Abstract
In spite of decades of malaria research and clinical trials, a fully effective and long-lasting preventive vaccine remains elusive. In the present study, 5370 proteins of Plasmodium falciparum genome were screened for the presence of signal peptide/anchor and GPI anchor motifs. Out of 45 screened surface-associated proteins, 22 were consensually predicted as antigens and had no orthologs in human and mouse except circumsporozoite protein (PF3D7_0304600). Among 22 proteins, 19 were identified as new antigens. In the next step, a total of 4944 peptides were predicted as CD8+ T cell epitopes from 22 probable antigens. Of these, the highest scoring 262 epitopes from each antigen were taken for optimization study in the malaria-endemic regions which covered a broad human population (~93.95%). The predicted epitope 13ILFYFFLWV21 of antigen 6-cysteine (PF3D7_1346800) was binding to the HLA-A*0201 allele with the highest fraction (26%) of immunogenicity in the target populations of North-East Asia, South-East Asia, and sub-Saharan Africa. Therefore, these epitopes are proposed to be favored in vaccine designs against malaria.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0947-7) contains supplementary material, which is available to authorized users.
Keywords: Malaria, Antigen, Vaccine, Epitope, Immunoinformatics
Introduction
Malaria is a life-threatening disease caused by Plasmodium parasites that are transmitted to people through the bites of infected female Anopheles mosquitoes (vector). Human malaria is caused by one of five species of Plasmodium, P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi. About 3.2 billion people (almost half of the world’s population) are at risk of malaria. Sub-Saharan Africa depicts a high share (90%) of the global malaria death (WHO 2016). The failure of malaria vector control, due to insecticidal resistance and development of anti-malarial drug resistance, has also resulted in an increase of malaria transmission worldwide, thereby currently receiving intensified attention toward the development of a malaria vaccine (Jones and Hoffman 1994). There are no licensed vaccines available globally so far against malaria. However, very recently, WHO confirmed phase-III clinical trials of the world’s first malaria vaccine known as RTS, S, which is partially effective against the most deadly malaria parasite, i.e., P. falciparum that is reported to be highly prevalent in Africa (Gosling and von Seidlein 2016; Vandoolaeghe and Schuerman 2016). Remarkably, the pathogenesis and clinical manifestations of malaria are dependent on the age and genetics of the human host including the transmission dynamics of the parasite (Cowman et al. 2016).
In the past decades, several approaches have been used for the development of malaria vaccines, ranging from the immunization with killed/live-attenuated pathogen to the formulation of safer subunit vaccines (Koganty 2003; McCarthy and Good 2010; Lu et al. 2016; Tan and Jiang 2016) together with genome editing (Singer and Frischknecht 2016). Malaria vaccines, presently under development, are based on a single or few antigens and offer too narrow immune response, i.e., neither optimal protection nor protection of genetically diverse backgrounds. Therefore, no vaccine candidate has been advanced with sufficient efficacy and population coverage for commercial production (Trieu et al. 2011; Davies et al. 2015). Several other investigators believe that an effective malaria vaccine will need to target multi-stage antigens that induce both antibody and cellular immune responses (De Sousa and Doolan 2016). Also, there are population-specific differences in the subset of T cells, which are expanded resulting in primary infection of Plasmodium, suggesting that malaria vaccine development may require optimization according to the target parasite as well as host (Good and Doolan 2010; Singh et al. 2015).
Conventionally, vaccines have been developed by cultivating disease-causing agents and isolating the killed/attenuated whole pathogen or its subunit selected on the basis of function, abundance, and immunogenicity. Nevertheless, these methods may have overlooked many potentially excellent candidates present with high abundance under the natural conditions during colonization and infection. These antigens may have been present only in lower amounts on the surface of the pathogen during in vitro expression. Interestingly, in the year 2000, a new approach called reverse vaccinology (RV), enabled novel vaccine discovery processes based on genomic information without growing pathogen in the laboratory (Sette and Rappuoli 2010). In the last two decades, an advancement in genome sequencing and annotation, coupled with the new and improved bioinformatics tools, has revolutionized the vaccine design strategy, which allows vaccines to be designed even for noncultivable pathogens. Rather than selecting individual proteins for in vitro/in vivo evaluation, the RV approach analyses the entire protein repertoire of the pathogen using bioinformatics tools to prioritize potential target proteins for their high-throughput expression and validation in animal models. These proteins are either surface components of the target pathogen or secreted by the pathogen which may be predicted as antigens (Goodswen et al. 2013). The predictions are based on protein structural features, such as integral transmembrane arrangements (alpha-helices and β-barrels), signal peptides/anchors, GPI anchor and secretary proteins. In reality, a licensed novel vaccine against MenB has been developed by applying the RV approach (Rappuoli et al. 2016). Thus, identifying novel candidate antigens is one way to boost up new malaria vaccine development (Dellagostin et al. 2017).
Moreover, vaccine development using native antigens is not always optimal and engineered constructs with only protective epitopes may perform better in eliciting the optimal immune response (Liljeroos et al. 2015). Hence, by using bioinformatics tools, all potential antigens of a pathogen could be screened virtually for the presence of their T cell epitopes that may lead to the design of improved vaccines by incorporating T cell immunity in addition to antibody-mediated immunity (Rappuoli et al. 2016). Cytotoxic T cell-mediated response in humans is elicited by a pathway comprising intracellular antigen processing and presentation. The epitopes selected for a vaccine must have binding affinity with more than one human leukocyte antigen (HLA) allele to cover a larger population. The antigens that contain many epitopes recognized by the multiple HLA alleles are known as promiscuous binders. However, the HLA supertype refers to a set of HLA alleles with overlapping peptide-binding specificities. The alleles in the given HLA supertype often represent the same epitope, which refers to the region on an antigen capable of eliciting T cell immunity (Davies et al. 2015; Terry et al. 2015; De Sousa and Doolan 2016). Very recently, Teo et al. (2016) have demonstrated the functional roles of antibodies to protect malaria. However, the targets of T cell immunity are largely unknown (Plebanski and Hill 2000). Thus, the identification of supertype CD8+ T cell epitopes in the antigenic proteins that promiscuously bind to several HLA alleles are prime targets for future malaria vaccine design. These epitopes are relevant to the larger sections of malaria-endemic human population (Singh and Mishra 2016). However, the determination of in vitro/in vivo binding specificities even for a single antigen and single major histocompatibility complex (MHC) allele are expensive, laborious, time-consuming and not possible to conduct studies for MHC supertypes (Reche and Reinherz 2007; Singh et al. 2010).
In the present study, we took the advantage of the partially annotated P. falciparum genome and conducted whole protein repertoires-based screening of probable antigens by using the RV approach (Doolan et al. 2014). To be potentially good vaccine candidates, the probable antigens must be surface exposed and able to be recognized by the cellular immune system (Lin et al. 2014). Thus, the study integrates analysis for supertype epitope prediction and optimization of selected antigens for the affected human population. These newly identified antigens/epitopes may help in designing fusion chimera for multi-component human malaria vaccine (Shamriz and Ofoghi 2016).
Materials and methods
Retrieval of genome sequence data
The protein sequences of P. falciparum genome were retrieved from the GeneDB/PlasmoDB database (Christiane Hertz-Fowler et al. 2004) and analyzed by the bioinformatics tools presented in Table 1 for screening of probable vaccine candidates. Three gold standard antigens (EXP-1, LSA-3 and AMA-1) were taken as control for validation of the present RV approach following Doolan et al. (2003).
Table 1.
Summary of the bioinformatics tools used in the study
| S. no. | Name of database/tool | Functional prediction | Web address |
|---|---|---|---|
| 1 | SignalP | Presence and location of signal peptide cleavage sites | http://www.cbs.dtu.dk/services/SignalP/ |
| 2 | TMHMM | Transmembrane helices in the integral membrane proteins | http://www.cbs.dtu.dk/services/TMHMM/ |
| 3 | VaxiJen | Prediction of protective antigens | http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html |
| 4 | SCRATCH protein predictor: ANTIGENpro, BETAWRAP, SOLpro and ABTMpro | Antigenicity, super secondary structural motif, solubility and transmembrane regions | http://scratch.proteomics.ics.uci.edu/ |
| 5 | NetCTL | CTL epitopes prediction | http://www.cbs.dtu.dk/services/NetCTL/ |
| 6 | OptiTope | Selection of optimal epitope set based on population coverage | http://etk.informatik.uni-tuebingen.de/optitope |
| 7 | BIMAS | MHC class I-binding affinity prediction | https://www-bimas.cit.nih.gov/molbio/hla_bind/ |
| 8 | PAComplex | Infer peptide antigen family and TCR–pMHC binding model | http://pacomplex.life.nctu.edu.tw/ |
Methodology of the work
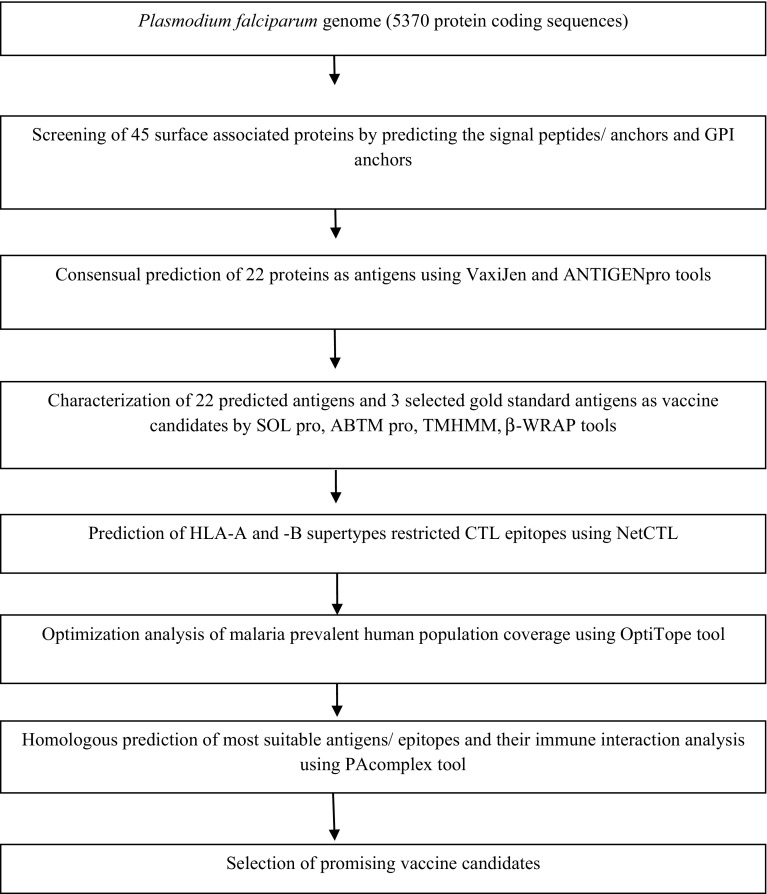
The present RV-based study involved two major steps. The first step was the screening of probable antigens from whole protein sequence of P. falciparum genome using computational tools such as SignalP, TMHMM, VaxiJen, SCRATCH suite (ANTIGENpro, BETAWRAP, SOLpro, ABTM pro) and OrthoMCL database linked BlastP (Chen et al. 2006). The second step covers prediction of immunogenic T cell epitopes using NetCTL program including population coverage analysis by OptiTope of the selected epitopes. Moreover, PAComplex tool was used to infer peptide homologous antigens and 3D structures available in PDB (Fig. 1). The summary of bioinformatics tools used in the present study is given in Table 1 and mostly default settings (parameters) were used in the analysis.
Fig. 1.
Schematic flow diagram for the screening of malaria vaccine candidate from the Plasmodium falciparum genome
Results and discussion
Screening of putative surface-exposed antigens
The screenings of VCs are performed by means of different bioinformatics tools that analyze the properties of each protein and their epitopes for possible human immune response (Table 1). The following are the properties of good VCs considered in the present study: (1) they do not show homology with human proteins to avoid the generation of a potential autoimmune response, (2) they lack transmembrane (TM) regions to facilitate their expression, (3) they possess antigenic and adhesin properties, which are important for the pathogenesis of the microorganism and for protection against the disease, (4) they are extracellular or cell surface localized to increase their accessibility to the immune system and (5) they possess optimal set of epitopes with respect to a target population (e.g. endemic regions of malaria) (El-Manzalawy et al. 2016; Singh and Mishra 2016). Furthermore, cellular location and expression level also influence the induction and magnitude of parasite-specific T cell responses (Lin et al. 2014).
In general, these aforementioned protein features such as signal peptides (SP) and GPI anchor motifs may serve to target intracellular proteins to the extracellular surface of plasma membranes/apical surfaces/outer membrane and/or activation of host–pathogen interaction (Varki et al. 1999; Gilson et al. 2006). Considering these, in the present study, whole protein of the P. falciparum genome was screened for the presence of a SP/anchor and GPI anchor using SignalP 3.0 and DGPI methods, respectively. Out of 5370 coding sequences, 45 proteins were predicted to contain the SP/anchor and GPI anchor (Fig. 1). Subsequently, their antigenicity was predicted using VaxiJen 2.0 and ANTIGENpro web servers with a threshold score ≥0.4, which provided a set of 22 potential antigens (Table 2). Furthermore, their characterizations as VCs were performed by a number of bioinformatics tools (Table 1) based on the prediction of signal peptides and transmembrane regions, which are broadly immunogenic with high CD8+ T cell epitope densities (Kovjazin et al. 2011). It has also been observed that bacterial and fungal proteins such as toxins, virulence factors, adhesins and surface proteins have parallel beta-helices that play an important role in human infectious disease (Cowen et al. 2002). Out of 22 predicted antigens, 17 (except nos. 1, 11, 13, 15, 17) were predicted to contain the right-handed parallel beta-helix (super secondary structural motif) using a program called BETAWRAP. Since it is difficult to clone, express and purify protein antigens with >1 TM spanning region, it might be better to ignore those proteins with multiple TM spanning regions as vaccine candidates. Furthermore, among these 22 proteins, 14 were predicted to contain ≤1 TM spanning region using the most reliable TMHMM web server, where three proteins (nos. 7, 11, 14) have alpha helical TM protein predicted by the ABTMpro program. As protein insolubility is another major obstacle for expression studies, a prediction method SOLpro was used to calculate the propensity of a protein to be soluble on overexpression and to identify mutations likely to increase the solubility of insoluble proteins. In this case, only seven proteins are predicted to be soluble on overexpression with the probability ≥0.5. For VCs, orthologous information gives an idea about the probable use of the vaccine in other related species (Kumar et al. 2015). Hence, an exhaustive search for potential orthologs was performed through the OrthoMCL database (Chen et al. 2006) using BLASTp homology prediction. In this context, among 22 probable antigens, only 4 (nos. 9, 14, 15, 16) had no orthologs on other related species such as P. y. yoelii, P. c. chabaudii, P. berghei, P. knowlesi and P. vivax and only one antigen (no. 17) showed similarity with the hosts. Thus, it could be excluded from the probable vaccine candidates (Table 2).
Table 2.
Computational characterization of the probable 22 P. falciparum antigens as vaccine candidates along with three control proteins
| Antigen number | PlasmoDB ID (amino acids) | Product | Predicted solubility upon overexpression using SOL pro | Predicted TM protein probabilities using ABTM pro | Number of TM regions predicted by TMHMM V. 2.0 | ||
|---|---|---|---|---|---|---|---|
| α helix | β barrel | Total | |||||
| 1 | PF3D7_0304600 (397) | Circumsporozoite protein (CSP) | 0.899185 soluble | 0.122872 | 0.0150902 | 0.862038 non TM | 1 |
| 2 | PF3D7_0317100 (969) | 6-Cysteine protein (B9) | 0.550358 soluble | 0.0647027 | 0.0492132 | 0.886084 non TM | 0 |
| 3 | PF3D7_0502400 (597) | Ring-stage membrane protein 1 (merozoite surface protein 8) | 0.602869 insoluble | 0.188048 | 0.0249447 | 0.787008 non TM | 1 |
| 4 | PF3D7_0502500 (594) | Conserved Plasmodium protein, unknown function | 0.743426 soluble | 0.218857 | 0.110808 | 0.670335 non TM | 2 |
| 5 | PF3D7_0514200 (867) | Conserved Plasmodium protein, unknown function | 0.736929 insoluble | 0.278051 | 0.0839772 | 0.637972 non TM | 3 |
| 6 | PF3D7_0612700 (347) | 6-Cysteine protein (Pf12) | 0.831126 insoluble | 0.0722029 | 0.0759844 | 0.851813 non TM | 1 |
| 7 | PF3D7_0612800 (371) | 6-Cysteine protein | 0.69182 soluble | 0.652834 alpha helical TM | 0.089623 | 0.257543 | 2 |
| 8 | PF3D7_0620400 (525) | Merozoite surface protein 10 | 0.628471 insoluble | 0.129475 | 0.0221658 | 0.848359 non TM | 2 |
| 9 | PF3D7_0702300 (594) | Sporozoite threonine and asparagine-rich protein (STRAP) | 0.99965 soluble | 0.0825028 | 0.0241054 | 0.893392 non TM | 0 |
| 10 | PF3D7_0811600 (1210) | Conserved Plasmodium protein, unknown function | 0.642934 insoluble | 0.232866 | 0.0357845 | 0.731349 non TM | 0 |
| 11 | PF3D7_0815700 (373) | Ubiquitin | 0.724588 insoluble | 0.774742 alpha helical TM | 0.0381447 | 0.187113 | 4 |
| 12 | PF3D7_0819200 (676) | Perforin-like protein 5 | 0.787379 insoluble | 0.221343 | 0.0583538 | 0.720303 non TM | 1 |
| 13 | PF3D7_0828800 (738) | GPI-anchored micronemal antigen (GAMA) | 0.598343 soluble | 0.258044 | 0.118927 | 0.623028 non TM | 2 |
| 14 | PF3D7_1000600 (361) | Rifin | 0.798433 insoluble | 0.757776 alpha helical TM | 0.00142111 | 0.240803 | 3 |
| 15 | PF3D7_1035300 (1233) | Glutamate-rich protein (GLURP) | 1.000000 insoluble | 0.0372566 | 0.00479055 | 0.957953 non TM | 0 |
| 16 | PF3D7_1035800 (712) | Probable protein, unknown function (M712) | 0.946569 soluble | 0.0460952 | 0.0253439 | 0.928561 non TM | 0 |
| 17 | PF3D7_1129000 (321) | Spermidine synthase | 0.750027 insoluble | 0.00193768 | 0.000100 | 0.997964 non TM | 0 |
| 18 | PF3D7_1346700 (448) | 6-Cysteine protein (Pfs48/45) | 0.739131 insoluble | 0.413481 | 0.116925 | 0.469594 non TM | 1 |
| 19 | PF3D7_1346800 (439) | 6-Cysteine protein (Pfs47) | 0.764391 insoluble | 0.47335 | 0.011404 | 0.515246 non TM | 2 |
| 20 | PF3D7_1364100 (796) | 6-cysteine protein (Pf92) | 0.732238 insoluble | 0.0783885 | 0.0691225 | 0.852489 non TM | 1 |
| 21 | PF3D7_1420700 (969) | Surface protein, Pf113 | 0.802372 insoluble | 0.307898 | 0.0247637 | 0.667338 non TM | 1 |
| 22 | PF3D7_1466000 (313) | Conserved Plasmodium protein, unknown function | 0.764391 insoluble | 0.0336363 | 0.0198285 | 0.946535 non TM | 0 |
| EXP-1 | PF3D7_1121600 (162) | Circumsporozoite-related antigen (exported protein 1) | 0.880007 soluble | 0.86553 alpha helical TM | 0.061304 | 0.0731663 | 1 |
| LSA-3 | PF3D7_0220000 (1558) | Liver stage antigen 3 | 1.000000 insoluble | 0.0346269 | 0.00134067 | 0.964032 non TM | 2 |
| AMA-1 | PF3D7_1133400 (622) | Apical membrane antigen 1 | 0.812039 insoluble | 0.00413516 | 0.000637273 | 0.995228 non TM | 1 |
Overall, in the first step, this study resulted in a list of predicted 22 antigens that consist of 19 new antigens and 3 known antigens (CSP, STRAP, GLURP) containing HLA class I binding peptide and/or CD8+ T cell response available in PlasmoDB-linked IEDB database, and the detailed immunogenicity information is presented in supplementary Table S1. These experimentally validated antigens have been successfully included as vaccine candidates (Kumar et al. 2006; Fidock et al. 1994; Antwi-Baffour et al. 2017). The only licensed malaria vaccine RTS,S is composed of truncated P. falciparum CSP which is a major surface protein expressed on sporozoites and contains CD8+ and CD4+ T cell epitopes as well as B cell epitopes (Supplementary Table S1).Thus, the currently used RV strategy for screening of novel antigens seems to be an effective approach (Holz et al. 2016).
CD8+ T cell epitope predictions by the NetCTL method
Malaria vaccines work mainly by inducing serum antibodies, a necessary and often sufficient constituent of vaccine efficacy that can inactivate pathogens directly or by cooperation with complement or immune cells (Doolan and Martinez-Alier 2006). Further, it is well known that CD8+ T cell responses are a key element of the immune reactivity elicited by several P. falciparum vaccines (Sette and Rappuoli 2010). Experimental studies of Doolan et al. (2003) have also demonstrated the protective immune responses induced by immunization with irradiated sporozoites that were broadly dispersed on a relatively large number of parasite antigens and against multiple epitopes on those antigens with variable potency. In light of the important role of cellular immunity in vaccine efficacy, it seems logical and necessary to also incorporate a cellular immunity dimension into the malaria vaccine design strategy. One approach for identifying targets of T cell responses is an antigen identification based on the prediction of high-affinity binding MHC-restricted T cell epitopes using computerized algorithms (Sette et al. 1994). Based on algorithms that predict binding to MHC molecules, measured as 50% inhibitory concentration (IC50) values expressed in nanomolars (nM), a meta-analysis using an affinity cutoff of 500 nM predicted that 52% of a panel P. falciparum peptides bound to HLA-A*0201 and led to the development of publicly available algorithms that are specific for MHC molecules (Doolan et al. 2003). The outcome of the similar studies led to the establishment of the IEDB database that contains open access data and analytical tools such as NetCTL for malaria and a wide range of other organisms (Fleri et al. 2017). To reduce experimental workload, HLA class I alleles have been grouped into nine major supertypes (clusters of alleles with similar peptide-binding motifs) and the majority of alleles fit these supertypes. Furthermore, analysis of the IEDB database suggests that >50% of HLA class I-restricted ligands bind to two or more HLA molecules often spanning different supertypes (Rao et al. 2011). In addition, HLA class I-restricted epitopes are well known to be promiscuous (binding to multiple HLA alleles) for malarial antigens including CSP (Doolan et al. 1996) and has been extended to include epitopes from other organisms (Frahm et al. 2007).
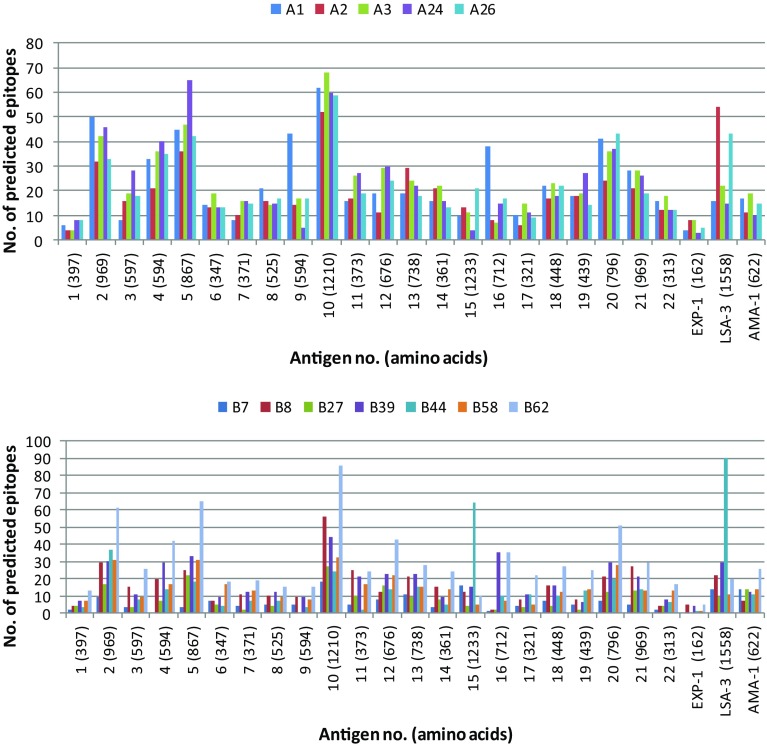
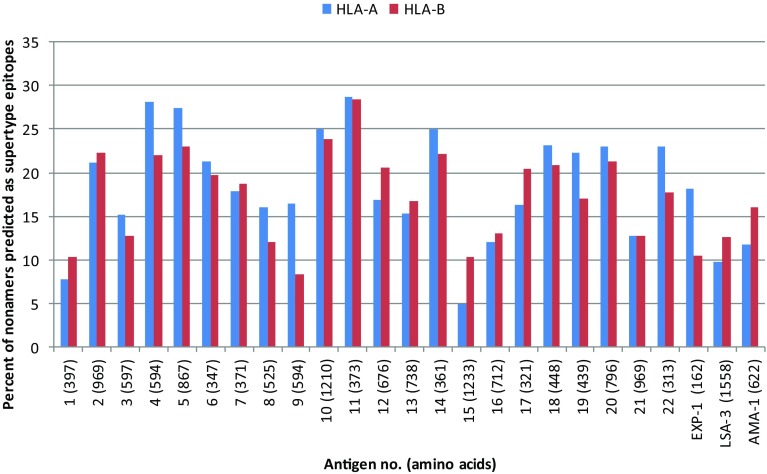
Keeping in view the above facts, the immunogenicity screening in the present study was limited to the predicted peptides that were able to bind HLA molecule with binding affinity (IC50) ≤500 nM using the NetCTL algorithm. The algorithm predicts MHC-binding peptides that are correctly processed from the native antigen using the combined epitope prediction score of 0.75, which includes predictions of proteasomal cleavage, TAP binding and HLA binding (Larsen et al. 2005). As a result, the amino acid sequences of the 22 predicted antigens and three control proteins were screened for their potential HLA class I-restricted supertype binding epitopes using the NetCTL algorithm. The control antigen proteins were included in the study to compare the epitope density/immunogenicity level of the predicted antigens. A total of 4944 CTL epitopes were predicted for 12 supertypes including 2523 HLA-A and 2421 HLA-B supertypes for the 22 predicted antigens (Fig. 2). For the control antigens, EXP-1 has 44 supertype epitopes (28 HLA-A and 16 HLA-B), LSA-3 has 345 supertype epitopes (150 HLA-A and 195 HLA-B) and AMA-1 has 170 supertype epitopes (72 HLA-A and 98 HLA-B). Furthermore, all the 22 probable antigens exhibited better level of immunogenicity as compared to threshold (more than 10% of HLA-A or -B supertype epitopes) defined by the selected three control antigens (Fig. 3). Out of 22 probable antigens, 13 (nos. 2, 4, 5, 6, 10, 11, 12, 14, 17, 18, 19, 20, 22) were predicted to contain more than 20% of HLA-A or -B supertype epitopes, which are higher than the epitope density of any control antigen (Fig. 3).
Fig. 2.
Distribution of predicted epitopes for HLA-A and -B supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, B62) by NetCTL from 22 probable antigens and 3 control antigens of P. falciparum malaria
Fig. 3.
Percent of nonamers predicted as epitopes (epitope density) of HLA-A (A1, A2, A3, A24, A26) and HLA-B (B7, B8, B27, B39, B44, B58, B62) supertypes against the selected panel of 22 probable antigens and 3 control antigens
Optimization of epitope set for malaria prevalent population coverage by OptiTope
In the past studies, it has been now accepted that all antigens or pathogen-derived epitopes are not equal in terms of their capacity to be recognized by the host immune system. This is called immunodominance, where pathogen-specific immune responses target only a small fraction of the full range of possible antigens or peptide epitopes (Sercarz et al. 1993). This may be due to properties intrinsic to the epitope(s), including efficiency of antigen processing and presentation, epitope abundance and MHC-binding affinity. Thus, the identification of epitopes, which are important targets of protective immune responses, will stimulate effective immunity against the target pathogen and is a key component of rational vaccine design (Rueckert and Guzmán 2012). Since MHC is highly polymorphic, each individual host possesses a set of MHC class I molecules of different binding specificities. Therefore, binding prediction of antigenic peptides to MHC is important for designing effective vaccine and hence activation of the immune system. The NetCTL predicts MHC-binding peptides that are correctly processed from the native antigen. In the design of a peptide-based specific and effective vaccine, selection of the set of epitopes which yields the best immune response in a given population or individual is an important criterion (Peters et al. 2013). Therefore, finally 262 predicted highest scoring CTL epitopes were screened from the selected panel of 22 probable antigens belonging to 5 HLA-A supertypes (Table 3a) and 7 HLA-B supertypes (Table 3b) for optimization study in malaria-endemic human population, viz. North-East (NE) Asia, South-East (SE) Asia, South-West (SW) Asia and sub-Saharan (SS) Africa. These epitopes were also searched in human protein reference database (HPRD) for similarity analysis, but none of the epitopes were found similar to human proteins. In this study, each of the 22 probable antigens was tested initially toward positive selection for each population and then the predicted highest scoring CTL epitopes by NetCTL of positive antigens were considered as a set of candidate epitopes. To compare the results of optimization of probable antigens with experimentally known antigens, the highest scoring distinct epitope set predicted by NetCTL of control antigens was also optimized for the same populations. For NE Asian population, ten probable antigens (nos. 1, 7, 9, 10, 13, 16, 17, 19, 20, 21) were tested for positive results. Similarly for the SE Asian population, 13 antigens (nos. 1, 4, 7, 9, 10, 11, 12, 13, 16, 17, 19, 20, 21) tested for positive selection. There are 11 antigens (nos. 1, 4, 7, 9, 10, 13, 16, 17, 19, 20, 21) tested for positive selection toward the SW Asian population and ten antigens (nos. 1, 4, 7, 9, 10, 16, 17, 19, 20, 21) tested positive for the SS African population. Among the control antigens, only LSA-3 showed a positive result in the SS African population. In OptiTope, the BIMAS program was selected as MHC class I-binding affinity prediction with threshold immunogenicity 1%. After the optimization of all the constraints under investigation (like MHC allele probabilities in the target population, peptide mutation rates and maximum number of selected peptides), the OptiTope algorithm provides an optimal set of epitopes along with additional information on their respective contribution to the overall immunogenicity including the alleles covered by the epitope in the selected population. Table 4 depicts the final optimization results of OptiTope for probable antigens and control antigen at 1% immunogenicity threshold. For the target population of NE Asia, out of the 119 candidate epitopes from ten probable antigens, OptiTope optimized 94 epitopes that covered maximally 98.26% population with 20 MHC class I alleles. However, 59 epitopes have fractional immunogenicity ≥0.01, where the most immunogenic epitope 13ILFYFFLWV21 from antigen no. 19 (6-cysteine protein-Pfs47), binds to the allele HLA-A*0201 and contributes a maximum of 6% to the overall immunogenicity. Similarly, for the target population of SE Asia, OptiTope provided 105 optimal epitopes set out of selected 154 epitopes from 13 antigens covered 96.25% of the population having all 12 MHC alleles, where the most immunogenic epitope 506FYFDINNKL514 from antigen no. 4 (conserved Plasmodium protein, unknown function) binded with alleles HLA-A*2402, -Cw*0401 and contributed maximally 5%, while the epitope the 13ILFYFFLWV21 from antigen no. 19 ranked at 20th position which binds with the allele HLA-A*0201 and contributed 2% of the overall immunogenicity. In case of South-West Asia, OptiTope selected 89 epitopes among 130 candidate epitopes from 11 probable antigens that covered 95.93% of the population with all 18 MHC alleles. However, 53 epitopes having fractional immunogenicity ≥0.01, where most immunogenic epitope 13ILFYFFLWV21 is from antigen no. 19, again binds with the allele HLA-A*0201 and contributes 8% to the overall immunogenicity. Likewise, in SS Africa population, OptiTope selected 67 epitopes out of 106 candidate epitopes of ten antigens covering a minimum of 85.37% of the population with 12 MHC class I alleles. Nevertheless, out of 67 optimal set of epitopes, 51 epitopes have fractional immunogenicity ≥0.01, in which the most immunogenic epitope 13ILFYFFLWV21 was found to bind with the alleles HLA-A*0201, -A*0205, -*0301 and again contributed 10% to the overall immunogenicity. Furthermore, in case of control antigen LSA-3 covered 64.12% SS African population. Overall, the combined analysis of the OptiTope results of 22 probable antigens for target populations where susceptibility to malaria is prevalent suggest that an epitope 13ILFYFFLWV21 from antigen no. 19 contributed most significantly: about 26% immunogenicity in all four populations having the major allele HLA-A*0201. Therefore, epitope 13ILFYFFLWV21 seems to be a novel vaccine candidate.
Table 3.
Highest scoring 262 CTL epitopes binding to HLA-A (a) and HLA-B (b) supertype as predicted by NetCTL from a panel of 22 probable and 3 control antigens of P. falciparum malaria
| (a) Antigen no. |
Epitope binding to HLA-A supertype (start position and CTL processing score) | ||||
|---|---|---|---|---|---|
| A1 | A2 | A3 | A24 | A26 | |
| 1 | PSDKHIKEY (311: 2.7000) | YLNKIQNSL (319: 1.2470) | KLRKPKHKK (85: 1.5478) | SFLFVEALF (12: 1.8273) | FVEALFQEY (15: 1.0471) |
| 2 | SSDIFDLSY (685: 3.7323) | YLAWSFNSL (329: 1.4288) | KVYTFYFYK (476: 1.6699) | KYILIYKNF (484: 1.9743) | DIFDLSYKY (687: 2.1903) |
| 3 | DSDIFLETY (400: 3.3753) | YIFFFLFLV (7: 1.4404) | KLLYPTLYY (465: 1.6781) | SYIFFFLFL (6: 1.8173) | ETYNLISGL (406: 2.1144) |
| 4 | DSDIFLETY (400: 3.3753) | YIFFFLFLV (7: 1.4404) | KLLYPTLYY (465: 1.6781) | SYIFFFLFL (6: 1.8173) | ETYNLISGL (406: 2.1144) |
| 5 | YSDFLILLF (807: 3.3260) | KLVYYNFIL (154: 1.4287) | YMYNIFLSY (394: 1.6305) | VWIFFFFNF (852: 1.9479) | YIFQKIKGY (187: 2.1707) |
| 6 | LTNIIMDHY (256: 3.2588) | FVLYILLSV (14: 1.4493) | VSAFNLSGK (238: 1.2369) | FYSRLPSLI (268: 1.8090) | FVIGSSMFM (100: 2.0667) |
| 7 | CTCDNSLTF (142: 1.7258) | FIFPFLYVI (360: 1.3715) | GIMKIHLKK (159: 1.5592) | FYFIFIFPF (356: 1.8206) | FIIFFFALF (6: 1.8150) |
| 8 | SSDHNLLGY (248: 3.4396) | YILFIVILL (510: 1.2959) | LLYFNNIVY (17: 1.4572) | YYATAVRNF (401: 1.9493) | YIVDMSVNY (445: 2.2900) |
| 9 | LSIWTTLLY (13: 2.7525) | ILAFALYML (584: 1.1966) | LLYSNKNLK (19: 1.5312) | TYVIKHNRF (39: 1.8533) | NIKSMINAY (509: 1.9343) |
| 10 | SADVYKSLY (667: 3.3520) | YLLLYTFDI (572: 1.4939) | RLHFYLLNK (331: 1.7244) | TYIYNVRTF (1125: 2.0610) | YIVKYLPYY (375: 2.4654) |
| 11 | SSSLYNNEY (172: 3.5241) | YMGDITTII (292: 1.4468) | ILFLISPFK (328: 1.7586) | FYKFSHFLF (338: 2.0231) | YLKNKLTYY (189: 2.0412) |
| 12 | FSTINSNLY (280: 3.3171) | YAFALFCTV (12: 1.1289) | KEYKIRMYK (266: 1.4592) | KYLNDHFVF (605: 2.1241) | DIKRNWAQY (434: 2.2182) |
| 13 | FSTKYVLIY (610: 3.5086) | FLFFSFILI (726: 1.3537) | KLIHKFTFK (134: 1.7568) | YYYFFPYFI (274: 1.9418) | DILNKLNAY (327: 2.0578) |
| 14 | VSNIYHLIY (198: 3.3669) | ALVMIIIYL (309: 1.3219) | KTAALAASK (145: 1.5142) | HYSNILLFF (4: 1.8934) | ETVLNVINY (187: 2.4988) |
| 15 | ETNIQEQLY (292: 2.4796) | NLFHITICL (3: 1.2816) | FITYISTKK (1185: 1.3069) | KFKKVSQTI (1193: 1.5103) | ETNIQEQLY (292: 1.6393) |
| 16 | YWDDFYHEY (408: 2.1577) | YILSISLFL (3: 1.4028) | FLILLNLYK (10: 1.4903) | KYILSISLF (2: 2.0677) | ELSDTENYY (372: 2.0208) |
| 17 | FADLKYYNY (293: 3.0794) | FLLKEIENI (313: 1.3871) | HLSQFCFSK (33: 1.4601) | NYENHSAAF (300: 1.6707) | GTIKNMIGY (238: 2.4826) |
| 18 | MTVTIDSAY (421: 2.8090) | FLAKTFIFL (432: 1.3097) | MMLYISAKK (1: 1.5931) | IYHKNLTIF (250: 1.9798) | SIFCTIHSY (68: 1.9156) |
| 19 | VLDTPNIEY (365: 3.0718) | ILFYFFLWV (13: 1.3530) | KLIPPYCFK (335: 1.5663) | KYISMFLIF (419: 1.9673) | SIINIILFY (8: 2.6122) |
| 20 | GTAMESLLY (661: 3.4765) | YLNTYHLAI (278: 1.4079) | KLFEVRLPK (399: 1.6583) | DYVEVQFHF (443: 1.90470) | DTVNKIYTY (360: 2.1999) |
| 21 | RMAVYNALY (640: 2.7522) | KLADNISLL (356: 1.4461) | KMSLLASLK (416: 1.6269) | KYAIMGNSF (669: 1.9124) | DSNDFLKKY (662: 1.7505) |
| 22 | FSTELNVEY (122: 2.8596) | FTYGYATFL (227: 1.3263) | IMASQICQK (92: 1.5831) | TYINLFFLL (3: 2.0372) | YTVEVKRAY (71: 2.3109) |
| EXP-1 | NTEKGRHPF (102: 0.8923) | GLLGVVSTV (83: 1.2151) | ALFFIIFNK (10: 1.7339) | VFFLALFFI (6: 1.4778) | EVNKRKSKY (66: 1.9752) |
| LSA-3 | LTDKMIDAV (936: 1.8323) | KLIEETQEL (1010: 1.3486) | ILNEAGGLK (871: 1.3939) | SYVVGFFTF (1448: 2.0792) | DIFKNLKHY (1493: 2.3066) |
| AMA-1 | TLDEMRHFY (194: 3.1764) | NLFSSIEIV (92: 1.2062) | SMFCFRPAK (272: 1.5246) | KYVKNLDEL (206: 1.4514) | EIVERSNYM (98: 2.1159) |
| (b) Antigen no. | Epitope binding to HLA-B supertype (start position and CTL processing score) | ||||||
|---|---|---|---|---|---|---|---|
| B7 | B8 | B27 | B39 | B44 | B58 | B62 | |
| 1 | MPNDPNRNV (285: 0.9327) | LRKPKHKKL (86: 1.5202) | TRVLNELNY (32: 1.2692) | NYDNAGTNL (39: 1.1377) | DELDYANDI (356: 0.9684) | ILSVSSFLF (7: 1.7560) | ALFQEYQCY (18: 1.3784) |
| 2 | IPSTRTIKI (644: 1.5871) | YLAWSFNSL (329: 1.6220) | FRFVLFKNF (778: 1.6170) | NYDEINFSL (425: 2.0832) | KETEVYFNL (217: 1.9571) | FSLIIPLSF (431: 1.8251) | TQKNIELSY (813: 1.4416) |
| 3 | LPGFNNIKI (516: 0.9995) | FIKSFVVEF (478: 1.7602) | HRNAFIKSF (474: 1.5987) | IKDDIYYIL (440: 1.9056) | KEFELINYL (387: 1.8518) | SSYIFFFLF (5: 1.7470) | KLLYPTLYY (465: 1.3747) |
| 4 | NA | FISIKYLSL (3: 1.5258) | YRIFYFINI (448: 1.2584) | FYFDINNKL (506: 1.6966) | EETKEYIPL (183: 1.8413) | KTNKSSILF (562: 1.7065) | FQNYISSFY (499: 1.3364) |
| 5 | PPSLNKGML (497: 1.2186) | FLILLFSPF (810: 1.8883) | YRADYKSNK (56: 1.5670) | YRELSEESL (385: 2.2779) | FEKNVHPIL (666: 1.7168) | LSYFSYLHY (400: 1.8124) | YMYNIFLSY (394: 1.5285) |
| 6 | HPNQQTSVT (42: 1.1563) | KLKLTNIIM (253: 1.3057) | SRNMMHLKK (88: 1.4165) | YCLGISFVL (8: 1.4636) | KEFVIGSSM (98: 1.4703) | ISNSSFLTL (320: 1.4146) | SSFLTLSSY (323: 1.3647) |
| 7 | RPVECFEYI (89: 1.2605) | YDKQKILPL (192: 1.5565) | LRFICPMRK (72: 1.3204) | MHIVSFIIF (1: 1.1613) | REHKLSEIL (102: 1.9382) | IVSFIIFFF (3: 1.4401) | VTIKKSQVY (247: 1.2871) |
| 8 | KPEVKNALL (318: 1.3426) | IMKGRYYAT (396: 1.6875) | GRYYATAVR (399: 1.6544) | YKDSLSNKL (94: 1.8128) | IEAFFPFIL (208: 1.9354) | TAIIDETVY (382: 1.1623) | LLYFNNIVY (17: 1.3749) |
| 9 | NPRNQITHL (542: 1.4070) | FIVVIYIAF (562: 1.7318) | NA | YKYVGKLIL (577: 2.4229) | NETTSDDEL (488: 1.2880) | VVIYIAFNW (564: 1.8080) | NQYVFANNY (479: 1.3857) |
| 10 | KPKYKHLKI (948: 1.5387) | YMKERYKNL (82: 2.1101) | QRLYFNIKK (922: 1.6416) | YHTEGINTL (753: 2.8654) | KEFKNIFQL (1099: 2.0999) | FSTPQNLKW (1143: 1.9155) | KQRKKYIEY (692: 1.4679) |
| 11 | KPFYKFSHF (336: 1.4554) | HIKKFFLYL (318: 1.7217) | KRIKQLFQY (282: 1.7683) | FFSVPNNIL (346: 1.7372) | IEEIYGIPL (101: 1.9058) | KSNIFCKSF (76: 1.5588) | FTNILSASY (358: 1.3693) |
| 12 | LPTNKNLLL (585: 1.4635) | YIRERNLIM (300: 2.0207) | RRNVLKKRK (74: 1.6360) | NRSEIHQIL (512: 1.9701) | WEQAKPVKL (375: 1.4091) | LASSSQTYF (572: 1.8982) | HQILTKNTF (517: 1.4490) |
| 13 | NPNNSSTPL (666: 1.7380) | YSKTNLSAL (638: 1.6769) | YRTKDMVNK (305: 1.4794) | IHEKDKISL (115: 2.2627) | REFLITGIL (224: 2.0900) | LSVGVQNTF (699: 1.8197) | QQVNNNNNY (442: 1.4448) |
| 14 | TPHHTTTTT (33: 1.0884) | LRYRRKKKM (319: 1.3152) | RRKKKMNKK (322: 1.6493) | PHHTTTTTL (34: 1.4569) | SEYGVHTSI (46: 1.7438) | VSNIYHLIY (198: 1.6676) | YSFKILNPF (342: 1.4249) |
| 15 | FPRQKHKKV (220: 1.5557) | FPRQKHKKV (220: 1.9607) | KRIGGPKLR (33: 1.3142) | FEDVHTEQL (800: 1.5889) | FESLSDLEL (151: 1.9433) | KSNKVQNHF (143: 2.0406) | IVSVMINAY (1201: 1.3909) |
| 16 | KPTYLNYHM (417: 1.2954) | GIKKFKNVF (196: 1.2073) | FQSYFNQSK (204: 0.8209) | THDEFNVPL (648: 2.7480) | LEGTYGENL (60: 1.4632) | TTNEESHNF (499: 1.5645) | TLYEPNNFY (428: 1.4407) |
| 17 | YPCGCIGIL (264: 1.2461) | FSKKWFSEF (39: 1.8660) | KKLFKKVEY (248: 1.0576) | YHLKNKFHL (26: 2.3134) | LESKEFADL (288: 1.5339) | YANISIPTY (256: 1.5275) | SIMWPGQAF (48: 1.4394) |
| 18 | KPKYEKKVI (287: 1.3798) | DMRERRSIF (62: 1.7436) | GRSAMVHVR (170: 1.0666) | FQEGKEKAL (235: 1.6916) | AEGDDKIKL (387: 1.7105) | KSSSPEFKI (89: 1.4990) | SIFCTIHSY (68: 1.3885) |
| 19 | CPKKNNGDF (77: 0.9266) | WVKKSISEL (20: 1.3686) | KKYAINSSF (120: 1.3434) | NQYNNIIEL (181: 1.8303) | KEGMYMLAL (378: 1.8096) | IINIILFYF (9: 1.5583) | TQYVCDFYF (32: 1.2525) |
| 20 | TPAVYSGSL (144: 1.6718) | VPRKIGCEL (348: 1.7022) | YRIVVEFDY (473: 1.5796) | YKYIYNNKL (370: 2.3093) | FETFDPQYL (302: 1.8799) | LSFDEGNNW (193: 1.7229) | VQFHFPIYY (447: 1.3709) |
| 21 | RRRKRITEL (523: 1.2293) | ELRKKTQSF (86: 2.0191) | RRMAVYNAL (639: 1.8852) | TTDEGTEEL (795: 1.6885) | KELAHRTAL (345: 1.7821) | ISSDIFFKY (479: 1.7725) | ALYEKAQSY (646: 1.4891) |
| 22 | NPHEKRATI (194: 1.3523) | YLIVRDQTL (153: 2.2147) | KRDNNLIKI (57: 0.9285) | HQGVTHKYL (146: 1.2382) | KENVYENMI (204: 1.1746) | KTYINLFFL (2: 1.2804) | YTVEVKRAY (71: 1.3755) |
| EXP-1 | NA | ILSVFFLAL (3: 1.6313) | NA | ILSVFFLAL (3: 1.0259) | GEPNAGPQV (131:1.0800) | LSVFFLALF (4: 1.2489) | FLALFFIIF (8: 1.1843) |
| LSA-3 | RPKLEEVLL (1260: 1.2460) | KLKELEKAL (1031: 1.8133) | KRIEKVKEK (1356: 1.5727) | SKNDVTNVL (1407: 1.8112) | LEELHENVL (663: 1.7718) | TTAESVTTF (634: 1.6572) | HMREKINKY (37: 1.4607) |
| AMA-1 | RASHTTPVL (608: 1.5105) | YLKDGGFAF (175: 1.7612) | KRKGNAEKY (568: 1.5485) | SSYIATTAL (423: 1.7190) | FEFTYMINF (13: 1.7547) | RSNYMGNPW (102: 1.7730) | YLKDGGFAF (175: 1.5466) |
NA not available
Table 4.
Population coverage analysis of the highest scoring 262 epitope set from 22 probable antigens and gold standard antigen LSA-3 of P. falciparum in the malaria-endemic regions
| Population selected | No. of optimized epitopes/candidate epitopes | No. of covered/target alleles | Locus coverage (%) | Population coverage (%) | ||
|---|---|---|---|---|---|---|
| A | B | C w | ||||
| Probable 22 antigens | ||||||
| NE Asia | 94/119 | 20/20 | 62.56 | 52.89 | 25.13 | 98.26 |
| SE Asia | 105/154 | 12/12 | 59.66 | 36.2 | 24.79 | 96.25 |
| SW Asia | 89/130 | 18/19 | 50.95 | 39.06 | 32.51 | 95.93 |
| SS Africa | 67/106 | 13/14 | 24.32 | 24.76 | 32.82 | 85.37 |
| LSA-3 | ||||||
| SS Africa | 4/12 | 7/14 | 13.65 | 18.89 | 14.48 | 64.12 |
Prediction of malarial homologous antigens
Keeping in view the immunodominance correlation with hydrophobicity of T cell receptor (TCR) contact residues, PAComplex web server was used for the detailed atomic interactions (Chowell et al. 2015). The most prominent antigen (no. 19) analyzed from the above work was further investigated for the presence of T cell epitopes by using PAComplex web server and five peptides were obtained above the threshold J z (√(ZMHC × ZTCR) value 4. The peptide 13ILFYFFLWV21 with J z value 4.77 was again ranked first and matched 3/9 residues to a template LLFGYPVYV (PDB id: 1bd2). The detailed residue interactions (e.g., hydrogen bonds and VDW forces) of peptide 13ILFYFFLWV21 with HLA-A*0201-TCR template are described below. The positions 3, 5 and 8 prefer aromatic residues. The third (Phe) residue of the top hit peptide forms strong VDW with Y159 and L156, and H-bond with Y99 of the MHC molecule. At position 5 (Phe) of the hit peptide, an H-bond is formed with D30 and strong VDWs are formed with M92, A95 and G94 of TCR molecule. The position 8 (Trp) of the hit peptide forms an H-bond (W147) and strong VDW interaction (Q72) with the MHC molecule. It also forms two strong VDW interactions (G97 and G98) with the TCR molecule. Conversely, the compositions of positions 4, 6 and 7 are diverse. However, the positions 1, 2, and 9 prefer non-polar residues. The first (Ile) residue forms H-bonds with Y159, Y7 and Y171 and strong VDW with W167 in MHC. The second (Leu) forms H-bonds with E63 and K66 and strong VDW (F9, M45 and V67) with HLA-A*02101. Similarly, the ninth (Val) forms three H-bonds (D77, Y84 and T143) and strong VDW (L81) with HLA-A*02101. The comparative results (J z value) with three control antigens suggest that the predicted 13ILFYFFLWV21 of antigen no. 19 could be considered as a potential immunodominant novel vaccine candidate. However, functional assays are required to define the protective efficacy of the epitope (Sedegah et al. 2016). A similar study toward the designing and computational analysis of a fusion protein has been also reported in P. falciparum (Shamriz and Ofoghi 2016) and Staphylococcus aureus (Hajighahramani et al. 2017) as a step toward developing a new vaccine candidate. If the sequence-based bioinformatics approaches fail to identify bona fide antigens, then an additional structure-based method may provide the rational design of next-generation subunit vaccines that correlate protective and broadly cross-reactive immune responses in vaccinated individuals with specific structural features (Doud et al. 2012; Liljeroos et al. 2015). Therefore, more efficient and high-throughput screening approaches are needed that can be accomplished on a whole genome scale to provide more empirical data to inform antigen selection (Davies et al. 2015; Kassegne et al. 2016; Zeng et al. 2017) and further plant-based transient expression system for the fast production of vaccine candidates (Boes et al. 2016).
Conclusions
In recent years, peptide-based subunit vaccines have attracted more attention because of their different advantages. In the present work, using several available bioinformatics resources, we tried to find out probable antigenic proteins from P. falciparum genome and design an efficient multi-epitope subunit vaccine, which could stimulate CD8+ T cell immune responses in malaria-endemic populations. Out of 22 probable antigens, 13 were predicted to contain more than 20% CD8+ epitopes, which is greater than the immunogenicity level (predicted epitope density) of any control antigen. The highest scoring CD8+ epitopes also covers better human population than the control antigens in the optimization study. These data provide the foundation for the development of an antigen/epitope map (immunosome) of P. falciparum with practical value to facilitate the design and discovery of multi-antigen and multi-epitope vaccines to mimic the complexity of responses elicited by natural infection. We expect that our designed vaccine shows promising results against P. falciparum malaria in practice. Moreover, the present approach offers the potential to overcome the deficiencies of the current ad hoc approach to antigen selection by using biological samples from humans or animals with immunity to malaria. However, the overall reliability of any in silico approach is strongly dependent on the accuracy of prediction, which in turn necessitates a robust definition of antigenic protein. There are limits to what in silico methods can achieve as well as immense opportunities to exploit its potential.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Dr. Akhilesh Kumar Singh and Dr. Brijesh Pandey from Amity University Uttar Pradesh Lucknow campus for their help in revising the present manuscript.
Compliance with ethical standards
Conflict of interest
We declare that we have no conflict of interest in the publication.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0947-7) contains supplementary material, which is available to authorized users.
References
- Antwi-Baffour S, Adjei JK, Agyemang-Yeboah F, et al. Proteomic analysis of microparticles isolated from malaria positive blood samples. Proteome Sci. 2017;15:5. doi: 10.1186/s12953-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes A, Reimann A, Twyman RM, et al. A plant-based transient expression system for the rapid production of malaria vaccine candidates. Methods Mol Biol. 2016;1404:597–619. doi: 10.1007/978-1-4939-3389-1_39. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Roos DS, et al. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34(D363–D368):2936–2943. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell D, Krishna S, Becker PD, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci USA. 2015;112(14):1754–1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiane Hertz-Fowler CH, Peacock CS, Wood V, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen L, Bradley P, Menke M, et al. Predicting the beta-helix fold from protein sequence data. J Comput Biol. 2002;9(2):261–276. doi: 10.1089/10665270252935458. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Healer J, Marapana D. Malaria: biology and disease. Cell. 2016;167(3):610–624. doi: 10.1016/j.cell.2016.07.055. [DOI] [PubMed] [Google Scholar]
- Davies DH, Duffy P, Bodmer JL. Large screen approaches to identify novel malaria vaccine candidates. Vaccine. 2015;33(52):7496–7505. doi: 10.1016/j.vaccine.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa KP, Doolan DL. Immunomics: a 21st century approach to vaccine development for complex pathogens. Parasitology. 2016;143(2):236–244. doi: 10.1017/S0031182015001079. [DOI] [PubMed] [Google Scholar]
- Dellagostin OA, Grassmann AA, Rizzi C, et al. Reverse vaccinology: an approach for identifying leptospiral vaccine candidates. Int J Mol Sci. 2017;18(1):E158. doi: 10.3390/ijms18010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Martinez-Alier N. Immune response to pre-erythrocytic stages of malaria parasites. Curr Mol Med. 2006;6:169–185. doi: 10.2174/156652406776055249. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Sedegah M, Hedstrom RC, et al. Circumventing genetic restriction of protection against malaria with multigene DNA immunization: CD8+ cell-, interferon gamma-, and nitric oxide-dependent immunity. J Exp Med. 1996;183:1739–1746. doi: 10.1084/jem.183.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Southwood S, Freilich DA, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan DL, Apte SH, Proietti C. Genome-based vaccine design: the promise for malaria and other infectious diseases. Int J Parasitol. 2014;44(12):901–913. doi: 10.1016/j.ijpara.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Doud MB, Koksal AC, Mi LZ, et al. Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc Natl Acad Sci USA. 2012;109(20):7817–7822. doi: 10.1073/pnas.1205737109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Manzalawy Y, Munoz EE, Lindner SE. PlasmoSEP: predicting surface-exposed proteins on the malaria parasite using semisupervised self-training and expert-annotated data. Proteomics. 2016;16(23):2967–2976. doi: 10.1002/pmic.201600249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Bottius E, Brahimi K, et al. Cloning and characterization of a novel Plasmodium falciparum sporozoite surface antigen, STARP. Mol Biochem Parasitol. 1994;64(2):219–232. doi: 10.1016/0166-6851(94)00012-3. [DOI] [PubMed] [Google Scholar]
- Fleri W, Paul S, Dhanda SK, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm N, Yusim K, Suscovich TJ, et al. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur J Immunol. 2007;37:2419–2433. doi: 10.1002/eji.200737365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, Neb T, Vukcevic D, et al. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteom. 2006;5(7):1286–1299. doi: 10.1074/mcp.M600035-MCP200. [DOI] [PubMed] [Google Scholar]
- Good MF, Doolan DL. Malaria vaccine design: immunological considerations. Immunity. 2010;33(4):555–566. doi: 10.1016/j.immuni.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Goodswen SJ, Kennedy PJ, Ellis JT. A guide to in silico vaccine discovery for eukaryotic pathogens. Brief Bioinform. 2013;14(6):753–774. doi: 10.1093/bib/bbs066. [DOI] [PubMed] [Google Scholar]
- Gosling R, von Seidlein L. The future of the RTS, S/AS01 malaria vaccine: an alternative development plan. PLoS Med. 2016;13(4):e1001994. doi: 10.1371/journal.pmed.1001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajighahramani N, Nezafat N, Eslami M, et al. Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect Genet Evol. 2017;48:83–94. doi: 10.1016/j.meegid.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Holz LE, Fernandez-Ruiz D, Heath WR. Protective immunity to liver-stage malaria. Clin Transl Immunol. 2016;5(10):e105. doi: 10.1038/cti.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Hoffman SL. Malaria vaccine development. Clin Microbiol Rev. 1994;7(3):303–310. doi: 10.1128/CMR.7.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassegne K, Abe EM, Chen JH, et al. Immunomic approaches for antigen discovery of human parasites. Expert Rev Proteom. 2016;13(12):1091–1101. doi: 10.1080/14789450.2016.1252675. [DOI] [PubMed] [Google Scholar]
- Koganty RR. Vaccine safety: a case for synthetic vaccine formulation. Expert Rev Vaccin. 2003;2:725–727. doi: 10.1586/14760584.2.6.725. [DOI] [PubMed] [Google Scholar]
- Kovjazin R, Volovitz I, Daon Y, et al. Signal peptides and trans-membrane regions are broadly immunogenic and have high CD8+ T cell epitope densities: implications for vaccine development. Mol Immunol. 2011;48(8):1009–1018. doi: 10.1016/j.molimm.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KA, Sano G, Boscardin S, et al. The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature. 2006;444:937–940. doi: 10.1038/nature05361. [DOI] [PubMed] [Google Scholar]
- Kumar H, Frischknecht F, Mair GR. In silico identification of genetically attenuated vaccine candidate genes for Plasmodium liver stage. Infect Genet Evol. 2015;36:72–81. doi: 10.1016/j.meegid.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Larsen MV, Lundegaard C, Nielsen M, et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC-I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35(8):2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- Liljeroos L, Malito E, Ferlenghi I, et al. Structural and computational biology in the design of immunogenic vaccine antigens. J Immunol Res. 2015;2015:156241. doi: 10.1155/2015/156241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Shaw TN, Annoura T, et al. The subcellular location of ovalbumin in Plasmodium berghei blood stages influences the magnitude of T-cell responses. Infect Immun. 2014;82(11):4654–4665. doi: 10.1128/IAI.01940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu T, Zhu F, et al. A whole-killed, blood-stage lysate vaccine protects against the malaria liver stage. Parasite Immunol. 2016;39(1):e12386. doi: 10.1111/pim.12386. [DOI] [PubMed] [Google Scholar]
- McCarthy JS, Good MF. Whole parasite blood stage malaria vaccines: a convergence of evidence. Hum Vaccin. 2010;6:114–123. doi: 10.4161/hv.6.1.10394. [DOI] [PubMed] [Google Scholar]
- Peters B, Sette A, Soisson L, et al. Identification of minimal human MHC-restricted CD8+ T-cell epitopes within the Plasmodium falciparum circumsporozoite protein (CSP) Malar J. 2013;12:185. doi: 10.1186/1475-2875-12-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebanski M, Hill AV. The immunology of malaria infection. Curr Opin Immunol. 2000;12:437–441. doi: 10.1016/S0952-7915(00)00117-5. [DOI] [PubMed] [Google Scholar]
- Rao X, Hoof I, Costa AI, et al. HLA class I allele promiscuity revisited. Immunogenetics. 2011;63:691–701. doi: 10.1007/s00251-011-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R, Bottomley MJ, D’Oro U, et al. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med. 2016;213(4):469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche PA, Reinherz EL. Definition of MHC supertypes through clustering of MHC peptide-binding repertoires. Methods Mol Biol. 2007;409:163–173. doi: 10.1007/978-1-60327-118-9_11. [DOI] [PubMed] [Google Scholar]
- Rueckert C, Guzmán CA. Vaccines: from empirical development to rational design. PLoS Pathog. 2012;8(11):e1003001. doi: 10.1371/journal.ppat.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedegah M, Peters B, Hollingdale MR, et al. Vaccine strain-specificity of protective HLA-restricted class 1 P. falciparum epitopes. PLoS One. 2016;11(10):e0163026. doi: 10.1371/journal.pone.0163026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercarz EE, Lehmann PV, Ametani A, et al. Dominance and crypticity of T cell antigenic determinants. Ann Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33(4):530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Vitiello A, Reherman B, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- Shamriz S, Ofoghi H. Design, structure prediction and molecular dynamics simulation of a fusion construct containing malaria pre-erythrocytic vaccine candidate, PfCelTOS, and human interleukin 2 as adjuvant. BMC Bioinform. 2016;17:71. doi: 10.1186/s12859-016-0918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Frischknecht F. Time for genome editing: next-generation attenuated malaria parasites. Trends Parasitol. 2016;33(3):202–213. doi: 10.1016/j.pt.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Singh SP, Mishra BN. Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol. 2016;77(3):295–306. doi: 10.1016/j.humimm.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Singh SP, Khan F, Mishra BN. Computational characterization of Plasmodium falciparum proteomic data for screening of potential vaccine candidates. Hum Immunol. 2010;71(2):136–143. doi: 10.1016/j.humimm.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Singh SP, Verma V, Mishra BN. Characterization of Plasmodium falciparum proteome at asexual blood stages for screening of effective vaccine candidates: an immunoinformatics approach. Immunol Immunogenet Insights. 2015;7:21–30. doi: 10.4137/III.S24755. [DOI] [Google Scholar]
- Tan M, Jiang X. Recent advancements in combination subunit vaccine development. Hum Vaccin Immunother. 2016;13(1):180–185. doi: 10.1080/21645515.2016.1229719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A, Feng G, Brown GV, et al. Functional antibodies and protection against blood-stage malaria. Trends Parasitol. 2016;32(11):887–898. doi: 10.1016/j.pt.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Terry FE, Moise L, Martin RF, et al. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert Rev Vaccin. 2015;4(1):21–35. doi: 10.1586/14760584.2015.955478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu A, Kayala MA, Burk C. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol Cell Proteom. 2011;10(9):M111.007948. doi: 10.1074/mcp.M111.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandoolaeghe P, Schuerman L. The RTS, S/AS01 malaria vaccine in children 5–17 months of age at first vaccination. Expert Rev Vaccin. 2016;15(12):1481–1493. doi: 10.1080/14760584.2016.1236689. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, et al. Essentials of glycobiology. New York: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- World Health Organization (WHO) (2016) World malaria report december
- Zeng L, Wang D, Hu N, et al. A novel pan-genome reverse vaccinology approach employing a negative-selection strategy for screening surface-exposed antigens against leptospirosis. Front Microbiol. 2017;8:396. doi: 10.3389/fmicb.2017.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.