Abstract
Deficiency of the cysteine protease inhibitor cystatin M/E (Cst6) in mice leads to disturbed epidermal cornification, impaired barrier function, and neonatal lethality. We report the rescue of the lethal skin phenotype of ichq (Cst6-deficient; Cst6−/−) mice by transgenic, epidermis-specific, reexpression of Cst6 under control of the human involucrin (INV) promoter. Rescued Tg(INV-Cst6)Cst6ichq/ichq mice survive the neonatal phase, but display severe eye pathology and alopecia after 4 mo. We observed keratitis and squamous metaplasia of the corneal epithelium, comparable to Cst6−/−Ctsl+/− mice, as we have reported in other studies. We found the INV promoter to be active in the hair follicle infundibulum; however, we did not observe Cst6 protein expression in the lower regions of the hair follicle in Tg(INV-Cst6)Cst6ichq/ichq mice. This result suggests that unrestricted activity of proteases is involved in disturbance of hair follicle biology, eventually leading to baldness. Using quenched activity-based probes, we identified mouse cathepsin B (CtsB), which is expressed in the lower regions of the hair follicle, as an additional target of mouse Cst6. These data suggest that Cst6 is necessary to control CtsB activity in hair follicle morphogenesis and highlight Cst6-controlled proteolytic pathways as targets for preventing hair loss.—Oortveld, M. A. W., van Vlijmen-Willems, I. M. J. J., Kersten, F. F. J., Cheng, T., Verdoes, M., van Erp, P. E. J., Verbeek, S., Reinheckel, T., Hendriks, W. J. A. J., Schalkwijk, J., Zeeuwen, P. L. J. M. Cathepsin B as a potential cystatin M/E target in the mouse hair follicle.
Keywords: proteases, cysteine protease inhibitor, epidermis, alopecia, activity-based probes
Cystatin M/E (CST6) is a cysteine protease inhibitor that is mainly expressed in the epidermis, more specifically in the upper layers of the stratum granulosum, sweat glands, sebaceous glands, and hair follicles (HFs) (1–3), suggesting an important role for CST6 in skin and hair physiology. Our previous studies have identified CST6 as a component in a novel biochemical pathway that controls skin barrier formation by regulating both cornification and desquamation of the stratum corneum (SC). CST6 plays a key role in this pathway, as it controls the activity of both the papain-like proteases cathepsin L (CtsL) and CTSV and the asparaginyl endopeptidase legumain (LGMN) (4). Furthermore, as human CTSL is the elusive enzyme that processes and activates human transglutaminase (TGM)-3 (4), CST6 may regulate cross-linking of structural proteins during the cornification process.
The importance of Cst6 and its target proteases in terminal differentiation of the epidermis is illustrated by the severe skin phenotype of the Cst6-deficient mouse (ichq, referred to herein as Cst6−/−), characterized by abnormal SC and HF formation, disturbed skin barrier function, and neonatal lethality (5–7). The lethal skin phenotype of the Cst6-deficient mouse makes it impossible to study the consequences of Cst6 deficiency in adult animals. To circumvent this problem, we generated Cst6−/−/Tgm3−/−, Cst6−/−/Lgmn−/−, and Cst6−/−Ctsl−/− double-knockout mice and have shown that a tightly regulated balance between the protease CtsL and Cst6 is essential for tissue integrity of the epidermis and for maintenance of corneal epithelium (8). Both Cst6−/−Ctsl−/− and Cst6−/−Ctsl+/− mice are completely bald, a phenomenon that thus could not be attributed to deleterious effects of free CtsL activity and rather suggests that there are hitherto unknown Cst6 targets in mice.
In the current study, we sought to identify possible new Cst6 targets by rescuing Cst6 deficiency in skin, using classic transgenesis and analyzing the consequences of Cst6 deficiency in other tissues. We found disturbance of HF biology in adult rescued mice, most likely related to unrestricted protease activity, as Cst6 is absent in the lower regions of the HF in these rescued mice. The data provide evidence that cathepsin (Cts)-B represents a novel and crucial Cst6 target in the mouse HF.
MATERIALS AND METHODS
Construction of the Cst6 expression vector
Total RNA from mouse tissues was isolated by using Trizol Reagent (Thermo Fisher Scientific, Waltham, MA, USA) (6). Oligo-dT-primed single-strand cDNA was synthesized from total RNA by Moloney murine leukemia virus RNase-H− reverse transcriptase (Boehringer, Mannheim, Germany). To amplify the full mouse Cst6 coding sequence, we used the following oligonucleotide primers: forward, 5′-CTGAATCCGCGGCTATGGAG-3′ and reverse, 5′-CCTGACTCTGTCACCCTGG-3′ (corresponding to nt 13–32 and 483–501, respectively, in the mouse Cst6 cDNA sequence; accession ID: MGI:1920970). PCR conditions (1 mM MgCl2) were 94°C for 6 min followed by 35 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 2 min. The resulting mouse Cst6 cDNA fragment was cloned into the pCR2.1-TOPO cloning vector (Thermo Fisher Scientific). The Cst6 cDNA vector, together with the human involucrin (INV) promoter-containing plasmid pBS-3700 (a kind gift from Dr. Joseph M. Carroll, State University of New York, Stony Brook, NY, USA), and the pIRES2-EGFP vector (BD Biosciences Clontech, Mountain View, CA, USA), were used for construction of the final Cst6 expression vector (pBS-INV-Cst6-IRES-eGFP). First, pIRES2-EGFP was modified by inserting a BamHI adaptor (5′-TTAACGGGATCCCG-3′) at the unique AflII site. Subsequently, the internal ribosomal entry site (IRES), enhanced green fluorescent protein (eGFP), and simian virus 40 poly(A) signal sequences were excised with BamHI and inserted into the unique BamHI site in pBS-3700. Finally, the Cst6 coding sequence was cloned into the intermediate vector with EcoRI, resulting in the expression vector pBS-INV-Cst6-IRES-eGFP. All cloning steps were examined by DNA sequence analysis.
Cell culture and transfection of human keratinocytes
Human epidermal keratinocytes, obtained from abdominal skin after surgical correction, were cultured in keratinocyte growth medium (Lonza, Basel, Switzerland) (9). Transfection of keratinocytes with pBS-INV-Cst6-IRES-eGFP or pIRES2-eGFP was performed at 90–100% confluence, followed by switching to keratinocyte growth medium/5% fetal calf serum to stimulate keratinocyte differentiation. Transfections were performed in OptiMEM I (Thermo Fisher Scientific),with Lipofectamine 2000 (LF2000; Thermo Fisher Scientific), according to the manufacturer’s protocol with a DNA:LF2000 ratio of 1 µg:5 µl. Finally, keratinocytes were harvested for flow cytometry or RNA isolation, and the culture supernatant was used for ELISA.
Detection of eGFP expression
Cultured keratinocytes were harvested and resuspended in PBS and subsequently sorted on an EPICS Elite flow cytometer (Coulter, Luton, United Kingdom) that detects fluorescent signals. Viable cells were separated from dead cells based on morphology and amount of propidium iodide uptake and analyzed for eGFP expression. In addition, keratinocyte suspensions were analyzed directly by fluorescence microscopy.
Detection of Cst6 transgenic transcripts
RNA was isolated from cultured keratinocytes, treated with DNase I (Thermo Fisher Scientific) to prevent DNA contamination, and used for cDNA synthesis as described above. Subsequently, the cDNA templates were used in a PCR reaction for amplification of the mouse Cst6 part of the construct. The PCR (35 cycles) contained 2.5 mM MgCl2 and was performed at a melting temperature of 61°C with the following primers: forward, 5′-ATGGAGCGTCCTCACTTCC-3′ and reverse, 5′-CCTGACTCTGTCACCCTGG-3′ (corresponding to nt 27–45 and 483–501, respectively, in the mouse Cst6 cDNA sequence; accession ID: MGI:1920970). PCR products were analyzed on ethidium bromide-stained 1.5% agarose gels.
ELISA for mouse Cst6
Recombinant mouse Cst6 and human CST6 (both from R&D Systems, Minneapolis, MN, USA) were used as a standard and as a negative control, respectively, in concentrations varying from 5 to 0.156 ng/ml. The wells of a 96-well plate were coated overnight at 4°C with polyclonal rabbit anti-human CST6 antibody (2), followed by incubation with 1% bovine serum albumin (ICN Biomedicals, Aurora, OH, USA) and 1% normal goat serum (Vector Laboratories, Burlingame, CA, USA) in PBS for 30 min. Subsequently, standards, controls, and samples (undiluted up to 32× diluted) were incubated for 1 h, followed by incubation with monoclonal rat anti-mouse Cst6 (R&D Systems) in PBS/1% normal rabbit serum/0.1% bovine serum albumin/0.05% Tween-20 for 30 min. Next, wells were incubated with goat anti-rat biotinylated antibody (Vector Laboratories) for 30 min, followed by a final incubation with avidin-biotinylated horseradish peroxidase complex (Vector Laboratories) for 30 min. The above incubation steps were performed at 37°C and separated by repeated washing steps with PBS/0.05% Tween-20. Chromogenic substrate 1-step Ultra TMB (Thermo Fisher Scientific) was used as substrate for detection and the reaction was stopped by adding 4 M H2SO4. Each well was measured for mouse Cst6 at an absorbance of 450 nm with an ELISA microplate reader (Bio-Rad, Hercules, CA, USA).
Microinjection of the transgene in mouse zygotes
The generation of transgenic mice was performed according to standard protocols (10) and the guidelines of the local Committee for Animal Welfare. The transgenic expression plasmid was isolated from bacteria with an endotoxin-free midi-prep kit (Sigma-Aldrich, St. Louis, MO, USA), followed by excision of the INV-Cst6-IRES-eGFP transgene [Tg(INV-Cst6-IRES-eGFP)] with SalI and SpeI restriction enzymes and subsequent purification of the linear DNA fragment. Superovulation was induced in female C57BL/6J mice and at 0.5 d after coitus the mice were euthanized and the fertilized eggs (zygotes) collected. The transgene was microinjected into the male pronucleus, and surviving zygotes were subsequently cultured in vitro overnight onto the 2-cell stage. Viable microinjected zygotes were then transferred into the oviduct of a 0.5 d postcoitus pseudopregnant female C57BL/6J mouse to resume embryonic development.
Mice
We have reported that a null mutation in the Cst6 gene (Cst6) causes the lethal skin phenotype of ichq/ichq mice, which harbor a spontaneous mutation first described in BALB/cJ mice (6). After they were backcrossed 6 times to C57BL/6J mice, the C57BL/6J Cst6+/− mice were cross-bred to the transgenic, epidermis-specific Cst6-expressing mice to obtain the rescued Tg(INV-Cst6)Cst6ichq/ichq mice. All mice were housed in specific-pathogen–free facilities at the Central Animal Laboratory (University of Nijmegen). All animal experiments were approved by the University of Nijmegen Institutional Animal Care and Use Committee.
Mouse genotyping
Tail tissue biopsies were collected, and samples were incubated overnight at 55°C for cell lysis in 200 µl DirectPCR solution (Viagen Biotech, Los Angeles, CA, USA) and 100 µl proteinase K (0.5 mg/ml; Roche, Mannheim, Germany). Genomic DNA was isolated by phenolchloroform extraction and alcohol precipitation. The genotyping PCR (42 cycles) contained 2.5 mM MgCl2 and was performed at a melting temperature of 53°C. The following primers were used to identify the transgene: forward primer (exon 2) 5′-TACTACCTGACTTTGGACATA-3′ and reverse primer (exon 3) 5′-AAAGTTGCAACGCAGTTT-3′ (corresponding to nt 285–305 and 393–410, respectively, in the mouse Cst6 cDNA sequence; accession ID: MGI:1920970). PCR primers used to distinguish between the Cst6 wild-type (WT) and mutant alleles were the following: forward primer 5′-GGATGGAAGTCCAGACAC-3′ for both alleles, reverse primer 5′-AGGCAGAATGCGAGCAG-3′ for the WT Cst6 allele; and reverse primer 5′-GAGGCAGAATGCGAGAA-3′ for the mutant Cst6 allele. PCR products were analyzed on ethidium bromide-stained 1.5% agarose gels.
Histopathology and immunofluorescence staining
We collected dorsal skin and eyes from the offspring of both founders in a time course, starting at wk 7 and continuing until 1 yr. Dorsal skin and eye were rinsed in PBS, fixed for 4 h in buffered 4% formalin, and embedded in paraffin wax, from which 7-µm sections were cut and stained with hematoxylin and eosin (H&E) or analyzed by immunofluorescence (11). Primary antibodies that were used in this study were rabbit anti-human/mouse loricrin and rabbit anti-human/mouse filaggrin (all from Covance, Richmond, CA, USA), goat anti-human/mouse Cst6, goat anti-mouse CtsB (both from R&D Systems), rabbit anti-mouse Ki-67, rat anti-mouse CD34, rabbit polyclonal anti-human/mouse eGFP, and chicken anti-human/mouse K15 (all from Abcam, Cambridge, United Kingdom). For immunofluorescence analysis, the following secondary reagents were used: Alexa-Fluor 488 goat anti-rabbit IgG, highly cross-absorbed, and Alexa-Fluor 594 goat anti-rat, goat anti-mouse, and chicken anti-goat IgG, highly cross-absorbed (Thermo Fisher Scientific).
Cell culture and fluorescent quenched activity-based probe labeling of cell lysates
Mouse monocyte macrophages (RAW cells) (12) and mouse skin melanoma cells (B16-F10) (13) were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Thermo Fisher Scientific). Some 2 × 105 cells per condition were harvested, washed with PBS, and resuspended in 10 µl citrate buffer per condition [50 mM citrate buffer (pH 5.5), 5 mM DDT, 0.5% CHAPS, and 0.1% Triton X]. After 15 min on ice and centrifugation at 4°C for 30 min, the supernatants were collected. The lysate was incubated with the following protease inhibitors; E-64, Z-FY (t-BU)DMK (Calbiochem, San Diego, CA, USA), CA-074 (Brunschwig Chemie, Amsterdam, The Netherlands), N-ethylmaleimide (Sigma-Aldrich), DCG-04 (kindly provided to M. V. by Prof. Hermen S. Overkleeft, Leiden University, Leiden, The Netherlands), and mouse Cst6 (R&D Systems) (20× DMSO in citrate buffer) for 1 h at 37°C, followed by incubation with 1 µM of the quenched activity-based probe (qABP) BMV109 (20× in DMSO) for 1 h at 37°C (14). In the heat inactivated control, the lysate was heated for 5 min at 95°C before the addition of BMV109. SDS-sample buffer (4×) was added, and the protein was denatured for 3 min at 95°C and resolved by SDS-PAGE (15%), and the labeled proteases were visualized by scanning the gel with a Typhoon Trio imager (GE Healthcare, Little Chalfont, United Kingdom). Labeling intensities were quantified with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Fluorometric enzyme assay
Protease inhibitor activity of recombinant mouse Cst6 (R&D Systems) against recombinant mouse CtsB (R&D Systems) was determined by measuring the hydrolysis of the fluorescent Z-Leu-Arg-AMC substrate (R&D Systems) (4). Assays were performed in a 25 mM 2-(N-morpholino)ethanesulfonic acid buffer (pH 5.0). The CtsB inhibitor CA-074 (Brunschwig Chemie) was used as the control.
RESULTS
Generation of Tg(INV-Cst6-IRES-eGFP) transgenic mice
To generate transgenic, epidermis-specific Cst6-expressing mice, we first constructed a vector in which the complete mouse Cst6 coding sequence was placed under the control of the INV promoter and in front of an IRES-eGFP cassette, to facilitate detection of transgene expression (Supplemental Fig. 1A). INV is a structural epidermal protein that is exclusively expressed in the suprabasal layers of stratified squamous epithelium (15). To validate the construct, we transiently transfected human keratinocytes, stimulated their differentiation, and analyzed the relative number of eGFP+ cells by flow cytometry and fluorescence microscopy (Supplemental Fig. 1B, C). These pBS-INV-Cst6-IRES-eGFP–transfected keratinocytes also expressed mouse Cst6, as determined by RT-PCR amplification of Cst6 coding sequences (Supplemental Fig. 1D) and ELISA detection of mouse Cst6 in the keratinocyte culture medium (Supplemental Fig. 1E).
Transgenic founder mice were created by microinjection of the transgene into C57BL/6J mouse zygotes. Genotyping of resulting litters for endogenous and transgenic Cst6 sequences was performed by PCR using exon-spanning primers (not shown). Overexpression of Cst6 protein in the epidermis of the founder mice did not cause an obvious skin phenotype (not shown). The resulting transgenic, epidermis-specific, Cst6-expressing founder was subsequently cross-bred with Cst6+/− mice (breeding schedule is depicted in Supplemental Fig. 2). The obtained Tg(INV-Cst6-IRES-eGFP)Cst6ichq/ichq rescued mice were viable, but developed hair and eye abnormalities as they matured (Fig. 1A). Further breeding with these transgenic rescued founders, from now on referred to as Tg(INV-Cst6)Cst6ichq/ichq, demonstrated stable germline transmission of the transgene in the resulting offspring. The mice all showed keratitis and a severe fur phenotype, with complete loss of the HFs at ∼16 wk of age. We noted with interest that the Tg(INV-Cst6)Cst6ichq/ichq mice developed eye lashes and whiskers.
Figure 1.
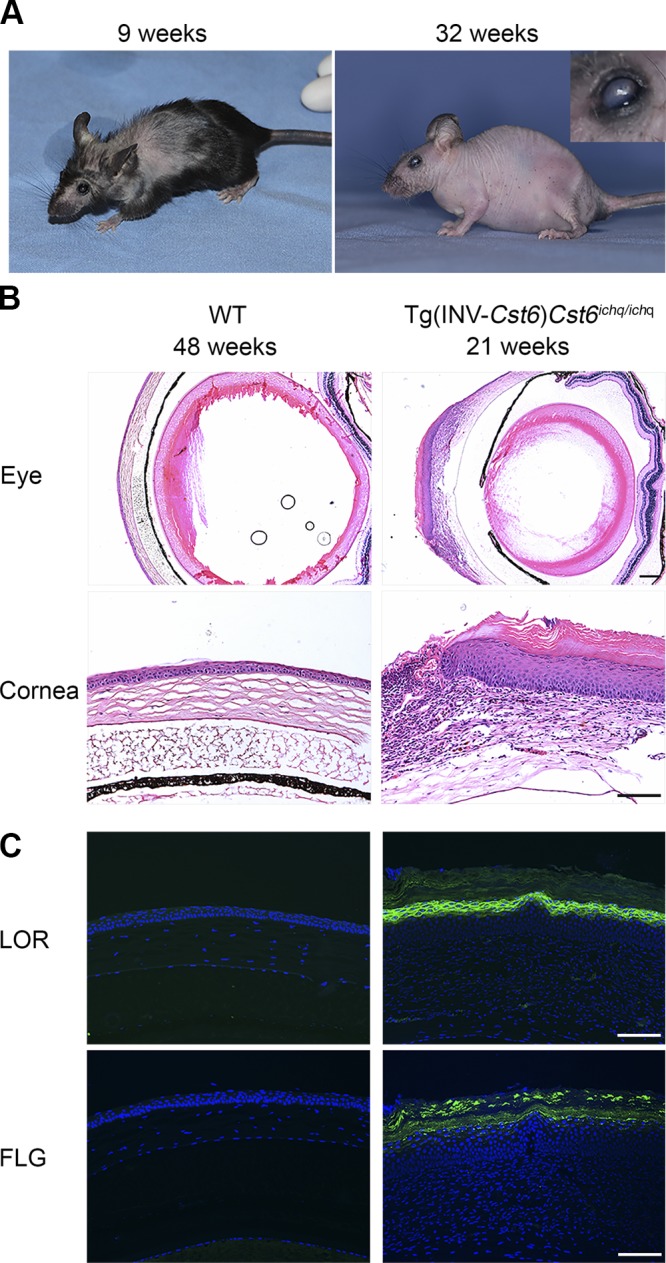
Phenotype of rescued Tg(INV-Cst6)Cst6ichq/ichq mice. A) Tg(INV-Cst6)Cst6ichq/ichq mice survived and showed periodic hair loss. After 4 mo, the progenies became completely bald. Keratitis and thickening of the cornea were observed in Tg(INV-Cst6)Cst6ichq/ichq mice from 4 to 5 mo. The mice shown are 9 and 32 wk old. Inset: magnified view of an affected eye. B) Keratitis and metaplasia of the corneal epithelium in Tg(INV-Cst6)Cst6ichq/ichq mice. H&E staining of the eye and the cornea in WT and Tg(INV-Cst6)Cst6ichq/ichq mice. C) Immunofluorescence staining for the expression of loricrin (LOR) and filaggrin (FLG) in the cornea. Scale bars, 100 µm.
Keratitis and metaplasia of the corneal epithelium in Tg(INV-Cst6)Cst6ichq/ichq mice
Microscopic analysis of affected eyes in Tg(INV-Cst6)Cst6ichq/ichq mice revealed that from wk 20 onward, the corneal epithelium became thickened, and keratitis was apparent (Fig. 1B), resembling the eye phenotype that we had observed in the Cst6−/−Ctsl+/− mice (8). The latter mice also developed severe keratitis, thickening of the corneal stroma, and squamous metaplasia of the corneal epithelium, a particular phenotype in which the cornea starts to resemble epidermis after about half a year. To investigate whether this was also the case in Tg(INV-Cst6)Cst6ichq/ichq mice, we analyzed expression of the cornified epithelium marker proteins loricrin and filaggrin. As in Cst6−/−Ctsl+/− mice, these proteins were aberrantly expressed in the corneal epithelium of adult Tg(INV-Cst6)Cst6ichq/ichq mice (Fig. 1C).
Disturbed HF biology in Tg(INV-Cst6)Cst6ichq/ichq mice
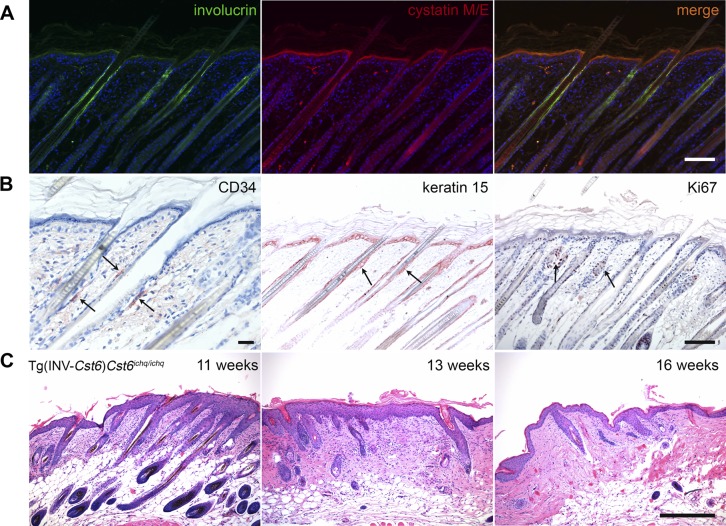
A cyclic phenotype of periodic loss and regrowth of hair was observed in Tg(INV-Cst6)Cst6ichq/ichq mice; however, after 4 mo, these rescued mice remained bald. Immunostaining for Cst6 and INV (Fig. 2A) revealed that, in WT mice, both proteins colocalized in the epidermis and in the proximal part of the HF, whereas Cst6 expression was also observed in lower regions of the HF, including the area around the bulge. Epithelial stem cells were readily detected in the HF bulge area of WT mice upon staining for the stem cell markers CD34 and keratin 15, and for the proliferation marker Ki67 (Fig. 2B). However, we were not able to detect cells that are positive for these markers in the HFs of Tg(INV-Cst6)Cst6ichq/ichq mice. Histologic analysis over a time course revealed that the HFs in Tg(INV-Cst6)Cst6ichq/ichq mice disappeared as early as from 11 wk onward (Fig. 2C). We did not observe an increase in immune cells in the lower region of the HFs.
Figure 2.
Destruction of the HFs in Tg(INV-Cst6)Cst6ichq/ichq mice. A) Immunofluorescence double labeling of INV and Cst6 in the epidermis and the HF of WT mice. Cst6 was expressed in the stratum granulosum and halfway up the HF, including the area around the bulge, whereas INV expression was also seen in the stratum granulosum, but remained only in the proximal part of the HF. B) Immunohistological labeling in WT mice to detect the location of the bulge area (arrows) using the stem cell markers CD34 and keratin 15 and the proliferation marker Ki67. C) H&E staining of Tg(INV-Cst6)Cst6ichq/ichq mice from 11, 13, and 16 wk showed disappearance of the HFs. Scale bar, 100 µm.
Mouse Cst6 targets CtsB in the HF
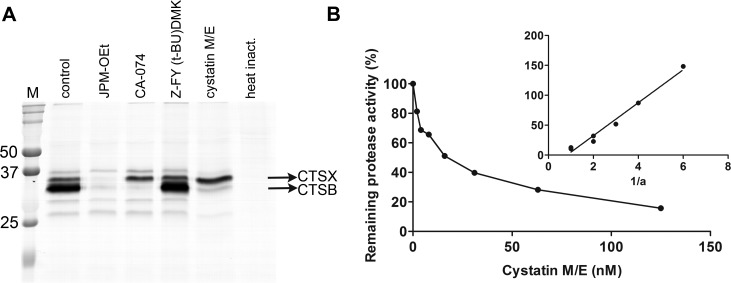
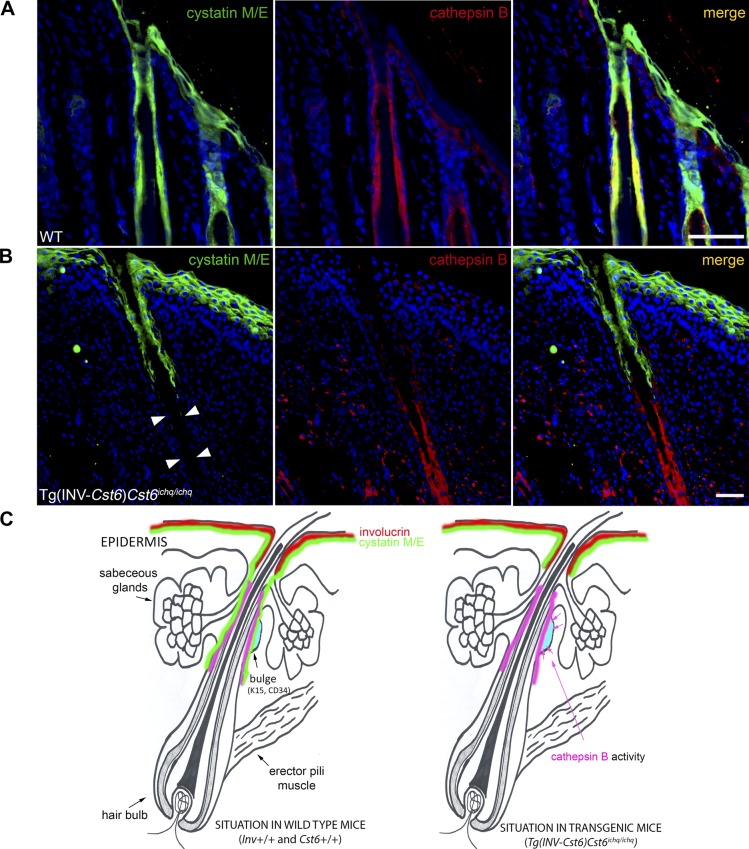
As complete baldness is also observed in Cst6−/−Ctsl−/− mice (8), the hair loss in Tg(INV-Cst6)Cst6ichq/ichq rescued mice cannot be caused by unrestricted CtsL activity. We hypothesized that there must be another target protease for Cst6 that is responsible for destruction of the HFs. To identify such candidate proteases we used a qABP, BMV109 (14), which becomes fluorescent upon covalently binding to active cysteine cathepsins. Cell lysates of RAW cells were preincubated with recombinant mouse Cst6 or other specific protease inhibitors and subsequently incubated with the probe. The resulting proteins were then denatured, resolved on an SDS-PAGE gel and scanned for fluorescence. We observed specific inhibition of mouse CtsB by mouse Cst6, suggesting that CtsB is another physiologic target of Cst6 (Fig. 3A). Similar results were obtained when using B16-F10 mouse melanoma cells (Supplemental Fig. 3). Furthermore, in a fluorometric protease inhibitor activity enzyme assay a strong inhibition of recombinant mouse CtsB by recombinant mouse Cst6 was observed, as reflected by Ki = 0.98 nM (Fig. 3B). Immunolocalization of CtsB in the skin of mice, in a double labeling with Cst6 revealed that CtsB is expressed in the HF, where it colocalizes with Cst6 (Fig. 4A). Together, these data suggest that the absence of Cst6 in the lower region of the HFs of Tg(INV-Cst6)Cst6ichq/ichq rescued mice might lead to uncontrolled CtsB activity (Fig. 4B) and, as a result, destruction of the surrounding tissue and HFs (Fig. 4C).
Figure 3.
Inhibition of mouse CtsB by mouse Cst6. A) BMV109 labels active cysteine cathepsins, such as CTSB and CTSX (14) in RAW cell lysates (murine macrophage cell line). Lane 1, no inhibitor; lane 2, JPM-OEt (pan-cathepsin inhibitor, 50 µM); lane 3, CA-074 (CtsB inhibitor, 10 µM); lane 4, Z-FY (t-BU)DMK (CtsL inhibitor, 10 µM); lane 5, mouse Cst6 (18 µM); and lane 6, heat inactivation. Note that CA-074 and Cst6 inhibited CtsB (lane 3 and 5). M = precision plus protein dual-color marker. B) The Ki for the inhibition of CtsB by Cst6 was determined by measuring the residual enzymatic activity of a fixed concentration of enzyme, incubated with increasing concentrations of the inhibitor. An Easson-Stedman plot (inset) was used to calculate the Ki, according to the equation [I]/1 − a = (Ki/a) + E0, were I is the inhibitor concentration, E0 is the enzyme concentration at time 0, and a is the fractional activity. The plot yielded a straight line with a Ki slope of 0.98 nM.
Figure 4.
CtsB and Cst6 colocalize in the mouse HF. A) Immunofluorescence double staining for Cst6 (green) and CtsB (red) in WT mice revealed that colocalization of both proteins was found in the proximal part of the HF as well as the lower region where the bulge area resides. Cst6 is also expressed in the epidermis, whereas no CtsB is observed. Scale bar, 100 µm. B) Cst6 and CtsB expression in mouse epidermis and HFs of Tg(INV-Cst6)Cst6ichq/ichq mice. No immunofluorescence staining for Cst6 was observed in the lower region of the HF (close to the bulge, arrowheads). Scale bar, 100 µm. C) Schematic presentation of INV, Cst6, and CtsB localization in the HF and epidermis of WT mice and the situation in rescued transgenic Tg(INV-Cst6)Cst6ichq/ichq mice.
DISCUSSION
In this report, we present in vivo evidence that Cst6 is not only a key molecule in a biochemical pathway that controls SC homeostasis but plays an additional role in maintaining HF integrity. Biochemical and histologic analyses suggest that unrestricted CtsB activity is responsible for disturbance of HF biology leading to the baldness phenotype of Tg(INV-Cst6)Cst6ichq/ichq transgenic mice. Our studies advance the understanding of the Cst6-controlled pathways that are critical for skin barrier function (4, 7, 8, 11) and HF maintenance (Fig. 5). Moreover, we provide evidence that CtsB represents a novel Cst6 target.
Figure 5.
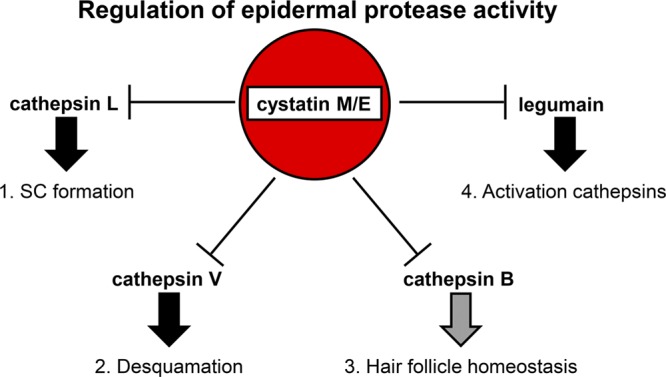
Regulation of epidermal protease activity by CST6. Model of the regulatory role of CST6 in processes that control epidermal cornification, desquamation and HF maintenance. 1) Inhibition of CTSL activity by CST6 is important in the cornification process, as CTSL is the elusive processing and activating enzyme for (TGM)-3. CTSL is also able to process CTSD, which in turn can activate TGM-1. 2) Inhibition of CTSV regulates desquamation, as CTSV is able to degrade desmosomal and corneodesmosomal proteins, such as desmoglein-1, desmocollin-1, and corneodesmosin. As CTSV is expressed only in humans, murine CTSL probably controls the specific functional enzymatic activities of both human CTSL and CTSV. 3) The findings in the present study suggest that inhibition of CtsB by Cst6 protects HF maintenance in mice. 4) Inhibition of human LGMN regulates the processing of (pro)-cathepsins; however mouse LGMN is not inhibited by mouse Cst6.
We have demonstrated, using a Cst6/Ctsl double-knockout approach in mice, that ablation of CtsL could rescue the lethal skin phenotype of Cst6-deficient mice, suggesting that CtsL is responsible for the ichthyosis phenotype and early lethality of these mice (8). Cst6−/−Ctsl+/−, but not Cst6−/−Ctsl−/−, mice developed keratitis and squamous metaplasia of the corneal epithelium. CstB-deficient mice display a similar eye phenotype (16), as well. As both CstB and Cst6 are high-affinity CtsL inhibitors, this finding strongly supports our conclusion that unrestricted CtsL activity is responsible for corneal pathology. In the current study, we observed a similar corneal phenotype (i.e., keratitis and expression of epidermal differentiation markers in the corneal epithelium) in rescued adult Tg(INV-Cst6)Cst6ichq/ichq mice. Apparently, Cst6 is not reexpressed in the cornea by the transgene, a finding corroborated by the absence of eGFP expression in the corneal epithelium of rescued transgenic mice (not shown), subsequently leading to an excess of CtsL activity. This observation is remarkable, as some reports suggest that INV is expressed in the normal human (17) and mouse (18) corneal epithelium. However, we and others could not detect INV protein expression in cornea of adult WT mice (8, 19). Loss of eyelashes, in itself, can lead to corneal irritation and is associated with ocular diseases (20). As the Tg(INV-Cst6)Cst6ichq/ichq mice develop normal eye lashes, the observed keratitis is probably not a secondary phenotype originating from eyelash alopecia.
The observed hair loss in adult Tg(INV-Cst6)Cst6ichq/ichq transgenic mice cannot be the consequence of unrestricted CtsL activity, as the Cst6−/−Ctsl−/− mouse is also bald (8). Thus, other protease activities are likely to be responsible for this phenotype. We used a qABP to identify candidate proteases. qABPs are small-molecule tools that allow for the monitoring and profiling of protease activities in complex biologic systems (14, 21, 22). They form activity-dependent covalent bonds with protease active site nucleophiles and, in this manner, can provide a readout of the levels of active protease in a cell, tissue or even whole organism (22). Protease abundance does often not directly correlate with activity. Therefore, the use of these qABPs is of great interest and provides a novel method of identifying new target proteases.
In WT mice, Cst6 and INV colocalize in the epidermis and the proximal part of the HF, whereas Cst6 expression is also present in lower regions of the HF. As INV protein expression could not be detected in this region in WT mice, this finding means that in Tg(INV-Cst6)Cst6ichq/ichq transgenic mice, Cst6 is not reexpressed. We hypothesize that in this part of the HF, Cst6 has another function or target. A plausible candidate target could be CtsB, for which human Cst6 showed a moderate inhibition (2, 23, 24). CtsB is a ubiquitously expressed cysteine protease, belonging to the papain family, similar to CtsL (25). It has been shown to act as a dominant protease in cellular apoptosis and senescence and to play a role in lysosomal cathepsin–dependent apoptosis pathways (26, 27). Whereas CtsL-deficient mice are known for their prominent periodic hair loss phenotype, manifesting together with epidermal hyperplasia, acanthosis, and hyperkeratosis (28), CtsB-deficient mice do not display any obvious skin or fur phenotype (29). In a recent study, the murine skin proteome and degradome of CtsB and CtsL-deficient mice were compared (30). Protein abundances of >1300 proteins were analyzed in skin specimens from WT and CtsB- and CtsL-deficient mice. Several proteins that are associated with skin phenotypes in knockout mice were more abundant in CtsL-deficient mice: CtsD, Cst6, periostin, and caspase 14. Also, CstB was significantly more abundant. The more pronounced impact of CtsL ablation on proteins important for mouse skin biology corresponds to the fact that CtsL-deficient mice show an overt skin phenotype, while CtsB-deficient mice do not. In comparison, our Tg(INV-Cst6)Cst6ichq/ichq transgenic mice model probably results in uncontrolled CtsB expression in the HFs. Altered CtsB activity correlates with several pathologic conditions in humans, including bone and joint disorders, pancreatitis, Alzheimer’s disease, and metastatic cancers (31–35). Comparison of CtsB deficiency and overexpression in 4 unrelated genetic mouse models of de novo tumorigenesis consistently describe CtsB as a tumor-promoting protease and thereby, a potential therapeutic target (36).
We speculate that unrestricted CtsB activity leads to disturbance of HF biology, which might explain the observed baldness in Tg(INV-Cst6)Cst6ichq/ichq transgenic mice. It appears that both CtsB and CtsL are essential for HF maintenance. Both proteases are expressed in the external root sheath of rat HFs (37). CTSB and CTSL are also present in human bulge cells and outer root sheath cells, as identified by cDNA microarrays (38). Both proteases can be regulated by CST6, but also by other cathepsins. The major, general extracellular cysteine protease inhibitor CSTC is a high-affinity inhibitor of CTSB (39), whereas CSTA is a moderate CTSB inhibitor (40). CSTA, -B, -C, and -F are all high-affinity CTSL inhibitors (4). Although CSTA, C, and M/E are all present in outer root sheath and HF bulge cells, only CST6 is significantly overrepresented in this area (7-fold change) (38). Therefore, CST6 is well suited to play an important role in HF maintenance, likely by controlling CTSB activity.
The present study further expands our understanding of the role of Cst6 in complex proteolytic pathways that maintain skin barrier function and HF morphogenesis. Based on the phenotypes of the knockout (Cst6−/− and Cst6−/−Ctsl−/−) and rescued (Tg(INV-Cst6)Cst6ichq/ichq) transgenic mice, Cst6 and the other components of proteolytic pathways in skin and HFs represent candidate genes for human disorders of cornification and HF integrity. Whether CST6 and its target proteases are involved in various congenital or acquired forms of hair loss in humans, like different forms of alopecia (41), remains to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Daphne Reijnen (Central Animal Laboratory, Radboud University Nijmegen Medical Center) for assistance with the animal experiments. This work was supported by Earth and Life Sciences (ALW) Grant 821.02.013 (to J.S. and P.L.J.M.Z.) from the Netherlands Organization for Health Research and Development (ZonMW), and Deutsche Forschungsgemeinschaft SFB 850 Project B7 (to T.R.). The authors declare no conflicts of interest.
Glossary
- CST6
cystatin M/E
- Cts
cathepsin
- eGFP
enhanced green fluorescent protein
- H&E
hematoxylin and eosin
- HF
hair follicle
- INV
involucrin
- IRES
internal ribosomal entry site
- LGMN
legumain
- qABP
quenched activity-based probe
- SC
stratum corneum
- TGM
transglutaminase
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
W. J. A. J. Hendriks, J. Schalkwijk, and P. L. J. M. Zeeuwen designed the research; M. A. W. Oortveld, I. M. J. J. van Vlijmen-Willems, F. F. J. Kersten, T. Cheng, P. E. J. van Erp, and P. L. J. M. Zeeuwen performed the experiments; M. A. W. Oortveld, J. Schalkwijk, and P. L. J. M. Zeeuwen wrote the paper; M. Verdoes provided reagents and analytic tools for activity-based probe assays; and S. Verbeek and T. Reinheckel provided tools and advice for the generation of the transgenic mouse model.
REFERENCES
- 1.Cheng T., van Vlijmen-Willems I. M. J. J., Hitomi K., Pasch M. C., van Erp P. E. J., Schalkwijk J., Zeeuwen P. L. J. M. (2009) Colocalization of cystatin M/E and its target proteases suggests a role in terminal differentiation of human hair follicle and nail. J. Invest. Dermatol. 129, 1232–1242 [DOI] [PubMed] [Google Scholar]
- 2.Zeeuwen P. L. J. M., Van Vlijmen-Willems I. M. J. J., Jansen B. J. H., Sotiropoulou G., Curfs J. H., Meis J. F., Janssen J. J., Van Ruissen F., Schalkwijk J. (2001) Cystatin M/E expression is restricted to differentiated epidermal keratinocytes and sweat glands: a new skin-specific proteinase inhibitor that is a target for cross-linking by transglutaminase. J. Invest. Dermatol. 116, 693–701 [DOI] [PubMed] [Google Scholar]
- 3.Zeeuwen P. L. J. M., Cheng T., Schalkwijk J. (2009) The biology of cystatin M/E and its cognate target proteases. J. Invest. Dermatol. 129, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 4.Cheng T., Hitomi K., van Vlijmen-Willems I. M. J. J., de Jongh G. J., Yamamoto K., Nishi K., Watts C., Reinheckel T., Schalkwijk J., Zeeuwen P. L. J. M. (2006) Cystatin M/E is a high affinity inhibitor of cathepsin V and cathepsin L by a reactive site that is distinct from the legumain-binding site: a novel clue for the role of cystatin M/E in epidermal cornification. J. Biol. Chem. 281, 15893–15899 [DOI] [PubMed] [Google Scholar]
- 5.Sundberg J. P., Boggess D., Hogan M. E., Sundberg B. A., Rourk M. H., Harris B., Johnson K., Dunstan R. W., Davisson M. T. (1997) Harlequin ichthyosis (ichq): a juvenile lethal mouse mutation with ichthyosiform dermatitis. Am. J. Pathol. 151, 293–310 [PMC free article] [PubMed] [Google Scholar]
- 6.Zeeuwen P. L. J. M., van Vlijmen-Willems I. M. J. J., Hendriks W., Merkx G. F. M., Schalkwijk J. (2002) A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet. 11, 2867–2875 [DOI] [PubMed] [Google Scholar]
- 7.Zeeuwen P. L. J. M., van Vlijmen-Willems I. M. J. J., Olthuis D., Johansen H. T., Hitomi K., Hara-Nishimura I., Powers J. C., James K. E., op den Camp H. J., Lemmens R., Schalkwijk J. (2004) Evidence that unrestricted legumain activity is involved in disturbed epidermal cornification in cystatin M/E deficient mice. Hum. Mol. Genet. 13, 1069–1079 [DOI] [PubMed] [Google Scholar]
- 8.Zeeuwen P. L. J. M., van Vlijmen-Willems I. M. J. J., Cheng T., Rodijk-Olthuis D., Hitomi K., Hara-Nishimura I., John S., Smyth N., Reinheckel T., Hendriks W. J. A. J., Schalkwijk J. (2010) The cystatin M/E-cathepsin L balance is essential for tissue homeostasis in epidermis, hair follicles, and cornea. FASEB J. 24, 3744–3755 [DOI] [PubMed] [Google Scholar]
- 9.Van Ruissen F., de Jongh G. J., Zeeuwen P. L. J. M., Van Erp P. E. J., Madsen P., Schalkwijk J. (1996) Induction of normal and psoriatic phenotypes in submerged keratinocyte cultures. J. Cell. Physiol. 168, 442–452 [DOI] [PubMed] [Google Scholar]
- 10.Behringer R., Gertsenstein M., Nagy K. V., Nagy A. (2014) Manipulating the Mouse Embryo: A Laboratory Manual, 4th ed., p. 298, Cold Spring Harbor Laboratory Press, Stony Brook, NY, USA [Google Scholar]
- 11.Zeeuwen P. L. J. M., Ishida-Yamamoto A., van Vlijmen-Willems I. M. J. J., Cheng T., Bergers M., Iizuka H., Schalkwijk J. (2007) Colocalization of cystatin M/E and cathepsin V in lamellar granules and corneodesmosomes suggests a functional role in epidermal differentiation. J. Invest. Dermatol. 127, 120–128 [DOI] [PubMed] [Google Scholar]
- 12.Ralph P., Nakoinz I. (1977) Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J. Immunol. 119, 950–954 [PubMed] [Google Scholar]
- 13.Fidler I. J. (1975) Biological behavior of malignant melanoma cells correlated to their survival in vivo. Cancer Res. 35, 218–224 [PubMed] [Google Scholar]
- 14.Verdoes M., Oresic Bender K., Segal E., van der Linden W. A., Syed S., Withana N. P., Sanman L. E., Bogyo M. (2013) Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J. Am. Chem. Soc. 135, 14726–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice R. H., Green H. (1979) Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: activation of the cross-linking by calcium ions. Cell 18, 681–694 [DOI] [PubMed] [Google Scholar]
- 16.Pennacchio L. A., Bouley D. M., Higgins K. M., Scott M. P., Noebels J. L., Myers R. M. (1998) Progressive ataxia, myoclonic epilepsy and cerebellar apoptosis in cystatin B-deficient mice. Nat. Genet. 20, 251–258 [DOI] [PubMed] [Google Scholar]
- 17.Tong L., Corrales R. M., Chen Z., Villarreal A. L., De Paiva C. S., Beuerman R., Li D. -Q., Pflugfelder S. C. (2006) Expression and regulation of cornified envelope proteins in human corneal epithelium. Invest. Ophthalmol. Vis. Sci. 47, 1938–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y., Senba E., Shirai K., Ueyama T., Reinach P., Saika S. (2008) Perturbed intraepithelial differentiation of corneal epithelium in c-Fos-null mice. Jpn. J. Ophthalmol. 52, 1–7 [DOI] [PubMed] [Google Scholar]
- 19.De Paiva C. S., Pangelinan S. B., Chang E., Yoon K. C., Farley W. J., Li D. Q., Pflugfelder S. C. (2009) Essential role for c-Jun N-terminal kinase 2 in corneal epithelial response to desiccating stress. Arch. Ophthalmol. 127, 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Andrade F. A., Giavedoni P., Keller J., Sainz-de-la-Maza M. T., Ferrando J. (2014) Ocular findings in patients with alopecia areata: role of ultra-wide-field retinal imaging. Immunol. Res. 60, 356–360 [DOI] [PubMed] [Google Scholar]
- 21.Blum G., von Degenfeld G., Merchant M. J., Blau H. M., Bogyo M. (2007) Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol. 3, 668–677 [DOI] [PubMed] [Google Scholar]
- 22.Edgington-Mitchell L. E., Bogyo M., Verdoes M. (2017) Live cell imaging and profiling of cysteine cathepsin activity using a quenched activity-based probe. Methods Mol. Biol. 1491, 145–159 [DOI] [PubMed] [Google Scholar]
- 23.Ni J., Abrahamson M., Zhang M., Fernandez M. A., Grubb A., Su J., Yu G. -L., Li Y., Parmelee D., Xing L., Coleman T. A., Gentz S., Thotakura R., Nguyen N., Hesselberg M., Gentz R. (1997) Cystatin E is a novel human cysteine proteinase inhibitor with structural resemblance to family 2 cystatins. J. Biol. Chem. 272, 10853–10858 [DOI] [PubMed] [Google Scholar]
- 24.Sotiropoulou G., Anisowicz A., Sager R. (1997) Identification, cloning, and characterization of cystatin M, a novel cysteine proteinase inhibitor, down-regulated in breast cancer. J. Biol. Chem. 272, 903–910 [DOI] [PubMed] [Google Scholar]
- 25.Turk V., Turk B., Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foghsgaard L., Wissing D., Mauch D., Lademann U., Bastholm L., Boes M., Elling F., Leist M., Jäättelä M. (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turk B., Stoka V., Rozman-Pungercar J., Cirman T., Droga-Mazovec G., Oresić K., Turk V. (2002) Apoptotic pathways: involvement of lysosomal proteases. Biol. Chem. 383, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 28.Roth W., Deussing J., Botchkarev V. A., Pauly-Evers M., Saftig P., Hafner A., Schmidt P., Schmahl W., Scherer J., Anton-Lamprecht I., Von Figura K., Paus R., Peters C. (2000) Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and perturbation of hair follicle cycling. FASEB J. 14, 2075–2086 [DOI] [PubMed] [Google Scholar]
- 29.Deussing J., Roth W., Saftig P., Peters C., Ploegh H. L., Villadangos J. A. (1998) Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc. Natl. Acad. Sci. USA 95, 4516–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tholen S., Biniossek M. L., Gansz M., Gomez-Auli A., Bengsch F., Noel A., Kizhakkedathu J. N., Boerries M., Busch H., Reinheckel T., Schilling O. (2013) Deletion of cysteine cathepsins B or L yields differential impacts on murine skin proteome and degradome. Mol. Cell. Proteomics 12, 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halangk W., Lerch M. M., Brandt-Nedelev B., Roth W., Ruthenbuerger M., Reinheckel T., Domschke W., Lippert H., Peters C., Deussing J. (2000) Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J. Clin. Invest. 106, 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berardi S., Lang A., Kostoulas G., Hörler D., Vilei E. M., Baici A. (2001) Alternative messenger RNA splicing and enzyme forms of cathepsin B in human osteoarthritic cartilage and cultured chondrocytes. Arthritis Rheum. 44, 1819–1831 [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto Y., Kakegawa H., Narita Y., Hachiya Y., Hayakawa T., Kos J., Turk V., Katunuma N. (2001) Significance of cathepsin B accumulation in synovial fluid of rheumatoid arthritis. Biochem. Biophys. Res. Commun. 283, 334–339 [DOI] [PubMed] [Google Scholar]
- 34.Hook V., Toneff T., Bogyo M., Greenbaum D., Medzihradszky K. F., Neveu J., Lane W., Hook G., Reisine T. (2005) Inhibition of cathepsin B reduces β-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate β-secretase of Alzheimer’s disease. Biol. Chem. 386, 931–940 [DOI] [PubMed] [Google Scholar]
- 35.Kos J., Mitrović A., Mirković B. (2014) The current stage of cathepsin B inhibitors as potential anticancer agents. Future Med. Chem. 6, 1355–1371 [DOI] [PubMed] [Google Scholar]
- 36.Reinheckel T., Peters C., Krüger A., Turk B., Vasiljeva O. (2012) Differential impact of cysteine cathepsins on genetic mouse models of de novo carcinogenesis: cathepsin B as emerging therapeutic target. Front. Pharmacol. 3, 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morioka K., Sato-Kusubata K., Kawashima S., Ueno T., Kominami E., Sakuraba H., Ihara S. (2001) Localization of cathepsins B, D, L, LAMP-1 and calpain in developing hair follicles. Acta Histochem. Cytochem. 34, 337–347 [Google Scholar]
- 38.Ohyama M., Terunuma A., Tock C. L., Radonovich M. F., Pise-Masison C. A., Hopping S. B., Brady J. N., Udey M. C., Vogel J. C. (2006) Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J. Clin. Invest. 116, 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett A. J., Rawlings N. D., Woessner J. F. (1998) Handbook of Proteolytic Enzymes, Academic Press, Amsterdam [Google Scholar]
- 40.Pavlova A., Krupa J. C., Mort J. S., Abrahamson M., Björk I. (2000) Cystatin inhibition of cathepsin B requires dislocation of the proteinase occluding loop: demonstration By release of loop anchoring through mutation of his110. FEBS Lett. 487, 156–160 [DOI] [PubMed] [Google Scholar]
- 41.Tong X., Coulombe P. A. (2003) Mouse models of alopecia: identifying structural genes that are baldly needed. Trends Mol. Med. 9, 79–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.