Abstract
microRNAs are important in cancer biogenesis and development. However, their underlying mechanisms in multiple myeloma (MM) are barely characterized. microRNA-20a (miR-20a) is a member of the microRNA-17-92 cluster. It has been implicated in various cancers, regulating the proliferation and invasion of cancer cells in vitro. Compared with healthy donors, it also has been reported to be elevated in plasma of MM patients. Here, we investigated the function of miR-20a. Our results showed that it promotes proliferation and inhibits apoptosis of MM cells in vitro by inhibiting early growth response protein 2. The effects of miR-20a were also evaluated in MM xenograft models of SCID/NOD mice. Apparent antitumor activity was achieved in xenograft mice injected with miR-20a inhibitor, while mimics of miR-20a significantly promoted tumor growth. These data indicate that miR-20a plays a crucial role in the biology of MM and represents a potential target for novel therapies for MM patients.
Keywords: miR-20a, EGR2, multiple myeloma
Introduction
Multiple myeloma (MM) is a malignant tumor characterized by clonal proliferation of plasma cells (PCs) in the bone marrow (BM), accounting for the leading cause of death in hematologic cancer patients.1–3 Despite recent and extensive progress in the understanding of the molecular mechanisms of MM pathogenesis and the advent of promising new therapies,4–9 MM is still an incurable disease. Additional research on MM and corresponding new therapeutic strategies are urgently required. Pathogenesis of MM was considered to be a multistep process. In each step, PCs undergo complicated molecular and cellular transformations. The human BM milieu, which supports the generation and growth of PCs, may be the key player in survival and drug resistance of neoplastic PCs as well.10–12 Increasing evidence has progressively disclosed the microRNA (miRNA) network in MM pathogenesis, providing potential new tools in clinical interventions in this important disease.13–16
miRNAs represent a class of small, noncoding RNAs with 19–25 nucleotides in length. miRNAs bind to specific targets through base pairing with partially or fully complementary sites of mRNA 3′ untranslated region (3′-UTR), leading to degradation or translational repression of the target, thereby inhibiting gene expression.17–19 Mature miRNAs play a pivotal role in a variety of normal biological processes such as development and cell proliferation, differentiation, and apoptosis.20 Moreover, they are known to be involved in pathogenesis of some solid tumors as well as hematologic malignancies.21–23 Depending on cellular contexts, miRNAs can serve as either oncogenes or tumor suppressors.24–26
microRNA-20a (miR-20a) is a member of the miR-17-92 cluster, which is located on chromosome 13 within a region of 1 kb. Both oncogenes and tumor suppressors have been reported among members of the miR-17-92 cluster.27 miR-20a, especially heavily researched, is upregulated in a range of cancers, such as thyroid cancer, gastric cancer, gliomas, cervical cancer, colon adenocarcinoma, and prostate cancer.28–33 Elevated plasma levels of miR-20a in MM patients were also observed.34 In the present study, we investigated the biological effects of miR-20a and its underlying mechanisms. Its function was further confirmed in mouse models. We suggest miR-20a could be a promising target for developing future miRNA-based MM therapy.
Materials and methods
Clinical samples and cells
BM samples were collected from patients with relapsed/refractory MM (n=18) and 18 healthy donors (HDs). All samples were obtained before clinical treatment. PCs were separated using CD138+ microbeads selection system (Miltenyi Biotec, Bergisch Gladbach, Germany) based on the manufacturer’s manual. Written informed consents were acquired from all patients and HDs. Human MM cell lines (H929, MM1S, RPMI-8226, U266) were purchased from the American Type Culture Collection. U266 cells were grown in Iscove’s modified Dulbecco’s Medium, while the other MM cells (H929, MM1S, and RPMI-8226) were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA). Cells were cultured with 10% fetal bovine serum plus 100 U/mL of penicillin and 100 μg/mL of streptomycin (all from Thermo Fisher Scientific) at 37°C with 5% CO2. Our experiments were approved by the Medical Ethics Committee of the Second Affiliated Hospital of Shandong University.
Quantitative real-time amplification (QPCR) of miRNAs and mRNAs
Total RNA from patient samples or HD or cell lines was extracted using TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. Total RNA (1 μg) was used to synthesize complementary DNA (cDNA) in 20 μL reactions. Reverse transcription kit was obtained from Thermo Fisher Scientific. Single-tube TaqMan miRNA assays (Thermo Fisher Scientific) were used to detect and quantify mature miR-20a after normalization to U6 small nuclear RNA. For analysis of early growth response protein 2 (EGR2), a SYBR green mix polymerase chain reaction (PCR) kit (Takara Bio Inc., Kusatsu, Japan) was used. Glyceraldehyde-3-phosphate dehydrogenase was used as internal control. The relative expression levels of miR-20a or EGR2 were determined using the 2−ΔΔCT method. The primers were synthesized by Sangon Biotech (Shanghai, China) and purified by polyacrylamide gels. Primer sequences were designed according to a previous report.32
Transfection of MM cells
Synthetic hsa-miR-20a mimics (miR-20a mimics), negative control, and miRNA inhibitors (anti-miR-20a and inhibitor negative control) were all purchased from Exiqon, Vedbæk, Denmark. Small interfering RNA (siRNA) against EGR2 and scrambled negative control were designed by Sangon Biotech. Cell transfection of these oligos was performed using Lipofectamine 2000 Reagent (Thermo Fisher Scientific). The final concentration of oligos was 100 nmol/L. Plasmids for luciferase assay were electro-transfected with Neon Transfection System (Thermo Fisher Scientific). A total of 2.5×105 cells was transfected with 2 pulses at 1,050 V, 30 ms.
Cellular viability assay
For cell proliferation analysis, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used. Briefly, MM cells were seeded in 96-well plates at a density of 3×103 cells per well. After 24 hours, cells were transfected with oligos. After indicated hours (0, 24, 48, 72, and 96 hours) of incubation at 37°C, 15 μL MTT (0.5 mg/mL; Sigma-Aldrich Co, St Louis, MO, USA) solution was added to each well. Another 4 hours of incubation at 37°C was performed before the culture medium was removed, and crystals were solubilized with 150 μL dimethyl sulfoxide (Sigma-Aldrich Co) at room temperature for 20 minutes. Optical density was measured at a wavelength of 450 nm. All samples were arranged in triplicate and experiments were repeated at least three times. Average of results was used to draw the growth curves. Growth inhibition rate was calculated relative to the control group.
Cell apoptosis assay
To examine apoptotic cell death, MM cells were stained using fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection Kit (Abcam, Cambridge, UK) based on the manufacturer’s instruction. Briefly, cells were centrifuged down, washed once with ice-cold phosphate-buffered saline (PBS), resuspended with binding buffer, and stained at room temperature for 10 minutes. Apoptotic cells were analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell cycle assay
For cell cycle analysis, cells were harvested by centrifugation. After washing once with PBS, cells were fixed with ice-cold 70% ethanol for 2 hours at 4°C. Fixed cells were washed with PBS twice and treated with RNase A (0.1 mg/mL) and 0.1% Triton X-100 for 10 minutes, stained with propidium iodide (PI, 50 μg/mL) for 30 minutes at room temperature, and analyzed with flow cytometry.
Luciferase reporter assay
EGR2 3′-UTR was amplified with PCR from cDNA reverse transcripted from RNA of MM cells, subcloned to pGEM-T vector. Putative miR-20a binding sites were mutated (Mut) with a site-directed mutagenesis kit (Stratagene, San Diego, CA, USA) according to the manufacturer’s instruction. Wild-type (WT) or Mut EGR2 3′-UTR were transferred to psiCHECK-2 vector via Xho I and Not I restriction sites. WT or Mut EGR2 3′-UTR inserted psiCHECK-2 was transfected together with hsa-miR-20a mimics or negative control into U266 cells. Dual-luciferase measurements (Promega Corporation, Fitchburg, WI, USA) were performed 48 hours post-transfection.
Western blot
Cells were rinsed twice with cold PBS and lysed in cell lysis buffer containing protease inhibitor cocktail (Hoffman-La Roche Ltd, Basel, Switzerland). Whole-cell lysates (30 μg per lane) were separated on 10% sodium dodecyl sulfate polyacrylamide gels, transferred to polyvinylidene fluoride membranes (GE Healthcare Biosciences Corp, Piscataway, NJ, USA). Primary antibody incubation was performed overnight at 4°C with suggested dilution of the manufacturer (Abcam). Peroxidase conjugated secondary antibody (Sigma-Aldrich Co) incubation was performed at room temperature for 1 hour with a dilution of 10,000.
Animals and in vivo model of human MM
Female SCID/NOD mice (6–8 weeks old) were maintained in our animal experimental center. Experimental content and animal manipulation protocols were approved by the Medical Ethics Committee of the Second Affiliated Hospital of Shandong University. Our MM xenografts were constructed according to previous reports.35,36 Mice were subcutaneously inoculated with 1×106 OPM2 cells. Treatment was started when tumors were palpable and reached an average volume of 100 mm3. Intra-tumor injection with miR-20a, miR-20a inhibitor, or negative controls was administered every 2 days for a total of seven injections. The tumor sizes were assessed as previously described.
Statistical analysis
All in vitro experiments were repeated at least three times. At the same time, QPCR, MTT assay, and luciferase assay were arranged in triplicate. A representative experiment or the average and standard deviation are shown in figures. Statistical significance of differences between two groups was determined using Student’s t-test or one-way analysis of variance (among three groups). p<0.05 was considered statistically significant.
Results
Expression of miR-20a in MM patients and cell lines
To understand the clinical pathologic significance of miR-20a in MM, we collected BM samples from 18 MM patients and 18 HDs, and detected miR-20a expression level by QPCR in these samples. The miR-20 expression in MM patients was significantly higher than that in HDs (Figure 1A). We further evaluated miR-20a expression in four MM cell lines, and observed that, compared to normal PCs, miR-20a is relatively highly expressed in MM cell lines (Figure 1B). Among these four cell lines, we selected NCI-H929 and U266 for further studies. NCI-H929 expresses relatively low levels while U266 expresses the highest levels of miR-20 among these four cell lines, which may be used in the study of the growth promoting role of miR-20a mimics or the antitumor activity of miR-20a inhibitor, respectively.
Figure 1.
miR-20a is upregulated in MM patient bone marrow and MM cell lines.
Notes: QPCR detection of miR-20a in bone marrow samples of HDs and MM patients (A) and in nPCs and MM cell lines (B). Data are mean ± SD of three independent experiments. *p<0.05.
Abbreviations: MM, multiple myeloma; HDs, healthy donors; SD, standard deviation; QPCR, quantitative polymerase chain reaction; nPCs, normal plasma cells; miR, microRNA.
MiR-20a promotes proliferation and inhibits apoptosis of MM cells
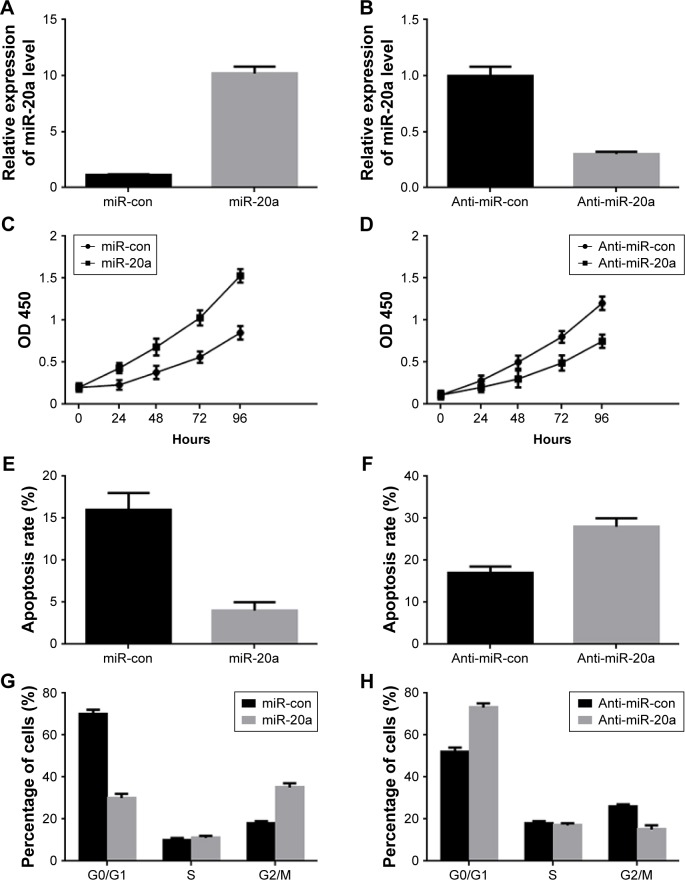
To determine the role of miR-20a in MM cells, we transfected miR-20a mimics and inhibitor (anti-miR-20a) to NCI-H929 cells and U266 cells, respectively. The transfection efficiency was confirmed by QPCR (Figure 2A and B), detecting miR-20a expression. MTT assay was then performed at different time points after transfection to investigate the role of miR-20a in cell growth. Enforced expression of the synthetic mimics of miR-20a indeed promoted NCI-H929 cell proliferation (Figure 2C), while transfection of miR-20a inhibitor in U266 cells inhibited proliferation of these cells (Figure 2D). Then, we hypothesized if the effect of miR-20a on NCI-H929 and U266 cells may be correlated with cell apoptosis and cell cycle distribution. To test this, these treated cells were subjected to FITC-annexin V/PI staining followed by flow cytometry analysis. Our results showed that, with miR-20a mimics transfection, the apoptosis rate was significantly decreased in NCI-H929 cells (Figure 2E); with miR-20a inhibitor transfection, the apoptosis rate was significantly promoted in U266 cells (Figure 2F). To determine if modulating cell apoptosis was a result of the cell cycle changes, transfected cells were fixed and subjected to PI staining followed by flow cytometry analysis. Compared with those transfected with the negative control, miR-20a mimics transfection decreased the proportion of NCI-H929 cells in the G0/G1 phase and increased the proportion of G2/M phase cells (Figure 2G). On the contrary, miR-20a inhibitor transfection in U266 increased the proportion of cells in the G0/G1 phase and decreased the proportion of cells in G2/M phase, compared to negative control transfection (Figure 2H). These results indicated that miR-20a promoted MM cell proliferation by inhibiting cell apoptosis in vitro.
Figure 2.
Role of miR-20a in MM cells proliferation and cell cycle progression.
Notes: (A and B) Relative expression of miR-20a in NCI-H929 cells after transfection with miR-20a mimics (miR-20a, A) or in U266 cells after transfection with miR-20a inhibitor (anti-miR-20a, B) determined by QPCR. (C–H) MM cells proliferation (C and D), apoptosis rate (E and F), cell cycle distribution (G and H) change after transfection with miR-20a mimics (C, E, and G, performed in NCI-H929 cells) or miR-20a inhibitor (D, F, and H, performed in U266 cells) determined by MTT assay or flow cytometry. Data are mean ± SD of three independent experiments.
Abbreviations: con, control; MM, multiple myeloma; SD, standard deviation; QPCR, quantitative polymerase chain reaction; OD, optical density; miR, microRNA; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide.
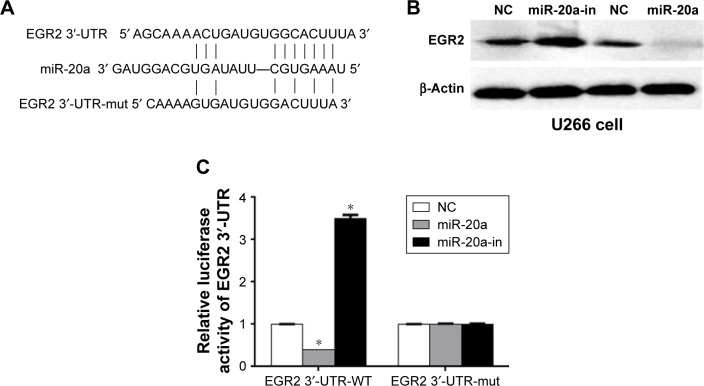
MiR-20a directly targets EGR2 by binding to its 3′-UTRin MM
It is generally believed that miRNAs inhibit mRNAs expression by targeting the 3′-UTR of mRNA. To find the target(s) of miR-20a, we performed a search on the TargetScan 6.2 website. Among >1,000 potential targets, EGR2, which is a C2H2 zinc finger transcription regulatory factor and regulates cell proliferation and cell cycle in aggressive bone derived cancer cells,37 caught our attention (Figure 3A). To determine if miR-20a truly inhibits EGR2 expression, we transfected U266 cells with miR-20a mimics, miR-20a inhibitors, or their negative controls, respectively. Western blot was then performed to determine EGR2 expression in these cells. As expected, miR-20a inhibitor increased EGR2 expression, while miR-20a mimics suppressed EGR2 expression in U266 cells (Figure 3B) compared to negative control transfection. To find whether EGR2 is a direct target of miR-20a or the regulation is mediated by other partners, we performed the luciferase assay. WT EGR2 3′-UTR or mutant 3′-UTR with nucleotides replacement in potential miR-20a target site (Figure 3A) was cloned to psiCHECK-2 vector, co-transfected with miR-20a mimics or miR-20a inhibitor in U266 cells, respectively, and dual-luciferase were then measured 24 hours later. As shown in Figure 3C, co-transfection of WT EGR2 3′-UTR-psiCHECK-2 vector and miR-20a mimics significantly repressed luciferase activity, whereas co-transfection of miR-20a inhibitor and WT EGR2 3′-UTR-psiCHECK-2 vector enhanced luciferase activity. Co-transfection of mutant vector showed no changes in luciferase activity. These results indicated that EGR2 is a direct target of miR-20a.
Figure 3.
miR-20a targets EGR2 3′-UTR.
Notes: (A) Sequence of mature miR-20a and its predicted target sequence of EGR2 3′-UTR and positions of mutation (EGR2 3′-UTR-mut). (B) EGR2 expression of U266 cells transfected with miR-20a mimics or inhibitor determined by Western blot. β-actin was used as the loading control. (C) Dual-luciferase assay of U266 cells co-transfected with the WT (or mut) EGR2 3′-UTR-psiCHECK2 reporter vector and miR-20a mimics or miR-20a-in. *p<0.05.
Abbreviations: miR-20a, microRNA-20a; WT, wild-type; EGR2, early growth response protein 2; 3′-UTR, 3′ untranslated region; mut, mutated; NC, negative control; miR, microRNA; in, inhibitor.
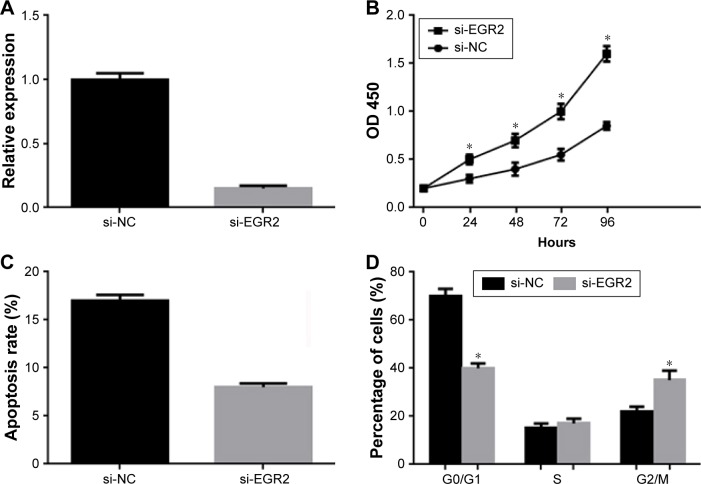
Knockdown of EGR2 promotes proliferation and inhibits apoptosis of U266 cells
As shown earlier, miR-20a promotes proliferation and inhibits apoptosis of MM cells, and miR-20a directly targets EGR2. We wonder if downregulation of EGR2 had a similar function to miR-20a overexpression. To address this question, we synthesized siRNA to EGR2 and transfected to U266 cells. QPCR was performed 48 hours post-transfection to validate RNA interference efficiency (Figure 4A). As shown by the MTT assay, knockdown of EGR2 significantly stimulated proliferation of U266 cells (Figure 4B) compared to control. Knockdown of EGR2 also inhibited U266 apoptosis (Figure 4C). Cell cycle analysis was also performed in EGR2-knockdown U266 cells by flow cytometry, and we found fewer cells arrested in G1/S phase (Figure 4D). In brief, these results showed that, similar to miR-20a mimics transfection, knockdown of EGR2 led to a remarkable promotion of proliferation and inhibition of apoptosis in U266 cells.
Figure 4.
Effect of EGR2 on U266 cells proliferation and apoptosis.
Notes: (A) Efficiency of small RNA interference to EGR2 determined by QPCR. Cell proliferation (B), apoptosis (C), and cell cycle distribution (D) of U266 cells with EGR2 knockdown measured by MTT assay or flow cytometry. Data are mean ± SD of three independent experiments. *p<0.05.
Abbreviations: si-NC, scrambled negative control of si-EGR2; si-EGR2, small RNA interference to EGR2; SD, standard deviation; OD, optical density; QPCR, quantitative polymerase chain reaction; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; EGR2, early growth response protein 2.
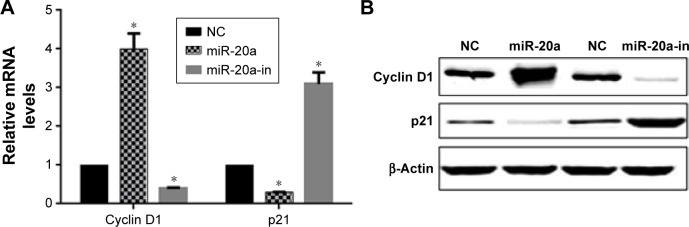
miR-20a alters p21 and cyclin D1 in U266 cells
To investigate the mechanism of miR-20a on cell proliferation and cell cycle, we investigated the effect of miR-20a on critical cell proliferation and cell cycle related regulators, cyclin D1 and cyclin-dependent kinase inhibitor p21, which negatively (p21) and positively (cyclin D1) regulate cell proliferation and cell cycle progression. The cyclin-dependent kinase inhibitor p21 and cyclin D1 are two miR-20a targeted proteins that negatively (p21) and positively (cyclin D1) regulate and control cell cycle progression and proliferation. U266 cells were transfected with miR-20a mimics or inhibitor; QPCR and Western blot were then performed to detect p21 and cyclin D1 expression. Our results showed that miR-20a mimics upregulated cyclin D1 mRNA (Figure 5A) and protein levels (Figure 5B) and downregulated those of p21. In contrast, miR-20a inhibitor promoted p21 expression and suppressed cyclin D1 expression (Figure 5A and B). Our results showed that, by regulating cell cycle genes, miR-20a promoted U266 cell proliferation and cell cycle progression.
Figure 5.
miR-20a alters p21 and cyclin D1 expression in U266 cells.
Notes: QPCR (A) and Western blot (B) detection of cyclin D1 and p21 expression in U266 cells as forced miR-20a overexpression or inhibition. β-Actin served as the loading control in Western blot. *p<0.05.
Abbreviations: QPCR, quantitative polymerase chain reaction; miR, microRNA; NC, negative control; in, inhibitor.
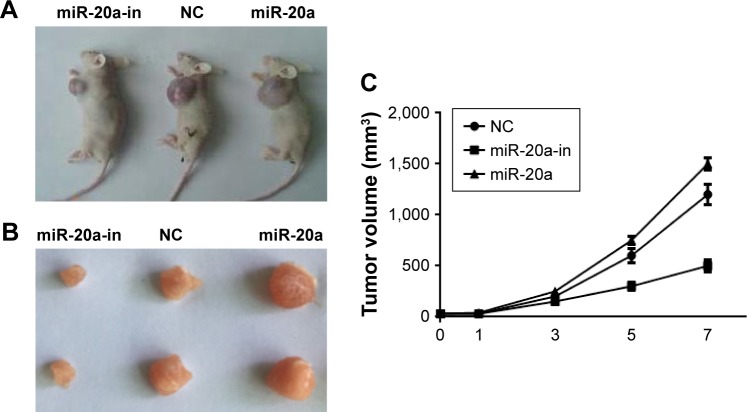
Antitumor activity of miRNA-20a inhibitor in vivo
We next investigated if our findings of miR-20a in vitro could be reproduced in a therapeutic model. The effect of miR-20a on MM xenografts in vivo was evaluated. When tumors became palpable, mice were randomized into three groups, with each group having 15 mice. Mice were treated intratumorally with synthetic miR-20a mimics, miR-20a inhibitor, or control. miRNA inhibitors were used in two different experimental systems. One is administered as lipidic formulation with neutral lipid emulsion particles that have been demonstrated to successfully deliver oligonucleotides in vivo; the other is used as unformulated agents. A total of seven injections were administered, and tumor volumes were measured each time; we found that miR-20a mimics significantly promoted tumor xenograft growth, while miR-20a inhibitor largely repressed tumor growth (Figure 6A and C). The same conclusion could be drawn when tumors were isolated from mice of each group at the end of therapy (Figure 6B). Our findings demonstrate that miR-20a promoted tumor growth in vivo, while delivery of its inhibitor exerts significant anti-MM xenograft activity.
Figure 6.
Oncogenic activity of miR-20a in MM xenograft SCID/NOD mice.
Notes: miR-20a mimics and inhibitor significantly promoted and suppressed xenograft tumor growth, respectively. Representative xenograft mice treated with miR-20 mimics or inhibitor or control oligos (A) and their isolated tumor nodule (B; the samples were separated in 3 groups, each group contained 2 mice). (C) Tumor volumes of xenograft mice treated with miR-20a mimics or miR-20a inhibitor.
Abbreviations: MM, multiple myeloma; miR, microRNA; NC, negative control; in, inhibitor.
Discussion
MM is a malignant monoclonal tumor of BM PCs often affecting old age people. Clonal proliferation and accumulation of myeloma cells in the BM caused bone destruction and affected normal BM function. Myeloma cells differ from normal PCs because they retain their self-renewing potential. Identifying new molecular techniques underlying myeloma cells proliferation is important for a better understanding of their biological behavior and for new treatment therapy. Growing evidence suggests that miRNAs play a pivotal role in cancer genesis and progression. miR-20a has been implicated in numerous cancers. In prostate cancer, it was reported that miR-20a was associated with Gleason score, a system to evaluate prognosis of prostate cancer, and it targets the ABL family non-receptor tyrosine kinases ABL2, and promoted prostate cancer cells invasion and migration.33 Antisense oligos to miR-20a treatment in lung cancer cells lead to apoptosis.38 However, the relationship between dysregulation of miR-20a and MM has been barely described. In the present work, we proved that miR-20a was highly expressed in BM samples of MM patients compared to HDs. It was also highly expressed in multiple MM cell lines with various degrees. The results suggest a potential role of miR-20a in MM. To determine if miR-20a is a novel tumor oncogene in MM and its role in MM cells proliferation, we tested the effect of miR-20a mimics and inhibitors on MM cell growth (Figure S1). We observed an increase in the growth rate of U266 cells which have a relatively low expression of miR-20a among four selected MM cells, when transfected with miR-20a mimics; and a decreased growth rate of NCI-H929 cells transfected with miR-20a, which has a higher baseline expression of miR-20a. Then, we supposed that the perturbation of miR-20a may be related to cell apoptosis and cell cycle redistribution. Flow cytometry analysis indeed confirmed a decreased apoptosis rate in U266 cells transfected with miR-20a mimics and an increased apoptosis rate in NCI-H929 cells transfected with miR-20a inhibitor, and these apoptosis rate changes were in line with cell cycle redistribution. These results indicated that miR-20a promotes MM cells proliferation by inhibiting cell apoptosis.
Previous studies with gastric, pancreatic, and breast cancer cells have confirmed that EGR2 inhibits tumor cell proliferation and metastasis.22–24 Previous studies have proven that EGR2 is a target of miR-20a.30 Luciferase reporter assay and Western blot indeed confirmed their relationship in MM cells. miR-20a mimics and inhibitor transfection induced and repressed EGR2 expression in MM cells, respectively; moreover, miR-20a mimics reduced luciferase expression bearing a EGR2 3′-UTR, and its inhibitor enhancing luciferase activity. Interfering EGR2 expression simulated the effect of miR-20a in MM cells. Together, these results indicated that miR-20a targets EGR2 to exert its cancer promoting activity.
Finally, we demonstrated the role of miR-20a in vivo. In SCID/NOD mice xenograft model, miR-20a mimics promoted tumor growth, while its inhibitors suppressed growth. The consistency of its in vitro and in vivo function potentiates miR-20a as a target for clinical applications. In conclusion, the current study revealed that miR-20a is upregulated in MM patient samples and MM cell lines. Fortifying miR-20a expression induces a marked promotion in MM cell proliferation and a decrease in apoptosis, and vice versa. The target of miR-20a in MM cells is EGR2 as determined by Western blot and luciferase assay. The anti-tumor activity of miR-20a inhibitors was further confirmed in mouse models. These results provide a foundation for the clinical usage of synthetic miR-20a inhibitors as novel MM therapeutics.
Supplementary material
U266 cells transfected with miR-20a have decreased sensitivity to bortezomib activity.
Abbreviations: bort, bortezomib; cont, control; miR, micro RNA; OD, optical density; scr, scrambled.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annu Rev Pathol. 2011;6:249–274. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Rajkumar SV. Treatment of multiple myeloma. Nat Rev Clin Oncol. 2011;8(8):479–491. doi: 10.1038/nrclinonc.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calimeri T, Battista E, Conforti F, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia. 2011;25(4):707–711. doi: 10.1038/leu.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tassone P, Neri P, Burger R, et al. Mouse models as a translational platform for the development of new therapeutic agents in multiple myeloma. Curr Cancer Drug Targets. 2012;12(7):814–822. doi: 10.2174/156800912802429292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuler J, Ewerth D, Waldschmidt J, Wasch R, Engelhardt M. Preclinical models of multiple myeloma: a critical appraisal. Expert Opin Biol Ther. 2013;13(Suppl 1):S111–S123. doi: 10.1517/14712598.2013.799131. [DOI] [PubMed] [Google Scholar]
- 7.Tassone P, Neri P, Carrasco DR, et al. A clinically relevant SCID-hu in vivo model of human multiple myeloma. Blood. 2005;106(2):713–716. doi: 10.1182/blood-2005-01-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tassone P, Tagliaferri P, Rossi M, et al. Challenging the current approaches to multiple myeloma-related bone disease: from bisphosphonates to target therapy. Curr Cancer Drug Targets. 2009;9(7):854–870. doi: 10.2174/156800909789760393. [DOI] [PubMed] [Google Scholar]
- 9.Munshi NC, Anderson KC. New strategies in the treatment of multiple myeloma. Clin Cancer Res. 2013;19(13):3337–3344. doi: 10.1158/1078-0432.CCR-12-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tassone P, Tagliaferri P, Rossi M, Gaspari M, Terracciano R, Venuta S. Genetics and molecular profiling of multiple myeloma: novel tools for clinical management? Eur J Cancer. 2006;42(11):1530–1538. doi: 10.1016/j.ejca.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 12.Tassone P, Tagliaferri P, Fulciniti MT, Di Martino MT, Venuta S. Novel therapeutic approaches based on the targeting of microenvironment-derived survival pathways in human cancer: experimental models and translational issues. Curr Pharm Des. 2007;13(5):487–496. doi: 10.2174/138161207780162872. [DOI] [PubMed] [Google Scholar]
- 13.Tagliaferri P, Rossi M, Di Martino MT, et al. Promises and challenges of microRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets. 2012;12(7):838–846. doi: 10.2174/156800912802429355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino MT, Leone E, Amodio N, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18(22):6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amodio N, Di Martino MT, Foresta U, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi M, Pitari MR, Amodio N, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228(7):1506–1515. doi: 10.1002/jcp.24306. [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Xie X, Lu J, Kulbokas EJ, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 24.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 26.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3(1):9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Y, Zhang L, Kebebew E. MiR-20a is upregulated in anaplastic thyroid cancer and targets LIMK1. PLOS One. 2014;9(5):e96103. doi: 10.1371/journal.pone.0096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Wang B, Shi Y, et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34(11):1407–1419. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zhang Z, Yu M, et al. Involvement of miR-20a in promoting gastric cancer progression by targeting early growth response 2 (EGR2) Int J Mol Sci. 2013;14(8):16226–16239. doi: 10.3390/ijms140816226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao S, Yao D, Chen J, Ding N, Ren F. MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS One. 2015;10(3):e0120905. doi: 10.1371/journal.pone.0120905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiang XF, Zhang ZW, Liu Q, et al. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. J Cell Biochem. 2014;115(7):1269–1276. doi: 10.1002/jcb.24778. [DOI] [PubMed] [Google Scholar]
- 34.Huang JJ, Yu J, Li JY, Liu YT, Zhong RQ. Circulating microRNA expression is associated with genetic subtype and survival of multiple myeloma. Med Oncol. 2012;29(4):2402–2408. doi: 10.1007/s12032-012-0210-3. [DOI] [PubMed] [Google Scholar]
- 35.Tassone P, Di Martino MT, Ventura M, et al. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol Ther. 2009;8(7):648–653. doi: 10.4161/cbt.8.7.7968. [DOI] [PubMed] [Google Scholar]
- 36.Neri P, Tagliaferri P, Di Martino MT, et al. In vivo anti-myeloma activity and modulation of gene expression profile induced by valproic acid, a histone deacetylase inhibitor. Br J Haematol. 2008;143(4):520–531. doi: 10.1111/j.1365-2141.2008.07387.x. [DOI] [PubMed] [Google Scholar]
- 37.Grunewald TG, Bernard V, Gilardi-Hebenstreit P, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47(9):1073–1078. doi: 10.1038/ng.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsubara H, Takeuchi T, Nishikawa E, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
U266 cells transfected with miR-20a have decreased sensitivity to bortezomib activity.
Abbreviations: bort, bortezomib; cont, control; miR, micro RNA; OD, optical density; scr, scrambled.