Abstract
In an effort to expand the genetic alphabet and create semi-synthetic organisms (SSOs) that store and retrieve increased information, we have developed the unnatural base pairs (UBPs) dNaM and d5SICS or dTPT3 (dNaM-d5SICS and dNaM-dTPT3). The UBPs form based on hydrophobic and packing forces, as opposed to complementary hydrogen bonding, and while they are both retained within the in vivo environment of an E. coli SSO, their development was based on structure-activity relationship (SAR) data generated in vitro. To address the likely possibility of different requirements of the in vivo environment, we screened 135 candidate UBPs for optimal performance in the SSO. Interestingly, we find that in vivo SARs differ from those collected in vitro, and most importantly, we identify four UBPs whose retention in the DNA of the SSO is higher than that of dNaM-dTPT3, which was previously the most promising UBP identified. The identification of these four UBPs further demonstrates that when optimized, hydrophobic and packing forces may be used to replace the complementary hydrogen bonding used by natural pairs and represents a significant advance in our continuing efforts to develop SSOs that store and retrieve more information than natural organisms.
Graphical abstract
INTRODUCTION
Natural organisms store genetic information in an alphabet of four nucleotide “letters,” and the replication and retrieval of this information is mediated by their selective pairing to form two base pairs. The addition of two new letters that form an unnatural base pair (UBP) would increase the potential information content and lay the foundation for semi-synthetic organisms (SSOs) capable of producing proteins with unnatural amino acids or even possessing novel forms and functions, which is the central goal of synthetic biology.1 We,2 as well as the Benner3 and Hirao4 groups, have worked towards this goal and have each identified UBPs that are well replicated in vitro. Our efforts have focused upon a family of UBPs represented by dNaM-d5SICS and dNaM-dTPT3 (Figure 1), which utilize hydrophobic and packing forces, as opposed to complementary hydrogen bonding, to stably pair within duplex DNA and during replication.5–10 This mode of pairing results in an edge-to-edge, Watson-Crick-like geometry during unnatural triphosphate insertion within the polymerase active site, but cross-strand intercalation once the UBP is synthesized,11–13 which likely mandates deintercalation for continued synthesis. Thus, the extended aromatic surface area of the unnatural nucleobases may be a liability. Nonetheless, with the unnatural triphosphates imported via transgenic expression of the nucleoside triphosphate transporter PtNTT2, we developed an Escherichia coli SSO that stably retains these UBPs in its DNA.10,14 While the retention of the UBPs in the DNA of the SSO is not equivalent to that of a natural base pair, retention may be raised to natural-like levels through the action of Cas9 targeted to degrade the DNA that has lost the UBP.14
Figure 1.
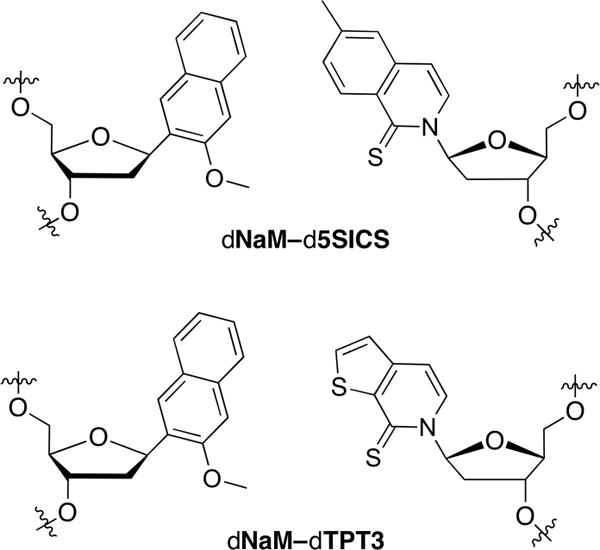
The dNaM-d5SICS and dNaM-dTPT3 UBPs.
Both the dNaM-d5SICS and dNaM-dTPT3 UBPs were identified from an intensive investigation of over 150 analogs that were evaluated in vitro, initially via steady-state kinetics and later by retention in PCR-amplified DNA.2 These studies provided the key structure-activity relationship (SAR) data used to optimize the unnatural nucleotides, but the in vivo environment of the SSO introduces additional constraints, such as toxicity, import, and polymerase availability. Thus, it is unclear whether UBPs optimized based on in vitro SAR are optimal for performance in vivo. Indeed, during in vitro optimization, we emphasized the importance of identifying multiple different UBPs whose constituent nucleotides possess varying physicochemical properties to provide flexibility during the effort to deploy them in an SSO.8,15
To extend our SAR data to include the restraints of the in vivo SSO environment, we conducted a screen for pairs of unnatural triphosphates that when added to growth media support high level UBP retention. From an examination of 135 candidate UBPs, we generate new SAR data that differs in several interesting ways from that generated in vitro, and remarkably, at least in some cases demonstrates that replication is more permissive in vivo than in vitro. Most importantly, we discover four new UBPs that are more efficiently retained in vivo than either dNaM-d5SICS or dNaM-dTPT3. One of the constituent nucleobases in each of the new UBPs is dTPT3, suggesting that it represents an at least currently optimal solution, but it is paired with different dNaM analogs. The most promising new UBP is retained in sequences where neither dNaM-d5SICS nor dNaM-dTPT3 is retained even with Cas9, and thus represents the most promising UBP identified to date for use in our continuing efforts to develop an SSO that stably stores increased information.
EXPERIMENTAL SECTION
General
All bacteria were cultured in 100 μL of liquid 2×YT media (casein peptone 16 g/L, yeast extract 10 g/L, NaCl 5 g/L) supplemented with potassium phosphate (50 mM, pH 7) in 96-well microwell plates. When noted, antibiotics were used at the following concentrations: chloramphenicol, 5 μg/mL; ampicillin, 100 μg/mL. Cell growth, indicated as OD600, was measured using a Perkin Elmer EnVision 2103 Multilabel Reader with a 590/20 nm filter. Unless otherwise stated, molecular biology reagents were purchased from New England Biolabs (Ipswich, MA) and were used according to the manufacturer’s protocols. As necessary, purification of nucleic acids was accomplished by microelution columns (Zymo Research Corp; Irvine, CA). All natural oligonucleotides were purchased from IDT (San Diego, CA), and oligonucleotides containing dNaM were synthesized by Biosearch Technologies (Petaluma, CA) with purification by reverse phase cartridge and were kindly provided by Synthorx (La Jolla, CA). Unnatural nucleotide triphosphates were prepared as previously described (Table S2), and confirmed by MALDI-TOF and UV/Vis.
Analysis of UBP Retention
Plasmids containing the dNaM-dTPT3 UBP were prepared and used to transform E. coli strain YZ3 as described previously.10,14 Following transformation, the SSO was allowed to recover at 37 °C for 1 h in media containing dNaMTP (125 μM) and dTPT3TP (25 μM). Cells were pelleted by centrifugation, resuspended in fresh media lacking unnatural triphosphates, and then used to inoculate cultures containing different pairs of unnatural triphosphates at the specified concentrations. When the cell density reached an OD600 of ~0.7, cells were pelleted and plasmids were recovered and PCR amplified with d5SICSTP and a biotinylated analog of dNaMTP, and UBP retention was determined by comparing the intensity of the streptavidin shifted and unshifted bands via PAGE as described previously10,14 and in Supporting Information. Plasmids with the dNaM-dTPT3 UBP were used as a starting point for the in vivo replication experiments to eliminate the need to construct separate plasmids for every candidate UBP examined and to eliminate differences in UBP retention during in vitro plasmid construction. We note that this requires each dNaM analog to pair with dTPT3 and each dTPT3 analog to pair with dNaM during the first round of replication, and similar pairing is required for the first round amplification during PCR analysis. It should also be noted that this assay detects bulk retention, and inversion of the pair by, for example, self-pairing, is not excluded.
RESULTS
To create a template for in vivo replication assays, Golden Gate assembly was used to construct a derivative of the pUC19 plasmid in which a single dNaM-dTPT3 UBP was embedded within the TK1 sequence (local sequence AXT, X = dNaM; referred to hereafter as sequence context 1), a context within which the dNaM-dTPT3 UBP is well replicated in our SSO.14 Plasmids were then used to transform the SSO, which was allowed to recover briefly in media containing dNaMTP and dTPT3TP. Upon resuspension in fresh media lacking triphosphates, the SSO culture was split into 100-μL aliquots and supplemented with varying concentrations of different pairs of unnatural triphosphates. Once the cultures reached an OD600 of ~0.7, plasmids were recovered and analyzed for UBP retention (Figure 2).
Figure 2.
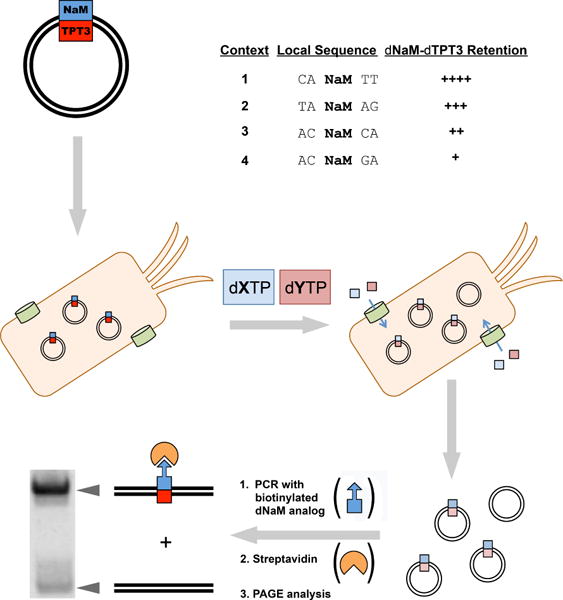
Graphical representation of screen workflow. For sequence contexts 1 – 4, X denotes NaM or a NaM analog and Y denotes TPT3 or a TPT3 analog. Retention levels of dNaM-dTPT3 are indicated with “+” symbols. See Ref 14 for original report of dNaM-dTPT3 retention data.
In a first phase of screening we explored the addition of 25 μM of dTPT3TP and one of 75 different dNaMTP analogs (structures shown in Figure 3A) added at a concentration of 125 μM or 10 μM. After plasmid recovery, we observed UBP retention of >90% with thirteen analogs (dMMO2TP, dDMOTP, dNaMTP, dClMOTP, dCNMOTP, d5FMTP, dFDMOTP, dFIMOTP, dZMOTP, dIMOTP, dMIMOTP, dFEMOTP, and dMMO2ATP) (Figure 4A and Supporting Information). Of the remaining analogs, four showed a retention of 50–90% (d2OMeTP, dTfMOTP dMEMOTP, dVMOTP), nine showed a retention of 20–50% (dDM5TP, d2MNTP, d45DMPyTP, dEMOTP, dDMTP, dTOK581TP, dTOK587TP, dPyMO2TP, d35DMPyTP), and the remainder showed a retention of less than 20%. Addition of the dNaMTP analogs at the lower concentration resulted in generally less efficient UBP retention, with only four, dMMO2TP, dClMOTP, dCNMOTP, and d5FMTP, resulting in high retention (>80%). Five, dFIMOTP, dIMOTP, dFEMOTP, dMMO2ATP, as well as dNaMTP itself, showed intermediate levels of retention (between 40–80%), and four, dFDMOTP, dVMOTP, d2OMeTP, and dZMOTP, showed slightly less retention (20–40%), with the remainder showing <20% retention.
Figure 3.
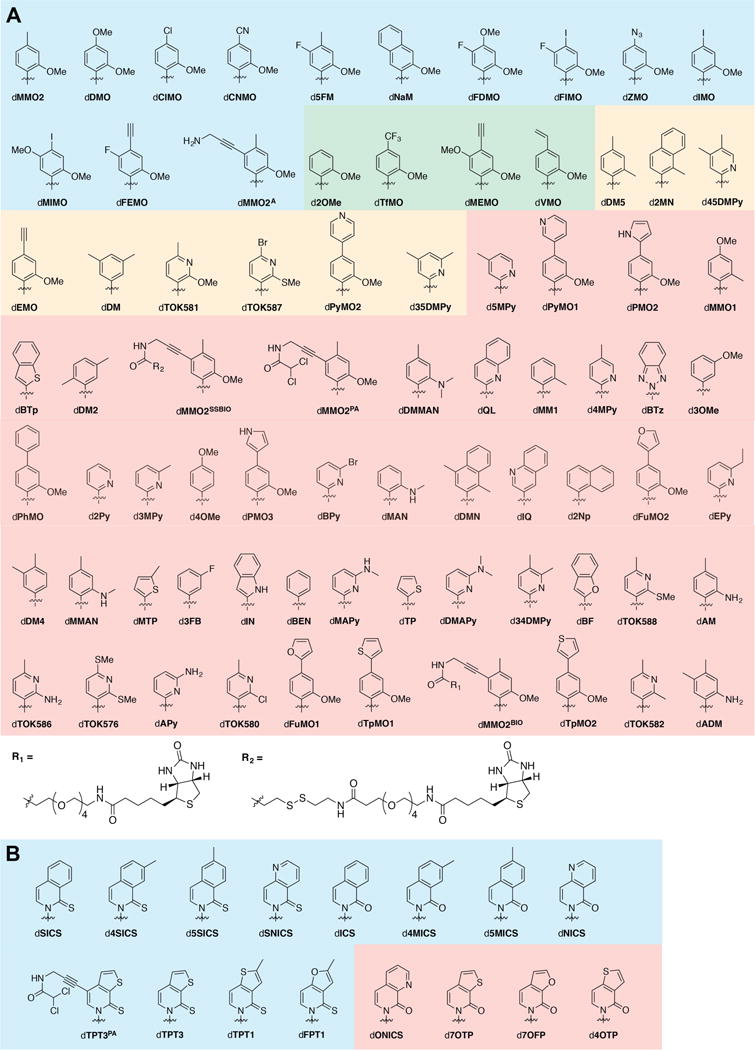
Structure of analogs used in current study. (A) dNaMTP analogs. Blue shading corresponds to analogs that at 125 μM showed greater than 90% retention in phase 1 of the screen. Green shading corresponds to those that at 125 μM showed retentions between 50% and 90% in phase 1. Orange shading corresponds to those that at 125 μM showed between 20% and 50% retention. Red shading corresponds to those that at 125 μM showed less than 20% retention. (B) dTPT3 analogs. Blue shading corresponds to those that at 10 μM showed greater than 90% retention in phase 1 of the screen and red shading corresponds to those that showed less than 10% retention at the same concentration. Ribose and phosphates omitted for clarity. For original references see Supporting Information.
Figure 4.
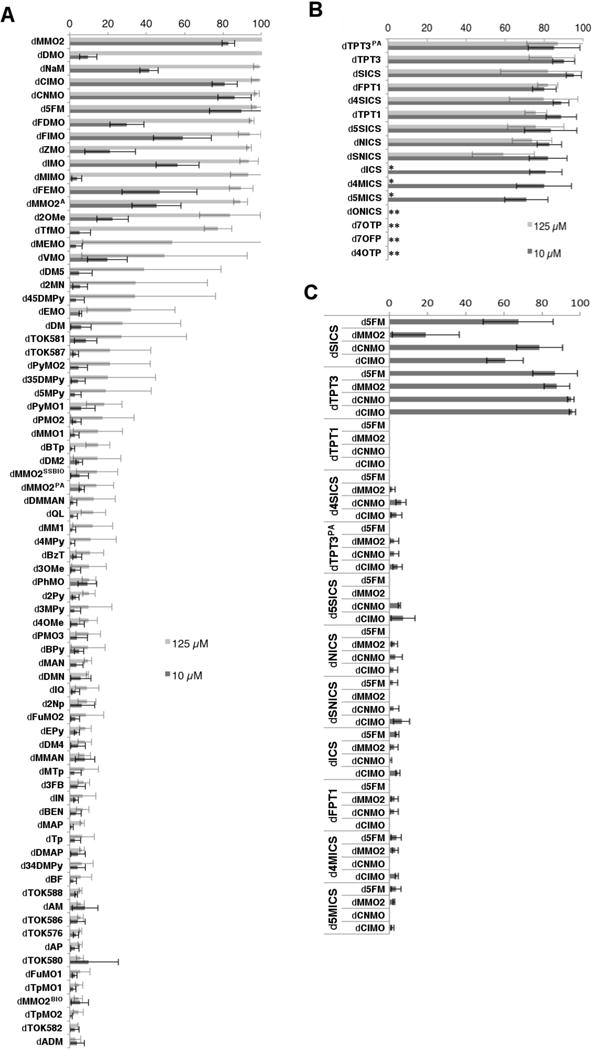
UBP retention (%). (A) dNaMTP analogs added at either 125 μM or 10 μM and dTPT3TP added at 25 μM. (B) dTPT3TP analogs added at either 125 μM or 10 μM and dNaMTP added at 125 μM. (C) Selected analogs screened against each other with dNaMTP analogs added at 25 μM and dTPT3TP analogs added at 10 μM. Data is an average of 3 independent trials, with error bars indicating standard deviation. A single asterisk indicates that no cell growth was observed at the higher concentration, and a double asterisk indicates that no cell growth was observed at either concentration.
We next explored UBP retention with the addition of 125 μM dNaMTP and one of 16 different dTPT3TP analogs (structures shown in Figure 3B) at a concentration of 125 μM or 10 μM. When provided at the higher concentration, nine of the dTPT3TP analogs, dTPT3PATP, dTPT3TP, dSICSTP, dFPT1, d4SICS, dTPT1, d5SICS, dNICS, and dSNICS, showed significant UBP retention upon plasmid recovery (Figure 4B and Supporting Information). Unlike with the dNaMTP analogs, these nine UBPs showed similar or better retention when provided at the lower concentration, while dICSTP, d4MICSTP, and d5MICSTP also showed significant retention (retention of these three analogs could not be determined at higher concentrations due to toxicity). Clearly UBP retention is more optimal with the lower concentration of these analogs, and under these conditions, when combined with dNaMTP, all triphosphate analogs examined except dONICSTP, d7OTPTP, d7OFPTP, and d4OTPTP (which were toxic at both concentrations) showed retention of the UBP in excess of 70%.
In a second phase of screening, we crossed the twelve most promising dTPT3TP analogs with the four most promising dNaMTP analogs identified in the first phase. We incorporated the UBP within the same plasmid, but embedded it within a local sequence of AXA (context 2, X = dNaM), a context in which we have found retention of dNaM-dTPT3 to be more challenging than sequence context 1.14 Based on the first phase of screening, we also focused on concentrations of the dNaMTP and dTPT3TP analogs of 25 μM and 10 μM, respectively, to increase the dynamic range of the screen. Significant retention was only observed with pairs containing dTPT3TP or dSICSTP, but each was found to yield at least moderate retention with each of the four dNaMTP analogs (Figure 4C and Supporting Information). Retention with dSICSTP was moderate when paired with dMMO2TP (19%), but more significant with d5FMTP, dCNMOTP, and dClMOTP, with 68%, 79%, and 61% retention, respectively. The highest retentions, however, were observed with dTPT3TP (all >87%).
Next we explored retention with the four most promising UBPs identified, d5FM-dTPT3, dMMO2-dTPT3, dCNMO-dTPT3, and dClMO-dTPT3, when embedded within context 2, but with unnatural triphosphate concentrations of 25 μM, 10 μM, or 2.5 μM (Figure 5). While it was clear that dMMO2TP and d5FMTP were retained best at 25 μM, UBP retention was too high to differentiate in the case of dCNMOTP and dClMOTP. Thus, we examined retention with the UBP positioned in the same plasmid, but within the local sequence context of CXC (context 3, X = dNaM), which is particularly challenging for dNaM-dTPT3 retention 14 (Figure 5). The data reveal that 25 μM is the most optimal concentration for both pairs and that dCNMOTP and dTPT3TP perform better than dClMOTP and dTPT3TP, with retentions of 42% and 21%, respectively. While d5FMTP and dMMO2TP resulted in slightly higher retention in this sequence context at high concentration (49% and 45%, respectively), they resulted in significantly less retention at the lower concentrations.
Figure 5.
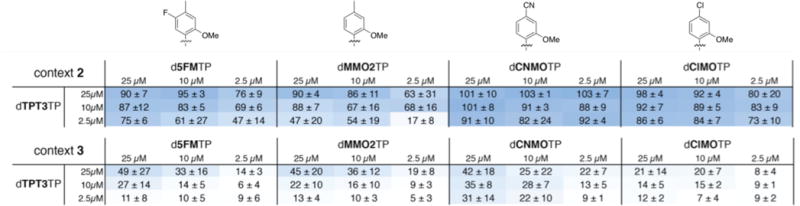
UBP retention (%) with dTPT3TP and different dNaMTP analogs added at varying concentrations to the media. Shading indicates level of UBP retention. Values are the average and standard deviation of three independent determinations.
Of the 135 candidate UBPs examined, the data reveal that dCNMO-dTPT3 is most efficiently replicated in the SSO. To directly and more thoroughly compare this UBP with dNaM-dTPT3, the most efficiently replicated UBP previously identified, we examined retention of both pairs in the three sequence contexts described above, as well as a fourth, which positions the UBP within the local sequence context of CXG (context 4), which is one of the most challenging sequences for dNaM-dTPT314 (Figure 6). dTPT3TP was added at a fixed concentration of 25 μM, while dNaMTP or dCNMOTP was added at a concentration of either 125 μM or 25 μM. At the higher concentration, we observed >99% retention in sequence context 1 with both dCNMOTP and dNaMTP, but while retention remained high with dCNMOTP added at the lower concentration (98%), it was decreased with dNaMTP (85%). In context 2, reduced retention was observed with dNaMTP at both the high concentration (73%) and the low concentration (36%), but retention at both concentrations remained high with dCNMOTP (>99%). In context 3, addition of dNaMTP at the higher concentration resulted in only moderate retention (26%), while addition at the lower concentration resulted in no retention. However, with dCNMOTP, significant retention was observed at both high (65%) and low concentrations (42%). Finally, with context 4, the UBP was not retained significantly at either concentration with dNaMTP, but remained moderate at high concentrations of dCNMOTP (24%).
Figure 6.
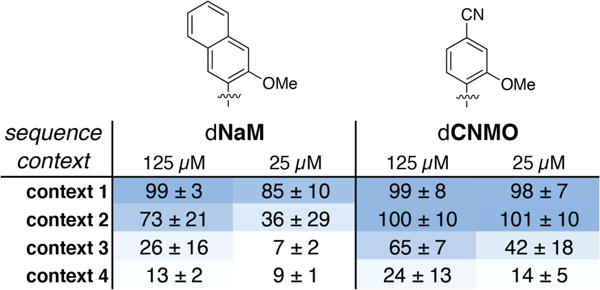
UBP retention (%) with 25 μM dTPT3TP and varying concentrations of dNaMTP or dCNMOTP added to the media. Shading indicates level of UBP retention. Values are the average and standard deviation of three independent determinations.
DISCUSSION
The discovery of dNaM-dTPT3 was driven by in vitro SARs that ultimately drew on over 150 unnatural nucleotides. While dNaM-dTPT3 was the most promising UBP discovered, and is clearly suitable for use within a living SSO,14 its retention is sequence context-dependent, with some sequences showing high retention and others less or none. During the in vitro discovery phase we also identified variants whose constituent nucleotides have distinct physicochemical properties that may differentiate performance in vivo. With these analogs, we have now examined 135 variant UBPs within the in vivo environment of our SSO. Interestingly, we find in vivo SARs that are both similar to and different from those collected in vitro. For example, dICS, and its methyl-derivatized analogs d4MICS and d5MICS, support UBP retention in vivo reasonably well, but only at low concentration, suggesting that they are misincorporated opposite natural nucleotides at high concentrations, thereby resulting in stalled replication forks and toxicity. Indeed, in vitro, steady state kinetic analyses suggest that these analogs can be misinserted opposite natural nucleotides in a template with reasonable efficiency.16 However, UBPs containing these analogs cannot be PCR amplified, suggesting that the observed retention is unique to the in vivo environment. Heteroatom derivatization of the dICS scaffold is generally deleterious and results in significant toxicity, again consistent with misincorporation, as we observed previously in vitro.17 An exception is dNICS, as this heteroatom-derivatized analog of dICS supports UBP retention reasonably well, and in fact, the additional sulfur substituent of dSNICS results in an analog that supports moderate retention at both low and high concentrations. The beneficial effect of the sulfur does not depend on aza substitution as retention is also increased with dSICS compared to dICS. Thus, the aza and sulfur substituents appear to independently reduce mispairing in vivo. While this was observed in vitro for the sulfur substituent, the opposite was observed with aza substitution, suggesting that its ability to reduce mispairing is unique to the in vivo environment.
The modification of unnatural nucleotides with linkers that allow for site-specific attachment of different functionalities is of particular interest for in vivo labeling experiments. Linker modification of dTPT3TP, resulting in dTPT3PATP, is well tolerated in vivo, as was also observed in vitro. However dMMO2ATP is reasonably well tolerated in vivo, while dMMO2PATP, dMMO2BIOTP and dMMO2SSBIOTP completely ablate retention, contrary to what is observed in vitro.18 While dMMO2ATP shows a decrease in retention at low concentration, which dTPT3PATP does not, its free amine linker should facilitate in vivo labeling or crosslinking experiments. Similarly, dZMOTP and dFEMOTP are well retained in vivo when supplemented at high concentrations, and provide an azide and alkyne moiety in the major groove, respectively, where they should also facilitate in vivo labeling or crosslinking.
A large body of in vitro SAR data demonstrates convincingly that an H-bond acceptor positioned ortho to the glycosidic bond, and thus oriented into the developing minor groove upon incorporation into DNA, is generally required for efficient continued primer elongation.19 The general requirement of an H-bond acceptor at this position is consistent with studies of natural base pairs,20–23 which invariably have a similarly disposed H-bond acceptor that is thought to engage in critical interactions with polymerase-based H-bond donors.24 An exception is the relatively efficient PCR amplification of DNA containing d2MN paired opposite dTPT3.8 In vivo, d2MNTP also supports retention with dTPT3TP, but when combined with dTPT3TP, dDM5TP does as well. Moreover, while dICSTP, dNICSTP, d4MICSTP, and d5MICSTP do not support PCR amplification when paired with any analog,8 they support reasonable retention in vivo when paired dNaMTP. Clearly, the requirements for the ortho group are somewhat different in vitro and in vivo, and at least in some cases, they are more permissive in vivo.
From a practical perspective, the most important results of the current study are the excellent in vivo performance of the d5FM-dTPT3, dMMO2-dTPT3, dCNMO-dTPT3, and dClMO-dTPT3 UBPs (Figure 7). Retention of each of these new UBPs in the SSO, in particular, dCNMO-dTPT3, is better than that of dNaM-dTPT3, the previously most promising UBP identified, and requires the addition of less nucleotide triphosphate to the growth media. In fact, dCNMO-dTPT3 shows at least moderate retention in sequence context 4, where dNaM-dTPT3 is retained so poorly that it cannot be rescued by Cas9, suggesting that it is lost immediately upon attempted replication. Interestingly, this contrasts with in vitro data where retention of dNaM-dTPT3 is better than retention of dCNMO-dTPT3,8,15 suggesting that E. coli provide a unique environment for which dCNMO-dTPT3 is more optimal. Possible contributing factors include PtNTT2-mediated uptake, stability within the cell, or recognition by different polymerases that can access the replication fork and actually mediate replication in vivo.
Figure 7.
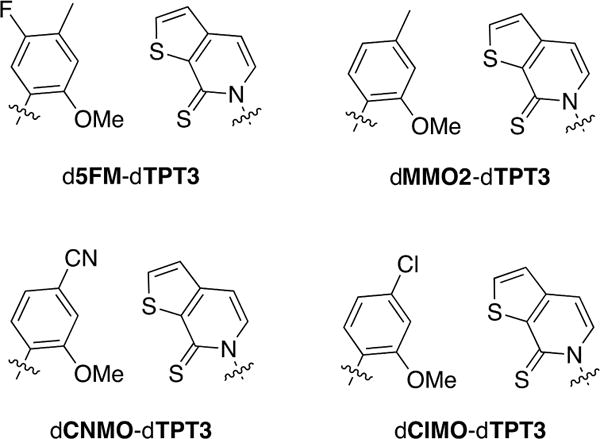
The four new optimal UBPs discovered.
Regardless of the specific properties that underlie their performance, it is clear that the present work has identified four new UBPs that now represent the most promising candidates for use in an SSO. These new UBPs further demonstrate the ability of hydrophobic and packing interactions to replace complementary H-bonding as the force underlying information storage. It is interesting that each of the new dNaM analogs bear smaller, single ring nucleobases. While this may have contributed to their in vivo performance by facilitating uptake, they are likely to be less prone to cross-strand intercalation, and more likely to adopt edge-to-edge structures. This may also contribute to their more optimal retention, and possibly even facilitate the replication of DNA with higher density UBPs. Only UBP loss was characterized in this study, as mutations involving natural nucleotides are predicted to be most problematic.5,25,26 However, cross-strand intercalation may also facilitate self-pairing, which even at low levels could cause UBP inversion (where the individual nucleotides switch strands). Whatever its level, self-pairing mediated UBP inversion may be less likely with these analogs. Finally, the performance of each of the new UBPs is likely to be even further improved by use of Cas9, and we are currently exploring this possibility. The availability of a family of UBPs that are well retained in the in vivo environment of the SSO, but that also possess distinct physicochemical properties, is of great significance as our efforts to retrieve the increased information via transcription and translation will likely introduce additional requirements and restraints.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (Grant Nos. GM060005 and GM118178 to F.E.R.). A.W.F. was supported by a National Science Foundation Graduate Research Fellowship (Grant No. NSF/DGE-1346837).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional methods, supporting tables and figures, and references (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
References
- 1.Leduc S. The Mechanisms of Life. Rebman Company; New York: 1911. [Google Scholar]
- 2.Malyshev DA, Romesberg FE. Angew Chem Int Ed. 2015;54:11930–11944. doi: 10.1002/anie.201502890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, et al. J Am Chem Soc. 2015;137:6734–6737. doi: 10.1021/jacs.5b02251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimoto M, Hirao I. In: Chemical Biology of Nucleic Acids: Fundamentals and Clinical Applications. Erdmann AV, Markiewicz TW, Barciszewski J, editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 131–148. [Google Scholar]
- 5.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavergne T, Malyshev DA, Romesberg FE. Chem Eur J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, Romesberg FE. Proc Natl Acad Sci USA. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhami K, Malyshev DA, Ordoukhanian P, Kubelka T, Hocek M, Romesberg FE. Nucleic Acids Res. 2014;42:10235–10244. doi: 10.1093/nar/gku715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Degardin M, Lavergne T, Malyshev DA, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2014;136:826–829. doi: 10.1021/ja408814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr, Romesberg FE. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A. Nat Chem Biol. 2012;8:612–614. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Romesberg FE, Marx A. J Am Chem Soc. 2013;135:18637–18643. doi: 10.1021/ja409609j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Chem Eur J. 2010;16:12650–12659. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Lamb BM, Feldman AW, Zhou AX, Lavergne T, Li L, Romesberg FE. Proc Natl Acad Sci U S A. 2017;114:1317–1322. doi: 10.1073/pnas.1616443114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavergne T, Degardin M, Malyshev DA, Quach HT, Dhami K, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2013;135:5408–5419. doi: 10.1021/ja312148q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Ogawa AK, Berger M, McMinn DL, Schultz PG, Romesberg FE. J Am Chem Soc. 2000;122:7621–7632. [Google Scholar]
- 17.Henry AA, Yu C, Romesberg FE. J Am Chem Soc. 2003;125:9638–9646. doi: 10.1021/ja035398o. [DOI] [PubMed] [Google Scholar]
- 18.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuda S, Leconte AM, Romesberg FE. J Am Chem Soc. 2007;129:5551–5557. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt TE. Biochemistry. 2001;40:2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- 21.Meyer AS, Blandino M, Spratt TE. J Biol Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 22.Morales JC, Kool ET. J Am Chem Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales JC, Kool ET. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Waksman G. Protein Sci. 2001;10:1225–1233. doi: 10.1110/ps.250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J Am Chem Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris SE, Feldman AW, Romesberg FE. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.7b00115. Published online 27 June. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


