Abstract
Avidocin-CDs are a new class of precision bactericidal agents that do not damage resident gut microbiota and are unlikely to promote the spread of antibiotic resistance. The precision killing properties result from the fusion of bacteriophage receptor binding proteins (RBPs) to a lethal contractile scaffold from an R-type bacteriocin. We recently described the prototypic Avidocin-CD, Av-CD291.2, that specifically kills C. difficile ribotype 027 strains and prevents colonization of mice. We have since selected two rare Av-CD291.2 resistant mutants of strain R20291 (RT027; S-layer cassette type-4, SCLT-4). These mutants have distinct point mutations in the slpA gene that result in an S-layer null phenotype. Reversion of the mutations to wild-type restored normal SLCT-4 S-layer formation and Av-CD291.2 sensitivity; however, complementation with other SCLT alleles did not restore Av-CD291.2 sensitivity despite restoring S-layer formation. Using newly identified phage RBPs, we constructed a panel of new Avidocin-CDs that kill C. difficile isolates in an SLCT-dependent manner, confirming the S-layer as the receptor in every case. In addition to bacteriophage adsorption, characterization of the S-layer null mutant also uncovered important roles for SlpA in sporulation, resistance to lysozyme and LL-37, and toxin production. Surprisingly, the S-layer-null mutant was found to persist in the hamster gut despite its completely attenuated virulence. Avidocin-CDs have significant therapeutic potential for the treatment and prevention of C. difficile Infection (CDI) given their exquisite specificity for the pathogen. Furthermore, the emergence of resistance forces mutants to trade virulence for continued viability and, therefore, greatly reduce their potential clinical impact.
Introduction
New antibacterial agents are needed to counteract the impending loss of effective treatment options for multi-drug resistant bacteria. Furthering this need is the realization that dysbiosis caused by broad-spectrum antibiotic use contributes to the prevalence of diseases/disorders such as inflammatory bowel disease, obesity and gastrointestinal infections (1). Strategies to overcome these threats include use of narrow spectrum or precision agents and the design of drugs that target virulence instead of in vitro viability (2, 3). One pathogen for which alternative treatment approaches are needed is C. difficile. This spore-forming, obligate anaerobe is the leading cause of nosocomial infections worldwide. Approximately 450,000 cases and 29,000 deaths each year are attributed to this pathogen in the US alone (4). As a result, the Centers for Disease Control and Prevention has identified C. difficile as an urgent threat to human health (5). This opportunistic pathogen exploits a reduction in gut microbiota diversity that often follows broad spectrum antibiotic use to proliferate, release toxins, and cause life-threatening colitis (6). Although the toxins have been studied in great detail, other aspects of C. difficile virulence, including colonization of the gut, are not well understood (6). The C. difficile cell surface is covered by a paracrystalline surface layer (S-layer) largely comprised of SlpA and sparsely interspersed by 28 related cell wall proteins (7). The S-layer precursor SlpA is proteolytically processed on the cell surface to generate the Low and High Molecular Weight S-layer Proteins (LMW and HMW SLPs). The SLPs interact with high-affinity to form a heterodimer, the basic unit of the mature S-layer (8). The slpA gene is located within a highly variable S-layer cassette consisting of 5 genes; 13 distinct S-layer cassette types (SLCTs) have been described to date (9). The variation that defines individual cassette types is largely confined to the LMW SLP-encoding region of slpA (4). The HMW SLP region is highly conserved and includes the cell wall binding motifs that anchor the S-layer to the cell wall (10). The S-layer and several associated cell wall proteins have been implicated in colonization of host tissues (7) and in stimulation of the host immune response via TLR4 signaling (11).
Whereas C. difficile is not significantly resistant to the frontline antibiotics used to treat CDIs (vancomycin, metronidazole and fidaxomicin), the use of these antibiotics causes further disruption of the resident microbiota leading to frequent CDI relapse (6). To be safe and effective, new agents to treat and prevent CDIs must not harm the diverse gut microbiota and the colonization resistance it provides. Avidocin-CDs represent one such potential agent (12). These bactericidal proteins are genetically modified versions of natural R-type bacteriocins (a.k.a. diffocins) produced by C. difficile to kill competing C. difficile strains (13). Diffocins resemble myoviridae phage tails and consist of a contractile sheath, nanotube core, baseplate and tail fiber structures. However, instead of delivering DNA across the bacterial membrane as does a bacteriophage, R-type bacteriocins function as killing machines by injecting a nanotube core through the bacterial cell envelope and creating a small pore that dissipates the cell’s membrane potential (14). Killing specificity is determined by the receptor binding proteins (RBPs) located at the tail fiber tips that trigger sheath contraction upon binding with a cognate receptor on the bacterial cell surface. Genetic replacement or fusion of the RBP gene with homologues from other strains or RBP sources (i.e. C. difficile bacteriophages and prophage insertions) make it possible to retarget killing (12, 15–17). These modified bacteriocins are known as Avidocin-CDs. An Avidocin-CD prototype, Av-CD291.2, constructed with a bacteriophage RBP identified within a prophage insertion was found to be more stable than the natural parent diffocin. Av-CD291.2 had a modified killing spectrum that included all hypervirulent RT027 C. difficile strains tested, blocked C. difficile colonization in a mouse model of spore transmission and did not disrupt the resident gut microbiota (12). These properties encourage the further development of Avidocin-CDs as oral human therapeutics.
Here we further characterize the Av-CD291.2 mechanism of action and describe an expanded panel of Avidocin-CDs that cover all clinically relevant C. difficile strain types using newly identified bacteriophage RBPs. Rare resistant mutants were isolated in vitro under Av-CD291.2 selection. Analysis of these mutants enabled the identification of SlpA as the cell surface receptor for all tested Avidocin-CDs and, by extension, the corresponding bacteriophage and prophage RBP-sources. We also identified previously unsuspected roles for SlpA in sporulation, resistance to lysozyme and the natural anti-microbial peptide LL-37, toxin production and virulence. Surprisingly, despite their complete attenuation of virulence, resistant mutants could colonize and persist in the hamster gut for the duration of a 14-day study. These findings imply that antimicrobial agents that force pathogens to trade virulence for continued viability would have clear clinical advantages should resistance emerge during treatment.
Results
Av-CD291.2-resistant mutants lack an S-layer
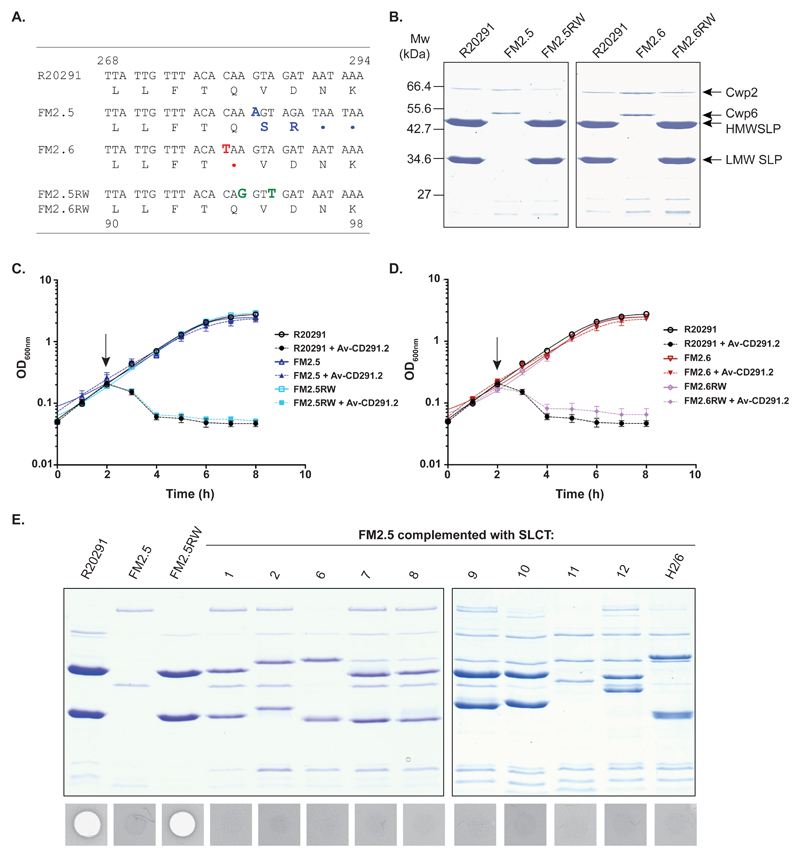
Resistance to any antimicrobial agent can occur and be exploited to understand its mode of action. We isolated two spontaneous mutants of C. difficile strain R20291 (ribotype 027) that were resistant to killing by Av-CD291.2. These mutants appeared at a frequency of < 1 x 10-9 and were found to encode independent point mutations in the slpA gene (Fig. 1A). Both mutations were predicted to truncate SlpA at a site N-terminal to the post-translational cleavage site and, thereby, prevent formation of an S-layer. Both FM2.5 and FM2.6 lacked detectable cell surface S-layer protein subunits as predicted but still expressed minor cell wall proteins including Cwp2 and Cwp6 (Fig. 1B). These mutations did not affect the growth rate of the bacteria in vitro (Fig. S4B); however, FM2.5 displayed a slight, but statistically significant earlier entry into stationary phase (maximum OD600nm: FM2.5 = 2.2; R20291 = 3.2, p = 0.000012). We attempted to complement the FM2.5 and FM2.6 mutations with a plasmid-borne wild-type R20291 slpA; however, unexpected homologous recombination restored the wild type slpA gene to the chromosome (Fig. S1). Accordingly, we created genetically identifiable recombinants “watermarked” with synonymous substitutions in the R20291 slpA allele (Fig. 1A). The resulting strains, FM2.5RW and FM2.6RW, were found to have LMW and HMW SLPs on their surface (Fig. 1B) and regained sensitivity to Av-CD291.2 (Fig. 1C and D). These results confirmed that an intact S-layer is required for Av-CD291.2 killing but did not determine whether SlpA itself or a protein dependent on the S-layer for surface localization was the receptor for Av-CD291.2. To address this question, slpA alleles from the most common non-R20291 SLCTs were individually cloned into an inducible expression plasmid and transferred into the S-layer deficient FM2.5 (Fig. 1E). Each resulting strain expressed the expected LMW and HMW SLPs on the cell surface, indicative of S-layer formation (9), but did not regain sensitivity to Av-CD291.2, thus, ruling out the possibility that the simple formation of an S-layer was responsible for sensitivity to Av-CD291.2. To address whether Av-CD291.2 directly interacts with SLCT-4 SlpA, plasmids encoding SlpA from 8 different SLCTs were introduced into the non-isogenic laboratory strain 630 (SLCT-7; ribotype 012), which is insensitive to Av-CD291.2 (Table S1). In this fully S-layer competent SLCT-7 wild type strain, only induction of SLCT-4 SlpA was sufficient to confer sensitivity to Av-CD291.2 (Fig. S3). Moreover, the degree of sensitivity was dependent on the level of induction with only 10 ng/mL of inducer required (Fig. S3A). Taken together these observations clearly demonstrate that the SLCT-4 variant of SlpA is the cell surface target of Av-CD291.2.
Fig. 1.
Mutations in slpA confer Av-CD291.2 resistance. (A) Alignment of the slpA sequence (nucleotides 268-294) from R20291, FM2.5, FM2.6, FM2.5RW and FM2.6RW. A nucleotide insertion at position 283 of FM2.5 slpA results in a frameshift and premature stop codon (in blue). A nucleotide substitution at position 280 of FM2.6 slpA results in a nonsense mutation (in red). To allow differentiation from the wild type sequence, two synonymous mutations were introduced into slpA in FM2.5RW and FM2.6RW (in green) (B) SDS-PAGE analysis of S-layer extracts from R20291, FM2.5, FM2.6, FM2.5RW and FM2.6RW. The positions of the LMW and HMW SLPs and minor cell wall proteins Cwp2 and Cwp6 are indicated. (C and D) The impact of Av-CD291.2 on exponentially growing R20291, FM2.5, FM2.5RW and FM2.6RW was monitored by measuring the optical density at 600 nm. Av-D291.2 addition is indicated with an arrow. Experiments were carried out in triplicate on biological duplicates. Means and standard deviations are shown. (E) SDS-PAGE analysis and Av-CD291.2 sensitivity of FM2.5 complemented with slpA alleles from multiple SLCTs following induction with anhydrotetracycline (20 ng/ml). R20291 and FM2.5RW are included as controls. A zone of clearance in the agar lawn indicates killing.
Sensitivity to all Avidocin-CDs is slpA allele-specific
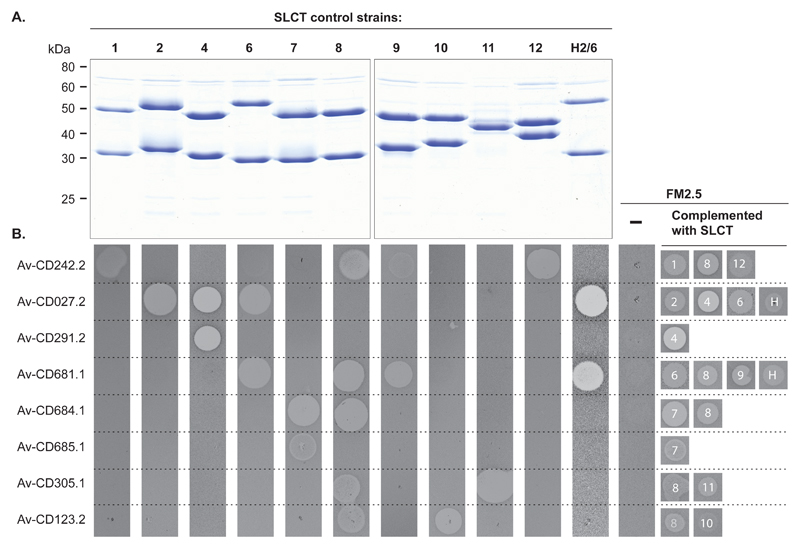
These observations suggested that each variant of SlpA may serve as a specific receptor for additional C. difficile bacteriophage RBPs. In an attempt to expand Avidocin-CD coverage beyond SLCT-4, we constructed ten new Avidocin-CDs using predicted bacteriophage RBPs mined from the genome sequences of C. difficile clinical isolates or newly isolated bacteriophages (Table S1). Source strains for RBP sequences were chosen based on their SLCT since strain typing information for the RBP source often correlates with sensitivity to the corresponding Avidocin (e.g. ribotype 027 and sensitivity to Av-CD291.2; SLCT-1 of phi-147 propagating strain and sensitivity to Av-CD147.1) (12, 15, 16). Preparations of Diffocin-4 (scaffold for all Avidocin-CDs), Av-CD291.2, and each of the new Avidocin-CDs were tested for killing activity on a panel of 62 C. difficile isolates containing all 13 known SLCTs and a newly identified 14th SLCT (Fig. S2). Only two strains, representing SLCTs 3 and 5, were not killed by a single Avidocin-CD (Fig S2); otherwise, every isolate from all 12 other SLCTs was killed by at least one Avidocin-CD. A near perfect correlation was observed between a strain’s SLCT and its sensitivity to each of the Avidocin-CDs. The only exceptions were SLCT-2 isolates with Av-CD027.2 and Av-CD685.1. A strong correlation was also observed between ribotype and sensitivity to a particular Avidocin-CD since all ribotypes in the panel, except 012, 014 and 015, were found to associate exclusively with a single SLCT. Similar correlations with strain sensitivity and SLCT or ribotype were not observed for killing with Diffocin-4, suggesting this natural R-type bacteriocin binds to C. difficile via another receptor.
Having made these observations, we wanted to determine if sensitivity to specific Avidocin-CDs was directly dependent upon the variant of SlpA present. The panel of isogenic FM2.5 strains complemented with slpA alleles from the 10 most common SLCTs was tested for sensitivity to a panel of the most potent Avidocin-CDs (Fig. 2). The corresponding parental strains from which the slpA alleles were obtained were used as controls. Each complemented strain became sensitive to the same Avidocin-CDs as the parental strain. To confirm that this engineered sensitivity did not result from altered cell surface architecture, an analogous experiment was performed in a second panel of isogenic strains created when plasmids encoding SlpAs from 8 SLCTs were introduced into the S-layer competent strain 630 (Fig. S3B and C). As before, the spectrum of killing was identical to that of the parental strains from which the slpA alleles were obtained. These data conclusively demonstrate that the polymorphic SlpA acts as the binding receptor for each of the Avidocin-CDs tested and, therefore, the corresponding bacteriophage RBPs.
Fig. 2.
Avidocin-CD sensitivity correlates with SLCT. (A) SDS-PAGE analysis of SLPs extracted from a panel of strains representing the 11 most commonly isolated SLCTs. (B) Spot bioassays with 8 Avidocin-CDs on the C. difficile strains used in panel A, as well as FM2.5 alone (-) and FM2.5 complemented with slpA alleles from 10 SLCTs following induction with anhydrotetracycline (20 ng/ml). The zone of clearance caused by each Avidocin-CD is shown along with SLCT (H = Hybrid 2/6).
S-layer null mutants are abnormally sensitive to innate immune effectors
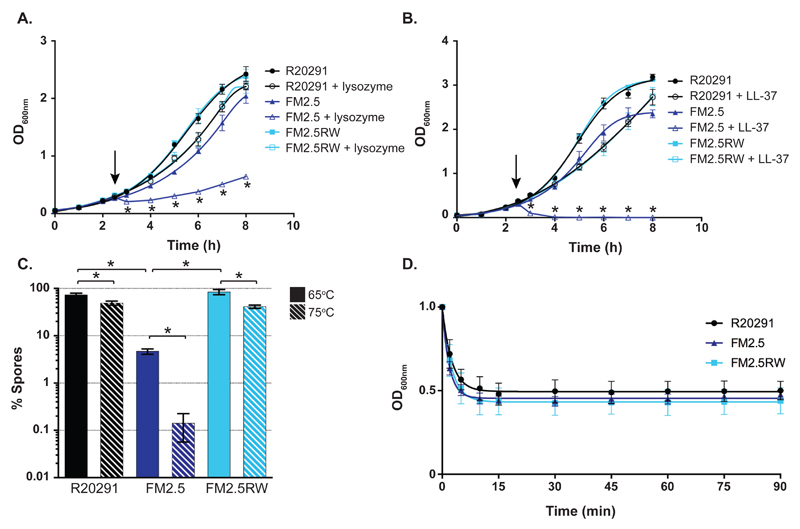
Bacterial S-layers serve many critical cellular functions (7). Given its location on the cell surface, the S-layer has been proposed to act as a molecular sieve to selectively limit exposure of the underlying cell envelope to large biomolecules such as the innate immune effector lysozyme (18). Until now, analysis of C. difficile S-layer function has been hampered by an inability to isolate slpA-deficient mutants (19). While C. difficile is highly resistant to killing by this enzyme, resistance has been attributed to extensive peptidoglycan deacetylation (20). To determine if the S-layer also plays a role in lysozyme resistance, we treated exponentially growing bacteria with a high concentration of lysozyme (500 μg/ml) and monitored the effects on growth (Fig. 3A). Upon addition of lysozyme an immediate decrease in optical density consistent with cell lysis, followed by slower growth, was observed for the S-layer mutant FM2.5. In contrast, R20291 and FM2.5RW displayed only a transient decrease in growth rate, consistent with natural resistance. Having confirmed a role for the S-layer in lysozyme resistance, we then tested for resistance to the human cathelicidin antimicrobial peptide LL-37 to determine if S-layer-mediated resistance extended to other innate immune effectors (Fig. 3B). LL-37 is found at mucosal surfaces at concentrations of up to 5 μg/ml in normal conditions with further expression induced in response to infection (21). Treatment with 5 μg/ml of LL-37 completely killed exponentially growing cultures of the S-layer mutant FM2.5; whereas, the same treatment caused only a slight reduction in growth rates for R20291 and FM2.5RW. Taken together, these data demonstrate a role for the S-layer in resistance to both lysozyme and LL-37.
Fig. 3.
Phenotypic characterization of FM2.5. (A and B) Cultures of R20291, FM2.5 and FM2.5RW were challenged with lysozyme (500 μg/ml; A) or LL-37 (5 μg/ml; B) in exponential phase after 2.5 h (indicated with arrows). Untreated control cultures were grown in parallel. Experiments were carried out in triplicate on biological duplicates. Means and standard deviations are shown. (C) Sporulation of R20291, FM2.5, and FM2.5RW after 5 days. Spore CFUs were determined following a standard 65°C heat treatment for 30 minutes or a harsher 75°C heat treatment for 30 minutes. Heat-resistant spore CFUs are expressed as a percentage of total viable CFUs (spores and vegetative cells). Experiments were carried out in duplicate on biological duplicates. Mean and standard deviation are shown. * = P<0.01, determined using using two-tailed t-tests with Welch's correction. (D) Germination of R20291, FM2.5, and FM2.5RW spores. Synchronous germination of purified spores was induced with the bile salt taurocholate. Germination initiation was monitored by measuring the resulting decrease in optical density at OD600nm.
S-layer null mutants display severe sporulation defects
It was noted that the S-layer mutant strains survived poorly in the standard charcoal medium used to transport C. difficile strains. The ability of C. difficile to survive in the environment and be transmitted to new hosts is reliant on the bacterium’s ability to produce a heat and chemical-resistant spore (22). We analyzed sporulation efficiency by measuring the numbers of heat-resistant spores as a percentage of total viable CFUs over 5 days (Fig. S4A). Spore production by wild type R20291 and FM2.5RW cultures was reproducible and equivalent and represented 73 and 85% of total viable counts on day 5, respectively (Fig. 3C). In contrast, spore production by FM2.5 was significantly lower (p = <0.00001). Spores only represented 4.3% of total viable counts on day 5, which is a 17-20-fold reduction compared to R20291 and FM2.5RW. These observed differences in FM2.5 spore formation were not due to an inability to germinate efficiently (Fig. 3D). When the bile salt germinant taurocholate was added to purified spore preparations, the speed and efficiency of germination initiation for FM2.5 spores was indistinguishable from that of R20291 and FM2.5RW. Analysis of bacterial cultures by phase contrast microscopy also pointed to a reduction in sporulation efficiency (Fig. S4C and D), as 5.2-fold fewer phase bright spores were observed in cultures of FM2.5 (FM2.5, 8.7% vs R20291, 44.9%; FM2.5RW, 45.4%). Interestingly, we noted a discrepancy between the magnitudes of the sporulation defect determined microscopically (5.2-fold less than R20291) compared with direct counting of viable spore CFUs (20-fold less than R20291). This suggests that many of the microscopically counted spores were nonviable following the 65°C heat treatment required to differentiate spores from vegetative cell CFUs. To test the FM2.5 spores for possible stress resistance defects, we exposed the cultures to a harsher heat treatment (75°C for 30 min). The 75°C heat treatment further reduced FM2.5 spore viability 33-fold compared with the standard 65°C (Fig. 3C). The same treatment only reduced spore viability by 1.5-fold for R20291 and 2-fold for FM2.5RW. Collectively, these data indicate that the production and quality of the infectious spores are severely impaired by the loss of the S-layer.
Transmission electron microscopy was employed to identify potential morphological changes associated with these defects (Fig. S5). In FM2.5 cultures we observed spores with disorganized material loosely attached to the electron-dense core that lacked discernible, well-organized protein coat layers (23)(Fig. S5A). To determine if these unusual spore morphologies were responsible for the observed thermal sensitivity, we repeated these analyses with spores purified on a Histodenz gradient. Following purification, spores of FM2.5 were morphologically indistinguishable from those of R20291 and FM2.5RW (Fig. S5C). Surprisingly, purified FM2.5 spores still displayed increased thermal sensitivity. A75°C heat treatment reduced FM2.5 spore viability 37.1-fold compared to 10- and 18.1-fold for R20291 and FM2.5RW respectively (Fig. S5B). Several distinct biochemical and structural features of the spore, including the concentric cortex peptidoglycan and protein layers and core dehydration, have been independently linked to heat resistance (24–26). Minor defects in any of these features could explain the observed thermal sensitivity of FM2.5 spores.
S-layer null mutants are completely avirulent despite persistent gut colonization
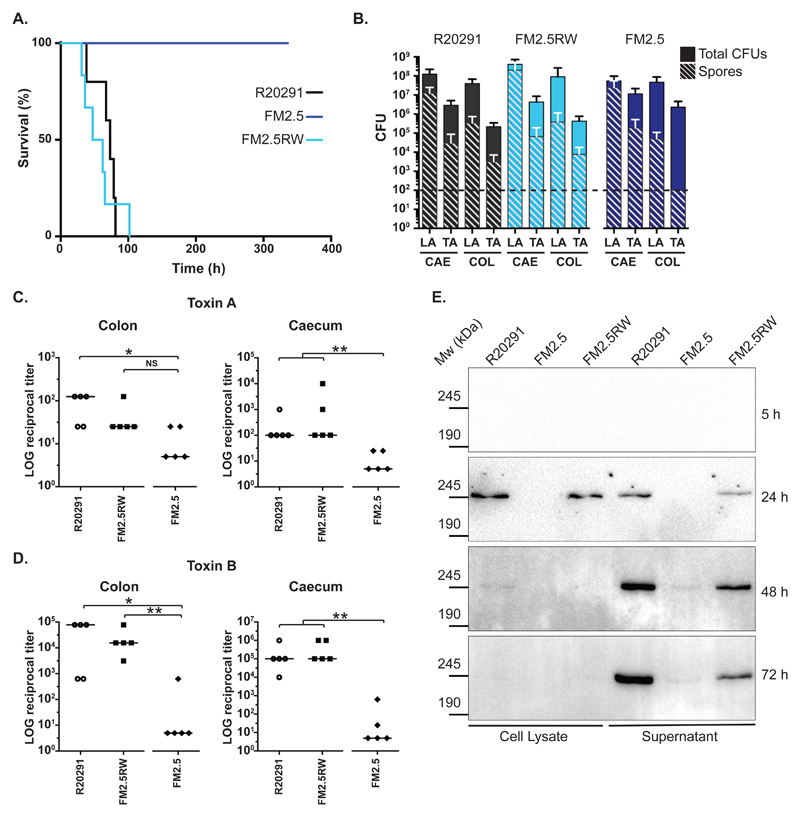
We tested the ability of the S-layer mutant FM2.5 to cause disease in the Golden Syrian hamster model of acute CDI. As expected, all animals inoculated with FM2.5RW behaved similarly to animals inoculated with the wild type R20291 strain and succumbed to C. difficile infection within 102 h of infection, with mean times to cull for R20291 and FM2.5RW of 67 h 36 min and 57 h 19 min, respectively (p = 0.52, Fig. 4A). Both groups of hamsters showed typical signs of disease including wet tail and drop in body temperature at experimental endpoint. In contrast, all animals inoculated with FM2.5 displayed no signs of disease and survived for the duration of the 14-day study (p = 0.0018; FM2.5 vs R20291, Fig. 4A). Very few described mutations located outside of the PaLoc locus have resulted in complete avirulence in this model (27). Surprisingly, the lack of virulence was not due to a colonization defect. FM2.5 was capable of persistent colonization; CFUs in the caecum and colon at the end of study (14 days) were not statistically different to those observed for R20291 and FM2.5RW in the same tissues taken at experimental endpoint some 10 days earlier (Fig. 4B). Toxin measurements from gut contents 14 days after inoculation with FM2.5 showed dramatic reductions in both toxin A or B activity compared to samples taken from hamsters that succumbed to infection with either the R20291or FM2.5RW (Fig. 4C and 4D).
Fig. 4.
In vivo analysis of slpA mutant in the Syrian Golden hamster. (A) Times to experimental endpoint of animals infected with R20291 (black line), FM2.5 (dark blue line) and FM2.5RW (light blue line) respectively. Each line represents 6 animals. (B) Total CFUs and spore CFUs (following heat treatment at 56°C for 20 min) were determined for Lumen- (LA) and tissue-associated (TA) bacteria recovered from caecum (CAE) and colon (COL) of infected animals and quantified at experimental endpoint (R20291 and FM2.5RW) or at 14 days post-infection (FM2.5). Shown are the mean and standard error. The horizontal dotted line indicates the limit of detection. None of the observed differences, including those for the TA-spore CFUs from the colon, are statistically significant. (C and D) Relative toxin activity of filtered gut samples on HT-29 (toxin A) and Vero cells (toxin B) respectively. Values represent the reciprocal of the first dilution in which cell morphology was indistinguishable from untreated wells. Samples were taken at experimental endpoint (R20291 and FM2.5RW) or at 14 days post-infection (FM2.5). (* = P <0.05, ** = P <0.01, NS = not significant, determined using using a two-tailed nonparametric Mann-Whitney test. (E) In vitro cell lysate and culture supernatant samples from R20291, FM2.5 and FM2.5RW were normalized to an equivalent optical density and separated on 6% SDS polyacrylamide gels. Toxin B was detected by Western immunoblot using an anti-Toxin B monoclonal antibody. Samples were taken at the indicated time points.
S-layer null mutants lack toxin production in vitro
Given the avirulence and low toxin activity observed in animals, we assayed the FM2.5 and control strains for toxin production in vitro (Fig. 4E). Toxin B was used as an indicator for both toxins since they are coordinately expressed and released (6). As expected, both R20291 and FM2.5RW produced toxin upon entry into stationary phase. A small amount of Toxin B was detected intracellularly at 24 hours; thereafter, Toxin B was exclusively detected in the culture supernatant. Cultures of FM2.5 produced less Toxin B at all time-points, consistent with in vivo observations.
Discussion
The bactericidal properties of Avidocin-CDs bode well for the clinical application of this new class of antimicrobial agents for combating CDI. Previous studies demonstrated that killing by the prototypic Avidocin-CD, Av-CD291.2, was highly specific for BI/NAP1/027-type strains and did not detectably alter the resident gut microbiota in mice (12). For production purposes fermentation of genetically modified B. subtilis expressing these agents is similar to many established industrial processes and scalable to thousands of liters. We selected, isolated and characterized rare C. difficile mutants resistant to Av-CD291.2 to better understand its mechanism of action. In the process we discovered the Avidocin-CD binding receptor, the C. difficile S-layer protein SlpA. Indeed, our 10 new Avidocin-CDs were all found to target variants of SlpA associated with different SLCTs. Given that the great majority of the sequence variation in SlpA is found within the surface exposed LMW subunit (9), this relationship between SLCT and Avidocin-CD sensitivity strongly suggests that the LMW SLP is the binding site for all the studied Avidocin-CDs. Further, it indirectly identifies this portion of the SlpA as the binding receptor for many C. difficile myophages since each Avidocin-CDs is constructed with a different bacteriophage-derived RBP. These findings help explain the limited host ranges observed for many C. difficile myophages (28, 29). It also suggests that antigenic variation observed between SLCTs may not be due to immune escape as previously proposed (9) but rather due to a molecular arms race between bacteria and bacteriophages. C. difficile is under selective pressure to change the bacteriophage receptor, which in turn puts selective pressure on the bacteriophages to evolve new RBPs.
Interestingly, patterns of strain sensitivity to each Avidocin-CD indicate killing was much broader than the parental bacteriophage’s host range (Fig. S2 and S7). A possible explanation for this observation is that a typical bacteriophage infection cycle is a 7-step process: attachment, genome injection, replication, transcription, translation, assembly and lysis. Bacteria can become resistant to a bacteriophage by blocking any stage of the infection cycle. For example, CRISPR-Cas or restriction-modification systems that prevent phage DNA from replicating (30). In contrast, Avidocin-CD killing is a 2-step process: attachment to the target bacterium followed by killing via creation of a small pore that dissipates the target bacterium’s membrane potential, importantly, without lysis or release of macromolecules such as exotoxin from the cytoplasm (Fig. S6). As observed with other Avidocins and R-type bacteriocins (16, 31), selected Avidocin-CD resistant mutants survived due to mutations that caused the loss or modification of the SlpA binding receptor and not due to mutations that directly disrupted the killing mechanism, such as improper pore formation, or caused proteolytic cleavage of the agent. The bias towards receptor mutations suggests the Avidocin-CD killing mechanism is simple and robust.
Modification of the binding receptor to avoid Avidocin-CD killing either through missense mutations or S-layer switching, as evidenced by the random association between clades and SLCTs (9), could also theoretically lead to resistance. While sequence variations between slpA alleles within each SLCT indicate missense mutations do occur (9), our findings suggest resistance due to this type of modification is unlikely since the bacteriophage RBPs used to construct the Avidocin-CDs have already evolved to counter this mode of potential escape. For instance, both Av-CD684.1 and Av-CD685.1 kill every SLCT-7 strain tested despite sequence identities between SLCT-7 SlpAs as low as 81% (Fig. S8). As for the emergence of resistance via horizontal transfer of the S-layer cassette, the administration of a cocktail of Avidocin-CDs that kill all the common SLCTs would make successful resistance via S-layer switching extremely unlikely. It appears that the only likely means of resistance to all the Avidocin-CDs is through complete loss of SlpA, as observed for resistance to Av-CD291.2. As a consequence the observed phenotypes for FM2.5 are germane to all likely Avidocin-CD-resistant mutants.
For a precision medicine agent to be successful, knowing the molecular target, in this case the binding receptor, is vital in designing accurate diagnostics to guide treatment decisions. If Avidocin-CDs were to be administered individually, a diagnostic determining SLCT of infecting C. difficile would be highly accurate for informing Avidocin-CD treatment decisions. However, an increase in the prevalence of hybrid cassettes, as found in ribotype 078 strains (9), would decrease the accuracy for this typing method. Strain ribotyping could also be employed to avoid development of new diagnostics, but predicting sensitivity to a particular Avidocin-CD may be challenging as ribotypes are not consistently linked to a single SLCT (i.e. 012, 014, and 015, as noted above). SlpA-typing would provide the most accurate diagnostic as well as prove more illuminating than ribotyping since SlpA is directly related to the physiology of C. difficile, whereas, ribotyping detects physiologically inconsequential ribosomal RNA gene polymorphisms. It may also be possible to administer a cocktail of 5-6 Avidocin-CDs that target 12 of the 14 C. difficile SLCTs. If such a cocktail of Avidocin-CDs were to be administered, a point of care diagnostic would only need to detect the presence of C. difficile to guide treatment decisions.
The strong selective pressure afforded by Av-CD291.2 allowed isolation of the first spontaneous C. difficile S-layer null mutants. In addition to enabling identification of the Avidocin-CD cell surface receptor, these mutants also provide an unprecedented opportunity to study S-layer function (Fig. 5). Given the ubiquity of S-layers in both Bacteria and Archaea, including many pathogenic species, surprisingly little is known about their function. It has been suggested that the S-layer could act as a molecular sieve to exclude certain large biomolecules from the cell envelope (18); however, this has not been confirmed in live cells. We have demonstrated that an intact S-layer is required for resistance to two components of the innate immunity system, lysozyme and the antimicrobial peptide LL-37. Assembled S-layers are highly symmetrical with regular repeating pores. The size of the pores in the C. difficile S-layer is not yet known but in other species pore sizes of between 2 and 6 nm have been reported. A pore size of 2 nm could conceivably exclude the16 kDa globular protein lysozyme but a small peptide such as LL-37 would experience no such steric hindrance. It is possible that charged surfaces on the assembled S-layer serve to sequester the cationic peptide away from the cell envelope in a manner analogous to capsular polysaccharides (32). Our data has identified other pleiotropic functions for the C. difficile S-layer. It is clear that the S-layer is the cell surface receptor for all of the Avidocin-CDs described here and, by extension, the receptor for the bacteriophages from which the RBP-encoding genes were cloned. Although a Bacillus bacteriophage has been found to bind S-layer protein Sap (33), this is the first time a receptor for a C. difficile bacteriophage has been identified. Furthermore, our data imply that S-layer recognition is a common feature of bacteriophages that infect this species.
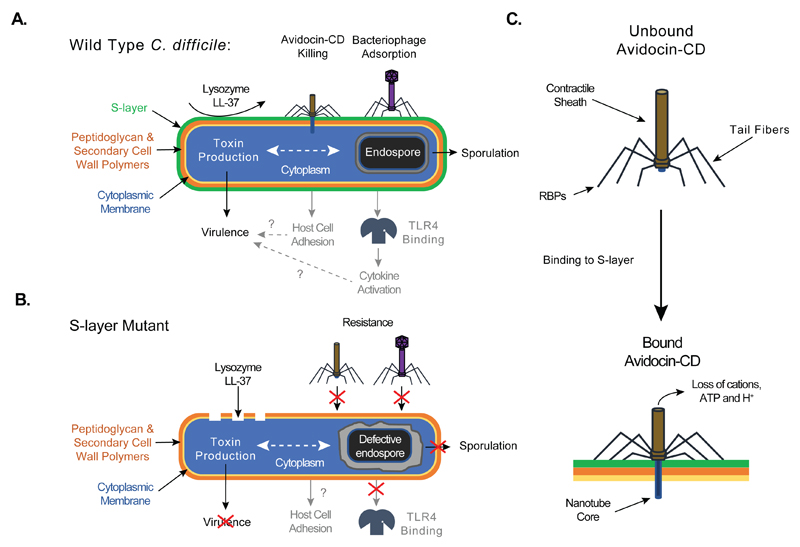
Fig. 5.
Schematic diagram depicting the phenotypes of Wild Type (A) and S-layer null mutant (B) cells in C. difficile biology and pathophysiology. Processes labeled in black indicate new functions discerned in this manuscript. Processes labeled in grey indicate previously identified functions. Dotted lines indicate relationships that need to be studied further. Question marks indicate previously described connections incompatible with observations made with the S-layer null mutant and warranting further investigation. (C) Schematic of Avidocin-CD structure and function. Binding to C. difficile by the Avidocin-CD receptor binding proteins (RBPs) triggers the sheath to contract and force the hollow nanotube core across the cell envelope. The resulting pore allows small metabolites, such as protons, ATP and cations, to escape from the cell cytoplasm, which, in turn, disrupts the cell’s membrane potential and kills the cell.
Surprisingly, the S-layer mutant also displayed severe sporulation defects, with fewer and morphologically defective spores produced. Despite a number of well-studied spore-forming organisms producing S-layers, including B. anthracis (33), S-layer biogenesis has not been reported previously to affect sporulation. There is currently no evidence to suggest that the SLPs are a structural part of the mature spore and the mechanisms by which the S-layer can influence sporulation are currently unknown. However, this observation has serious ramifications for the ability of an Avidocin-CD-resistant, S-layer defective mutant to survive and be transmitted to other hosts. The spore is an absolute requirement for C. difficile survival in the aerobic environment and is critical for transmission (22). In addition to sporulation defects, the S-layer mutant was also found to produce less toxin in vitro. Interestingly, there have been previous suggestions of feedback between sporulation and the complex regulatory network controlling toxin production (22). Although the mechanism by which these processes are affected in SlpA null mutants is far from clear, it is possible that the S-layer feeds into a point of crosstalk between regulation of virulence and transmission. Given the poor toxin production and other diverse phenotypes identified for the S-layer mutant, it is probably not surprising that the mutant was entirely avirulent in the hamster model of acute infection. What did come as a surprise, however, was the ability of the S-layer mutant to stably colonize and persist in the hamster gut for the 14-day duration of the experiment. Previous reports have pointed to a role for the S-layer in epithelial cell adhesion; however, these earlier studies were performed without access to an slpA-defective strain (34). The S-layer defective mutants and isogenic controls expressing the SLCT-specific slpA alleles provide the ideal controls to test these conflicting findings and better define the effect SlpA type has on these functions.
There are several study limitations that should be considered when evaluating the data presented. The activity of each new Avidocin-CD was observed in vitro and needs to be confirmed in an animal treatment model, as done for Av-CD291.2. Similarly, the exquisite specificity of each agent for distinct C. difficile SLCTs also needs be tested in vivo to confirm that treatment with these agents will not alter the diversity of the gut microbiota. The avirulent phenotype and long-term persistence of the Avidocin-CD-resistant slpA mutants in the hamster model should also be confirmed in other animal species. Finally, the implications of the mutants’ observed sporulation defects on transmission and the spread of resistance remain to be tested in vivo. In all cases, we do not anticipate different outcomes from those described here.
After accounting for these limitations, analysis of the data clearly demonstrates that acquisition of Avidocin-CD resistance results in loss of toxin production and complete loss of virulence. Virulence factors make attractive targets for new antimicrobials as they tend to be species-specific, and emergence of resistance is likely to reduce virulence. The ability of the S-layer mutant to stably colonize the gut in the absence of clinical disease reveals that the in vivo lifestyle of the organism is independent of toxin production and virulence. The prevalence of non-toxigenic, avirulent C. difficile strains in the general population (35) supports this hypothesis. As a result, we predict that there will be no competing selective pressure to restore virulence in the context of Avidocin-CD resistance.
In summary, we have developed and characterized multiple new Avidocin-CDs, providing crucial insights into their potential advantages in the clinic. The precise killing activity of Avidocin-CDs makes them attractive agents for both treatment and prevention since they can be administered to patients without altering the diversity of the complex gut microbiota. In addition, when resistance does emerge, Avidocin-CDs force the pathogen to sacrifice virulence for viability - making the potential clinical impact of resistance inconsequential.
Materials and Methods
Study design
The objective of this study was to characterize a panel of Avidocin-CDs, anti-bacterial protein complexes constructed to specifically target and kill C. difficile. During these experiments, the isolation of Avidocin-CD resistant slpA mutants allowed detailed analysis of S-layer function for the first time. Starting from the prototypic Av-CD291.2 (12) we constructed a panel of new Avidocin-CDs using bacteriophage RBPs we identified. Each new Avidocin-CD displayed a unique spectrum of killing activity with a strong correlation to SLCT. Sensitization of two insensitive C. difficile strains by heterologous expression of a cognate SLCT SlpA alone allowed identification of the S-layer as the receptor for all described Avidocin-CDs. Analysis of the slpA mutant identified previously unsuspected in vitro roles for the S-layer in resistance to the immune effectors lysozyme and LL-37, and in the production of mature heat-resistant spores. Finally, use of the Golden Syrian hamster model of acute infection demonstrated that the slpA mutant was entirely avirulent despite persistent infection. Greatly reduced toxin activity was detected in intestinal contents from animals colonized with the slpA mutant and this observation was supported by identification of a toxin production defect in vitro. The design and execution of these animal experiments is described in detail in Supplementary Methods.
Strains, bacteriophage and culture conditions
Bacterial strains used in this study are described in Figure S2. DNA oligonucleotides are described in Table S2. E. coli strains were routinely grown in LB broth and on LB agar (VWR). C. difficile strains were routinely grown under anaerobic conditions on BHI or BHI-S agar and in TY broth (36) except where otherwise stated. Cultures were supplemented with chloramphenicol (15 μg/ml), thiamphenicol (15 μg/ml) or anhydrotetracycline (20 ng/ml) as required. C. difficile SLCTs were determined by analyzing the nucleotide sequence of the slpA gene. When necessary, the slpA gene was sequenced using oligonucleotide primers previously described (37, 38) or primers 023-F and 023_010-R or 014+++-F and 014_002+-R (nucleotide sequences - Table S2). The variable region of strain 19142 (ribotype 046) slpA gene did not display high sequence identity with other slpA alleles, and has been designated SLCT-13. A partial strain 19142 slpA sequence was deposited in Genbank (accession: KX610658). Details of plasmid and strain construction are given in Supplementary Methods.
Bioassays to determine Avidocin-CD killing activity
Avidocin-CD bactericidal activity was assayed by a semi-quantitative spot method as previously described (12, 13). For broth-based killing assays, C. difficile strains were grown overnight in TY broth and then sub-cultured to an OD600nm of 0.05 in 1 ml fresh TY supplemented with 1mM CaCl2. Av-CD291.2 (50 μl) was added to each culture after 2.5 h. Growth was monitored by measuring the OD600nm hourly.
Extraction of S-layer and associated proteins
S-layer proteins were extracted using low pH glycine as previously described (8) and analyzed by SDS-PAGE using standard methods.
Quantitative analysis of sporulation and germination
Quantitative analysis of sporulation was carried out as previously described (19) and monitored by phase contrast and transmission electron microscopy as described in Supplementary Methods.
Analysis of resistance to lysozyme and LL-37
Broth based killing assays were carried out as described above but with lysozyme (500 μg/ml) or LL-37 (5 μg/ml) added after 2.5 h growth. Cell density was monitored by measuring the OD600nm hourly. Assays were carried out in triplicate on biological duplicates.
Animal experiments
The Golden Syrian hamster model was performed as previously described (39). All procedures were performed in strict accordance with the Animals (Scientific Procedures) Act 1986 with specific approval granted by the Home Office, U.K. (PPL60/4218). Further detail is given in Supplementary Methods.
Quantification of toxin expression
Quantification of toxin activity was performed using Vero cells as described previously (39) and by Western immunoblot using anti-Toxin B antibody. Further details are given in Supplementary methods.
Statistical analyses
Data were analyzed using GraphPad Prism software (GraphPad Software Inc.). Toxin production was compared using a two-tailed nonparametric Mann-Whitney test, and animal survival curves were analyzed using a Log-rank (Mantel-Cox) test. All other statistical analyses were performed using two-tailed t-tests with Welch's correction.
Supplementary Material
One Sentence Summary.
We have constructed a series of antimicrobial agents called Avidocin-CDs that specifically bind to the highly polymorphic S-layer and kill C. difficile; rare escape mutants lacked Surface layer protein A (SlpA) and exhibited severe phenotypic defects that fully compromised virulence without affecting colonization of the hamster gut.
Acknowledgments
We thank Fred Tenover, Tom Riley, Trevor Lawley, Vince Young, and Kate Dingle for providing C. difficile isolates; Neil Fairweather for a plasmid containing SLCT-11 slpA; Hilary Browne and Trevor Lawley for providing strain CD305 genome sequence; Chris Hill and the University of Sheffield Electron Microscopy Unit for TEM analysis.
Funding:
Supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R21AI121692. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support from the MRC (grant number MR/N000900/1; R.P.F.), AvidBiotics Corp. and the University of Sheffield via the Higher Education Innovation Fund 2011-2015 (R.P.F.), and the Wellcome Trust (grant number 086418; G.R.D.).
Footnotes
Author contributions:
D.S., G.R.D., G.R.G. and R.P.F. designed and coordinated the study. J.A.K, D.G., A.M.B., S.L., G.R.G. conducted the experiments. G.R.D., G.R.G. and R.P.F. wrote the manuscript with contributions from all co-authors.
Competing interests:
D.G., S.L., D.S., and G.R.G. are current or past employees of and own stock in AvidBiotics Corp. R.P.F. received a research grant from AvidBiotics Corp. AvidBiotics Corp. hold the following patents: US8206971 (Modified bacteriocins and methods for their use), US8673291 (Diffocins and methods of use therof), US9115354 (Diffocins and methods of use therof), and EP2576604 (Diffocins and methods of use therof).
Data and materials availability:
Nucleotide sequences have been deposited in Genbank with accession identifiers: KX610658, KX557294, KX592438, KX592434, KX592441. KX592442, KX592443, KX592444, KX592439, KX592435, KX592437, KX592436, KX592440. Avidocin-CDs are available from AvidBiotics Corp. subject to a material transfer agreement.
References
- 1.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 3.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 4.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, et al. Burden of Clostridium difficile Infection in the United States. New Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ANTIBIOTIC RESISTANCE THREATS in the United States, 2013, Threat Report 2013. Centers for Disease Control and Prevention; Atlanta: 2013. [Google Scholar]
- 6.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 8.Fagan RP, Albesa-Jove D, Qazi O, Svergun DI, Brown KA, Fairweather NF. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol Microbiol. 2009;71:1308–1322. doi: 10.1111/j.1365-2958.2009.06603.x. [DOI] [PubMed] [Google Scholar]
- 9.Dingle KE, Didelot X, Ansari MA, Eyre DW, Vaughan A, Griffiths D, Ip CL, Batty EM, Golubchik T, Bowden R, Jolley KA, et al. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J Infect Dis. 2013;207:675–686. doi: 10.1093/infdis/jis734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willing SE, Candela T, Shaw HA, Seager Z, Mesnage S, Fagan RP, Fairweather NF. Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol Microbiol. 2015;96:596–608. doi: 10.1111/mmi.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan A, Lynch M, Smith SM, Amu S, Nel HJ, McCoy CE, Dowling JK, Draper E, O'Reilly V, McCarthy C, O'Brien J, et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 2011;7:e1002076. doi: 10.1371/journal.ppat.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio. 2015;6:e02368. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhart D, Williams SR, Bishop-Lilly KA, Govoni GR, Willner KM, Butani A, Sozhamannan S, Martin D, Fortier LC, Scholl D. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J Bacteriol. 2012;194:6240–6247. doi: 10.1128/JB.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol. 2015;22:377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholl D, Cooley M, Williams SR, Gebhart D, Martin D, Bates A, Mandrell R. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob Agents Chemother. 2009;53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl D, Gebhart D, Williams SR, Bates A, Mandrell R. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One. 2012;7:e33637. doi: 10.1371/journal.pone.0033637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams SR, Gebhart D, Martin DW, Scholl D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol. 2008;74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sara M, Sleytr UB. Molecular sieving through S layers of Bacillus stearothermophilus strains. J Bacteriol. 1987;169:4092–4098. doi: 10.1128/jb.169.9.4092-4098.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho TD, Williams KB, Chen Y, Helm RF, Popham DL, Ellermeier CD. Clostridium difficile extracytoplasmic function sigma factor sigmaV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection. Infect Immun. 2014;82:2345–2355. doi: 10.1128/IAI.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowdish DM, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock RE. Impact of LL-37 on anti-infective immunity. J Leukoc Biol. 2005;77:451–459. doi: 10.1189/jlb.0704380. [DOI] [PubMed] [Google Scholar]
- 22.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barra-Carrasco J, Olguin-Araneda V, Plaza-Garrido A, Miranda-Cardenas C, Cofre-Araneda G, Pizarro-Guajardo M, Sarker MR, Paredes-Sabja D. The Clostridium difficile exosporium cysteine (CdeC)-rich protein is required for exosporium morphogenesis and coat assembly. J Bacteriol. 2013;195:3863–3875. doi: 10.1128/JB.00369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 26.Strong PC, Fulton KM, Aubry A, Foote S, Twine SM, Logan SM. Identification and characterization of glycoproteins on the spore surface of Clostridium difficile. J Bacteriol. 2014;196:2627–2637. doi: 10.1128/JB.01469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ransom EM, Williams KB, Weiss DS, Ellermeier CD. Identification and characterization of a gene cluster required for proper rod shape, cell division, and pathogenesis in Clostridium difficile. J Bacteriol. 2014;196:2290–2300. doi: 10.1128/JB.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepinski P, Douce GR, Clokie MR. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016;60:968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekulovic O, Garneau JR, Neron A, Fortier LC. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Appl Environ Microbiol. 2014;80:2555–2563. doi: 10.1128/AEM.00237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samson JE, Magadan AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie JM, Greenwich JL, Davis BM, Bronson RT, Gebhart D, Williams SR, Martin D, Scholl D, Waldor MK. An Escherichia coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob Agents Chemother. 2011;55:5469–5474. doi: 10.1128/AAC.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154:3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 33.Plaut RD, Beaber JW, Zemansky J, Kaur AP, George M, Biswas B, Henry M, Bishop-Lilly KA, Mokashi V, Hannah RM, Pope RK, et al. Genetic evidence for the involvement of the S-layer protein gene sap and the sporulation genes spo0A, spo0B, and spo0F in Phage AP50c infection of Bacillus anthracis. J Bacteriol. 2014;196:1143–1154. doi: 10.1128/JB.00739-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BA, Gerding DN, Viswanathan VK, Vedantam G. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One. 2013;8:e78404. doi: 10.1371/journal.pone.0078404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351:633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 36.Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 37.Kato H. Typing by sequencing the slpA gene of Clostridium difficile strains causing multiple outbreaks in Japan. J Med Microbiol. 2005;54:167–171. doi: 10.1099/jmm.0.45807-0. [DOI] [PubMed] [Google Scholar]
- 38.Karjalainen T, Saumier N, Barc MC, Delmee M, Collignon A. Clostridium difficile genotyping based on slpA variable region in S-layer gene sequence: an alternative to serotyping. J Clin Microbiol. 2002;40:2452–2458. doi: 10.1128/JCM.40.7.2452-2458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckley AM, Spencer J, Candlish D, Irvine JJ, Douce GR. Infection of hamsters with the UK Clostridium difficile ribotype 027 outbreak strain R20291. J Med Microbiol. 2011;60:1174–1180. doi: 10.1099/jmm.0.028514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011;286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–1936. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peltier J, Shaw HA, Couchman EC, Dawson LF, Yu L, Choudhary JS, Kaever V, Wren BW, Fairweather NF. Cyclic diGMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through ZmpI protease-mediated cleavage. J Biol Chem. 2015;290:24453–24469. doi: 10.1074/jbc.M115.665091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.