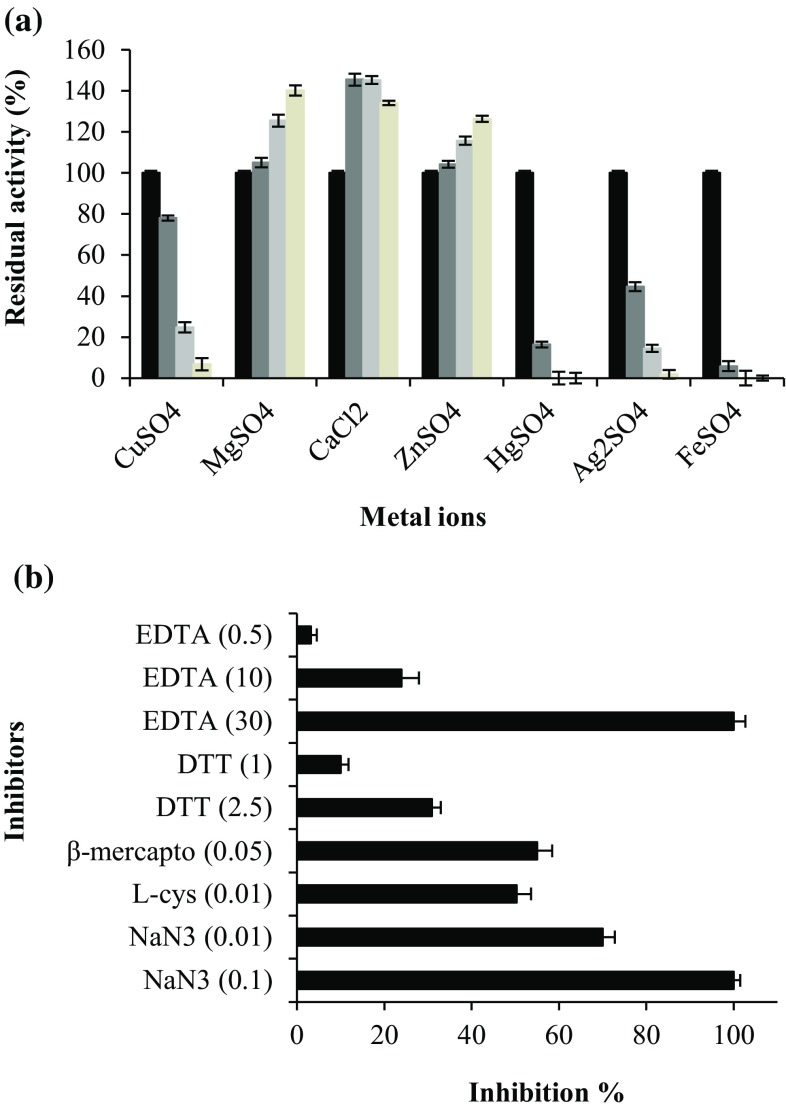
Enzymes for industrial purposes should display a higher tolerance and stability in the presence of inhibitors and metal ions present massively in industrial processes (Pan et al. 2014). The effect of metal ions on the Chaetomium sp. laccase activity was tested at three concentrations (10, 100 and 200 mM). As shown in Fig. 6a, a strong inhibition resulted after the addition of Hg2+, Ag+ and Fe2+ at a concentration of 10 mM, similarly to those reported from other fungal laccases (Lu et al. 2007, 2013; Neifar et al. 2010; Forootanfar et al. 2011; Yang et al. 2014; Castaño et al. 2015). Previous studies showed that Fe2+ inhibit strongly laccase activity, this feature was similar for most other laccases (Lu et al. 2007; Yang et al. 2013). In contrast, Niladevi et al. (2008) showed that Fe2+ could stimulate the laccase from Streptomyces psammoticus. In the presence of 10 mM of Ag+, Yang et al. (2013) reported that relative activity of laccase from Shiraia sp.SUPER-H168 decreased to 8.13%. The purified laccase was highly sensitive to Hg2+, similarly to the results obtained by Lu et al. (2013) and Mukhopadhyay and Banerjee (2015) (total inhibition occurred with Hg2+ concentration higher than 5 mM). The activity of the purified laccase in the presence of copper ions decreased slightly and remained around 78% of its relative activity in the presence of 10 mM Cu2+. However, similar result was found on laccase from Pycnoporus sp. SYBC-L1 in the presence of 10 mM Cu2+, which retained 71.52% of its relative activity (Wang et al. 2010). These results are not in agreement with previous reports that showed the enhancing effect of copper ions on laccase activity (Baldrian and Gabriel 2003; Niladevi et al. 2008; Castaño et al. 2015). It was found that for concentration of 10 mM of Mg2+ and Zn2+, laccase activity remained stable. Similar results were found by Wang et al. (2010) and Yang et al. (2013) and showed that around 99% of relative activity was retained. The presence of Mg2+, Ca2+ and Zn2+ at high concentrations (200 mM), showed a significant stimulation of laccase activity. Further increase of Ca2+ concentrations seems to have no effect on laccase activity. These observations indicated that metal ions had different effects on laccase catalytic activity depending on its type and changed within strains of the same species. The effect of some typical laccase inhibitors was tested in the presence of sodium azide (NaN3), the metal ion chelator EDTA and the thiol compounds; l-cysteine (l-cys), DTT and β-mercaptoethanol (β-mercapto), as shown in Fig. 6b. The purified Chaetomium sp. laccase was completely inhibited by 0.1 mM of NaN3, which is agree with those observed in other fungal laccases (Chakroun et al. 2010; Si et al. 2013; Castaño et al. 2015). Similar results reported a complete inhibition of laccase activity by the same concentration 0.1 mM NaN3 (Lu et al. 2007; Niladevi et al. 2008). This strong inhibition could be explained by the decrease of the oxygen consumption by laccase during the catalysis (Baldrian 2006) due to the binding of NaN3 with type 2 and 3 copper sites which block the electron transfer (Ryan et al. 2003; Chakroun et al. 2010; Lu et al. 2013). 1 mM DTT showed no inhibitory effect on the Chaetomium sp. laccase. The enzyme retained around 90% of its relative activity at this concentration, in contrast to that obtained for laccase from Pycnoporus sanguineus (Lu et al. 2007) which showed 90% of inhibition at the same concentration. The laccase was quite resistant to EDTA where 80% of the initial activity was retained at 10 mM. At a concentration of 30 mM EDTA, a complete inhibition was obtained, similarly to that reported for other laccases, where only high concentrations of EDTA (>25 mM) inhibited the enzyme, such as laccases from Xylaria sp. (Castaño et al. 2015), Pleurotus ostreatus (Palmieri et al. 1997), Streptomyces sp. C1 (Lu et al. 2013) as well as the aquatic ascomycete Phoma sp. UHH 5-1-03 (Junghanns et al. 2009). With 0.01 mM l-cysteine and 0.05 mM β-mercaptoethanol, laccase activity was reduced by 50% of its initial activity. Compared to previous studies, l-cysteine and β-mercaptoethanol were efficient inhibitors to the purified laccase at low concentrations (<0.1 mM). Similar results, reported by Sondhi et al. (2014), showed that the enzyme activity was completely inhibited when reducing agents l-cysteine and β-mercaptoethanol were added to the reaction mixture. It was reported that l-cysteine at a concentration of 0.1 mM has different effects on laccases from Lentinus squarrosulus MR13 (30% inhibition) (Mukhopadhyay and Banerjee 2015), Bacillus licheniformis LS04 (40% inhibition) (Lu et al. 2012) and Trichoderma atroviride (58% inhibition) (Chakroun et al. 2010). Yang et al. (2013) reported that 10 mM of l-cysteine inhibits 50% of laccase activity produced by Shiraia sp. SUPER-H168. At higher concentration, β-mercaptoethanol was able to inhibit only 7% (Halaburgi et al. 2011) and 14% (Mukhopadhyay and Banerjee 2015) of enzyme activity at 10 and 0.1 mM, respectively. The purified Chaetomium sp. laccase was tolerant to common environmental metal ions and typical inhibitors presenting in the industrial effluents. This tolerance makes it an advantageous tool for practical applications since many other laccases are active in the presence of the same metal ions.
Fig. 6.
a Effect of metal ions on the purified laccase activity [control (filled square), 10 mM (filled square), 100 mM (filled square), 200 mM (filled square)]. b Effect of typical laccase inhibitors on purified laccase activity
Kinetic parameters of the purified laccase
The apparent kinetic constants (K m, k cat) of the Chaetomioum sp. laccase were determined for the small phenolic substrate 2,6-DMP and for the artificial substrate ABTS, at different concentrations. A comparison of the catalytic properties of the enzyme against both substrates is showed in Table 2. The highest catalytic efficiency was observed with ABTS, which showed the lower K m value (5.6 µM). The affinity of Chaetomium sp. laccase for 2,6-DMP was higher (10.2 µM), which agree with data reported for most of the fungal laccases showing highest activity towards the non-phenolic compound ABTS (Kiiskinen et al. 2002; Morozova et al. 2007). These K m values were considered to be in the same range of fungal laccases using ABTS (4–770 µM) and 2,6-DMP (8–14,720 µM) as substrate (Baldrian 2006). The purified laccase showed high affinity against ABTS similar to many previous studies. Laccases from Melanocarpus albomyces (Kiiskinen et al. 2004) and from Chaetomium thermophilum (Chefetz et al. 1998) demonstrated K m values of 280 and 190 μM, respectively, towards ABTS. It is also other factor to consider the potential biotechnological interest of the new laccase to be used in delignification, textile dye treatment and detoxification of contaminated soils.
Table 2.
Kinetic properties of the purified Chaetomium sp. laccase
| Substrate | Wavelength (nm) | ɛ (M−1 cm−1) | K m (mM) | k cat (s−1) | k cat /K m (mM−1 s−1) |
|---|---|---|---|---|---|
| ABTS | 436 | 29.3 | 0.0056 | 68.91 | 12306.32 |
| 2,6-DMP | 469 | 27.5 | 0.0102 | 58.78 | 5762.74 |
Decolourization of synthetic dyes
The decolourization based on the use of enzymatic methods is shown as a potential alternative to the chemical methods, and it is investigated in many studies using fungal laccase systems as a promising tool (Chen and Yien Ting 2015; Manai et al. 2016). Multiple studies have shown the treatment of industrial synthetic dyes by using purified laccases (Wong and Yu 1999; Wesenberg et al. 2003; Zouari-Mechichi et al. 2006; Koyani et al. 2013; Xu et al. 2016; Wang et al. 2016). In this work, we demonstrated the ability of the purified Chaetomium sp. laccase to degrade or transform various synthetic dyes including, triarylmethane type (Malachite Green), anthraquinonic (RBBR) and azoic (Direct Red 75, Direct Blue 86 and Acid Orange 51) dyes type (Table 3). The obtained results showed that the purified Chaetomium sp. laccase was able to decolorize all the tested dyes except Malachite Green and Direct Red 75, in the absence of any mediator. When hydroxy-benzotriazole (HBT) was used as mediator (0.5 mM) in the reaction, a small increase in the level of the decolourization was detected, which is consistent with the previous reports (Zouari-Mechichi et al. 2006; Grassi et al. 2011). The decolourization of Direct Blue 86 and Acid Orange 51 was slightly affected by the presence of HBT. By contrast, in the case of Malachite Green, the presence of HBT produced an increase of the decolourization rate from 0 to around 37%, after 24 h of incubation. Thus, the presence of HBT or other mediators was shown to be necessary to improve the laccase decolourization ability, which is in agreement to those reported in other fungal laccases from Trametes versicolor (Wong and Yu 1999), Trametes hirsuta (Abadulla et al. 2000) and Aspergillus sp. (Soares et al. 2001). Although laccase-mediator systems are considered as responsible for the removal of most recalcitrant dyes, mediators are expensive and can affect the enzymatic activity caused by the generation of toxic unstable radicals (Moldes and Sanromán 2006; Pan et al. 2014). For this reason, our laccase could be used for the decolourization of industrial wastewater without mediator although more work to optimize dye decolourization is needed.
Table 3.
Decolourization of different synthetic dyes by the purified Chaetomium sp. laccase
| Dyes | Class | Decolourization (%) | |||
|---|---|---|---|---|---|
| −HBT | +HBT | ||||
| 4 h | 24 h | 4 h | 24 h | ||
| Acid Orange 51 | Azo | 26.46 ± 1.71 | 31.85 ± 0.45 | 34.43 ± 1.35 | 34.79 ± 2.34 |
| RBBR | Anthraquinone | 32.27 ± 2.05 | 38.42 ± 0.57 | 35.51 ± 1.25 | 55.6 ± 2.12 |
| Direct Red 75 | Azo | 3.46 ± 1.92 | 4.64 ± 1.74 | 26 ± 2.0 | 59.75 ± 2.25 |
| Direct Blue 86 | Azo | 50.42 ± 1.10 | 54.16 ± 0.85 | 46.2 ± 1.3 | 56.56 ± 1.68 |
| Malachite Green | Triphenylmethane | 0.0 | 0.0 | 4.88 ± 1.52 | 36.92 ± 1.27 |
Conclusion
The search for laccase in ascomycete fungi, where its presence is poorly known, could provide new proteins with catalytic properties of biotechnological and industrial interest. For this reason, this study started with the screening of a newly thermo-tolerant ascomycete fungal strain isolated in Tunisia. The laccase of the most promising fungi, identified as a Chaetomium sp. strain, was purified and characterized and its biochemical and kinetic properties were studied. The purified laccase was highly stable at neutral pH, showing considerable tolerance to the presence of sodium chloride (NaCl). This novel laccase was investigated to test its ability in the decolourization of azoic and anthraquinonic dyes under acid conditions, with and without HBT as mediator. It showed an important ability to decolorize all used synthetic dyes without mediator, except for Malachite Green, although the presence of this mediator played an interesting role as an enhancer of dye decolourization rates. The results suggest that the new laccase could be a potential and suitable tool for further biotechnological applications.
Acknowledgements
We thank Marίa Jesùs Martίnez and Jorge Barriuso for their help in carrying out this work.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Abadulla E, Tzanov T, Costa S, Robra KH, Cavaco-Paulo A, Gübitz GM. Decolorization and detoxification of textile dyes with a laccase from Trametes hirsuta. Appl Environ Microbiol. 2000;66:3357–3362. doi: 10.1128/AEM.66.8.3357-3362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El-Azeem AM, Salem FM. Biodiversity of laccase producing fungi in Egypt. Mycosphere. 2012;3:900–920. [Google Scholar]
- Ademakinwa AN, Agboola FK. Biochemical characterization and kinetic studies on a purified yellow laccase from newly isolated Aureobasidium pullulans NAC8 obtained from soil containing decayed plant matter. J Genet Eng Biotechnol. 2016;14:143–151. doi: 10.1016/j.jgeb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal R, Sharma V, Kharbikar LL. Molecular characterization of Chaetomium species using URP-PCR. Genet Mol Biol. 2008;31:943–946. doi: 10.1590/S1415-47572008005000011. [DOI] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrian P. Fungal laccases—occurrence and properties. FEMS Microbiol Rev. 2006;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Gabriel J. Lignocellulose degradation by Pleurotus ostreatus in the presence of cadmium. FEMS Microbiol Lett. 2003;220:235–240. doi: 10.1016/S0378-1097(03)00102-2. [DOI] [PubMed] [Google Scholar]
- Ben Younes S, Sayadi S. Purification and characterization of a novel trimeric and thermotolerant laccase produced from the ascomycete Scytalidium thermophilum strain. J Mol Catal B Enzym. 2011;73:35–42. doi: 10.1016/j.molcatb.2011.07.014. [DOI] [Google Scholar]
- Ben Younes S, Mechichi T, Sayadi S. Purification and characterization of the laccase secreted by the white rot fungus Perenniporia tephropora and its role in the decolourization of synthetic dyes. J Appl Microbiol. 2007;102:1033–1042. doi: 10.1111/j.1365-2672.2006.03152.x. [DOI] [PubMed] [Google Scholar]
- Berka RM, Schneider P, Golightly EJ, Brown SH, Madden M, Brown KM, Halkier T, Mondorf K, Xu F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl Environ Microbiol. 1997;63:3151–3157. doi: 10.1128/aem.63.8.3151-3157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, Grigoriev IV, Otillar R, Salamov A, Grimwood J, Reid I, Ishmael N, John T, Darmond C, Moisan MC, Henrissat B, Coutinho PM, Lombard V, Natvig DO, Lindquist E, Schmutz J, Lucas S, Harris P, Powlowski J, Bellemare A, Taylor D, Butler G, de Vries RP, Allijn IE, van den Brink J, Ushinsky S, Storms R, Powell AJ, Paulsen IT, Elbourne LD, Baker SE, Magnuson J, Laboissiere S, Clutterbuck AJ, Martinez D, Wogulis M, de Leon AL, Rey MW, Tsang A. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- Boonlue S, Aimi T, Morinaga T. Molecular characterization of a xylanase-producing thermophilic fungus isolated from Japanese Soil. Curr Microbiol. 2003;47:0119–0124. doi: 10.1007/s00284-002-3918-z. [DOI] [PubMed] [Google Scholar]
- Castaño JD, Cruz C, Torres E. Optimization of the production, purification and characterization of a laccase from the native fungus Xylaria sp. Biocatal Agric Biotechnol. 2015;4:710–716. [Google Scholar]
- Chakroun H, Mechichi T, Martinez MJ, Dhouib A, Sayadi S. Purification and characterization of a novel laccase from the ascomycete Trichoderma atroviride: application on bioremediation of phenolic compounds. Process Biochem. 2010;45:507–513. doi: 10.1016/j.procbio.2009.11.009. [DOI] [Google Scholar]
- Chefetz B, Chen Y, Hadar Y. Purification and characterization of laccase from Chaetomium thermophilium and its role in humification. Appl Environ Microbiol. 1998;64:3175–3179. doi: 10.1128/aem.64.9.3175-3179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Yien Ting AS. Biodecolorization and biodegradation potential of recalcitrant triphenylmethane dyes by Coriolopsis sp. isolated from compost. J Environ Manage. 2015;150:274–280. doi: 10.1016/j.jenvman.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Claus H. Laccases: structure, reactions, distribution. Micron. 2004;35:93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- D’Souza-Ticlo D, Sharma D, Raghukumar C. A thermostable metal-tolerant laccase with bioremediation potential from a marine-derived fungus. Mar Biotechnol. 2009;11:725–737. doi: 10.1007/s10126-009-9187-0. [DOI] [PubMed] [Google Scholar]
- Daâssi D, Zouari-Mechichi H, Prieto A, Martínez MJ, Nasri M, Mechichi T. Purification and biochemical characterization of a new alkali-stable laccase from Trametes sp. isolated in Tunisia: role of the enzyme in olive mill waste water treatment. World J Microbiol Biotechnol. 2013;29:2145. doi: 10.1007/s11274-013-1380-7. [DOI] [PubMed] [Google Scholar]
- Damm U, Verkley GJM, Crous PW, Fourie PH, Haegi A, Riccioni L. Novel Paraconiothyrium species on stone fruit trees and other woody hosts. Persoonia Mol Phylogeny Evol Fungi. 2008;20:9–17. doi: 10.3767/003158508X286842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer JK, Patel NJ, Dhawale SW, Dhawale SS. Production of multiple laccase isoforms by Phanerochaete chrysosporium grown under nutrient sufficiency. FEMS Microbiol Lett. 1997;149:65–70. doi: 10.1111/j.1574-6968.1997.tb10309.x. [DOI] [Google Scholar]
- Dwivedi UN, Singh P, Pandey VP, Kumar A. Structure–function relationship among bacterial, fungal and plant laccases. J Mol Catal B Enzym. 2011;68:117–128. doi: 10.1016/j.molcatb.2010.11.002. [DOI] [Google Scholar]
- Fan X, Zhou Y, Xiao Y, Xu Z, Bian Y. Cloning, expression and phylogenetic analysis of a divergent laccase multigene family in Auricularia auricula-judae. Microbiol Res. 2014;169:453–462. doi: 10.1016/j.micres.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Fang Z, Li T, Wang Q, Zhang X, Peng H, Fang W, Hong Y, Ge H, Xiao Y. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl Microbiol Biotechnol. 2011;89:1103–1110. doi: 10.1007/s00253-010-2934-3. [DOI] [PubMed] [Google Scholar]
- Farnet AM, Gil G, Ferre E. Effects of pollutants on laccase activities of Marasmius quercophilus, a white-rot fungus isolated from a Mediterranean schlerophyllous litter. Chemosphere. 2008;70:895–900. doi: 10.1016/j.chemosphere.2007.06.086. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fernaud JRH, Marina A, González K, Vázquez J, Falcón MA. Production, partial characterization and mass spectrometric studies of the extracellular laccase activity from Fusarium proliferatum. Appl Microbiol Biotechnol. 2006;70:212–221. doi: 10.1007/s00253-005-0221-5. [DOI] [PubMed] [Google Scholar]
- Forootanfar H, Faramarzi MA, Shahverdi AR, Yazdi MT. Purification and biochemical characterization of extracellular laccase from the ascomycete Paraconiothyrium variabile. Bioresour Technol. 2011;102:1808–1814. doi: 10.1016/j.biortech.2010.09.043. [DOI] [PubMed] [Google Scholar]
- Frasconi M, Favero G, Boer H, Koivula A, Mazzei F. Kinetic and biochemical properties of high and low redox potential laccases from fungal and plant origin. Biochim Biophys Acta BBA Proteins Proteom. 2010;1804:899–908. doi: 10.1016/j.bbapap.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Garzillo AMV, Colao MC, Caruso C, Caporale C, Celletti D, Buonocore V. Laccase from the white-rot fungus Trametes trogii. Appl Microbiol Biotechnol. 1998;49:545–551. doi: 10.1007/s002530051211. [DOI] [PubMed] [Google Scholar]
- Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. Laccases: a never-ending story. Cell Mol Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi E, Scodeller P, Filiel N, Carballo R, Levin L. Potential of Trametes trogii culture fluids and its purified laccase for the decolorization of different types of recalcitrant dyes without the addition of redox mediators. Int Biodeterior Biodegrad. 2011;65:635–643. doi: 10.1016/j.ibiod.2011.03.007. [DOI] [Google Scholar]
- Hakulinen N, Kiiskinen LL, Kruus K, Saloheimo M, Paananen A, Koivula A, Rouvinen J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat Struct Mol Biol. 2002;9:601–605. doi: 10.1038/nsb823. [DOI] [PubMed] [Google Scholar]
- Halaburgi VM, Sharma S, Sinha M, Sinha M, Singh TP, Karegoudar TB. Purification and characterization of a thermostable laccase from the ascomycetes Cladosporium cladosporioides and its applications. Process Biochem. 2011;46:1146–1152. doi: 10.1016/j.procbio.2011.02.002. [DOI] [Google Scholar]
- Jaouani A, Guillén F, Penninckx MJ, Martίnez AT, Martίnez MJ. Role of Pycnoporus coccineus laccase in the degradation of aromatic compounds in olive oil mill waste water. Enzyme Microb Technol. 2005;36:478–486. doi: 10.1016/j.enzmictec.2004.11.011. [DOI] [Google Scholar]
- Junghanns C, Pecyna MJ, Böhm D, Jehmlich N, Martin C, Von Bergen M, Schauer F, Hofrichter M, Schlosser D. Biochemical and molecular genetic characterisation of a novel laccase produced by the aquatic ascomycete Phoma sp. UHH 5-1-03. Appl Microbiol Biotechnol. 2009;84:1095–1105. doi: 10.1007/s00253-009-2028-2. [DOI] [PubMed] [Google Scholar]
- Kiiskinen LL, Viikari L, Kruus K. Purification and characterisation of a novel laccase from the ascomycete Melanocarpus albomyces. Appl Microbiol Biotechnol. 2002;59:198–204. doi: 10.1007/s00253-002-1012-x. [DOI] [PubMed] [Google Scholar]
- Kiiskinen LL, Rättö M, Kruus K. Screening for novel laccase-producing microbes. J Appl Microbiol. 2004;97:640–646. doi: 10.1111/j.1365-2672.2004.02348.x. [DOI] [PubMed] [Google Scholar]
- Kittl R, Mueangtoom K, Gonaus C, Khazaneh ST, Sygmund C, Haltrich D, Ludwig R. A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J Biotechnol. 2012;157:304–314. doi: 10.1016/j.jbiotec.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Koyani RD, Sanghvi GV, Sharma RK, Rajput KS. Contribution of lignin degrading enzymes in decolourisation and degradation of reactive textile dyes. Int Biodeterior Biodegrad. 2013;77:1–9. doi: 10.1016/j.ibiod.2012.10.006. [DOI] [Google Scholar]
- Krishnamurthy YL, Naik SB, Jayaram S. Fungal communities in herbaceous medicinal plants from the malnad region, southern India. Microbes Environ. 2008;23:24–28. doi: 10.1264/jsme2.23.24. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunamneni A, Ballesteros A, Plou FJ, Alcalde M. Fungal laccase: a versatile enzyme for biotechnological applications. In: Mendez-Vilas A, editor. Communicating current research and educational topics and trends in applied microbiology. Microbiology book series. Spain: Formatex Publishers; 2007. pp. 233–245. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levasseur A, Saloheimo M, Navarro D, Andberg M, Pontarotti P, Kruus K, Record E. Exploring laccase-like multicopper oxidase genes from the ascomycete Trichoderma reesei: a functional, phylogenetic and evolutionary study. BMC Biochem. 2010;11:32. doi: 10.1186/1471-2091-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liers C, Ullrich R, Pecyna M, Schlosser D, Hofrichter M. Production, purification and partial enzymatic and molecular characterization of a laccase from the wood-rotting ascomycete Xylaria polymorpha. Enzyme Microb Technol. 2007;41:785–793. doi: 10.1016/j.enzmictec.2007.07.002. [DOI] [Google Scholar]
- Lineweaver H, Burk D. The determination of enzyme dissociation constants. J Am Chem Soc. 1934;56:658–666. doi: 10.1021/ja01318a036. [DOI] [Google Scholar]
- Lu L, Zhao M, Zhang BB, Yu SY, Bian XJ, Wang W, Wang Y. Purification and characterization of laccase from Pycnoporus sanguineus and decolorization of an anthraquinone dye by the enzyme. Appl Microbiol Biotechnol. 2007;74:1232–1239. doi: 10.1007/s00253-006-0767-x. [DOI] [PubMed] [Google Scholar]
- Lu L, Zhao M, Wang TN, Zhao LY, Du MH, Li TL, Li DB. Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresour Technol. 2012;115:35–40. doi: 10.1016/j.biortech.2011.07.111. [DOI] [PubMed] [Google Scholar]
- Lu L, Wang TN, Xu TF, Wang JY, Wang CL, Zhao M. Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its characterization. Bioresour Technol. 2013;134:81–86. doi: 10.1016/j.biortech.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Maheshwari R, Bharadwaj G, Bhat MK. Thermophilic fungi: their physiology and enzymes. Microbiol Mol Biol Rev. 2000;64:461–488. doi: 10.1128/MMBR.64.3.461-488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manai I, Miladi B, El Mselmi A, Smaali I, Ben Hassen A, Hamdi M, Bouallagui H. Industrial textile effluent decolourization in stirred and static batch cultures of a new fungal strain Chaetomium globosum IMA1 KJ472923. J Environ Manage. 2016;170:8–14. doi: 10.1016/j.jenvman.2015.12.038. [DOI] [PubMed] [Google Scholar]
- Marwah RG, Fatope MO, Deadman ML, Al-Maqbali YM, Husband J. Musanahol: a new aureonitol-related metabolite from a Chaetomium sp. Tetrahedron. 2007;63:8174–8180. doi: 10.1016/j.tet.2007.05.119. [DOI] [Google Scholar]
- Michniewicz A, Ullrich R, Ledakowicz S, Hofrichter M. The white-rot fungus Cerrena unicolor strain 137 produces two laccase isoforms with different physico-chemical and catalytic properties. Appl Microbiol Biotechnol. 2006;69:682–688. doi: 10.1007/s00253-005-0015-9. [DOI] [PubMed] [Google Scholar]
- Moldes D, Sanromán MÁ. Amelioration of the ability to decolorize dyes by laccase: relationship between redox mediators and laccase isoenzymes in Trametes versicolor. World J Microbiol Biotechnol. 2006;22:1197–1204. doi: 10.1007/s11274-006-9161-1. [DOI] [Google Scholar]
- Morozova OV, Shumakovich GP, Gorbacheva MA, Shleev SV, Yaropolov A. “Blue” laccases. Biochem Mosc. 2007;72:1136–1150. doi: 10.1134/S0006297907100112. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Banerjee R. Purification and biochemical characterization of a newly produced yellow laccase from Lentinus squarrosulus MR13. 3. Biotech. 2015;5:227. doi: 10.1007/s13205-014-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz C, Guillén F, Martίnez AT, Martίnez MJ. Induction and characterization of laccase in the ligninolytic fungus Pleurotus eryngii. Curr Microbiol. 1997;34:1–5. doi: 10.1007/s002849900134. [DOI] [PubMed] [Google Scholar]
- Neifar M, Jaouani A, Ellouze-Ghorbel R, Ellouze-Chaabouni S. Purification, characterization and decolourization ability of Fomes fomentarius laccase produced in solid medium. J Mol Catal B Enzym. 2010;64:68–74. doi: 10.1016/j.molcatb.2010.02.004. [DOI] [Google Scholar]
- Niladevi KN, Jacob N, Prema P. Evidence for a halotolerant-alkaline laccase in Streptomyces psammoticus: purification and characterization. Process Biochem. 2008;43:654–660. doi: 10.1016/j.procbio.2008.02.002. [DOI] [Google Scholar]
- Ogel ZB, Yüzügüllü Y, Mete S, Bakir U, Kaptan Y, Sutay D, Demir AS. Production, properties and application to biocatalysis of a novel extracellular alkaline phenol oxidase from the thermophilic fungus Scytalidium thermophilum. Appl Microbiol Biotechnol. 2006;71:853–862. doi: 10.1007/s00253-005-0216-2. [DOI] [PubMed] [Google Scholar]
- Palmieri G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G. A novel white laccase from Pleurotus ostreatus. J Biol Chem. 1997;272:31301–31307. doi: 10.1074/jbc.272.50.31301. [DOI] [PubMed] [Google Scholar]
- Palonen H, Saloheimo M, Viikari L, Kruus K. Purification, characterization and sequence analysis of a laccase from the ascomycete Mauginiella sp. Enzyme Microb Technol. 2003;33:854–862. doi: 10.1016/S0141-0229(03)00247-3. [DOI] [Google Scholar]
- Pan K, Zhao N, Yin Q, Zhang T, Xu X, Fang W, Hong Y, Fang Z, Xiao Y. Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour Technol. 2014;162:45–52. doi: 10.1016/j.biortech.2014.03.116. [DOI] [PubMed] [Google Scholar]
- Park JH, Choi GJ, Jang KS, Lim HK, Kim HT, Cho KY, Kim JC. Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol Lett. 2005;252:309–313. doi: 10.1016/j.femsle.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Phonkerd N, Kanokmedhakul S, Kanokmedhakul K, Soytong K, Prabpai S, Kongsearee P. Bis-spiro-azaphilones and azaphilones from the fungi Chaetomium cochliodes VTh01 and C. cochliodes CTh05. Tetrahedron. 2008;64:9636–9645. doi: 10.1016/j.tet.2008.07.040. [DOI] [Google Scholar]
- Qasemian L, Guiral D, Ziarelli F, Dang TKV, Farnet AM. Effects of anthracene on microbial activities and organic matter decomposition in a Pinus halepensis litter from a Mediterranean coastal area. Soil Biol Biochem. 2012;46:148–154. doi: 10.1016/j.soilbio.2011.12.002. [DOI] [Google Scholar]
- Ramírez-Cavazos LI, Junghanns C, Ornelas-Soto N, Cárdenas-Chávez DL, Hernández-Luna C, Demarche P, Enaud E, García-Morales R, Agathos SN, Parra R. Purification and characterization of two thermostable laccases from Pycnoporus sanguineus and potential role in degradation of endocrine disrupting chemicals. J Mol Catal B Enzym. 2014;108:32–42. doi: 10.1016/j.molcatb.2014.06.006. [DOI] [Google Scholar]
- Rich JO, Leathers TD, Anderson AM, Bischoff KM, Manitchotpisit P. Laccases from Aureobasidium pullulans. Enzyme Microb Technol. 2013;53:33–37. doi: 10.1016/j.enzmictec.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Riva S. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 2006;24:219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Rodgers CJ, Blanford CF, Giddens SR, Skamnioti P, Armstrong FA, Gurr SJ. Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 2010;28:63–72. doi: 10.1016/j.tibtech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Rodríguez Couto S, Toca Herrera JL. Industrial and biotechnological applications of laccases: a review. Biotechnol Adv. 2006;24:500–513. doi: 10.1016/j.biotechadv.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Rodríguez Couto S, Rodríguez A, Paterson RR, Lima N, Teixeira JA. Laccase activity from the fungus Trametes hirsuta using an air-lift bioreactor. Lett Appl Microbiol. 2006;42:612–616. doi: 10.1111/j.1472-765X.2006.01879.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez K, Stchigel A, Guarro J. Three new species of Chaetomium from soil. Mycologia. 2002;94:116–126. doi: 10.1080/15572536.2003.11833254. [DOI] [PubMed] [Google Scholar]
- Rodríguez E, Ruiz-Dueñas FJ, Kooistra R, Ram A, Martínez AT, Martínez MJ. Isolation of two laccase genes from the white-rot fungus Pleurotus eryngii and heterologous expression of the pel3 encoded protein. J Biotechnol. 2008;134:9–19. doi: 10.1016/j.jbiotec.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Ruijssenaars HJ, Hartmans S. A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl Microbiol Biotechnol. 2004;65:177–182. doi: 10.1007/s00253-004-1571-0. [DOI] [PubMed] [Google Scholar]
- Ryan S, Schnitzhofer W, Tzanov T, Cavaco-Paulo A, Gübitz GM. An acid-stable laccase from Sclerotium rolfsii with potential for wool dye decolourization. Enzyme Microb Technol. 2003;33:766–774. doi: 10.1016/S0141-0229(03)00162-5. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem Rec. 2007;7:220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- Santhanam N, Vivanco JM, Decker SR, Reardon KF. Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol. 2011;29:480–489. doi: 10.1016/j.tibtech.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Schückel J, Matura A, Van Pée KH. One-copper laccase-related enzyme from Marasmius sp.: purification, characterization and bleaching of textile dyes. Enzyme Microb Technol. 2011;48:278–284. doi: 10.1016/j.enzmictec.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Sharma P, Goel R, Capalash N. Bacterial laccases. World J Microbiol Biotechnol. 2007;23:823–832. doi: 10.1007/s11274-006-9305-3. [DOI] [Google Scholar]
- Si J, Peng F, Cui B. Purification, biochemical characterization and dye decolorization capacity of an alkali-resistant and metal-tolerant laccase from Trametes pubescens. Bioresour Technol. 2013;128:49–57. doi: 10.1016/j.biortech.2012.10.085. [DOI] [PubMed] [Google Scholar]
- Siroosi M, Amoozegar MA, Khajeh K. Purification and characterization of an alkaline chloride-tolerant laccase from a halotolerant bacterium, Bacillus sp. strain WT. J Mol Catal B Enzym. 2016;134:89–97. doi: 10.1016/j.molcatb.2016.10.001. [DOI] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Soares GMB, Costa-Ferreira M, Pessoa de Amorim MT. Decolorization of an anthraquinone-type dye using a laccase formulation. Bioresour Technol. 2001;79:171–177. doi: 10.1016/S0960-8524(01)00043-8. [DOI] [PubMed] [Google Scholar]
- Sondhi S, Sharma P, Saini S, Puri N, Gupta N. Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from Bacillus tequilensis SN4. PloS One. 2014;9:e96951. doi: 10.1371/journal.pone.0096951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni SK, Soni R. Regulation of cellulase synthesis in Chaetomium erraticum. BioResources. 2010;5:81–98. [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhulle S, Trovaslet M, Enaud E, Lucas M, Sonveaux M, Corbisier AM. Cytotoxicity and genotoxicity evolution during decolorization of dyes by white rot fungi. World J Microbiol Biotechnol. 2008;24:337–344. doi: 10.1007/s11274-007-9475-7. [DOI] [Google Scholar]
- Wang X, Zheng R. Chaetomium ampulliellum sp. nov. (Chaetomiaceae, Ascomycota) and similar species from China. Nova Hedwig. 2005;81:247–256. doi: 10.1127/0029-5035/2005/0081-0247. [DOI] [Google Scholar]
- Wang Z, Cai Y, Liao X, Zhang F, Zhang D, Li Z. Production and characterization of a novel laccase with cold adaptation and high thermal stability from an isolated fungus. Appl Biochem Biotechnol. 2010;162:280–294. doi: 10.1007/s12010-009-8801-y. [DOI] [PubMed] [Google Scholar]
- Wang B, Yan Y, Tian Y, Zhao W, Li Z, Gao J, Peng R, Yao Q. Heterologous expression and characterisation of a laccase from Colletotrichum lagenarium and decolourisation of different synthetic dyes. World J Microbiol Biotechnol. 2016;32:40. doi: 10.1007/s11274-015-1999-7. [DOI] [PubMed] [Google Scholar]
- Wesenberg D, Kyriakides I, Agathos SN. White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol Adv. 2003;22:161–187. doi: 10.1016/j.biotechadv.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Wijeratne EMK, Turbyville TJ, Fritz A, Whitesell L, Gunatilaka AA. A new dihydroxanthenone from a plant-associated strain of the fungus Chaetomium globosum demonstrates anticancer activity. Bioorg Med Chem. 2006;14:7917–7923. doi: 10.1016/j.bmc.2006.07.048. [DOI] [PubMed] [Google Scholar]
- Wong Y, Yu J. Laccase-catalyzed decolorization of synthetic dyes. Water Res. 1999;33:3512–3520. doi: 10.1016/S0043-1354(99)00066-4. [DOI] [Google Scholar]
- Wu YR, Luo ZH, Kwok-Kei Chow R, Vrijmoed LLP. Purification and characterization of an extracellular laccase from the anthracene-degrading fungus Fusarium solani MAS2. Bioresour Technol. 2010;101:9772–9777. doi: 10.1016/j.biortech.2010.07.091. [DOI] [PubMed] [Google Scholar]
- Xu F. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry (Mosc) 1996;35:7608–7614. doi: 10.1021/bi952971a. [DOI] [PubMed] [Google Scholar]
- Xu Y, Lu Y, Zhang R, Wang H, Liu Q. Characterization of a novel laccase purified from the fungus Hohenbuehelia serotina and its decolourisation of dyes. Acta Biochim Pol. 2016;63:273–279. doi: 10.18388/abp.2015_1091. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ding Y, Liao X, Cai Y. Purification and characterization of a new laccase from Shiraia sp. SUPER-H168. Process Biochem. 2013;48:351–357. doi: 10.1016/j.procbio.2012.12.011. [DOI] [Google Scholar]
- Yang J, Lin Q, Ng TB, Ye X, Lin J. Purification and characterization of a novel laccase from Cerrena sp. HYB07 with dye decolorizing ability. PLoS One. 2014;9:e110834. doi: 10.1371/journal.pone.0110834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang Q. Expressed sequence tags-based identification of genes in the biocontrol agent Chaetomium cupreum. Appl Microbiol Biotechnol. 2007;74:650–658. doi: 10.1007/s00253-006-0701-2. [DOI] [PubMed] [Google Scholar]
- Zouari-Mechichi H, Mechichi T, Dhouib A, Sayadi S, Martίnez AT, Martίnez MJ. Laccase purification and characterization from Trametes trogii isolated in Tunisia: decolorization of textile dyes by the purified enzyme. Enzyme Microb Technol. 2006;39:141–148. doi: 10.1016/j.enzmictec.2005.11.027. [DOI] [Google Scholar]