Abstract
Here I give a brief history of my scientific career, beginning with my early interest in natural history and my introduction to the microscope and the wonderful world of the cell. My studies have focused on chromosomes, nucleoli, and other nuclear structures, with a few forays into the cytoplasm. In each case, I have tried to understand how proteins and nucleic acids are physically organized to give rise to the structures seen under the microscope. I describe how studies in my laboratory on amplified ribosomal RNA genes led to the development of in situ hybridization, a technique that permitted us to localize specific nucleic acid sequences with high precision. My early exposure to the diversity of animals and plants made it seem natural to choose organisms best suited to a particular problem, hence the use of salamanders, frogs, and mice, as well as protozoa, fruit flies, and other invertebrates.
Keywords: lampbrush chromosome, nucleolus, germinal vesicle, ribosomal DNA, in situ hybridization, Xenopus, Tetrahymena
BEGINNINGS
I was born in Washington, D.C., in 1928 and spent my early childhood in northern Virginia, which at that time was mostly rural. Some of my earliest memories are of walking along a stream looking for insects, frogs, tadpoles, or anything else that might be moving in the water or on the plants growing near the water. That scene was repeated over and over as I grew older, becoming a biologist without knowing there was such a thing as a biologist. My father was a lawyer who was often away from home, so it was left to my mother to nurture my interests. She let me bring all sorts of live things into the house, she found the appropriate books to encourage my interests, and above all she participated in what I was doing. Children had lots of free time in those days, before television and organized sports, and I spent many hours outdoors with my butterfly net, roaming over the fields and woods near our home. Insects were brought back, identified in one of my reference books, and then pinned and mounted in old cigar boxes. Somewhere along the line I became interested in drawing these insects, resulting in meticulous copies in full detail. In retrospect, I realize that most of my knowledge of the outside world was coming through my eyes, and that has been the story of my life.
LIFE ON THE FARM
In 1942, when I was 14 years old, our family moved to a 500-acre farm in northern Virginia. Although neither of my parents grew up on a farm, they both had a fondness for the countryside and a desire to get further from Washington, D.C., which even at that time was beginning to encroach on the surrounding suburbs. Moving to the farm had major consequences for me as a teenager. Because rural schools were not really an option for my education, I was sent away to a boarding school near Charlottesville. Woodberry Forest, as it was called, was a small school founded shortly after the Civil War and modeled after the British “public school” (i.e., private school for the well to do). Emphasis was on English, Latin, French, math, history, and to a lesser extent science, not to mention athletics, which was required every afternoon except for weekends. Because I was already bookish by any ordinary standard, this school suited me perfectly, an attitude not shared by many of my classmates. Summers were spent on the farm, where I had plenty of time to channel my early interest in natural history into more disciplined science. Just how I convinced my parents to buy me a real microscope, I do not remember. It could not have been easy for them to find one, because we were in the middle of the Second World War and items of that sort were scarce. Not only did I get a microscope, but I also converted my bedroom at home into a laboratory (Figure 1). With only books as my guide, I managed to make permanent slides of all sorts of things, from fleas and lice (kindly provided by the farm animals) to algae and protozoa from the farm pond. Eventually, I wanted to see more. I had a copy of E.B. Wilson’s classic The Cell in Development and Heredity (Wilson 1925), and several other books that described cells and their parts. So, in what seemed a natural progression for me at the time, I learned how to fix and embed tissues, and section them for observation with my microscope. I made free-hand sections at first, but this obviously did not work too well. Again, I prevailed on my parents and miraculously they found and bought a second-hand microtome for me to use. My parents must have worried a little when my interests progressed from tadpoles and grasshoppers to the internal organs of rabbits and groundhogs. But I do not think they were too concerned—after all, we lived on a farm where we knew how the animals in the barnyard became food on the table.
Figure 1.
The author at his microscope, about 1943.
YALE
After three years at prep school, it was time to consider college. Today this is a huge decision, involving tests upon tests, interviews, essays, and descriptions of extracurricular activities. I can honestly say that I do not know how the decision was made that I should go to Yale. I do know that it involved very little thought on my part. Most likely, my prep school wanted to send an occasional student to the Ivy League, just to prove that it was more than a finishing school for the sons of Southern industrialists. In any case, I found myself in New Haven in early summer of 1945. The War in Europe had just finished, and veterans had not yet returned from Europe or the Pacific. So my class consisted almost entirely of those who were too young to have been drafted. Prep school had fulfilled its name for me and fully prepared me for college courses that were a challenge to those who came from high school backgrounds. As a result, I did not feel stressed, as many of my classmates did, and I had time to continue my interests in biology.
I majored in zoology—botany was a separate subject at that time—and like nearly all majors in zoology, I considered myself headed for medical school. The curriculum included systematics, comparative anatomy, histology, embryology, genetics, and physiology. There was little biochemistry and what recognition there was of DNA took place in the genetics course. There was essentially no career counseling, so it was not until near the end of my undergraduate work that I realized that one could get a Ph.D. and have a career doing research and teaching in a university setting. With the help of my future advisor, Donald Poulson, I was admitted to the zoology graduate program at Yale, where I finally felt that my career was headed in the right direction.
Poulson was a young Drosophila geneticist who had trained with Alfred Sturtevant at CalTech. From his time at CalTech and elsewhere, he knew many of the early giants of genetics, including T.H. Morgan, Hermann Muller, George Beadle, and Barbara McClintock. His major interest was in the effect of mutations on early embryological development, and for his doctoral work, he had shown the dramatic effects of the Notch mutant on gastrulation and formation of the nervous system. As a mentor he was completely nondirective, allowing his few students to work on essentially anything they wanted to. By then I had developed an intense interest in chromosomes, particularly the giant polytene chromosomes of Drosophila with their enigmatic banded structure. Like many others, I felt that an understanding of these bands could help us learn more about genes and how they function. Unfortunately, I could not think of anything to do with the polytene chromosomes that had not already been done, and by the end of my first year of graduate work I was beginning to despair of finding a suitable research problem.
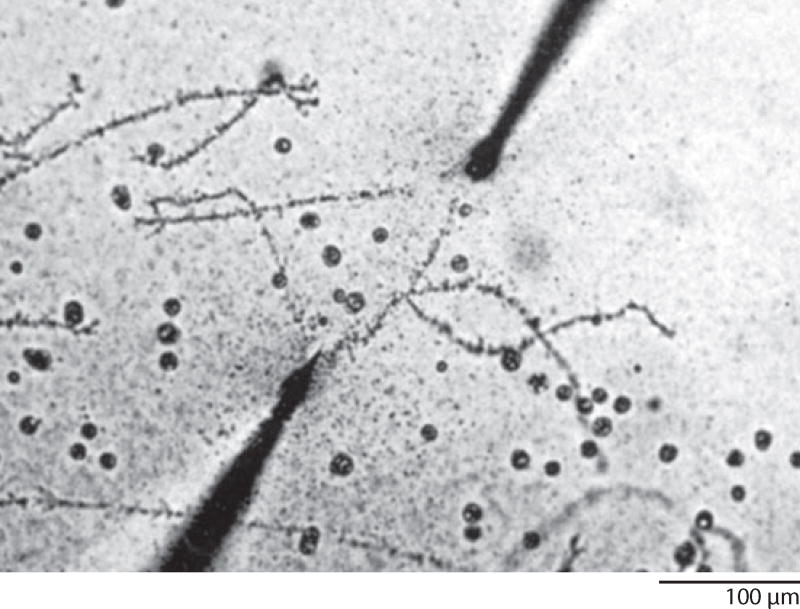
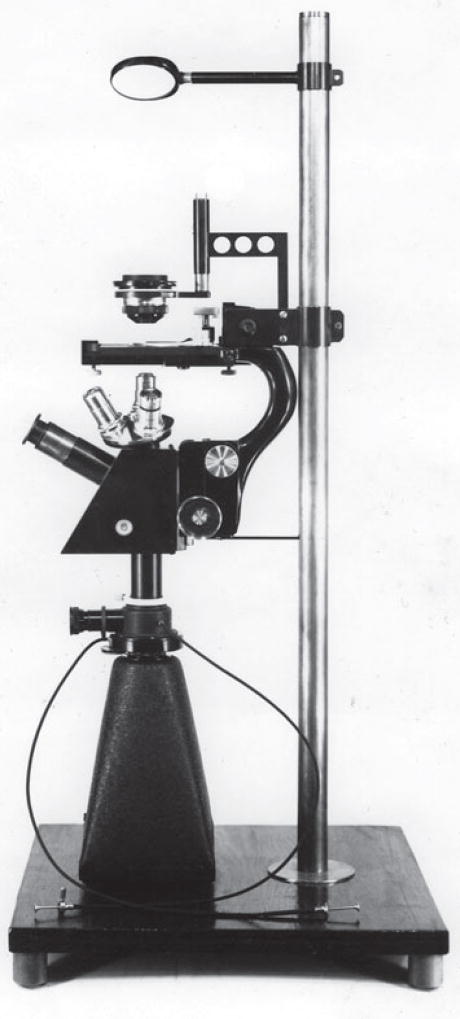
Then I made a chance discovery—not in the lab, but in a book—that affected the entire future course of my career. In C.H. Waddington’s An Introduction to Modern Genetics (Waddington 1939), I ran across an image of a lampbrush chromosome (LBC) from a salamander oocyte. The chromosome was being stretched between microneedles, and I felt sure that the stated magnification was wrong, because the chromosome was nearly half a millimeter in length. When I went to the original paper, written by William R. Duryee (1937), I discovered that the magnification was indeed correct and that giant chromosomes had been described from salamander oocytes several times since the 1880s (Figure 2). They had received the name lampbrush (Lampencylinderputzer) because they reminded an early German worker (Rückert 1892) of the brushes used to clean soot from kerosene lamp chimneys. Digging further, I found that these marvelous chromosomes had been more or less ignored except for Duryee’s studies. Almost immediately, I knew I had a thesis project. There was no consensus on how these chromosomes were related to the much smaller chromosomes in other cells of the salamander or to the smaller chromosomes of other organisms. I decided to study the LBCs of the newt Notophthalmus viridescens with the aid of the newly invented phase contrast microscope, one of which had just been acquired by the Zoology Department. It soon became apparent that I needed an inverted microscope stand if I wanted to study the LBCs without fixation and attachment to a microscope slide. Because there were no commercially available inverted microscopes at that time, it was up to me to design and build one, which I did with the help of my mother’s brother, who happened to be an accomplished machinist. The final product was partly homemade and partly scavenged from my personal microscope (Figure 3). In my thesis, I began to work out the morphology of these interesting giant chromosomes (Gall 1954a), but in fact I missed a number of important features that only became clear over the next few years.
Figure 2.
A pair of lampbrush chromosomes being stretched between microneedles. These chromosomes, from an oocyte nucleus of the newt Cynops (Triturus) pyrrhogaster, measure 435 µm in length. Reproduced from Duryee (1941) with permission from the University of Pennsylvania Press.
Figure 3.
The author’s inverted microscope, used in early studies on lampbrush chromosomes.
MINNESOTA
After completing my Ph.D., I was anxious to set up my own laboratory, and I elected not to do a postdoc, which in any case was not considered mandatory at that time. So at the tender age of 24 years, I took up my first position as an instructor in the Zoology Department at the University of Minnesota in Minneapolis. I was hired to teach and, if time permitted, carry out research. Like most new faculty members, I did not realize how time-consuming teaching could be. Fortunately, the faculty was supportive, and I was able to find time for research, especially during the summers. At first, I continued to study LBCs, clearing up structural points that I had gotten wrong in my thesis. At about this time I began correspondence with an investigator in Scotland, H. G. “Mick” Callan, who was also interested in LBCs. Our overlapping scientific interests led first to a professional relationship and eventually a close personal friendship that lasted for more than 40 years. Although we published only one joint paper during the early part of our collaboration, we felt that we were working together on problems of mutual interest. We always shared new information long before it was published, and in this way benefited from each other’s input.
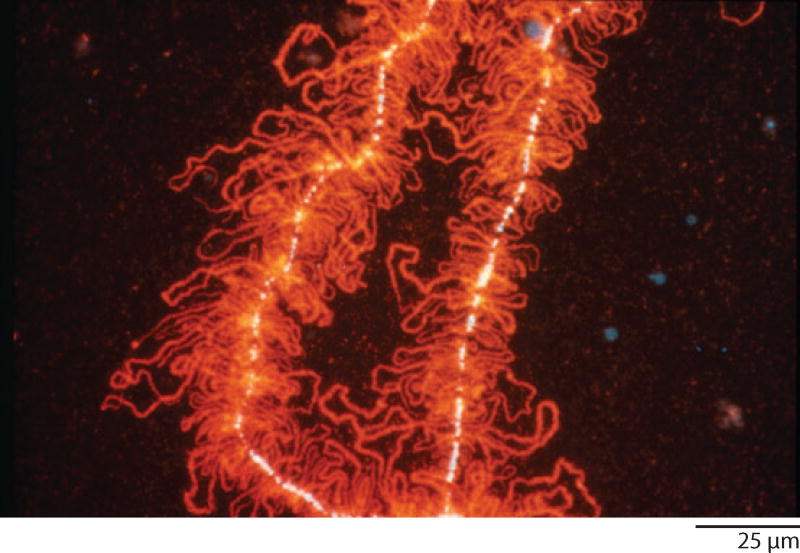
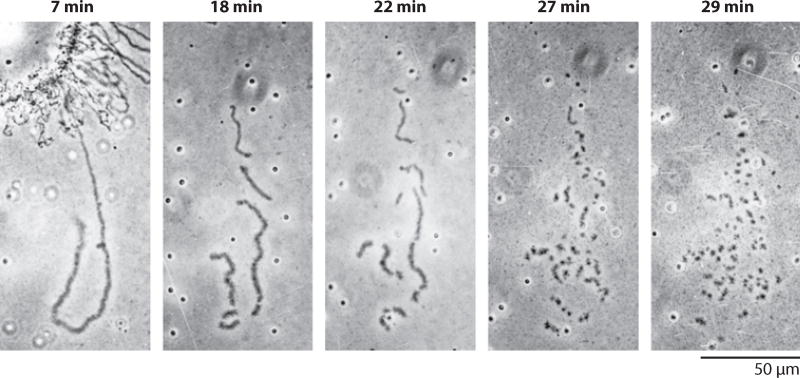
During the years I was at Minnesota, Mick and I clarified two major features of newt LBCs, one structural and the other functional. Structurally, we showed that each LBC consists of only two chromatids, which extend laterally from the chromosome axis as hundreds of pairs of loops (Figure 4). The loop pairs alternate with condensed regions (chromomeres) where the chromatids are intimately associated (Callan 1954; Gall 1955, 1956, 1958; Callan & Lloyd 1960). Callan’s student, Herbert Macgregor, showed that the loops were cut into short fragments when a LBC was placed in a DNase solution, whereas they became thinner but did not fragment when treated with RNase or various proteases (Callan & Macgregor 1958). When I repeated Macgregor’s observations, it immediately struck me that the number of breaks in a loop did not increase as a linear function of time, but at some higher rate (Figure 5). By counting the number of loop fragments over time, I showed that a loop breaks with two-hit kinetics, whereas the main axis (where the chromatids are paired) breaks with four-hit kinetics (Gall 1963). The obvious conclusion from these observations was that a chromatid consisted of one double helix of DNA, the so-called unineme model of chromosome structure. Herbert Taylor (Taylor et al. 1957) had earlier shown that newly incorporated thymidine was distributed in a semiconservative fashion when plant chromosomes replicated. With evidence from a plant and a vertebrate, it was now clear that chromosomes of higher organisms contained one long DNA molecule, just as Meselson & Stahl (1958) had found for the bacterial chromosome.
Figure 4.
A portion of a lampbrush chromosome from the newt Notophthalmus viridescens. Each chromosome consists of a central axis of condensed chromatin (white, stained with the DNA-specific dye DAPI) from which loops of transcribing chromatin extend laterally. Each loop consists of an invisible DNA axis with associated ribonucleoprotein transcripts, here stained red with a fluorescent antibody. Reproduced from Roth & Gall (1987) with permission from Rockefeller University Press.
Figure 5.
A time series showing DNase digestion of a single loop of a lampbrush chromosome from the newt Notophthalmus viridescens. Note that the number of cuts per minute increases as time progresses. The kinetics are 2-hit, demonstrating that the axis of the loop consists of one double helix of DNA. Reproduced from Gall (1966b).
The second insight gained from LBCs was that the loops are sites of active RNA synthesis. This was shown by their incorporation of radioactive RNA precursors (Gall 1958, Gall & Callan 1962). Subsequent studies amply confirmed this conclusion, showing that a given loop may contain one to several transcription units (Figure 6).
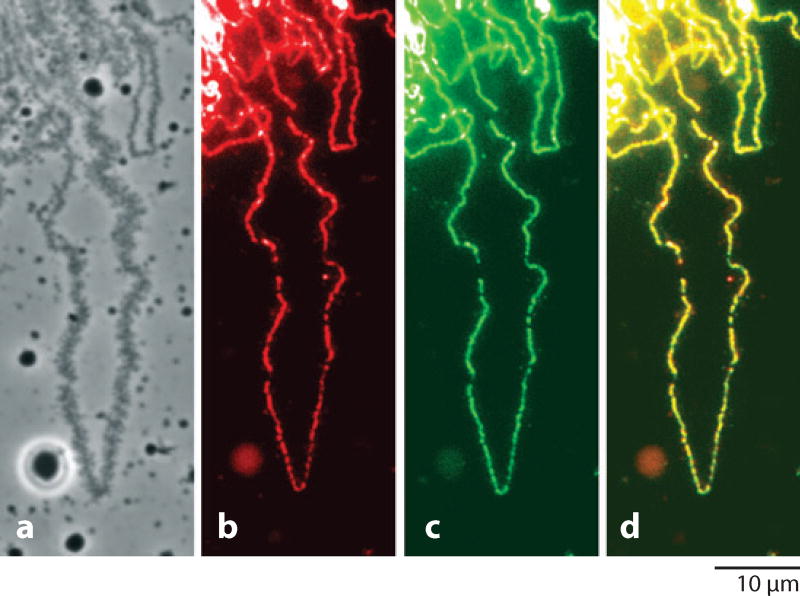
Figure 6.
A single loop of a lampbrush chromosome from the newt Notophthalmus viridescens. (a) Phase contrast showing the thin-thick distribution of ribonucleoprotein matrix. (b) Immunofluorescent staining of RNA polymerase II along the central axis of the loop. (c) Incorporation of bromouridine (BrU) along the axis. (d) Merge of polymerase and BrU images. From Gall et al. (1999) with permission from Molecular Biology of the Cell.
Like other cell biologists in the 1950s, I was fascinated by the increased resolving power of the electron microscope and all the new information it provided about cell structure. At that time the NIH was anxious to install electron microscopes in various laboratories around the country, and it was not difficult for me to obtain one for the Zoology Department at Minnesota, along with the newly invented Porter-Blum microtome for thin sectioning. Following Mick Callan’s lead, I studied the nuclear pore complexes in isolated nuclear envelopes, eventually demonstrating their octagonal symmetry (Gall 1954b, 1967). Studies on centriole replication showed how newly formed centrioles arise from a small procentriole situated at right angles to the mother (Gall 1961, Mizukami & Gall 1966).
RETURN TO YALE
In 1963, Don Poulson invited me to spend a year at Yale as a visiting professor, and I readily accepted his offer. What started out as a sabbatical leave ended up as a permanent position in the recently formed Biology Department. This was an opportune time because the new Kline Biology Tower was being constructed for the department, and I was able to design a floor for myself and two other investigators. The 1960s were a time of transition for biology as a science, and especially for cell biology. The American Society for Cell Biology had just been organized. People were beginning to realize that the old disciplines of cytology, embryology, genetics, and biochemistry were highly arbitrary—that a new synthesis was taking place in which these fields would eventually be understood at the molecular level. Molecular biology was the catchword for this change in attitude, and many of us felt an urgent need to become molecular biologists. Because of my interest in chromosomes, I decided to concentrate on nucleic acids, and so I spent several years learning how to isolate, characterize, quantitate, and otherwise deal with RNA and DNA. This was the time when CsCl gradients were developed for separating DNA fractions, denaturation and renaturation of DNA became standard techniques, and RNA/DNA hybrids let one make the first forays into molecular analysis of specific sequences.
ODD NUCLEOLI
Not surprisingly, I chose the amphibian oocyte nucleus as the subject for my molecular studies. People who work on the oocyte nucleus call it the germinal vesicle (GV), the name given to it by Purkinje, who first saw it in a chicken egg in 1825. I found that the GV made a lot of RNA, almost all of which was ribosomal RNA (Gall 1966a). This was a fortunate turn of events, since ribosomal RNA was one of the very few specific types of RNA that could be studied at that time (the revolution in cloning and sequencing was still another decade in the future). Recent studies on cultured cells had shown that ribosomal RNA was synthesized in the nucleolus (Perry 1962). Moreover, the genes coding for ribosomal RNA were almost certainly located in the nucleolus organizer, the chromosomal locus to which the nucleolus is attached (McClintock 1934, Ritossa & Spiegelman 1965). All of this posed a puzzle for the amphibian GV. I knew from my Ph.D. thesis—and indeed from classical studies going back to the nineteenth century—that the GV contained 1000 or more nucleoli. But these nucleoli were floating freely in the nucleus, not attached to the chromosomes. I had assumed (incorrectly) that the multiple nucleoli were being shed from the nucleolus organizer on a specific chromosome, and that they accumulated as some kind of storage product over the many weeks of oogenesis. Now we had to think more radically. Perhaps the large amount of ribosomal RNA in the GV was being synthesized in the free nucleoli themselves. But if this were true, then these nucleoli had to contain ribosomal DNA (rDNA) to act as the template for synthesis. In fact, many years earlier, Brachet (1940) and Painter & Taylor (1942) reported that amphibian oocyte nucleoli contain DNA. Their claims were based on staining with the Feulgen reagent, a highly specific and quantitative stain for DNA. Callan and I had been unsuccessful in attempts to stain oocyte nucleoli of salamanders with the Feulgen reagent, and had more or less discounted these earlier reports. But several people now began to reexamine this question. Most notably, elegant observations by Jim Keser (Peacock 1965) and Oscar Miller (1966) showed that the multiple oocyte nucleoli of salamanders could take the form of beaded rings, and these rings broke into smaller units when treated with DNase. In my lab, we showed that the multiple nucleoli could bind 3H-actinomycin, an antibiotic that was known to inhibit RNA synthesis by binding to DNA (Ebstein 1967).
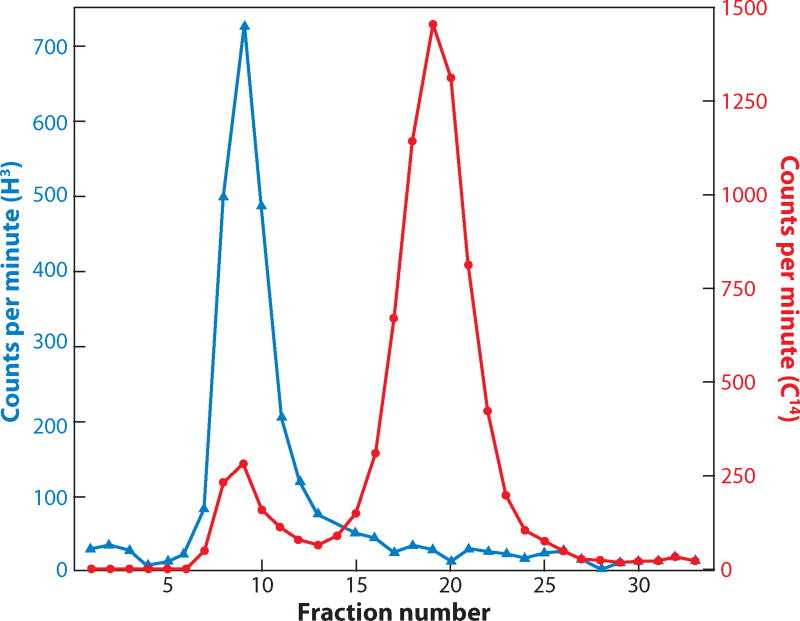
We still had to demonstrate that this nucleolar DNA was rDNA. Here, I was helped by Painter and Taylor’s account. They had not only described a dot of DNA in each nucleolus in late stage oocytes but had shown that the extra DNA accumulated very early in oogenesis as a prominent cap on one side of the nucleus. It turned out to be quite easy to see the extra DNA by staining Xenopus oocytes during the early stages of oogenesis, especially at pachytene. Furthermore, incorporation experiments with 3H-thymidine showed that the extra DNA was being synthesized during prophase, not at the premeiotic S-phase, when the genome is normally replicated (Figure 7) (Gall 1968). The next logical step was to show biochemically that the extra DNA was, indeed, rDNA. This was possible by isolating total DNA from “baby” ovaries. We demonstrated that such ovary DNA contained a large excess of rDNA compared with DNA from somatic tissues (Figure 8). At about the same time, Don Brown and Igor Dawid at the Carnegie Institution approached this issue by hand-isolating thousands of GVs from Xenopus and two species of salamander. Unlike somatic DNA examined at the same time, DNA extracted from the GVs had a prominent satellite peak, which they showed was amplified rDNA (Brown & Dawid 1968).
Figure 7.
Autoradiograph of two pachytene oocyte nuclei of the toad Xenopus after incorporation of radioactive thymidine. Black silver grains lie above the cap of darkly stained amplified rDNA. Note that the chromosomes are not labeled. Thymidine is normally incorporated into DNA during the S phase of mitosis, not during prophase, as in these nuclei.
Figure 8.
A graph showing hybridization of radioactive ribosomal RNA (blue line) to Xenopus ovarian DNA (red line) after centrifugation on a CsCl density gradient. Note the detectable peak of DNA in fractions 6–11, which represents amplified rDNA. Such a peak is not seen in DNA isolated from somatic tissue. Redrawn from Gall (1968).
The extrachromosomal amplification of rDNA in amphibian oocytes posed a number of interesting questions. What was the molecular organization of the amplified genes? How did the rDNA get out of the chromosome? How did it replicate? What was its ultimate fate at the end of oogenesis? Did other organisms have rDNA amplification in their oocytes? Did other genes undergo amplification? My lab and others continued to work on these questions for several years, but space does not permit further discussion here.
IN SITU HYBRIDIZATION
From my earliest observations on giant chromosomes, particularly the polytene chromosomes of Drosophila, I had been intrigued by the possibility of identifying specific genes by some sort of staining test. Coons & Kaplan had shown as early as 1950 that specific protein molecules could be visualized in tissue sections by fluorescent antibodies (Coons & Kaplan 1950). While still at Minnesota, I was so excited by their results that I tried to stain lampbrush chromosomes with fluorescent antibodies—with very little luck—I might add. The thought remained with me that something similar for nucleic acids should be possible. When the melting and reassociation of DNA was demonstrated in the early 1960s and particularly when RNA/DNA hybridization was described, I became even more convinced that some sort of cytological test for gene sequences should be possible. Like others at the time, my lab used 32P-labeled rRNA for hybridization to denatured DNA on filters. Why not try the same on tissues, but use autoradiography as the method of detection and 3H as the label for high resolution? We knew where to look for a positive signal, because we knew that the genes coding for rRNA resided in the nucleolus. I tried several times to hybridize 3H-labeled rRNA to tissue squashes on slides, but each time the resulting autoradiographs were completely negative.
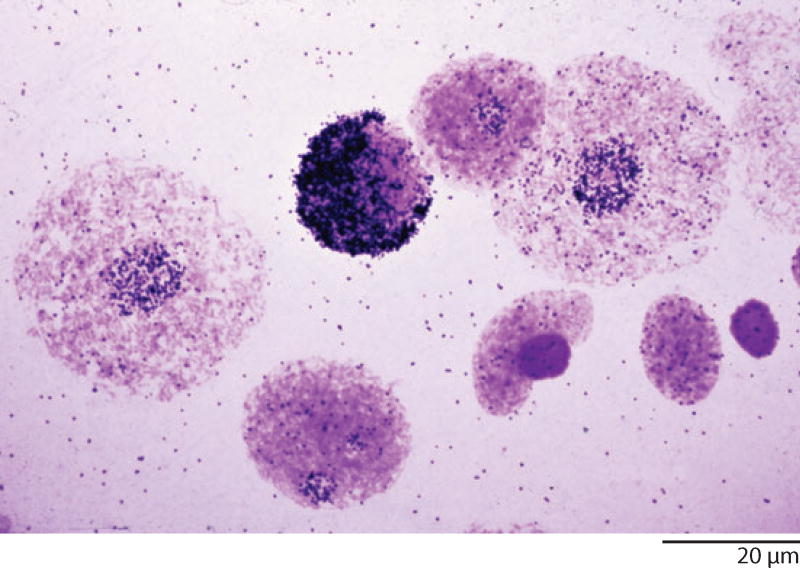
When I first saw the amplified rDNA in young oocytes of Xenopus, I knew I had the perfect test system for a hybridization technique. In these nuclei, the rDNA was amplified roughly 1000 times over that in a diploid cell, so that more than half the DNA in the nucleus was one specific sequence. At this time, I was worried that adequate denaturation of the nuclear DNA would lead to loss of material from the slide, so I decided to use a somewhat cumbersome technique in which oocyte nuclei were suspended in melted agar and then spread as a thin layer on a microscope slide. After hybridization with 3H-labeled rRNA, the slide was covered with autoradiographic emulsion and exposed in the dark for a week or two. The method was not optimal, because only nuclei on the very surface of the agar were close enough for 3H disintegrations to reach the emulsion. Nevertheless, there were hopeful signs that some of the nuclei were radioactive, and that was enough to encourage further experiments. At this point, my student Mary Lou Pardue joined the project, and together we optimized the technique, eventually eliminating the agar altogether (Gall & Pardue 1969, 1971; Pardue & Gall 1969, 1972). In retrospect it became clear that the major problem in the earlier experiments had been the low specific activity of the probes, too low to detect signals in diploid nuclei. Our new in situ hybridization technique allowed us to examine the amplification process in detail. We learned that the major period of amplification was during pachytene of meiosis, but that a low level of amplification was, in fact, detectable as far back as the oogonial stage (Figure 9). The technique was used to examine the distribution of rDNA in other situations as well, such as in the giant polytene nuclei of Drosophila, Sciara, and Rhynchosciara (Pardue et al. 1970, Eckhardt & Gall 1971, Gerbi 1971).
Figure 9.
Autoradiograph of nuclei from the ovary of the toad Xenopus after in situ hybridization with 3H-labeled ribosomal RNA. The most heavily labeled nucleus is in the pachytene stage of meiosis, when the amount of amplified rDNA reaches its maximum. The two largest round nuclei are from oogonia. They show a low level of rDNA amplification compared with the essentially unlabeled diploid follicle nuclei and dense red blood cell nuclei at lower right.
ODD CHROMATIN
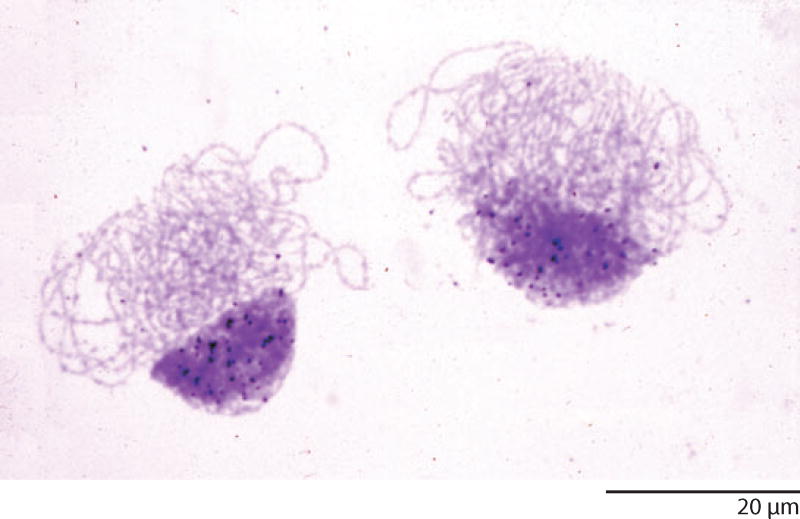
Our new in situ hybridization technique provided a great way to detect specific sequences at the subcellular level, but we were limited to sequences that could be obtained by the relatively crude fractionation procedures available at the time. Cloning and sequencing were still a few years in the future. Among the most obvious candidates were the so-called satellite DNAs, named for the fact that they formed smaller satellite bands near the main band when DNA was centrifuged to equilibrium on a CsCl gradient. Mouse satellite DNA had been characterized in some detail. It constituted approximately 10% of the total mouse genome, and from its reassociation kinetics was known to consist of a relatively simple repeated sequence, but its origin was obscure. Mary Lou Pardue and I made 3H-labeled probes and hybridized them to metaphase chromosome spreads of the mouse (Pardue & Gall 1970). We were delighted to see that hybridization occurred in a distinct region of each chromosome next to the centromere, which was referred to as the centromeric or pericentromeric heterochromatin (Figure 10). Heterochromatin was, and still is, a word with multiple meanings. Originally, it referred to parts of chromosomes that remain condensed and stain darkly during the interphase of mitosis, when the rest of the chromosome, the euchromatin, becomes highly decondensed and stains weakly. Evidence from several organisms, but particularly from Drosophila, suggested that there were very few genes in the heterochromatin. Our observations on mouse satellite provided a straightforward explanation for the lack of genes in heterochromatin—the DNA in those regions was too simple and monotonously repetitive to code for proteins.
Figure 10.
Metaphase chromosomes from cultured cells of the mouse after in situ hybridization with 3H-labeled mouse satellite DNA. Label is evident over the heterochromatin at one end of each chromosome.
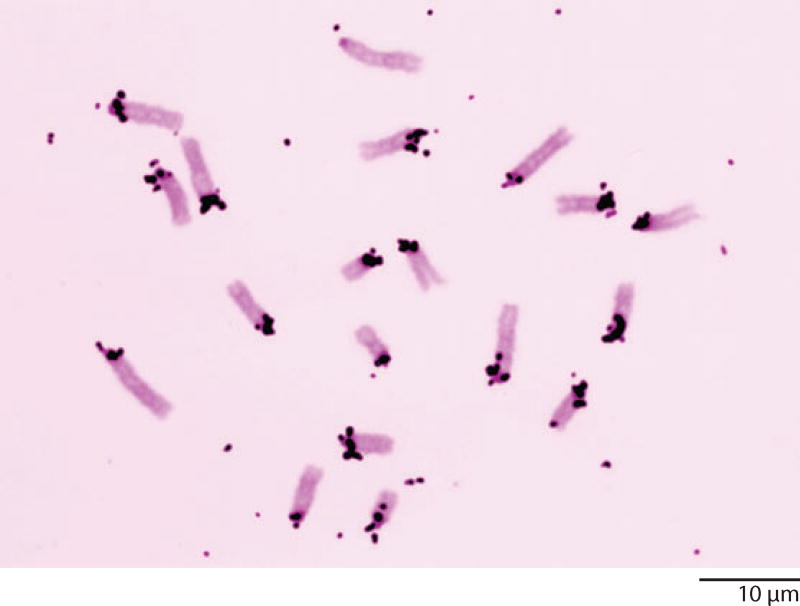
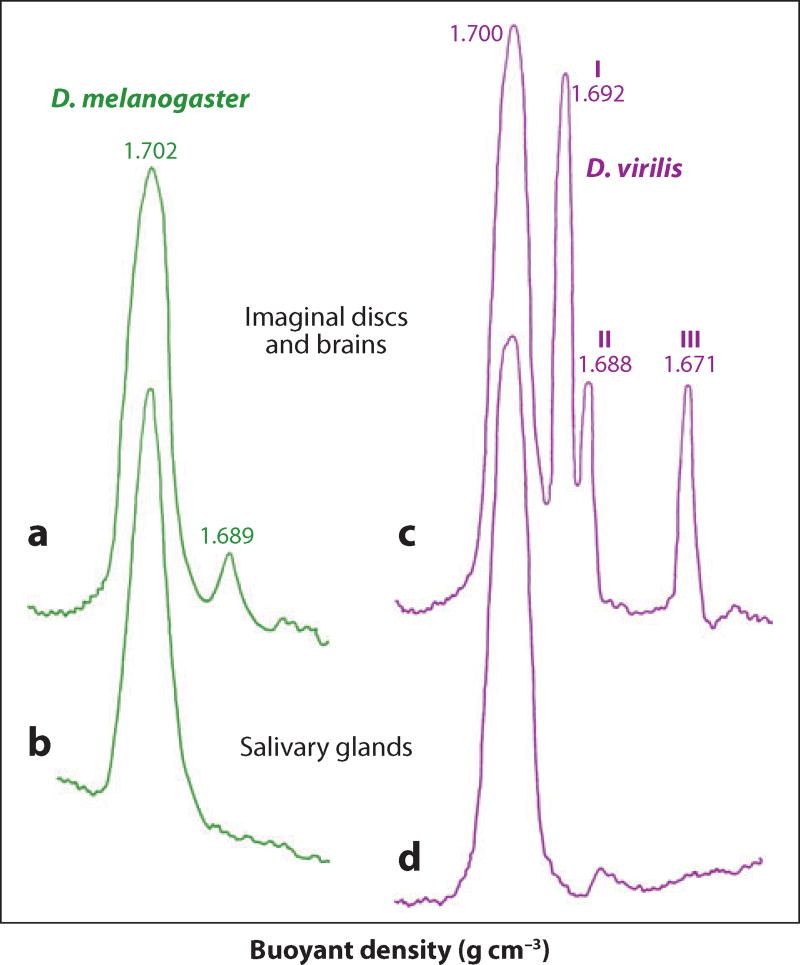
We knew from the genetics and cytology of Drosophila that it could provide an even more compelling case for the nature of heterochromatin. Each chromosome arm of Drosophila melanogaster contained a large block of heterochromatin next to the centromere (the Y chromosome consisted entirely of heterochromatin). Detailed cytogenetic studies had shown that essentially all genes were located in the euchromatin, and that crossing over did not occur in the heterochromatin. The heterochromatin had still another unusual feature. It failed to replicate during formation of the giant polytene chromosomes in the salivary glands—the banded regions corresponded to euchromatin, whereas the unreplicated (or possibly under-replicated) heterochromatin formed a small chromocenter from which the euchromatic arms extended. Studies on another species of Drosophila, D. virilis, showed an even more dramatic situation in which at least half of each chromosome arm consisted of heterochromatin. So we predicted that the DNA from diploid cells of D. melanogaster, and particularly from D. virilis, should have prominent satellite peaks, and that these peaks should not be present or should be much reduced in DNA isolated from the polytene salivary glands.
We were pleased when ultracentrifugation studies confirmed these predictions (Gall et al. 1971). D. melanogaster had one major satellite and D. virilis had three huge satellites that made up approximately 60% of the total DNA (Figure 11). Melting studies suggested that the satellites consisted of short, highly repeated sequences. More detailed analysis later provided the actual sequences (Gall & Atherton 1974, Endow et al. 1975). The satellites were missing from DNA extracted from the salivary glands, suggesting very strongly that the satellites were derived from the heterochromatin. In situ hybridization studies showed that this was, indeed, the case. Satellite probes hybridized prominently to the heterochromatic regions of the chromosomes. The situation in the polytene nuclei of the salivary glands was particularly revealing. Here, hybridization was limited to the chromocenter region, and the amount of hybridization in one giant polytene nucleus was roughly the same as in each nearby diploid cell on the same slide. In other words, during polytenization the heterochromatin failed to replicate, but it did not disappear. The studies from Drosophila nicely complemented those from the mouse, and clearly demonstrated a correlation between heterochromatin, lack of genes, and simple sequence satellite DNA.
Figure 11.
CsCl gradients of DNA isolated from Drosophila melanogaster and D. virilis. The DNA from imaginal discs and brains comes largely from diploid cells. In D. melanogaster there is a small satellite peak on the light side of the main peak (1.689 g cm−3), whereas in D. virilis there are three unusually large peaks (1.671, 1.688, and 1.692 g cm−3). In DNA isolated from salivary glands, which comes largely from the polytene chromosomes, the satellites are essentially absent. The absence of satellites correlates with the failure of the heterochromatin to replicate during polytenization. Redrawn from Gall et al. (1971) with permission from Springer Verlag.
ODD DNA FROM AN ODD ORGANISM
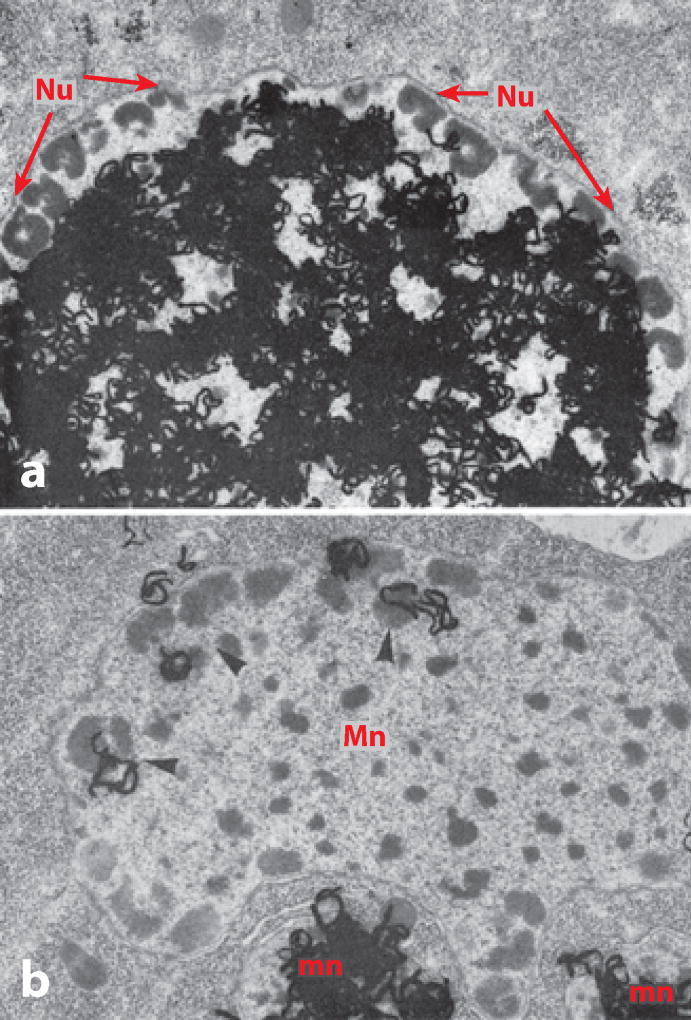
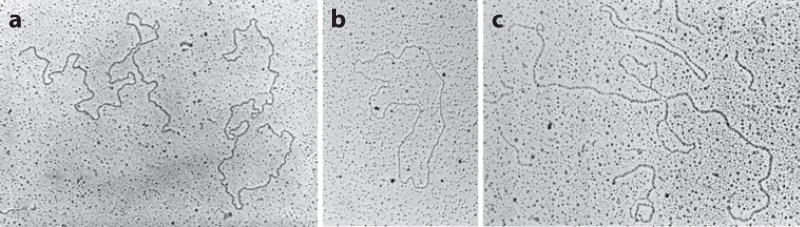
Sometimes research takes an unexpected turn, based on a chance observation or a hunch. This was certainly the case when I started work on the rDNA of Tetrahymena. Tetrahymena is a ciliated protozoan with an unusual genetic system consisting of two nuclei, a large macronucleus, and a small micronucleus. The micronucleus is a normal diploid nucleus. It undergoes mitosis at each cell division and meiosis at the time of conjugation. The macronucleus, however, is polyploid, and unlike almost all other known nuclei, it simply pinches in half at the time of cell division. Furthermore, all transcription takes place in the macronucleus. In other words, Tetrahymena segregates the hereditary and functional aspects of the genome into two separate nuclei. Protozoa had always fascinated me. I collected and observed many species as a teenager, and at Minnesota I carried out research on a distant relative of Tetrahymena called Euplotes (Gall 1959). In 1969 a short paper appeared on DNA replication in Tetrahymena by a French investigator named Charret (1969). Charret fed Tetrahymena 3H-thymidine and then made autoradiographs showing the distribution of label in the nuclei. Most individuals showed uniform label throughout the macronucleus exclusive of the periphery, whereas a few showed label only at the periphery where there were multiple nucleoli (Figure 12). When I first ran across this paper, early in 1974 as I recall, I was struck by the uncanny resemblance to an amphibian oocyte nucleus, and especially by the independent replication of nucleolar DNA. Could there be amplified rDNA molecules in Tetrahymena? To test this hypothesis, I grew up a large batch of Tetrahymena, isolated the DNA, and ran it on a CsCl gradient. The rDNA formed a tiny satellite peak that was well separated from the bulk of the DNA. A second round of centrifugation provided essentially pure rDNA. I then spread the rDNA molecules for observation in the electron microscope. To my delight, the rDNA consisted of linear and circular molecules, most of which had the same length (Figure 13a,b). The whole experiment took less than two weeks, but it provided the starting point for roughly five years of investigation by my laboratory (Gall 1974).
Figure 12.
Autoradiographs of 3H-thymidine incorporation into the DNA of Tetrahymena macronuclei. These are electron micrographs in which individual silver grains in the emulsion are visible as intensely black squiggles. Two patterns are seen in the macronuclei, depending on the point in the cell cycle when labeling took place. (a) The macronucleus is uniformly labeled except for the nucleoli around the periphery. (b) The converse is true: label is limited to the peripheral nucleoli. This pattern suggests that the nucleoli might contain free rDNA molecules. Adapted from Charret (1969) with permission from Elsevier.
Figure 13.
Individual rDNA molecules from the macronucleus of Tetrahymena, viewed by electron microscopy. (a) Two linear molecules, each approximately 22 kb in length. (b) A circular molecule of the same length. (c) A molecule that was partially denatured and then allowed to reassociate. Because the molecule is a palindrome, various configurations are possible after reassociation. In this case, the original center of the molecule now exists as two unpaired loops, connected to the rest of the molecule by two short paired regions. Above the rDNA molecule is a smaller circle of ΦX174 DNA, used as an internal size marker.
PALINDROMES
The first order of business was to characterize the rDNA in more detail. A rough quantitative hybridization experiment suggested that each rDNA molecule contained one 18S and one 28S rRNA coding region. However, this was not correct, as my student Kathy Karrer demonstrated (Karrer & Gall 1976). Instead, each molecule contained two coding regions that were transcribed divergently from the center. In other words, each rDNA molecule was a giant palindrome (Figure 13c). At the time Kathy made this discovery, I was in England visiting Herbert Macgregor’s lab in Leicester, and she conveyed the news in a telegram written as a palindrome. It took me several hours to decipher her meaning and nearly two days to compose a suitable palindromic response! Kathy’s thesis provides an example of an observation made before its time. To determine where the coding regions lay, she hybridized rRNA to partially denatured rDNA molecules, so-called R-loop mapping. In some of her images, there is an extra loop corresponding to what we now know is a short intron in the 28S gene. It was another two years before introns and splicing were discovered in adenovirus (Berget et al. 1977, Chow et al. 1977). The intron in the Tetrahymena gene later led to a Nobel Prize, when Tom Cech showed that it is self-splicing (Zaug & Cech 1980).
DNA ENDS
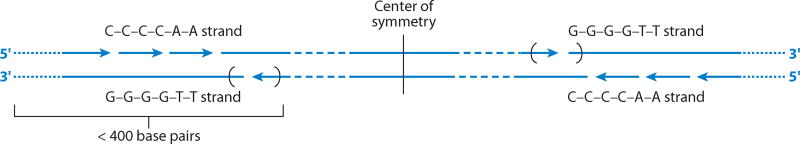
The Tetrahymena rDNA molecule provided still another sequence that turned out to have far-reaching significance. This was the repeated hexanucleotide at the ends of the molecule discovered by Liz Blackburn. Liz came to my laboratory in 1975 as a postdoctoral fellow, after completing a thesis with Fred Sanger at the MRC Laboratory in Cambridge. For her thesis, she had learned the brand-new technique of DNA sequencing and had sequenced 48 bases from phage ΦX174 (Blackburn 1975, 1976). From our first electron microscopic observations on Tetrahymena rDNA, we suspected that the ends of the molecules were unusual, because in the same preparation we found both linear and circular molecules, as well as star-shaped configurations consisting of several molecules held together by their ends. Liz decided to analyze these ends to find out what gave them their unusual sticky property. Eventually, she showed that the ends were heterogeneous, consisting of 20–70 repeats of a simple hexanucleotide sequence CCCCAA/GGGGTT (Blackburn & Gall 1978). Single-stranded nicks and a probable fold-back at the very end of the molecule contributed to the unusual structure and behavior of the rDNA palindromes (Figure 14).
Figure 14.
Hypothetical arrangement of the CCCCAA/GGGGTT repeats at the ends of a Tetrahymena rDNA molecule. Adapted from Blackburn & Gall (1978).
Liz Blackburn’s description of the hexanucleotide CCCCAA/GGGGTT in Tetrahymena is sometimes cited as the discovery of the telomere sequence. This is only a partial truth. In our original paper, there is no mention of telomeres for the simple reason that we knew nothing about telomere sequences in other organisms at that time. We were more concerned with rDNA amplification in Xenopus and how the hexanucleotide repeat might be important for what we considered a similar amplification process in Tetrahymena. It was several years later that Liz and Jack Szostak showed that Tetrahymena repeats could be transferred to the end of a yeast plasmid, where they functioned as a telomere (Szostak & Blackburn 1982). Still later, hexanucleotide sequences were demonstrated at the ends of chromosomes in higher eukaryotes. Some of these were identical to the sequence in Tetrahymena, whereas others differed by only one or two bases. Thus, although in the end (no pun intended) Tetrahymena rDNA termini turned out to be telomeres, this knowledge came more as hindsight than as our research objective at the time.
Later, Liz and her student Carol Greider made the exciting discovery that telomere sequences are added to the ends of chromosomes by templating from the RNA part of a ribonucleoprotein, which they named telomerase (Greider & Blackburn 1987). The role of telomerase in maintaining chromosome ends continues to be an exciting area of study with implications for many important biological problems, including senescence and cancer.
THE CARNEGIE INSTITUTION
By the early 1980s, I had been at Yale for nearly 20 years. Research had gone well, and I enjoyed my colleagues in both the Biology Department and the Molecular Biophysics and Biochemistry Department, where I held a joint appointment. The undergraduates and especially the graduate students were outstanding. Why, then, was I feeling restless and looking for a change? With seniority came many responsibilities that took increasing amounts of my time. I was Director of Graduate Studies, I managed two training grants for graduate students, my lab was full, and I had been teaching a large undergraduate cell biology course for many years. None of these were onerous tasks, and I could not really complain, because I had successfully finessed heavier administrative responsibilities. Still, my real love was working in the lab with my own two hands (and eyes), and I found less and less time for such enjoyable activity. In a chance conversation with Don Brown, I learned that a position might be open in the Embryology Department of the Carnegie Institution, where he was Director. I had spent a sabbatical leave in Don’s lab at the Carnegie in 1981, but at that time I had not been thinking seriously about a change. Nevertheless, it had given me a good chance to see how things operated at the Carnegie, and when the opportunity arose, I jumped at it. The Carnegie provided an extraordinary environment for research free from the teaching and administrative duties that were beginning to weigh on me at Yale.
BACK TO THE GERMINAL VESICLES AND LAMPBRUSH CHROMOSOMES
I always tell my Ph.D. students not to continue work on their thesis problem after they get their own lab. This is generally good advice, but I did not practice what I preached—I continued to work on the amphibian oocyte at Minnesota, at Yale, and again at Carnegie. My excuse is that this cell, and particularly its spectacular GV, is not simply a thesis problem, but a model system for cells and nuclei in general. In any case, at Carnegie I discontinued work on Tetrahymena and returned to oocytes.
Most earlier studies on the amphibian GV, both in my lab and elsewhere, had concentrated on the LBCs and nucleoli. We knew there were hundreds to thousands of other extrachromosomal bodies in each GV, but little attention had been paid to them. Our first task was to define how many different kinds of bodies there were, based on their molecular composition. Things turned out to be simpler than expected when we applied various in situ hybridization and immunofluorescent probes. The vast majority of bodies in the Xenopus GV fell into just two categories—a relatively small number of objects that were already known by the name spheres, and a larger number of nameless bodies that we dubbed B-snurposomes (Wu et al. 1991). Both stained conspicuously with antibodies against the Sm proteins, and both gave in situ hybridization signals with probes against snRNAs (hence the term snurposome, derived from snRNP, which is pronounced “snurp”). The B-snurposomes remain to be studied in detail. We think they correspond more or less to the structures called speckles or interchromatin granule clusters in other cell types. The spheres attracted my attention for a variety of reasons and became the focus of my research for several years.
SPHERES AND OTHER BODIES
From our earliest descriptions of LBCs, both Callan and I realized that spheres occurred not only as free bodies in the nucleoplasm, but also were attached to a few specific sites on two or three chromosomes, depending on the species under study. By chance, we discovered that these sites were the histone gene loci (Gall et al. 1981, Callan et al. 1991). We suspected that the machinery for processing histone pre-mRNA might be localized in the spheres, but the U7 snRNP, which carries out this reaction, had not been described for any amphibian. When probes became available for Xenopus, we showed that the U7 snRNP is indeed a unique component of spheres (Wu & Gall 1993). The association of spheres with the histone genes seemed a reasonable way to provide processing factors to sites where they were needed. But it did not explain why most spheres should be far distant from those sites, and this remains a puzzle to this day.
Spheres took on much greater interest with the discovery of a protein called coilin, which derived its name from its unique localization in small nuclear structures known as coiled bodies (Andrade et al. 1991, Raska et al. 1991). Coiled bodies were described in the late 1960s on the basis of their morphology in electron micrographs of mammalian nuclei, but they remained largely unknown to most cell biologists. Working independently, Mark Roth (a former postdoc) and I found coilin in the spheres of Xenopus, which we promptly renamed coiled bodies (Tuma et al. 1993, Wu et al. 1994). As more and more organisms were studied, it became clear that coiled bodies, as defined by coilin, were present in a wide variety of cell types in mammals, amphibians, insects, and even plants. Like the spheres in the GV, the bodies in other cell types had their own specific names (endobodies, nucleolus-associated bodies, etc.). The very first description came from the great neuroanatomist and Nobel laureate, Ramón y Cajal, who called them accessory bodies (in Spanish, cuerpo accesorio) because he thought they were somehow accessory to the nucleolus (Cajal 1903). Given the plethora of names, the time was ripe for a rationalization of nomenclature. So I suggested the name Cajal body (CB) for all of the previously described coilin-positive bodies (Gall et al. 1999). Fortunately, the field was relatively small at the time and my suggestion was accepted.
CBs in the Xenopus GV contain a wide variety of macromolecules in addition to those concerned with histone pre-mRNA processing. These include the splicing snRNPs, RNA polymerase, and factors involved in rRNA processing (Figure 15). Some of these are well-known constituents of CBs in other organisms. At the same time, however, it remains unclear whether there are major differences in CB composition and function in different organisms and cell types.
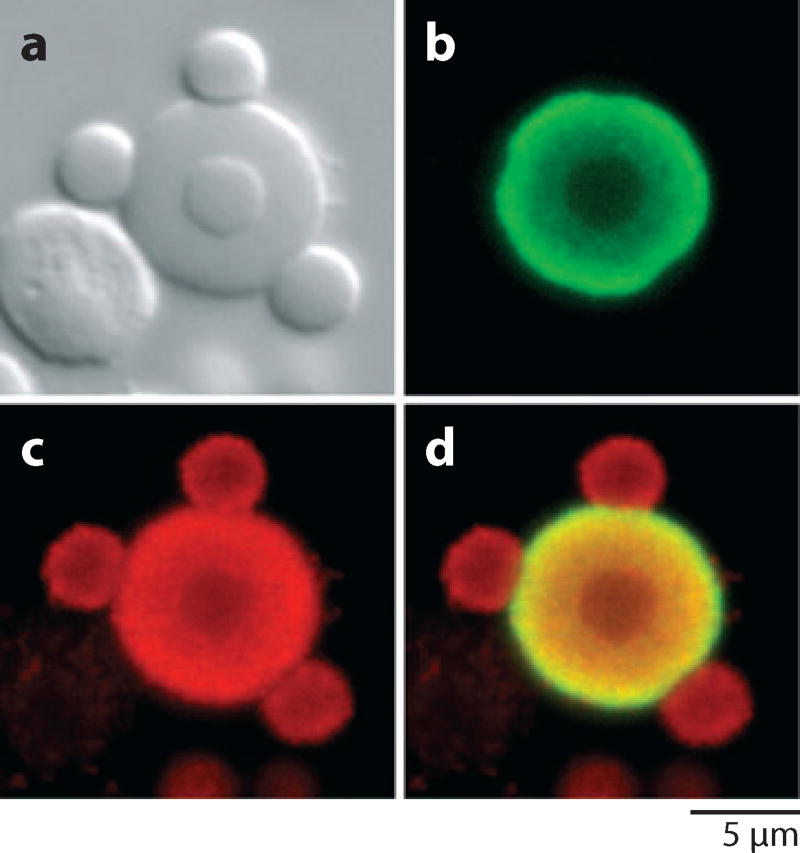
Figure 15.
A single Cajal body from a Xenopus germinal vesicle visualized by (a) differential interference contrast and (b–d ) confocal laser scanning microscopy.
(b) Stained with an antibody against coilin.
(c) Stained with an antibody against the Sm epitope, now known to be symmetric dimethylarginine.
(d ) Merge of (b) and (c). These images show three speckles on the surface of the Cajal body and one inside. The unstained object to the lower left of the Cajal body in (a) is a free nucleolus. Modified from Bellini & Gall (1998) with permission from Molecular Biology of the Cell.
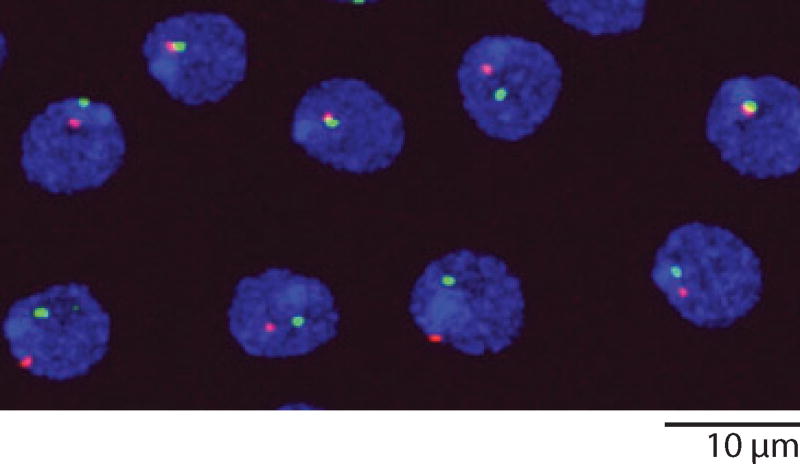
Several years ago, I decided to make a concerted effort to characterize coilin and the CB in Drosophila, an organism in which one might approach the function of CBs through genetics. A major problem with Drosophila was the fact that standard antibodies against vertebrate coilin did not give reliable cross-reactions with Drosophila tissues, and more importantly no one had found Drosophila coilin by sequence analysis. We decided to look for the Drosophila CB using markers other than coilin, especially the U7 snRNP and the small CB-associated (sca) RNA, U85 scaRNA. We were surprised to find these two components in separate bodies (Liu et al. 2006). The body that contained the U7 snRNP was invariably attached to the histone gene locus on the second chromosome. For that reason we called it the histone locus body (HLB). U85 scaRNA defined a second body. Because it also contained several other conventional CB components, we designated it the CB. In our most recent studies, we have at last identified the elusive Drosophila coilin. Not surprisingly, it is a prominent component of the previously identified CB, but it is also present in HLBs, usually at a lower concentration (Liu et al. 2009) (Figure 16). Now that we have identified Drosophila coilin, the coilin gene, and two types of bodies that contain coilin, we hope to use fly genetics, cell biology, and developmental biology to gain better insight into the role that CBs play in cellular physiology.
Figure 16.
Nuclei from the ejaculatory duct of a male Drosophila melanogaster, stained with antibodies against coilin (green) to show Cajal bodies and Lsm 11 (red ) to show histone locus bodies. Lsm11 is a component of the U7 snRNP.
EPILOGUE
In reviewing my scientific history, I am struck by how lucky I have been in ways that do not appear in the official record. I was privileged from the outset growing up in a well-to-do family under circumstances that shielded me from the violence of the Second World War. My formal education at Yale assured me access to the best scientific minds in the country. Moreover, the organization of science, and indeed the whole educational system in the United States, diverged sharply from the hierarchical European model. Shortly after the end of the Second World War, both the NIH and the NSF began to make research grants directly to investigators, not to departments or to powerful heads of institutes. Private foundations did the same. I am forever grateful that I have had both public and private support to run my own laboratory from the beginning of my career. We need to preserve this system, confident that innovation comes from individuals who have the freedom to pursue their own agendas.
At the same time, everything I have accomplished has been done with the help of dedicated graduate students, postdoctoral fellows, technicians, colleagues, and even a few undergraduates and high school students. I regret there is not space to acknowledge them all in this article. However, a complete list can be found on my Web site (http://www.ciwemb.edu/labs/gall/index.php).
Acknowledgments
Research from my laboratory has been supported continuously since the 1950s by grants from the National Institutes of Health, currently by GM33397 from the National Institute of General Medical Sciences. Most of our studies on Tetrahymena were supported by the American Cancer Society, which also appointed me American Cancer Society Professor of Developmental Genetics in 1984.
Biography

Footnotes
DISCLOSURE STATEMENT
The author is not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 1991;173:1407–19. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol. Biol. Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA. Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. 1977;74:3171–75. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Transcription by Escherichia coli RNA polymerase of a single-stranded fragment by bacteriophage ΦX174 DNA48 residues in length. J. Mol. Biol. 1975;93:367–74. doi: 10.1016/0022-2836(75)90283-1. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Transcription and sequence analysis of a fragment of bacteriophage ΦX174 DNA. J. Mol. Biol. 1976;107:417–31. doi: 10.1016/s0022-2836(76)80075-7. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Brachet J. La localisation de l’acide thymonucléique pendant l’oogenèse et la maturation chez les amphibiens. Arch. Biol. 1940;51:151–65. [Google Scholar]
- Brown DD, Dawid IB. Specific gene amplification in oocytes. Science. 1968;160:272–80. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- Cajal SRy. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. (Madrid) 1903;2:129–221. [Google Scholar]
- Callan HG. Fine Structure of Cells: Symposium of the VIIIth Congress in Cell Biology, Leiden 1954, Int. Union Biol. Sci. Publ, Ser. B. Groningen: Noordhof; 1954. Recent work on the structure of cell nuclei; pp. 89–109. [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–51. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Callan HG, Lloyd L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti) Philos. Trans. R. Soc. Lond. B. 1960;243:135–219. [Google Scholar]
- Callan HG, Macgregor HC. Action of deoxyribonuclease on lampbrush chromosomes. Nature. 1958;181:1479–80. doi: 10.1038/1811479a0. [DOI] [PubMed] [Google Scholar]
- Charret R. L’ADN nucléolaire chez Tetrahymena pyriformis: chronologie de sa réplication. Exp. Cell Res. 1969;54:353–61. doi: 10.1016/0014-4827(69)90214-6. [DOI] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Coons AH, Kaplan MH. Localization of antigen in tissue cells; improvements in a method for the detection of antigen by means of fluorescent antibody. J. Exp. Med. 1950;91:1–13. doi: 10.1084/jem.91.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duryee WR. Isolation of nuclei and non-mitotic chromosome pairs from frog eggs. Arch. exp. Zellforsch. 1937;19:171–76. [Google Scholar]
- Duryee WR. Cytology, Genetics, and Evolution. Univ. Penn. Bicentennial Conf. Philadelphia, PA: Univ. Penn; 1941. The chromosomes of the amphibian nucleus; pp. 129–41. [Google Scholar]
- Ebstein BS. Tritiated actinomycin D as a cytochemical label for small amounts of DNA. J. Cell Biol. 1967;35:709–13. doi: 10.1083/jcb.35.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt RA, Gall JG. Satellite DNA associated with heterochromatin in Rhynchosciara. Chromosoma. 1971;32:407–27. doi: 10.1007/BF00285252. [DOI] [PubMed] [Google Scholar]
- Endow SA, Polan ML, Gall JG. Satellite DNA sequences of Drosophila melanogaster. J. Mol. Biol. 1975;96:665–92. doi: 10.1016/0022-2836(75)90145-x. [DOI] [PubMed] [Google Scholar]
- Gall JG. Lampbrush chromosomes from oocyte nuclei of the newt. J. Morphol. 1954a;94:283–352. [Google Scholar]
- Gall JG. Observations on the nuclear membrane with the electron microscope. Exp. Cell Res. 1954b;7:197–200. doi: 10.1016/0014-4827(54)90054-3. [DOI] [PubMed] [Google Scholar]
- Gall JG. Fibrous Proteins and Their Biological Significance. Symp. Soc. Exp. Biol. No. 9. London: Soc. Exp. Biol; 1955. Problems of structure and function in the amphibian oocyte nucleus; pp. 358–70. [Google Scholar]
- Gall JG. Mutation. Brookhaven Symp. Biol. No. 8. Upton, NY: Brookhaven Natl. Lab; 1956. On the submicroscopic structure of chromosomes; pp. 17–32. [PubMed] [Google Scholar]
- Gall JG. Chromosomal differentiation. In: McElroy WD, Glass B, editors. A Symposium on the Chemical Basis of Development. Baltimore, MD: Johns Hopkins; 1958. pp. 103–35. [Google Scholar]
- Gall JG. Macronuclear duplication in the ciliated protozoan Euplotes. J. Biophys. Biochem. Cytol. 1959;5:295–308. doi: 10.1083/jcb.5.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Centriole replication. A study of spermatogenesis in the snail Viviparus. J. Biophys. Biochem. Cytol. 1961;10:163–93. doi: 10.1083/jcb.10.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Kinetics of deoxyribonuclease action on chromosomes. Nature. 1963;198:36–38. doi: 10.1038/198036a0. [DOI] [PubMed] [Google Scholar]
- Gall JG. Nuclear RNA of the salamander oocyte. In: Vincent WS, Miller OL, Drets ME, Saez FA, editors. The Nucleolus—Its Structure and Function. National Cancer Institute Monograph 23. Bethesda, MD: Natl. Cancer Inst; 1966a. pp. 475–88. [PubMed] [Google Scholar]
- Gall JG. Methods in Cell Physiology. New York: Academic; 1966b. Techniques for the study of lampbrush chromosomes; pp. 37–60. [Google Scholar]
- Gall JG. Octagonal nuclear pores. J. Cell Biol. 1967;32:391–99. doi: 10.1083/jcb.32.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oogenesis. Proc. Natl. Acad. Sci. USA. 1968;60:553–60. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc. Natl. Acad. Sci. USA. 1974;71:3078–81. doi: 10.1073/pnas.71.8.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Atherton DD. Satellite DNA sequences in Drosophila virilis. J. Mol. Biol. 1974;85:633–64. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Callan HG. H3 Uridine incorporation in lampbrush chromosomes. Proc. Natl. Acad. Sci. USA. 1962;48:562–70. doi: 10.1073/pnas.48.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Cohen EH, Polan ML. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–44. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Gall JG, Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. USA. 1969;63:378–83. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Pardue ML. Nucleic acid hybridization in cytological preparations. In: Grossman L, Moldave K, editors. Methods in Enzymology. Vol. 21. New York: Academic; 1971. pp. 470–80. Nucleic Acids, Pt. D. [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–71. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gerbi SA. Localization and characterization of the ribosomal RNA cistrons in Sciara coprophila. J. Mol. Biol. 1971;58:499–511. doi: 10.1016/0022-2836(71)90367-6. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–98. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Karrer KM, Gall JG. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J. Mol. Biol. 1976;104:421–53. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- Liu J-L, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell. 2009;20:1661–70. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006;172:875–84. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch. mikrosk. Anat. 1934;21:294–328. [Google Scholar]
- Meselson M, Stahl FW. The replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1958;44:671–82. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL. Structure and composition of peripheral nucleoli of salamander oocytes. In: Vincent WS, Miller OL, Drets ME, Saez FA, editors. International Symposium on The Nucleolus—Its Structure and function. Bethesda, MD: Natl. Cancer Inst; 1966. pp. 53–66. [PubMed] [Google Scholar]
- Mizukami I, Gall JG. Centriole replication II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter TS, Taylor AN. Nucleic acid storage in the toad’s egg. Proc. Natl. Acad. Sci. USA. 1942;28:311–17. doi: 10.1073/pnas.28.8.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl. Acad. Sci. USA. 1969;64:600–4. doi: 10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, Gall JG. Chromosomal localization of mouse satellite DNA. Science. 1970;168:1356–58. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Pardue ML, Gall JG. Molecular cytogenetics. In: Sussman M, editor. Molecular Genetics and Developmental Biology. Englewood Cliffs, NJ: Prentice-Hall; 1972. pp. 65–99. [Google Scholar]
- Pardue ML, Gerbi SA, Eckhardt RA, Gall JG. Cytological localization of DNA complementary to ribosomal RNA in polytene chromosomes of Diptera. Chromosoma. 1970;29:268–90. doi: 10.1007/BF00325943. [DOI] [PubMed] [Google Scholar]
- Peacock WJ. Chromosome replication. In: Valencia JI, Grell RF, editors. International Symposium on Genes and Chromosomes—Structure and function. Bethesda, MD: Natl. Cancer Inst.; 1965. pp. 101–31. Natl Cancer Inst. Monogr. 18. [Google Scholar]
- Perry RP. The cellular sites of synthesis of ribosomal and 4S RNA. Proc. Natl. Acad. Sci. USA. 1962;48:2179–86. doi: 10.1073/pnas.48.12.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, et al. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Ritossa FM, Spiegelman S. Localization of DNA complementary to ribosomal RNA in the nucleolus organizer region of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1965;53:737–45. doi: 10.1073/pnas.53.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Gall JG. Monoclonal antibodies that recognize transcription unit proteins on newt lampbrush chromosomes. J. Cell Biol. 1987;105:1047–54. doi: 10.1083/jcb.105.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert J. Zur Entwickelungsgeschichte des Ovarialeies bei Selachiern. Anat. Anz. 1892;7:107–58. [Google Scholar]
- Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–55. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Taylor JH, Woods PS, Hughes WL. The organization and duplication of chromosomes as revealed by autoradiographic studies using tritium-labeled thymidine. Proc. Natl. Acad. Sci. USA. 1957;43:122–28. doi: 10.1073/pnas.43.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J. Cell Biol. 1993;122:767–73. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. An Introduction to Modern Genetics. New York: Macmillan; 1939. p. 441. [Google Scholar]
- Wilson EB. The Cell in Development and Heredity. New York: Macmillan; 1925. p. 1232. [Google Scholar]
- Wu C-HH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. USA. 1993;90:6257–59. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J. Cell Biol. 1991;113:465–83. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol. Biol. Cell. 1994;5:1119–27. doi: 10.1091/mbc.5.10.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. In vitro splicing of the ribosomal RNA precursor in nuclei of Tetrahymena. Cell. 1980;19:331–38. doi: 10.1016/0092-8674(80)90507-3. [DOI] [PubMed] [Google Scholar]