Abstract
In China, H9N2 subtype avian influenza outbreak is firstly reported in Guangdong province in 1992. Subsequently, the disease spreads into vast majority regions nationwide and has currently become endemic there. Over vicennial genetic evolution, the viral pathogenicity and transmissibility have showed an increasing trend as year goes by, posing serious threat to poultry industry. In addition, H9N2 has demonstrated significance to public health as it could not only directly infect mankind, but also donate partial or even whole cassette of internal genes to generate novel human-lethal reassortants like H5N1, H7N9, H10N8 and H5N6 viruses. In this review, we mainly focused on the epidemiological dynamics, biological characteristics, molecular phylogeny and vaccine strategy of H9N2 subtype avian influenza virus in China to present an overview of the situation of H9N2 in China.
Introduction
Avian influenza (AI) is initially reported in 1878 in Italy to describe the disease resulted in massive poultry death, which was then termed as “Fowl plague” to distinguish from fowl cholera in 1880 [1]. Although had being identified as filterable virus in 1901, the causative agent is formally designated as influenza A virus until 1955 [1, 2]. Apart from the highly pathogenic forms, less virulent AI viruses have been successively detected in various countries since the mid-1900s that started with the first isolate from chickens in Germany in 1949 [A/chicken/Germany/1949(H10N7)] without being recognized and defined the specific subtype till 1960. As for the H9N2 subtype, with distinguished characteristics to challenge animal industry and even human health among the low pathogenic AI forces, the protovirus is generally considered as the early isolate from turkey flocks in Wisconsin in America in 1966 [A/turkey/Wisconsin/1/1966(H9N2)] [3]. The virus spread becomes more and more extensively at about 1990s, resulting continuous viral circulation in several countries in Asia, Middle East and North Africa [4]. On one hand, H9N2 AI virus could cause damage to birds with direct pathology, coinfection and immunosuppression [5, 6]. On the other hand, H9N2 viruses not only infect mankind directly, but also provide partial or even whole set of internal genes to emerging human-lethal H5N1, H7N9, H10N8 and H5N6 reassortants [7–11], posing a substantial threat to public health. Therefore, the study of H9N2 AI virus deserves great attention.
The etiology of AI
Avian influenza virus affiliates to the genus of type A influenza virus in the Orthomyxoviridae family, packaged with eight negative-sense and single-strand RNA segments in sequence of PB2, PB1, PA, HA, NP, NA, M and NS according to gene length [12]. Each viral gene encodes at least one protein, in which the three polymerase proteins (PB2, PB1 and PA) plus the nucleoprotein (NP) consist the minimal protein unit in forming the functional RNP structure essential for viral transcription and replication. Hemagglutinin (HA) and neuraminidase (NA) are the two major envelope glycoproteins indispensable in mediating influenza A virus to invade host cells and promoting matured newborn virions to disaggregate from cell surface, respectively [13]. Both M and NS genes utilize RNA splicing to synthesize two protein forms of matrix protein (M1) and ion channel protein (M2), nonstructural protein (NS1) and nuclear export protein (NS2), respectively. Through ribosomal frameshift, PB1 and PA genes can also be edited to generate additional PB1-F2 and PA-X proteins, effecting on virus pathogenicity [14, 15]. Based on the antigenic diversity, AI virus can be classified into 16 HA subtypes (H1–H16) and 9 NA subtypes (N1–N9), resulting in various subtype combinations. The criteria to discriminate highly pathogenic avian influenza virus (HPAIV) and low pathogenicity avian influenza virus (LPAIV) were defined at the First International Symposium on Avian Influenza in Beltsville in 1981 [1]. HPAIV only restricts to partial proportions of H5 or H7 subtype, whereas LPAIV covers all the remaining viruses. In particular, H9N2 is currently the most widely circulating and damaging LPAIV subtype in the world.
Outbreaks and prevalence of H9N2 in China
Isolation of AI virus in China has been documented since 1970s [16]. During November 1975 to October 1979, several different subtypes of AI viruses had been isolated from imported live poultry (duck, goose, chicken) in Guangdong and Guangxi provinces, of which the most prevalent subtype is H4N6 [17, 18]. In addition, domestic scholars also described type A influenza virus from duck flocks in some meat processing enterprise in Nanjing in 1980 [19]. However, those above mentioned AI viruses were all identified from apparently healthy birds, therefore insufficient to certify the actual existence of disease outbreaks.
Till 1992, Chen et al. isolated the first H9N2 subtype LPAIV strain AID93-1 (once erroneously identified as H9N3 subtype then), also the earliest published report of AI outbreaks in mainland China [20]. During November 1992 to May 1994, a total of 17 chicken farms and two minor poultry farms had suffered from AI outbreaks in regions of Guangdong province [20, 21]. A few years afterwards, several other parts in China intermittently reported sporadic disease outbreaks caused by H9N2 [22–24]. However, a massive H9N2 epizootics occurred from fall to winter in 1998, initially starting from Hebei province and rapidly spreading to majority of poultry raising areas nationwide in only 2 months [25, 26]. According to the statistics, the ratio of chicken flocks subjected to H9N2 subtype AI infection accounted for 93.89% in the period of 1996–2000, thereby demonstrating that H9N2 was the predominant subtype affecting poultry farming from the end of twentieth century to the beginning of twenty first century [27]. Even to this day, H9N2 is still one of the three primary AI subtypes devastating poultry industry other than the notorious H5N1 and emerging rookie of H7N9.
Theoretically, emerging diseases could possibly be effectively controlled by a stamping-out policy before disseminating into vast areas [28]. However, the optimal eradication opportunity for H9N2 through timely culling of infected poultry was missed during 1992–1998 in China, as the disease has remarkably spread into large regions especially since 1998 and the vaccination strategy has been extensively executed since then [29]. Presently, H9N2 has become stably established in chicken flocks to acquire the endemicity in vast majority of China, accompanied with the substantial implementation of vaccination programs [5]. Moreover, the virus is yet prevalent in wild birds, live poultry markets, backyard flocks and environment [30, 31]. Generally, the inherited complex breeding and trading patterns of poultry industry contributed critically to the current epidemiological situation of H9N2 in China. On one hand, traditional small-scale and backyard-level raisings such as free ranging and mixed ranging still occupy certain ratio in poultry production nationwide, while their biosecurity condition and vaccination coverage are relatively unsatisfactory as compared with typical intensive operations. On the other hand, live poultry markets (LPMs) as a distinctive manifestation of the consumption style that freshly-killed poultry meat is much more preferred rather than chilled or frozen meat, has provided a tremendous gene pool of avian influenza viruses which is evidenced by the continued high virus detection rate including multiple HA/NA subtypes [32]. It is worth noting that interventions involving implementation of one or two rest days per month in the wholesale and retail LPMs could significantly reduce the H9N2 isolation rates [33]. As China is still located on the important flyways for migration, the huge amount of domestic waterfowls which frequently contact the ecointerface with wild waterfowls when sharing common water or makeshift inhabitance also facilitated the persistence and evolution of H9N2 viruses in environment by means like inter-transmission and gene reassortment between birds [30]. Ecologically, at least those above mentioned intricate factors jointly shaped the enzootic status of H9N2 in China.
Genetic evolution of H9N2
H9N2 subtype AI virus is extensively distributed worldwide, generally divided into two major lineages of North-American lineage and Eurasian lineage. Specifically, the Eurasian lineage further blooms into various virus clusters, as represented by A/chicken/Beijing/1/1994(BJ/94-like) or A/duck/Hong Kong/Y280/1997(Y280-like), A/quail/Hong Kong/G1/1997(G1-like), A/duck/Hong Kong/Y439/1997(Y439-like), A/chicken/Shanghai/F/1998(F/98-like) and so on [34–36]. Comparing with the H9N2 viruses in Central Asia and the Middle East, Chinese isolates clustered independently as referred from the phylogenetic trees of HA and NA genes [36]. In China, G1-like circulated mainly in quails is of geography superiority in southern regions, whereas BJ/94-like and F/98-like prevailed in chicken flocks are regnant in northern and eastern areas, respectively [26, 35].
HA phylogenetic clades
To further systematically understand the evolutionary dynamics of H9N2 subtype AI virus globally, four stem evolutionary clades of h9.1–h9.4 have been designated by Jiang et al. to map the HA gene phylogeny through comparing more than 1000 HA sequences retrieved from GenBank, as referred to the nomenclature of the Asian H5N1 HPAIV defined by the WHO/OIE/FAO H5N1 working group [37, 38]. Particularly, h9.1 and h9.2 just corresponded to early North-American isolates in 1966 and the nineties, respectively. H9.3 covered the widest temporal span including Asia, Europe, Africa, Pacific and North America, so did expand the longest spatial range from 1976 until now. The most vast clade h9.4 included two subclades of h9.4.1 and h9.4.2, which coordinated to the G1-like (h9.4.1.1) and Y280-like (h9.4.2.4) H9N2 viruses prevailing in most Asian countries ever since 1994, respectively. In more detail, h9.4.1 contained isolates from Pakistan, India, Iran and Israel, whereas h9.4.2 accommodated exclusively Chinese strains. Chronologically, domestic H9N2 viruses before 2007 generally belonged to clades h9.4.2.1–h9.4.2.4, in which h9.4.2.1 equaled to the above mentioned F/98-like viruses. Thereafter, h9.4.2.5 represented by A/chicken/Guangxi/55/2005(H9N2) has become predominant step by step, whilst h9.4.2.6 distinguished by A/chicken/Guangdong/FZH/2011(H9N2) mainly in southern China has also acquired establishment and tended to spread readily across the country from about 2010. Hence, currently, h9.4.2.5 and h9.4.2.6 have co-circulated in China, while of which the former H9N2 viruses are yet superior over the latter ones.
Genotypic diversity
Owing to the segmented nature of AI virus genome, when two or more virus strains concurrently infect a single cell, exchange of gene segments would occur among different virus particles via gene reassortment to generate a series of newborn viral descendants inheriting parental components. Certainly, H9N2 subtype AI virus is also without exception that distinct virus clusters could reassort with each other or with other AI subtypes to produce various genotypes, which is defined on the basis of the combination of each individual gene phylogenies. For instance, virus harbored all the gene constellation from BJ/94-like is designated as genotype A, variant of three polymerase genes and NP gene from F/98-like while the remaining four genes from BJ/94-like is assigned for genotype H. Thus far, H9N2 subtype AI virus in China has evolved into diversified clusters and genotypes (A–W), showing clear spatio-temporal divergence (Table 1; Figure 1) [7, 26, 29, 39, 40]. Among the rest, three major genotypes of H9N2 subtype AI virus containing A, H and S, have predominated in chicken flocks during different periods since the nineties [6, 7, 29, 41]. In particular, early genotype A prevailing in the nineties had gradually been replaced by genotype H, evident of better adaptation in poultry and easier reassortment with other AI viruses, after 2000 [6]. However, genotype S with exogenous G1-like PB2 and M genes on the genetic backbone of F/98-like viruses emerged around 2007 and had become increasingly established in chickens afterwards, especially in the Yangtze River Delta region in eastern China [6, 7]. Updated epidemiological studies in more recent years also suggest the supreme of genotype S there [40]. Consistently, the additionally categorized genotype G57 (generally equivalent to genotype S) demonstrated greater infectivity than the other genotypes, and had been dominating ever since 2010 across China to cause severe damages to poultry farming [42].
Table 1.
Genotypes of H9N2 subtype avian influenza viruses in China.
| Genotype | Emerged year | Gene constellation | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | |||
| A | 1994 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | [29] |
| B | 1997 | G1/97 | G1/97 | BJ/94 | BJ/94 | BJ/94 | G9/97 | BJ/94 | BJ/94 | |
| C | 1999 | G1/97 | G1/97 | G1/97 | BJ/94 | BJ/94 | G9/97 | BJ/94 | G1/97 | |
| D | 1999 | G1/97 | G1/97 | G1/97 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | G1/97 | |
| E | 2000 | G1/97 | G1/97 | G1/97 | TY/WI/66 | TY/WI/66 | G9/97 | BJ/94 | BJ/94 | |
| F | 2000 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | G9/97 | BJ/94 | BJ/94 | |
| G | 2000 | G1/97 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | |
| H | 1998 | F/98 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | BJ/94 | BJ/94 | |
| I | 2001 | F/98 | F/98 | F/98 | BJ/94 | F/98 | G9/97 | BJ/94 | BJ/94 | |
| J | 1999 | F/98 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | BJ/94 | d73/76 | [26] |
| K | 2003 | BJ/94 | BJ/94 | Kor/323/96 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | |
| L | 2005 | F/98 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | BJ/94 | Kor/323/96 | |
| M | 1998 | BJ/94 | BJ/94 | F/98 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | BJ/94 | [39] |
| N | 2007 | BJ/94 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | BJ/94 | BJ/94 | |
| O | 2007 | F/98 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | G1/97 | BJ/94 | |
| P | 2008 | F/98 | F/98 | F/98 | BJ/94 | F/98 | G9/97 | G1/97 | BJ/94 | |
| Q | 2008 | F/98 | BJ/94 | Y439/97 | BJ/94 | F/98 | G9/97 | G1/97 | BJ/94 | |
| R | 2007 | F/98 | F/98 | Y439/97 | BJ/94 | F/98 | BJ/94 | G1/97 | BJ/94 | [7] |
| S | 2007 | G1/97 | F/98 | F/98 | BJ/94 | F/98 | BJ/94 | G1/97 | BJ/94 | |
| T | 2008 | F/98 | BJ/94 | F/98 | BJ/94 | F/98 | G9/97 | G1/97 | BJ/94 | |
| U | 2009 | G1/97 | BJ/94 | Y439/97 | BJ/94 | F/98 | G9/97 | G1/97 | BJ/94 | |
| V | 2014 | G1/97 | F/98 | F/98 | BJ/94 | F/98 | G9/97 | G1/97 | BJ/94 | [40] |
| W | 2014 | Wild Waterfowls | F/98 | F/98 | BJ/94 | F/98 | F/98 | G1/97 | BJ/94 | |
Genotypes were defined according to the array mode of the eight gene phylogenies, those which have or had persisted for a long time in China are labeled in bold
BJ/94: A/chicken/Beijing/1/1994(H9N2)-like; G1/97: A/quail/Hong Kong/G1/1997(H9N2)-like; G9/97: A/chicken/Hong Kong/G9/1997(H9N2)-like; Y439/97: A/duck/Hong Kong/Y439/1997(H9N2)-like; TY/WI/66: A/turkey/Wisconsin/1/1966(H9N2)-like; F/98: A/chicken/Shanghai/F/1998(H9N2)-like; Kor/323/96: A/chicken/Korea/38349-p96323/1996(H9N2)-like; d73/76: A/duck/Hong Kong/d73/1976(H6N1)-like
Figure 1.
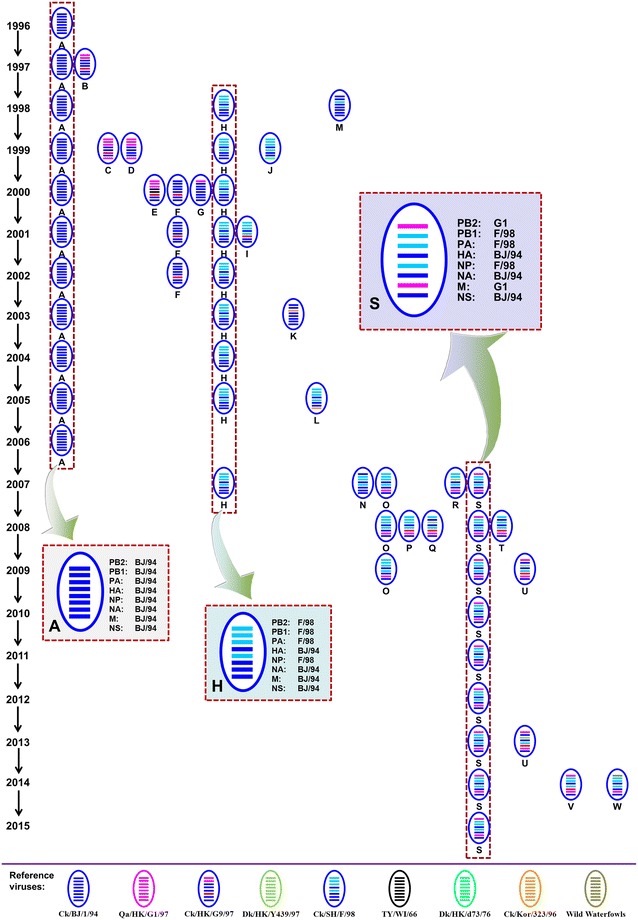
Genotypic diversity of H9N2 subtype avian influenza viruses in China during 1996–2015. The eight horizontal bars in oval (from top to bottom) represent PB2, PB1, PA, HA, NP, NA, M and NS genes, respectively. Each color represents a virus lineage. The resulting genotype designation is depicted below.
Biological property variation of H9N2
The premier isolates of H9N2 just infected turkeys, rarely encroached on chickens, but have gradually adapted to chickens and acquired pathogenicity after years of evolution [1, 3]. Since the initial isolation of H9N2 virus in China, its host range and virulence have become increasingly wider and stronger, respectively [5, 26, 35, 42, 43]. As revealed by a continuous surveillance on H9N2 subtype AI virus in eastern China from 1999 to 2008, most viruses before 2000 were competent to propagate in inoculated chickens but inadequate to be transmissible through respiratory droplets [26]. In contrast, variants after 2001 not only replicated well in vivo but also transmitted efficiently by respiratory droplets in chickens [44]. Comparing with the ones prior to 2010, H9N2 isolates circulating during 2010–2013 showed an obviously higher isolation rate and titers, as well as a longer period of virus shedding especially from cloaca in challenged chickens [42]. It was recently demonstrated that such improved viral fitness was resulted from the substitution of BJ/94-like M gene with the G1-like [45]. Specifically, H9N2 viruses containing G1-like M gene not only exhibited significantly efficient early augment of viral mRNA and vRNA to increase the amount of produced protein and benefit the release of progeny virions, but also conferred extrapulmonary virus spread in chickens [45]. Moreover, characterization of H9N2 viruses ranging from 2009 to 2013 in southern China indicated that natural H9N2 isolates of chicken origin had gradually acquired the preference for human-type α-2,6 sialic acid receptors, and several variants even developed the airborne transmissibility in ferrets [46].
Internal gene cassette reassortment of H9N2
It is acknowledged that the variation mechanism of AI virus mainly includes antigenic drift and genetic shift, with the former featured by point mutation of key amino acids in major immunoprotective proteins whereas the latter resulted from genomic reassortment [47]. As compared with genetic drift, gene reassortment poses a more radical effect on influenza virus by generating totally brand-new viruses with competitive advantage to spread widely such as those causing influenza pandemics in history [48–51]. According to literatures, H9N2 not merely donate partial gene segments but also the whole set of internal genes to reassort with other influenza A viruses [52, 53]. Especially in the past few years, the phenomenon that the six internal genes of H9N2 constituting a relatively stable community to transfer into other emerging reassortants as a whole cassette seems more distinguished (Figure 2). For example, the newly detected chicken H7N7 viruses in Wenzhou city of Zhejiang province, the human-infecting H7N9 and H10N8 viruses initially reported in 2013, and the more recent clade 2.3.4.4 human-lethal H5N6 viruses, were all generated on the basis of complete internal genes from H9N2 subtype AI viruses [9, 10, 54–56]. In addition, H9N2 even dedicated all the other seven gene segments except HA to the clade 7.2 HPAI H5N2 natural reassortants in recent years [57, 58]. Despite diversity, those H9N2 donor viruses all pertain to the unique S genotype prevailing in chicken flocks in China since 2007 [7]. As influenza A virus proved to choose gene segments specifically for package when more than one kind of viruses co-infect the same host cell, whether the intrinsic vRNA–vRNA interaction contributed crucially to the molecular mechanism of this particular internal-gene-cassette re-assortment deserves further exploration [59–61].
Figure 2.
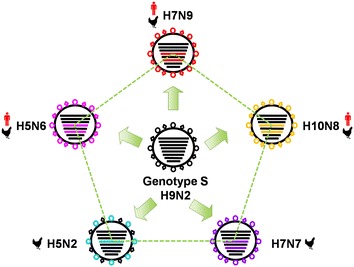
Illustrative scheme of the events induced by reassortment of whole set of internal genes from H9N2 to generate novel influenza A reassortants. The eight horizontal bars in circle (from top to bottom) represent PB2, PB1, PA, HA, NP, NA, M and NS genes, respectively. The ring and prismatic shape on outer surface of circle represent the HA and NA proteins, respectively. Each color represents a specific virus subtype. The black poultry label indicates that the generated influenza A virus challenges birds and the red figure label indicates that the reassortant can also infect human beings.
Vaccine strategy for control of H9N2
Presently, H9N2 subtype AI virus has been widely spread in China, and has established stable lineages in commercial chicken flocks with endemicity [40]. Despite that the mortality caused by H9N2 generally not exceed 20%, it usually leads to respiratory and egg-drop symptom, as well as sever secondary infection of other respiratory diseases, affecting poultry productivity [5, 29]. Therefore, at current stage, vaccination is still one of the principle strategies to control H9N2 AI in China apart from biosecurity.
Conventional whole-virus inactivated vaccines
The majority of commonly used AI vaccines are killed whole-virus vaccines, prepared from formaldehyde inactivation of virus-containing allantoic fluids proliferated via chicken embryos and accompanied with adjuvants, manifesting favorable immune efficacy [62]. Domestically, various H9N2 strains have been used for inactivated vaccine development, wherein the F strain and SS strain respectively belonging to genotype H and genotype A are the two typical representatives [63, 64]. The F strain is a natural reassortant of chicken origin isolating from Shanghai in 1998, with its polymerase genes being replaced with counterpart gene segments from distinct H9N2 clusters in ducks. This F/98-like virus entirety had existed over a long time in chicken flocks in China, and even served as the donor to provide internal genes for further reassortants until recently [65, 66]. As for SS vaccine, it is developed from the seed isolated from Guangdong province in 1994, which is also the first commercial vaccine for control of H9N2 subtype AI in China. However, as the ongoing evolution of H9N2 viruses, vaccination failure due to infection with prevailing antigenic variants evidently challenges the efficacy of the vaccines in China, like that in many other countries such as Iran and Korea [40, 67–72]. Therefore, updated vaccine seed strains based on continuous surveillance data have gradually been preparing and permitting for clinical practice. To simplify the immune procedure to reach an ideal goal of “one injection preventing multiple diseases”, a massive of double or multiple combined vaccines have been designed such as the triplex inactivated vaccines simultaneously against AI (H9 subtype), Newcastle disease and infectious bronchitis [73].
Recombinant and vector virus vaccines
Inactivated whole virus vaccine mainly elicits humoral immune response, deficient in inducing effective mucosal and cellular immunity. Furthermore, it also interferes with immunological surveillance and epidemiological investigation of AI virus under the condition of current technology. Therefore, novel DIVA (differentiating infected and vaccinated animals) vaccines against H9N2 come into being, including recombinant live virus vectored vaccine, subunit vaccine, DNA vaccine, VLPs (virus like particles) vaccine and so on. They could supplement certain shortages of traditional vaccines and are popular for AI vaccine development nowadays. Frequently used live virus vectors contain recombinant fowlpox virus, Newcastle disease virus, Marker’s disease virus, etc [74–76]. Subunit vaccine is generally developed based on the extraction of immunogenic proteins (usually HA) of AI virus, without introducing viral particles. Large amounts of HA protein could be acquired by ligation of HA gene with expressing plasmid vector for amplification, such as in the baculovirus expression system [77]. As for DNA vaccine, the exogenous gene encoding for protective antigen is initially cloned to eukaryotic expressing vector, followed by administrating the constructed DNA plasmids into animals to get expressed in vivo and to stimulate specific humoral and cellular immunity [78]. VLPs are self-assembled hollow protein particles by one or more viral structural proteins, containing no viral genetic materials but resembling integral viruses in appearance. Despite without infectivity, VLPs could still retain immunogenicity to provoke effective immune response and to serve as safe vaccines [79]. So far, a great number of novel genetically engineered AI vaccines have been designed in China, however, many of which are still in the stage of technical research and reserve, immature for clinical usage yet.
Interspecies transmission of H9N2
H9N2 in pigs
Apart from various kinds of poultry, H9N2 subtype AI viruses could also infect pigs, the long considered mixing vessel for mammalian and avian influenza variants. It is revealed by epidemiological survey that H9N2 viruses were isolated from pigs naturally when transported from southern China to Hong Kong for sale, as early as in 1998 [80]. Subsequently during 2001–2008, H9N2 had been detected incessantly in swine herds in several provinces covering Shandong, Fujian, Henan, Jiangxi, Guangdong, Guangxi, Hebei and so on [81–84]. In addition, the identified swine H9N2 isolates exhibited evident genetic and antigenic complexity with diversified genotypes [85]. Serological investigation also manifested the infection of H9N2 viruses in Chinese pig population [86–88].
H9N2 in humans
What’s more noteworthy, H9N2 subtype AI viruses have already acquired the ability to break through species barrier and directly invade human beings without intermediate hosts. The first documentation of human-infecting H9N2 viruses in China traced back to 1998, as described that five H9N2 strains were cultured from laryngopharyngeal mucus of flu-like outpatients and inpatients in southern regions [89]. Further gene sequence analysis indicated that those H9N2 human isolates probably derived from local chicken flocks [90]. In March 1999 in Hong Kong, another two children were confirmed infection with H9N2 viruses, with their genomic sequences highly homologous with the quail strain A/quail/Hong Kong/G1/1997 [91, 92]. Therefore, quails had also been suggest to play important roles in cross-species transmission of H9N2 viruses [93]. Still in 1999, A/chicken/Hong Kong/G9/1997-like H9N2 virus repeatedly isolated from human population in November in southern China [94]. Again in December 2003, Hong Kong reported a second human infection event of H9N2 virus, of which all the eight gene segments were of avian origin and clustered most intimately with those extensively distributed in live poultry market there [95]. Yet recently, laboratory-confirmed human infection of H9N2 virus have continuously been reporting sporadically from WHO, with an apparently higher rate in the last few years and even one fatal case additionally suffering from chronic underlying conditions in 2016 [45]. Besides, quite a number of people prove to have been exposed to H9N2 viruses by serological data, especially those poultry workers [89, 96–98]. Distinct from HPAI H5N1 infection, the overall human symptoms induced by H9N2 are analogous to seasonal flu with rapid recovery and no lethality. However, just such mild infection has made H9N2 easily be negligible in clinical, facilitating to adapt further in the body by reassortment with other human influenza viruses to yield potential variants with high reproductivity and even efficient interpersonal transmissibility.
Conclusion
Although being classified as LPAIV, H9N2 subtype AI virus is extensively distributed in chicken flocks to pose a persistent challenge. In China, traditional raising system of livestock including free-ranging and polyculture, continuously occupies a crucial status yet. It is inevitable for chicken to contact with domestic or wild waterfowl, which harbored large amount of H9N2 viruses. These apparently healthy latent birds could serve as the “Trojan horses” in chicken flocks to cause the circulation of H9N2. Furthermore, the LPMs extending throughout China still played an indispensable role in hosting and disseminating of H9N2 AI virus, as evidenced by significant higher rates of virus isolation than other locations. However, focusing on LPMs management, innovative control measures targeting principally against the emerging avian influenza A(H7N9) virus such as closure of LPMs or other more sustainable but yet effective interventions including washing and cleaning once a day, disinfecting once a week, having rest days once a month and banning live poultry overnight, as well as separating of aquatic and non-aquatic live poultry, would certainly simultaneously reduce the risk of H9N2 contamination at source and deserve high priority in implementation. On account of incessant viral mutation and reassortment, natural variants with increased pathogenicity have been emerging periodically. Even though vaccination remains one of the primary strategies to control H9N2 subtype AI in China, the majority of vaccine recipients are actually still under siege of wild-type variants. Therefore, disease outbreaks would still occur in vaccinated flocks in case of descended protection level or different kinds of immune failure. It is more prior important to establish favorable biosecurity management and take all practicable measures to control infection source, preventing virulent variants from intruding the poultry flocks. On the other hand, the human-infecting events of H9N2 AI virus deserve to be treated scientifically and rationally. Once animal influenza is controlled, should the risk of emerging human pandemic influenza be decreased to minimum level.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MG and LX drafted and revised the manuscript; XW helped in revision; XL designed the structure of the review. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31702245), the Jiangsu Provincial Natural Science Fund for Excellent Young Scholars (BK20170073), the National Natural Science Foundation of China (31772755), the National Key Research and Development Program of China (2017YFD0500101 and 2016YFD0500202), the Special Financial Grant from the China Postdoctoral Science Foundation (2017T100410), the Earmarked Fund for Modern Agro-Industry Technology Research System (nycytx-41-G07) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AI
avian influenza
- PB2
polymerase basic protein 2
- PB1
polymerase basic protein 1
- PA
polymerase acidic protein
- HA
hemagglutinin
- NP
nucleoprotein
- NA
neuraminidase
- M
matrix protein
- NS
nonstructural protein
- HPAIV
highly pathogenic avian influenza virus
- LPAIV
low pathogenicity avian influenza virus
- LPMs
live poultry markets
- WHO
World Health Organization
- OIE
World Organisation for Animal Health
- FAO
Food and Agriculture Organization of United Nations
- BJ/94
A/chicken/Beijing/1/1994(H9N2)
- F/98
A/chicken/Shanghai/F/1998(H9N2)
- G1
A/quail/Hong Kong/G1/1997(H9N2)
- SA
sialic acid
- DIVA
differentiating infected and vaccinated animals
- VLPs
virus like particles
Footnotes
Min Gu and Lijun Xu contributed equally to this work
Contributor Information
Min Gu, Email: gumin@yzu.edu.cn.
Lijun Xu, Email: xulijun011@126.com.
Xiaoquan Wang, Email: wxq@yzu.edu.cn.
Xiufan Liu, Phone: +86 514 8799 1416, Email: xfliu@yzu.edu.cn.
References
- 1.Lupiani B, Reddy SM. The history of avian influenza. Comp Immunol Microbiol Infect Dis. 2009;32:311–323. doi: 10.1016/j.cimid.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. doi: 10.1016/S0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 3.Homme PJ, Easterday BC. Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin-1966 virus. Avian Dis. 1970;14:66–74. doi: 10.2307/1588557. [DOI] [PubMed] [Google Scholar]
- 4.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, Liu J. H9N2 influenza virus in China: a cause of concern. Protein Cell. 2015;6:18–25. doi: 10.1007/s13238-014-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002) J General Virol. 2008;89:3102–3112. doi: 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 7.Gu M, Chen H, Li Q, Huang J, Zhao M, Gu X, Jiang K, Wang X, Peng D, Liu X. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China. Vet Microbiol. 2014;174:309–315. doi: 10.1016/j.vetmic.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 8.Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. doi: 10.1128/JVI.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen YY, Ke CW, Li Q, Yuan RY, Xiang D, Jia WX, Yu YD, Liu L, Huang C, Qi WB, Sikkema R, Wu J, Koopmans M, Liao M. Novel reassortant avian influenza A(H5N6) viruses in Humans, Guangdong, China, 2015. Emerg Infect Dis. 2016;22:1507–1509. doi: 10.3201/eid2208.160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Li R, Jiang L, Xiong C, Chen Y, Zhao G, Jiang Q. The complexity of human infected AIV H5N6 isolated from China. BMC Infect Dis. 2016;16:600. doi: 10.1186/s12879-016-1932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.RahimiRad S, Alizadeh A, Alizadeh E, Hosseini SM. The avian influenza H9N2 at avian-human interface: a possible risk for the future pandemics. J Res Med Sci. 2016;21:51. doi: 10.4103/1735-1995.187253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 13.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci U S A. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep. 2015;5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamal RP, Kumar A, Davis CT, Tzeng WP, Nguyen T, Donis RO, Katz JM, York IA. Emergence of highly pathogenic avian influenza A(H5N1) virus PB1-F2 variants and their virulence in BALB/c Mice. J Virol. 2015;89:5835–5846. doi: 10.1128/JVI.03137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shortridge KF. Pandemic influenza: a zoonosis? Semin Respir Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- 17.Shortridge KF. Avian influenza A viruses of southern China and Hong Kong: ecological aspects and implications for man. Bull World Health Organ. 1982;60:129–135. [PMC free article] [PubMed] [Google Scholar]
- 18.Shortridge KF, Butterfield WK, Webster RG, Campbell CH. Diversity of influenza A virus subtypes isolated from domestic poultry in Hong Kong. Bull World Health Organ. 1979;57:465–469. [PMC free article] [PubMed] [Google Scholar]
- 19.Han C, Xu W, Du N. Orthomyxoviruses and paramyxoviruses isolated from apparently healthy ducks. J Nanjing Agric Univ. 1982;2:87–100. [Google Scholar]
- 20.Chen B, Zhang Z, Chen W. The study of avian influenza: I. The isolation and preliminary serological identification of avian influenza virus in chicken. Chin J Vet Med. 1994;20:3–5. [Google Scholar]
- 21.Zhang Z, Chen B, Chen W. The study of avian influenza: II. The incidence and serological survey of avian influenza. Chin J Vet Med. 1994;20:6–7. [Google Scholar]
- 22.Tang X, Tian G, Zhao C. Isolation and characterization of prevalent strains of avian influenza viruses in China. Chin J Prev Vet Med. 1998;37:100–102. [Google Scholar]
- 23.Chen F, Xia C. Cloning and analysis of avian influenza nucleoprotein gene from A/Chicken/Beijing/1/96(H9N2) Chin J Prev Vet Med. 1999;167:1–28. [Google Scholar]
- 24.Liu JH, Okazaki K, Shi WM, Wu QM, Mweene AS, Kida H. Phylogenetic analysis of neuraminidase gene of H9N2 influenza viruses prevalent in chickens in China during 1995–2002. Virus Genes. 2003;27:197–202. doi: 10.1023/A:1025736829103. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Liu X, Cheng J, Peng D, Jia L, Huang Y. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996–2001. Avian Dis. 2003;47:116–127. doi: 10.1637/0005-2086(2003)047[0116:PAOTHG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Tang Y, Liu X, Liu W, Zhang X, Liu H, Peng D, Gao S, Wu Y, Zhang L, Lu S, Liu X. A novel genotype H9N2 influenza virus possessing human H5N1 internal genomes has been circulating in poultry in eastern China since 1998. J Virol. 2009;83:8428–8438. doi: 10.1128/JVI.00659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X, Fu C, Feng J (2000) Prevalence and control of subtype H9 of avian influenza. In: Proceedings of the 10th symposium on avian diseases, Hangzhou, September 2000, Chinese Association of Animal Science and Veterinary Medicine, pp 125–128
- 28.Moennig V. Eradication versus vaccination strategies to control infectious diseases–some lessons to be learned from terrestrial animals. Dev Biol. 2005;121:13–19. [PubMed] [Google Scholar]
- 29.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Zhang Z, Chen Z, Zhang Y, Lv Q, An X, Tong Y, Carr MJ, Sun S, Shi W. High genetic diversity and frequent genetic reassortment of avian influenza A (H9N2) viruses along the East Asian–Australian migratory flyway. Infect Genet Evol. 2016;39:325–329. doi: 10.1016/j.meegid.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Chen LJ, Lin XD, Guo WP, Tian JH, Wang W, Ying XH, Wang MR, Yu B, Yang ZQ, Shi M, Holmes EC, Zhang YZ. Diversity and evolution of avian influenza viruses in live poultry markets, free-range poultry and wild wetland birds in China. J General Virol. 2016;97:844–854. doi: 10.1099/jgv.0.000399. [DOI] [PubMed] [Google Scholar]
- 32.Chen LJ, Lin XD, Tian JH, Liao Y, Ying XH, Shao JW, Yu B, Guo JJ, Wang MR, Peng Y, Shi M, Holmes EC, Yang ZQ, Zhang YZ. Diversity, evolution and population dynamics of avian influenza viruses circulating in the live poultry markets in China. Virology. 2017;505:33–41. doi: 10.1016/j.virol.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Peiris JS, Cowling BJ, Wu JT, Feng L, Guan Y, Yu H, Leung GM. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. Lancet Infect Dis. 2016;16:252–258. doi: 10.1016/S1473-3099(15)00502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu JH, Liu XF, Shao WX, Liu YL, Wei DP, Liu HQ. Phylogenetic analysis of eight genes of H9N2 subtype influenza virus: a mainland China strain possessing early isolates’ genes that have been circulating. Virus Genes. 2005;31:163–169. doi: 10.1007/s11262-005-1790-1. [DOI] [PubMed] [Google Scholar]
- 35.Xu KM, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, Webster RG, Chen H, Peiris JS, Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fusaro A, Monne I, Salviato A, Valastro V, Schivo A, Amarin NM, Gonzalez C, Ismail MM, Al-Ankari AR, Al-Blowi MH, Khan OA, Maken Ali AS, Hedayati A, Garcia Garcia J, Ziay GM, Shoushtari A, Al Qahtani KN, Capua I, Holmes EC, Cattoli G. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol. 2011;85:8413–8421. doi: 10.1128/JVI.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Liu S, Hou G, Li J, Zhuang Q, Wang S, Zhang P, Chen J. Chinese and global distribution of H9 subtype avian influenza viruses. PLoS One. 2012;7:e52671. doi: 10.1371/journal.pone.0052671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.WHO/OIE/FAO H5N1 Evolution Working Group Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:e1. doi: 10.3201/eid1407.071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Hu B, Wen X, Cao S, Gavrilov BK, Du Q, Khan MI, Zhang X. Diversified reassortant H9N2 avian influenza viruses in chicken flocks in northern and eastern China. Virus Res. 2010;151:26–32. doi: 10.1016/j.virusres.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Liu YF, Lai HZ, Li L, Liu YP, Zhang WY, Gao R, Huang WK, Luo QF, Gao Y, Luo Q, Xie XY, Xu JH, Chen RA. Endemic variation of H9N2 avian influenza virus in China. Avian Dis. 2016;60:817–825. doi: 10.1637/11452-061616-Reg. [DOI] [PubMed] [Google Scholar]
- 41.Xu KM, Li KS, Smith GJ, Li JW, Tai H, Zhang JX, Webster RG, Peiris JS, Chen H, Guan Y. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol. 2007;81:2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Chen Q, Chen Z. Complete genome sequence of an H9N2 avian influenza virus isolated from egret in Lake Dongting wetland. J Virol. 2012;86:11939. doi: 10.1128/JVI.02042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong L, Wang X, Li Q, Liu D, Chen H, Zhao M, Gu X, He L, Liu X, Gu M, Peng D, Liu X. Molecular mechanism of the airborne transmissibility of H9N2 avian influenza A viruses in chickens. J Virol. 2014;88:9568–9578. doi: 10.1128/JVI.00943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pu J, Sun H, Qu Y, Wang C, Gao W, Zhu J, Sun Y, Bi Y, Huang Y, Chang KC, Cui J, Liu J. M Gene reassortment in H9N2 influenza virus promotes early infection and replication: contribution to rising virus prevalence in chickens in China. J Virol. 2017;91:e02055-16. doi: 10.1128/JVI.02055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog. 2014;10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uyeki TM, Cox NJ. Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med. 2013;368:1862–1864. doi: 10.1056/NEJMp1304661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178:63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antonovics J, Hood ME, Baker CH. Molecular virology: was the 1918 flu avian in origin? Nature. 2006;440:E9. doi: 10.1038/nature04824. [DOI] [PubMed] [Google Scholar]
- 52.Gu M, Liu WB, Cao JP, Cao YZ, Zhang XR, Peng DX, Liu XF. Genome sequencing and genetic analysis of a natural reassortant H5N1 subtype avian influenza virus possessing H9N2 internal genes. Bing Du Xue Bao. 2010;26:298–304. [PubMed] [Google Scholar]
- 53.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 56.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 57.Zhao G, Gu X, Lu X, Pan J, Duan Z, Zhao K, Gu M, Liu Q, He L, Chen J, Ge S, Wang Y, Chen S, Wang X, Peng D, Wan H, Liu X. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. PLoS One. 2012;7:e46183. doi: 10.1371/journal.pone.0046183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Yuan X, Qi L, Zhang Y, Xu H, Yang J, Ai W, Qi W, Liao M, Wang D, Song M, Li F. H9N2 avian influenza virus-derived natural reassortant H5N2 virus in swan containing the hemagglutinin segment from Eurasian H5 avian influenza virus with an in-frame deletion of four basic residues in the polybasic hemagglutinin cleavage site. Infect Genet Evol. 2016;40:17–20. doi: 10.1016/j.meegid.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 59.Octaviani CP, Goto H, Kawaoka Y. Reassortment between seasonal H1N1 and pandemic (H1N1) 2009 influenza viruses is restricted by limited compatibility among polymerase subunits. J Virol. 2011;85:8449–8452. doi: 10.1128/JVI.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JI, Lee I, Park S, Bae JY, Yoo K, Lemey P, Park MS, Song JW, Kee SH, Song KJ. Reassortment compatibility between PB1, PB2, and HA genes of the two influenza B virus lineages in mammalian cells. Sci Rep. 2016;6:27480. doi: 10.1038/srep27480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Essere B, Yver M, Gavazzi C, Terrier O, Isel C, Fournier E, Giroux F, Textoris J, Julien T, Socratous C. Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proc Natl Acad Sci U S A. 2013;110:3840–3848. doi: 10.1073/pnas.1308649110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapczynski DR, Swayne DE. Influenza vaccines for avian species. Curr Top Microbiol Immunol. 2009;333:133–152. doi: 10.1007/978-3-540-92165-3_6. [DOI] [PubMed] [Google Scholar]
- 63.Lu J, Liu X, Shao W, Zhang P, Wei D. Genetic characterization of the entire genome of an H9N2 avian influenza virus A/Chicken/Shanghai/F/98. Wei Sheng Wu Xue Bao. 2003;43:434–441. [PubMed] [Google Scholar]
- 64.Guo X, Liao M, Xin C. Sequence of HA gene of avian influenza A/Chicken/Guangdong/SS/1994 (H9N2) virus. Avian Dis. 2003;47:1118–1121. doi: 10.1637/0005-2086-47.s3.1118. [DOI] [PubMed] [Google Scholar]
- 65.He J, Liu LP, Hou S, Gong L, Wu JB, Hu WF, Wang JJ. Genomic characteristics of 2 strains of influenza A (H9N2) virus isolated from human infection cases in Anhui province. Zhonghua Liu Xing Bing Xue Za Zhi. 2016;37:708–713. doi: 10.3760/cma.j.issn.0254-6450.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010;146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Wei Y, Xu G, Zhang G, Wen C, Anwar F, Wang S, Lemmon G, Wang J, Carter R, Wang M, Sun H, Sun Y, Zhao J, Wu G, Webster RG, Liu J, Pu J. Antigenic evolution of H9N2 chicken influenza viruses isolated in China during 2009–2013 and selection of a candidate vaccine strain with broad cross-reactivity. Vet Microbiol. 2016;182:1–7. doi: 10.1016/j.vetmic.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia J, Cui JQ, He X, Liu YY, Yao KC, Cao SJ, Han XF, Huang Y. Genetic and antigenic evolution of H9N2 subtype avian influenza virus in domestic chickens in southwestern China, 2013–2016. PLoS One. 2017;12:e0171564. doi: 10.1371/journal.pone.0171564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge F, Li X, Ju H, Yang D, Liu J, Qi X, Wang J, Yang X, Qiu Y, Liu P, Zhou J. Genotypic evolution and antigenicity of H9N2 influenza viruses in Shanghai, China. Arch Virol. 2016;161:1437–1445. doi: 10.1007/s00705-016-2767-1. [DOI] [PubMed] [Google Scholar]
- 70.Sun Y, Pu J, Fan L, Sun H, Wang J, Zhang Y, Liu L, Liu J. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Vet Microbiol. 2012;156:193–199. doi: 10.1016/j.vetmic.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Bahari P, Pourbakhsh SA, Shoushtari H, Bahmaninejad MA. Molecular characterization of H9N2 avian influenza viruses isolated from vaccinated broiler chickens in northeast Iran. Trop Animal Health Prod. 2015;47:1195–1201. doi: 10.1007/s11250-015-0848-x. [DOI] [PubMed] [Google Scholar]
- 72.Lee DH, Song CS. H9N2 avian influenza virus in Korea: evolution and vaccination. Clin Exp Vaccine Res. 2013;2:26–33. doi: 10.7774/cevr.2013.2.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin QP, Chen RA, Huang WK, Yan JZ. Immunization program of newcastle disease, infectious bronchitis and avian influenza (H9 Subtype) vaccine, inactivated (Strain La Sota + Strain M41 + Strain SS/94) Chin Anim Husb Vet Med. 2012;39:192–196. [Google Scholar]
- 74.Zhang Z, Chen W, Ma C, Zhao P, Duan L, Zhang F, Sun A, Li Y, Su H, Li S, Cui H, Cui Z. Construction of recombinant Marek’s disease virus (MDV) lacking the meq oncogene and co-expressing AIV-H9N2 HA and NA genes under control of exogenous promoters. J Biotechnol. 2014;181:45–54. doi: 10.1016/j.jbiotec.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 75.Ge J, Tian G, Zeng X, Jiang Y, Chen H, Bua Z. Generation and evaluation of a newcastle disease virus-based H9 avian influenza live vaccine. Avian Dis. 2010;54:294–296. doi: 10.1637/8731-032509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 76.Chen HY, Shang YH, Yao HX, Cui BA, Zhang HY, Wang ZX, Wang YD, Chao AJ, Duan TY. Immune responses of chickens inoculated with a recombinant fowlpox vaccine coexpressing HA of H9N2 avain influenza virus and chicken IL-18. Antivir Res. 2011;91:50–56. doi: 10.1016/j.antiviral.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 77.Lin W, Fan H, Cheng X, Ye Y, Chen X, Ren T, Qi W, Liao M. A baculovirus dual expression system-based vaccine confers complete protection against lethal challenge with H9N2 avian influenza virus in mice. Virol J. 2011;8:273. doi: 10.1186/1743-422X-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan Z, Zhang X, Geng S, Cheng N, Sun L, Liu B, Huang J, Jiao X. Priming with a DNA vaccine delivered by attenuated Salmonella typhimurium and boosting with a killed vaccine confers protection of chickens against infection with the H9 subtype of avian influenza virus. Vaccine. 2009;27:1018–1023. doi: 10.1016/j.vaccine.2008.11.111. [DOI] [PubMed] [Google Scholar]
- 79.Lee DH, Park JK, Lee YN, Song JM, Kang SM, Lee JB, Park SY, Choi IS, Song CS. H9N2 avian influenza virus-like particle vaccine provides protective immunity and a strategy for the differentiation of infected from vaccinated animals. Vaccine. 2011;29:4003–4007. doi: 10.1016/j.vaccine.2011.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Yu K, Yang H, Xin X, Chen J, Zhao P, Bi Y, Chen H. Isolation and characterization of H5N1 and H9N2 influenza viruses from pigs in China. Chin J Prev Vet Med. 2004;26:1–6. [Google Scholar]
- 82.Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect. 2004;6:919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 83.Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 84.Zhang R, Cui H, Xu M, Li K, Chen H, Wang C, Wei D, Li C, Xu T. Molecular characterization and pathogenicity of swine influenza H9N2 subtype virus A/swine/HeBei/012/2008/(H9N2) Acta Virol. 2011;55:219–226. doi: 10.4149/av_2011_03_219. [DOI] [PubMed] [Google Scholar]
- 85.Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol. 2011;149:254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 86.Yuan Z, Zhu W, Chen Y, Zhou P, Cao Z, Xie J, Zhang C, Ke C, Qi W, Su S, et al. Serological surveillance of H5 and H9 avian influenza A viral infections among pigs in Southern China. Microb Pathog. 2013;64:39–42. doi: 10.1016/j.micpath.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 87.Liu W, Wei MT, Tong Y, Tang F, Zhang L, Fang L, Yang H, Cao WC. Seroprevalence and genetic characteristics of five subtypes of influenza A viruses in the Chinese pig population: a pooled data analysis. Vet J. 2011;187:200–206. doi: 10.1016/j.tvjl.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 88.Ninomiya A, Takada A, Okazaki K, Shortridge KF, Kida H. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet Microbiol. 2002;88:107–114. doi: 10.1016/S0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- 89.Guo Y, Li J, Cheng X. Discovery of men infected by avian influenza A (H9N2) virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999;13:105–108. [PubMed] [Google Scholar]
- 90.Guo Y, Dong J, Wang M, Zhang Y, Guo J, Wu K. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chin Med J. 2001;114:76–79. [PubMed] [Google Scholar]
- 91.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 92.Saito T, Lim W, Suzuki T, Suzuki Y, Kida H, Nishimura SI, Tashiro M. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2001;20:125–133. doi: 10.1016/S0264-410X(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 93.Perez DR, Lim W, Seiler JP, Yi G, Peiris M, Shortridge KF, Webster RG. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J Virol. 2003;77:3148–3156. doi: 10.1128/JVI.77.5.3148-3156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gou Y, Xie J, Wang M. A strain of influenza A H9N2 virus repeatedly isolated from human population in China. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2000;14:209–212. [PubMed] [Google Scholar]
- 95.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang R, Wang AR, Liu ZH, Liang W, Li XX, Tang YJ, Miao ZM, Chai TJ. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis. 2013;32:1347–1351. doi: 10.1007/s10096-013-1888-7. [DOI] [PubMed] [Google Scholar]
- 97.de Bruin E, Zhang X, Ke C, Sikkema R, Koopmans M. Serological evidence for exposure to avian influenza viruses within poultry workers in southern China. Zoonoses Public Health. 2017 doi: 10.1111/zph.12346. [DOI] [PubMed] [Google Scholar]
- 98.Li X, Tian B, Jianfang Z, Yongkun C, Xiaodan L, Wenfei Z, Yan L, Jing T, Junfeng G, Tao C, Rongbao G, Dayan W, Shu Y. A comprehensive retrospective study of the seroprevalence of H9N2 avian influenza viruses in occupationally exposed populations in China. PLoS One. 2017;12:e0178328. doi: 10.1371/journal.pone.0178328. [DOI] [PMC free article] [PubMed] [Google Scholar]


