Summary
Cell fate decisions are fundamental for development, but we do not know how cells select between alternate fates. Here, we asked how mouse embryonic stem (ES) cells leave the pluripotent state and choose between germ layer fates. By analyzing the dynamics of the transcriptional circuit that maintains the pluripotent state during lineage choice, we found that the proteins that maintain ES cell identity also orchestrate germ layer fate selection. Core pluripotency factors, Oct4 and Sox2, each suppress differentiation into a single germ layer fate. Differentiation signals continuously and asymmetrically modulate Oct4 and Sox2 protein levels, altering their binding pattern in the genome, and leading to cell fate choice. The same factors that maintain pluripotency, thus, also integrate external signals and control lineage selection. Our study provides a framework for understanding how complex transcription factor networks control cell fate decisions in progenitor cells.
Introduction
How progenitor cells decide their fate is a question that underlies all of developmental biology but is poorly understood. While complex regulatory networks are known to maintain cells in distinct cell fates (Davidson et al., 2002; Novershtern et al., 2011; Odom et al., 2004), we know little about how cells integrate signals and reorganize these networks to allow fate transitions.
Mouse embryonic stem cells (ES) provide a model system for studying cell fate choice (Nishikawa et al., 2007; Niwa, 2010). The cells integrate signals in their environment and choose whether to remain pluripotent or to differentiate into progenitors of the mesendoderm (ME) or neural ectoderm (NE) (Figure 1A) (Greber et al., 2010; Nishikawa et al., 2007; Niwa, 2007, 2010; Niwa et al., 2000; Tesar et al., 2007; Yamaguchi et al., 1999; Ying et al., 2003b). A complex circuit of transcription factors and epigenetic regulators (including Oct4, Sox2, Nanog, Klf4, Klf5, Tbx3; Jarid2, Suz12) holds the ES cell in a pluripotent state (Figure 1B) by repressing genes required for ME and NE differentiation (Ema et al., 2008; Han et al., 2010; Jiang et al., 2008; Pasini et al., 2010; Peng et al., 2009; Schuettengruber and Cavalli, 2009; Silva and Smith, 2008). High-throughput experiments have provided a complex but static picture of the pluripotency circuit (Chen et al., 2008; Lu et al., 2009; Marson et al., 2008; Wang et al., 2006) a part of which is shown in Figure 1B), but we do not know how an ES cell leaves the pluripotent state and selects between the ME and NE cell fate.
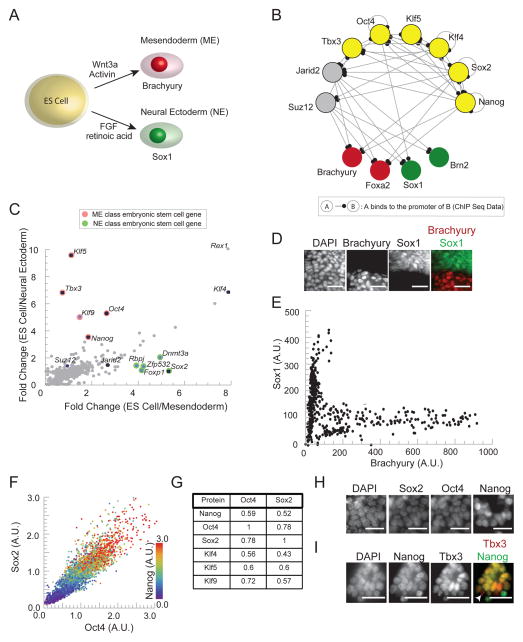
Figure 1. ES cells, defined by correlated expression of pluripotency factors, select between NE and ME fate in vitro.
(A) ES cells lose pluripotency and differentiate into mesendodermal (ME) progenitor cells to express Brachyury in response to Wnt3a or Wnt agonist, CHIR. They differentiate into neural ectoderm (NE) progenitors to express Sox1 in response to FGF signals or retinoic acid. (B) Diagram of interactions between the pluripotency factors (yellow), key epigenetic regulators (grey), and regulators of the ME (red) and NE (green) lineage inferred from ChIP-seq data in the literature (C) Plot of fold change in expression levels (obtained from microarray data in (Shen et al., 2008)) of genes expressed in ES cells compared to their expression in ME progenitors (x axis) versus the same fold change comparison in NE progenitors (y-axis). Pluripotency factors and key epigenetic regulators are indicated with black dots, ME class genes in red and NE class genes in green. (D) Fluorescence images of Sox1-GFP ES cells exposed to 3μM CHIR and stained with DAPI and immunostained for Brachyury show cytoplasmic GFP expression (green, NE marker) or nuclear Brachyury expression (red, ME marker). Scale bar 32μm (E) Scatter plot of Sox1 levels vs. Brachyury levels in differentiated cells. Each point represents Brachyury and Sox1-GFP signal in a single cell. (F) Scatter plot of protein levels in single cells for Oct4, Sox2, and Nanog derived from immunofluorescence measurements for n > 1000 ES cells growing in LIF and BMP. Protein levels measured in units normalized to the population mean. Figure S1F shows population distributions of Oct4 and Sox2 protein levels. (G) Pearson correlation coefficients for pairs of pluripotency factors measured using immunofluorescence and FACS. (H) Fluorescence images of ES cell nuclei stained simultaneously for DAPI, Sox2, Oct4 and Nanog. Scale bars 32 μm. (I) Fluorescence images of ES cells stained for DAPI, Nanog and Tbx3. White arrow points to punctate Tbx3 expression (red) in a nucleus with low Nanog (green). Intensity of the delocalized nuclear Tbx3 and Nanog have a correlation of R = .65. Scale bars 32 μm. See also Figure S1.
Since pluripotency circuit expression is sufficient to block all differentiation (Kim et al., 2008; Mitsui et al., 2003; Takahashi and Yamanaka, 2006), the circuit must be reorganized during germ layer differentiation so that the cells can release the gene expression program associated with either ME or NE lineage. Here, we study the dynamic regulation of pluripotency circuit components during in vitro differentiation to gain insight into the regulatory mechanisms underlying cell fate selection in this system.
By analyzing circuit dynamics during lineage selection, we are able to disentangle the complex network (Figure 1B) and focus on key factors that both regulate pluripotency and control germ layer differentiation. While most pluripotency circuit factors are down regulated or variably expressed during differentiation, Oct4 and Sox2 are not. Oct4 is up regulated in cells choosing the ME fate but repressed in cells choosing the NE fate. Conversely, Sox2 protein level is up regulated in cells choosing the NE and repressed in those choosing the ME fate. Oct4 and Sox2 protein levels provide continuous temporal markers of the cell’s progression towards lineage selection before lineage specific markers are activated. The lineage specific regulation of Oct4 and Sox2 is necessary for germ layer fate choice and alters their binding pattern in the genome. The intimate involvement of these key nodes of the pluripotency circuit in initiating differentiation enables the cell to integrate signals and choose between different fates.
Results
Microarray analysis suggests that transcription factors expressed in ES cells fall into three classes based on their modulation during differentiation
We identified transcription factors and DNA binding proteins that are expressed in ES cells and studied their regulation in ME and NE progenitor cells (Figure 1C) using published microarray data (Shen et al., 2008). We found that many genes (diagonal points in Figure 1C) present in the ES cell are down regulated in both ME and NE cells including Klf4, a pluripotency circuit factor, and Rex1, a commonly used ES cell marker (Masui et al., 2008) (see Supplemental Experimental Procedures). However, several genes were present in the ES cell and differentially present in either ME or NE progenitors and fell near either axis in Figure 1C.
We identified a class of genes that was expressed in ES cells and ME progenitors but specifically down-regulated in NE progenitor cells (Figure 1C, red points). This ME class of genes contained the pluripotency genes Oct4, Nanog, Klf5, Tbx3, as well as Klf9. Complementarily, a class of genes, we call the NE class, was expressed in ES cells and in NE progenitor cells but down-regulated in ME progenitor cells (Figure 1C, green points). This class contained the pluripotency circuit gene Sox2 as well as Foxp1, Rbpj, Dnmt3a, and Zfp532. We classified genes as belonging to the ME or NE class based upon their distance from the diagonal in Figure 1C (table with distances and p-values in Figure S1C, see Supplemental Experimental Procedures for details). Surprisingly, many of the core pluripotency circuit genes including Nanog, Oct4, Sox2, Klf5, and Tbx3 had signatures of lineage specific regulation, suggesting a deeper connection between pluripotency maintenance and lineage choice.
We then developed an experimental system for studying the expression of the ME and NE class genes during lineage choice in single cells.
Embryonic stem cells make a discrete choice between neural ectodermal and mesendodermal lineages in vitro
We reconstituted the NE versus ME cell fate decision in vitro and established an experimental system in which we could differentiate a cell population into either the ME or NE lineage by adding Wnt or retinoic acid.
We maintained ES cells in the pluripotent state using LIF and BMP in defined conditions without serum or feeder cells using methods described previously (Ying et al., 2003a; Ying et al., 2008), Supplemental Experimental Procedures). Cells removed from pluripotency promoting conditions did not immediately respond to NE or ME inducing signals, and this effect has been reported in the literature (Jackson et al., 2010). However, after 48 hours in N2B27, a defined medium that lacks differentiation signals (Supplemental Experimental Procedures), cells became competent to respond to signals.
After 48 hours in N2B27, cells responded to Wnt3a or CHIR, a Wnt agonist, by activating the core mesendodermal regulators Brachyury and Foxa2 (Figure 1D, S1D), with more than 70% of cells expressing Brachyury within 36 hours of signal addition at high levels of signal (eg, 3μM of CHIR). After 48 hours in N2B27, retinoic acid (RA) or FGF drove NE differentiation and triggered the activation of NE markers, Sox1, Brn2, and Nestin (Figure 1D, S1E). Consistent with the published literature, 70% of cells activated Sox1 within 36 hours of signal addition (Abranches et al., 2009; Ying et al., 2003b). In this way, Wnt or retinoic acid signals drove high efficiency differentiation of ES cells to the ME or NE cell fate, respectively.
Even in the presence of Wnt3a or CHIR, a population of cells activated Sox1, the NE regulator and not Brachyury, the ME regulator. This is presumably due to the paracrine FGF signaling between the cells leading some cells to adopt the NE fate (Ying et al., 2003b). Using a previously validated Sox1-GFP reporter cell line (Ying et al., 2003b), and Brachyury immunofluorescence, we could detect Sox1 and Brachyury activation in the same cell population (Ying et al., 2003b). In cell populations treated with Wnt or CHIR, Sox1-GFP expression and Brachyury staining were mutually exclusive (Figure 1D, 1E). There are no points on the diagonal of the scatter plot in Figure 1E, illustrating the absence of simultaneous high Sox1 and Brachyury expression in single cells. The “L” shape of the scatter plot shows that cells make a discrete decision to activate either Sox1 or Brachyury in vitro.
Our ability to induce and detect Sox1 positive and Brachyury positive cells under identical conditions provided a defined experimental system for studying the regulation of the pluripotency circuit factors during ME versus NE lineage selection. We validated the differentiation protocol with a variety of cell lines.
Nanog down regulation is necessary for lineage selection
We used immunofluorescence to measure the levels of the proteins identified in Figure 1C in single cells as we took the cells through the differentiation protocol described in the previous section. We performed immunofluorescence staining under identical conditions in all experiments so that protein levels could be quantitatively compared between different cell populations (Sachs et al., 2005).
Consistent with the dense network of positive regulatory interactions that have been inferred between pluripotency factors (Chew et al., 2005; Ivanova et al., 2006; Masui et al., 2007), in populations of cells growing under pluripotency promoting conditions, pluripotency circuit protein levels were strongly correlated across the cell population (Figures 1F,G,H,I)
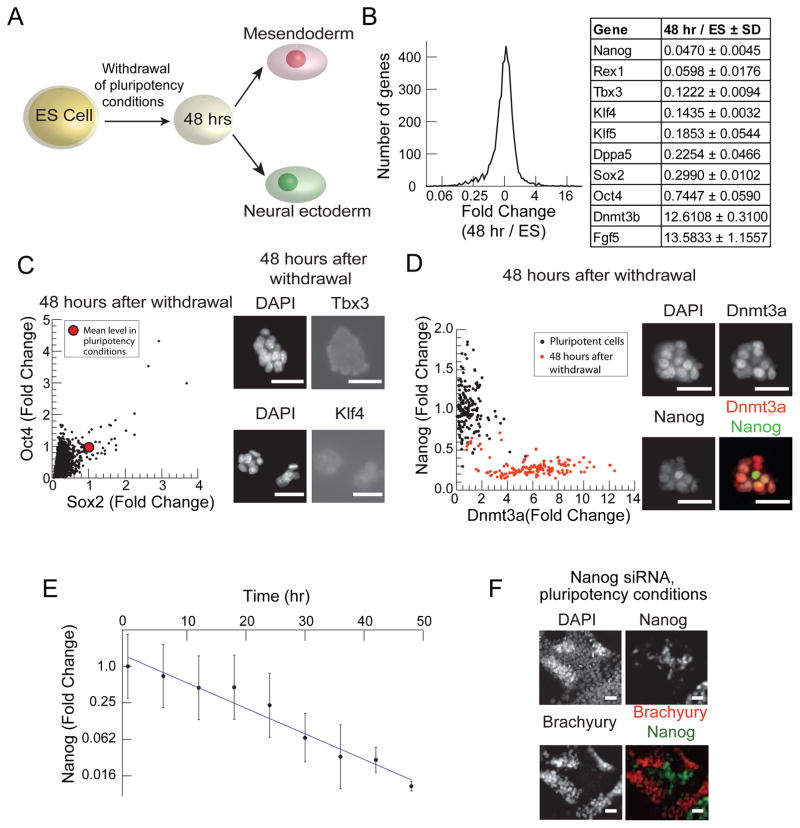
Cells responded to differentiation signals only 48 hours after the withdrawal of pluripotency promoting conditions (Figure 2A). After 48 hours in N2B27, microarrays showed that 87% of genes changed by less than 2 fold, while 9% (including many pluripotency factors) were down regulated by more than 2 fold, and 4% were induced by more than 2 fold after 48 hours, as shown by the histogram in Figure 2B. Nanog, Oct4, Sox2, Klf4, Klf5, and Tbx3 mRNA levels were 5%, 74%, 30%, 14%, 19% and 12% of their levels in ES cells (Figure 2B). A small number of genes were induced including Dnmt3b (Figure 2B, (Lu et al., 2009). Dnmt3b is a de-novo DNA methyl-transferase that acts with Dnmt3a to methylate the promoters of the pluripotency circuit factors including Nanog during germ layer differentiation (Li et al., 2007).
Figure 2. Down regulation of ES cell specific factors is necessary for differentiation.
(A) Schematic of ES cell differentiation protocol. 48 hours after the withdrawal of pluripotency promoting conditions (LIF and BMP), cells are exposed to differentiation signals. Cells then respond to ME inducing signals (Wnt3a or CHIR) by activating Brachyury and to NE inducing signals (retinoic acid or endogenous FGF) by activating Sox1. (B) Histogram of fold change of mRNA expression levels in the cell population 48 hours after withdrawal of LIF and BMP as measured by microarray. Table shows fold changes for a set of pluripotency factors as well as for Dnmt3b and Fgf5. (C) The scatter plot (left) of Oct4 vs. Sox2 expression in n > 1000 single cells 48 hours after withdrawal of pluripotency conditions. The red dot indicates mean expression level of these proteins in pluripotency conditions. Fluorescent images of cells immunostained for DAPI, Tbx3 (above) and Klf4 (below) 48 hours after withdrawal of pluripotency conditions. Scale bar 32μm (D) Scatter plot of Nanog vs. Dnmt3a levels in single cells under pluripotency supporting conditions (black) and 48 hours after the withdrawal of pluripotency conditions (orange). Images of cells immunostained for DAPI, Dnmt3a (red) and Nanog (green) 48 hours after pluripotency condition withdrawal. Scale bars 32 μm. (E) Plot of the fold changes of mean Nanog protein levels (> 20,000 cells) in cell populations fixed every 6 hours following withdrawal of LIF and BMP. Error bars indicate ±SD in Nanog protein levels in the cell population. Solid line shows Nanog decay fit to an exponential with a half-life of 7.5 hours. (F) Fluorescence images of cells in pluripotency conditions, LIF and BMP, stained for DAPI, Nanog, and Brachyury following CHIR addition for 24 hours and siRNA knock-down of Nanog. siRNA construct was added to cells for 24 hours prior to CHIR exposure. Scale bars 32μm.
Consistent with the microarray data, the protein levels of Oct4, Sox2, Nanog, Klf4, Klf5, Tbx3, and Klf9 fell during the transition, while Dnmt3a and Dnmt3b were induced up to 3-fold (Figure 2C,D, S2A). In Figure 2C the mean levels of Oct4 and Sox2 in more than 99% of the cells are below the mean expression levels of these proteins in pluripotency supporting conditions. Cell images (Figure 2C) show that Tbx3 and Klf4 protein levels are severely down regulated upon withdrawal of pluripotency sustaining conditions (Compare Figure S2A).
Thus, while in ES cells Nanog is expressed at high levels and Dnmt3a at low levels (black scatter plot in Figure 2D), after 48 hours, the cells show a complementary pattern of expression with high levels of Dnmt3a and low levels of Nanog (red scatter plot in Figure 2D, Images in Figure 2D), consistent with an antagonistic regulatory relationship between these factors (Li et al., 2007).
Next, we asked whether a fall in Nanog levels might drive the ES cell into the responsive state. We found that mean Nanog protein level in the cell population fell exponentially in time (half-life 7.5 hours) by 100 fold after 48 hours in N2B27 (Figure 2E), suggesting that Nanog down regulation might be an early and causal event for driving the ES cell into the responsive state.
Consistently, siRNA mediated knock down of Nanog in ES cells growing in pluripotency promoting conditions led to a correlated decrease in Oct4 and Sox2 protein levels (Figure S2). Further, in these conditions, CHIR drove Brachyury activation only in cells that had lost Nanog (Figure 2F). In the presence of Nanog, activation of the Wnt pathway with CHIR or other pathway agonists is known to promote pluripotency (Sato et al., 2004; Ying et al., 2008). Thus, Nanog knock down was necessary and sufficient for Brachyury activation by CHIR.
Oct4 and Sox2 are differentially regulated as cells choose their germ layer fate
The correlated expression pattern of the pluripotency factors in ES cells (Figure 1F) contrasts sharply with the lineage specific expression pattern that we observed in our microarray analysis (Figure 1C). We asked whether this change in correlation pattern is reflected in protein levels of pluripotency factors in individual cells during differentiation.
After 48 hours of pluripotency condition withdrawal (LIF and BMP), cells were not committed to a lineage and could be driven to either cell fate by signal addition (Figure 2A). We added differentiation signals to cells 48 hours after withdrawal of pluripotency conditions. Upon addition of signal (3uM CHIR, Figure 1D, 1E), some cells adopted the ME (Brachyury expression) and others the NE lineage (Sox1 expression).
We studied the regulation of genes in the ME class (Oct4, Nanog, Klf5, Tbx3, Klf9) and NE class (Sox2, Foxp1, Dnmt3a, Rbpj, Zfp532) defined above to ask how the pluripotency circuit is regulated during lineage selection. In addition to the ME and NE class, we studied the pluripotency factor, Klf4, and the key epigenetic regulator, Jarid2, which controls the targeting of repressive epigenetic modifications to the genome in pluripotent and differentiating ES cells(Peng et al., 2009).
Of the genes identified from the microarray analysis as belonging to the ME or NE class, Tbx3, Klf4, Klf9, and Rbpj proteins were absent or present at very low levels in cells entering both the NE and ME lineages and were not reactivated during differentiation (Figure 2C, Figure S3). This suggests that these factors play a role in pluripotency maintenance but not in lineage choice. In contrast, Oct4, Sox2, Nanog, Jarid2, Klf5, Foxp1, and Dnmt3a proteins were present in the nucleus of differentiating cells (Figure 3), and we classified these factors into groups based on their expression pattern.
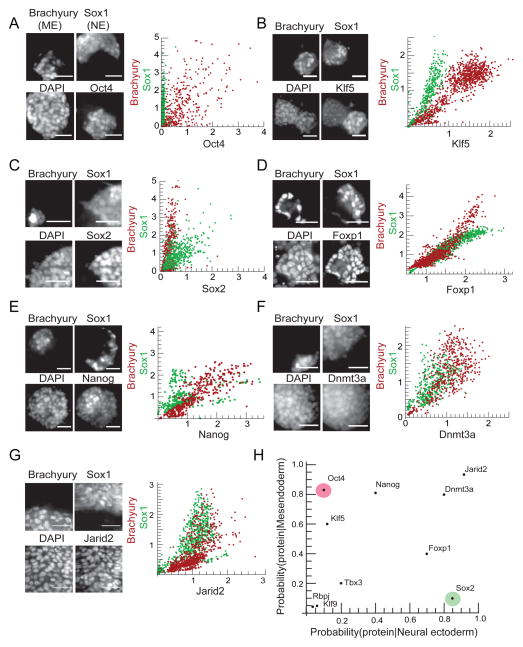
Figure 3. Key ES cell transcription factors are differentially expressed in ME and NE cells.
Cells were differentiated in conditions (see Experimental Procedures) where some cells adopted the ME fate (Brachyury positive) and others adopted the NE fate (Sox1-GFP positive). Images of a field of cells immunostained 36 hours after signal addition for ME marker Brachyury, NE marker Sox1, DAPI, and a specific factor (left panel); scatter plots (right panel) of the expression level of that factor against Brachyury in n > 300 Brachyury positive cells (red points) and against Sox1, n > 300 Sox1 positive cells (green points) for (A) Oct4, (B) Klf5, (C) Sox2, (D) Foxp1, (E) Nanog, (F) Dnmt3a and (G) Jarid2. All scale bars 32μm. (H) The data from A–G are summarized in a conditional probability plot. Each point in the plot represents the probability of finding the expression of the specific protein in ME progenitors (y-axis) versus NE progenitors (x-axis). See also Figure S3.
Oct4 and Klf5 constituted the first group of proteins (Figure 3A,B). Oct4 protein was present exclusively in Brachyury positive (ME) cells but absent in Sox1 positive (NE) cells. Consistently, scatter plots of the protein levels of Oct4 versus Brachyury in single cells correlated positively (Figure 3A, red scatter plot). The scatter plots of Oct4 versus Sox1 levels showed that cells with Sox1 have very low levels of Oct4 (Figure 3A, green scatter plot). Klf5 had a similar pattern of protein expression to Oct4. However, rather than being absent in Sox1 positive cells, Klf5 was confined to a sub-cellular compartment distinct from the nucleus (Figure 3B).
The second group of proteins included only Sox2, which was expressed in a complementary pattern to Oct4 and Klf5 (Figure 3C). In cells undergoing ME differentiation, Sox2 and Brachyury were mutually exclusive (Figure 3C, red scatter plot). In contrast, Sox2 and Sox1 (NE marker), expression correlated strongly during NE lineage induction (Figure 3C, green scatter plot).
As the final class of factors, Foxp1, Nanog, Dnmt3a, and Jarid2 were present variably in cells that had activated either Sox1 or Brachyury (Figure 3D, E, F, G). For example, scatter plots of Dnmt3a protein levels quantify this observation, and illustrate that the distribution of Dnmt3a protein is similar in both in Sox1 and Brachyury expressing cells (Figure 3F).
We quantified the data in the scatter plots by measuring the probability of observing a given protein conditioned on the presence of each lineage marker (Figure 3H, Supplemental Experimental Procedures) On this plot, variably expressed proteins like Dnmt3a lie on the diagonal and are present with high probability in both Sox1 and Brachyury expressing cells. Lineage specific proteins like Oct4 and Sox2 fall on the extreme off-diagonal of the plot and are present with high probability in either ME or NE progenitor cells but not both, consistent with the images in Figures 3A,C (see Supplementary Experimental Procedures).
To determine if the above patterns were established prior to lineage choice, we used FACS and live cell imaging to study the dynamics of protein regulation in response to signal. We did not study Klf5 further because Klf5 knock-out ES cell lines can differentiate into all three germ layers (Ema et al., 2008).
Dynamics of Oct4 and Sox2 protein levels reveal that differential regulation of the pluripotency circuit precedes cell fate choice
We asked how the relative levels of Oct4 and Sox2 protein change in time during ME and NE differentiation. We added CHIR or RA to cell populations to induce ME or NE differentiation as described (Experimental Procedures), and determined the levels of both factors in individual cells (n>40,000) using FACS.
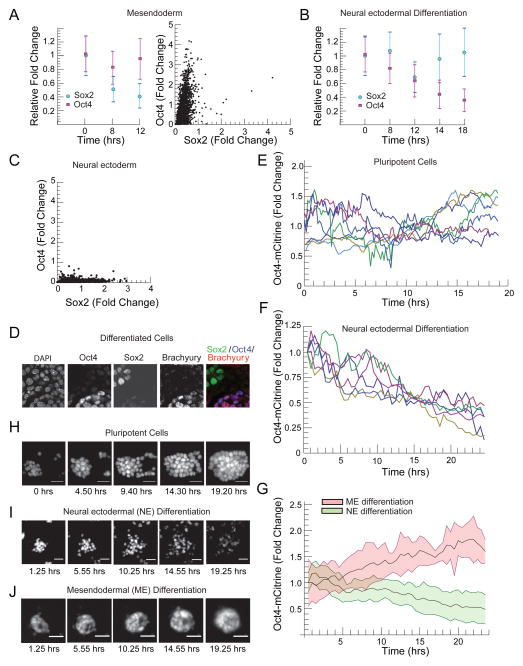
Following CHIR addition, the mean level of Sox2 protein fell by 72% after 8 hours and by 77% at 12 hours (Figure 4A). The mean Oct4 protein level, on the other hand, fell by 5%, over the first 12 hours. At 12 hours after CHIR addition, 42% of cells had activated Oct4 to levels greater than the mean level in the cell population prior to signal addition, while only 1.7% of cells had Sox2 protein levels exceeding the Sox2 mean in the initial cell population. After 12 hours, the fold change in relative Oct4 and Sox2 protein levels differed by more than the sum of the standard deviations of either protein in the cell population. The activation of ME regulator Brachyury occurred between 12 and 15 hours after CHIR addition. By 24 hours, Oct4 levels were up regulated and Sox2 repressed in cells that had activated Brachyury (Figure 4A) as shown in the scatter plot in Figure 4A, where cells adopting the ME fate showed high Oct4 and low Sox2 expression.
Figure 4. Oct4 and Sox2 protein levels diverge continuously during lineage selection.
(A) Left Panel, plot of mean ± SD of Oct4 (Purple squares) and Sox2 (blue circles) protein levels in n>40000 cells at 0,8, and 12 hours following CHIR addition, measured simultaneously in single cells using immunofluorescence and FACS. Protein levels normalized to the mean protein level in the cell population prior to signal addition. Right panel, scatter plot of Oct4 and Sox2 protein levels 24 hours after signal addition in single ME progenitor cells (~2400 cells), levels normalized to the mean level of these proteins in pluripotent cells. (B) Plot of mean ± SD of Oct4 (purple squares) and Sox2 (blue circles) levels in populations of n > 40,000 cells following RA addition (C) Scatter plot of Oct4 vs. Sox2 protein levels in n > 2000 cells 24 hours after RA addition. Protein levels normalized to the mean protein level in the cell population prior to signal addition. (D) Confocal microscopy images of cells undergoing ME differentiation stained for DAPI, Oct4, Sox2 and Brachyury. (E) Single cell trajectories of Oct4-mCitrine levels obtained using time lapse microscopy for ES cells growing under pluripotency promoting conditions. (F) Single cell trajectories of Oct4-mCitrine in cells differentiating into the neural ectodermal lineage. Conditions supporting pluripotency were removed for 48 hours and then retinoic acid (500nM) was added at t=0 to induce differentiation. (G) Temporal trajectories of Oct4-mCitrine in n >100 cells differentiating into NE (mean in solid line, standard deviation as a light green background) and ME progenitors (mean in solid line, standard deviation as a light red background) obtained by fluorescence time-lapse microscopy of the Oct4-mCitrine cell line (t=0 corresponds to time of signal addition, Figure 2A). (H–J) Fluorescence images from a time lapse imaging experiment following a field of Oct4-mCitrine cells (H) under pluripotency promoting conditions, (I) during retinoic acid driven NE differentiation, and (J) during CHIR driven ME differentiation. All scale bars 32 μm. See also Figure S4
Following RA addition, Oct4 and Sox2 protein levels diverged but on a longer time scale than after CHIR addition (Figure 4B). At 14 hours after RA addition, the mean level of Sox2 protein had fallen by 5% while Oct4 had fallen by 56%. At 18 hours, Sox2 protein levels were 100% of their initial level while Oct4 had fallen by 64%. At the same time point, 37% of cells had Sox2 levels greater than the mean level in the cell population prior to signal addition, while less than 1% of cells had Oct4 levels exceeding the mean in the initial cell population. At 18 hours, the relative fold change in Oct4 and Sox2 differed by more than the sum of the standard deviations of either protein in the cell population. The NE maker Sox1 was activated between 17 and 19 hours following RA addition. By 24 hours, Sox2 was activated and Oct4 repressed in Sox1 expressing cells as seen in Figure 4C.
Thus, following the addition of differentiation signals, Oct4 and Sox2 protein levels diverged in time, and the correlated pattern of Oct4 and Sox2 expression of the ES cell (Figure 1F) broke continuously into either a high Oct4 and low Sox2, or low Oct4 and high Sox2 pattern during differentiation (Figure 4A,C,D)..
To validate our FACS results, we measured Oct4 expression dynamics in single live cells during ME and NE differentiation. We created and validated (Experimental Procedures, Figure S4, (Tesar et al., 2007)) an Oct4-mCitrine fusion reporter cell line (Experimental Procedures, Figure S4). We used time lapse epifluorescence microscopy to track Oct4-mCitrine expression in hundreds of single cells both under pluripotency promoting conditions and during differentiation. Under pluripotency promoting conditions, Oct4 levels in single cells varied during normal rounds of cell division (Figure 4E,H). After 48 hours in N2B27, Oct4-mCitrine levels in single cells decreased to 60 ± 12 % of ES cell levels.
NE differentiation with retinoic acid drove a rapid down regulation of Oct4-mCitrine level that was detectable in single cells within 6 hours of signal addition (Figure 4F,I). On average, individual cells decreased their Oct4 level to half of its initial value by 16 hours after RA addition (Figure 4F). At the population level, Oct4 level decreased linearly in time (Figure 4G).
ME differentiation drove a complementary response. Cells responded to CHIR by up-regulating Oct4 (Figure 4J,G). In the cell population, Oct4 level increased linearly in time within 6 hours of CHIR addition (Figure 4G).
In this way, cells differentiating into the ME or NE lineage showed a detectable response in Oct4 protein levels within 6 hours of signal addition, a time that is on average 10 hours before the earliest detectable lineage specific marker expression. Further, the Oct4 temporal trajectories in cells differentiated with CHIR diverged from those differentiated with RA within 6 hours (Figure 4G). These results show that the differential modulation of Oct4 occurs prior to the expression of ME or NE lineage-specific markers.
We next determined the functional roles of Oct4 and Sox2 in lineage choice. The observed Oct4 and Sox2 protein expression patterns (Figure 3A,C; 4A,c suggested that Oct4 might specifically inhibit the NE lineage while promoting ME differentiation and that that Sox2 might specifically inhibit ME differentiation while promoting NE lineage choice. Our analysis of published ChIP-Seq data (Chen et al., 2008) provided further evidence for the asymmetric role for Oct4 and Sox2 in regulating ME and NE differentiation (Supplemental Data, Figure S5).
Oct4 and Sox2 bind together in ES cells but bind asymmetrically in lineage specific patterns in ME and NE progenitor cells
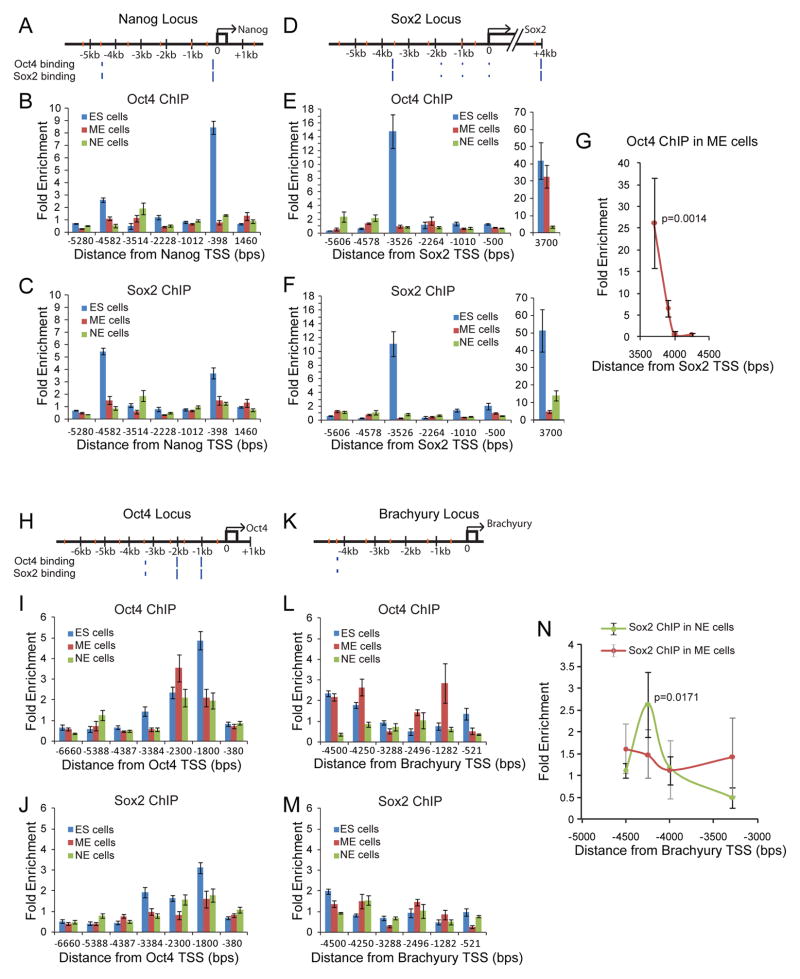
To ask how Oct4 and Sox2 DNA binding patterns change as their relative levels are modulated during differentiation, we used ChIP-qPCR to probe the binding of Oct4 and Sox2 along five genomic regions (Oct4, Sox2, Nanog, Brachyury, and Sox1) in ES cells and during ME and NE differentiation. We could not obtain reproducible ChIP-qPCR results along the Sox1 locus in ES cells, ME, or NE progenitors (see Experimental Procedures).
We measured the spatial distribution of binding enrichment along each genomic locus using tiled qPCR primers on ChIP samples for each factor Oct4 and Sox2, in the three lineages: ES, ME and NE. Experiments were performed on 3 biological replicates, and in 3 technical replicates for each primer pair. We validated enrichment peaks of interest using multiple primers targeted to and around the region of interest.
We validated our experimental system by comparing the results of our Oct4 and Sox2 ChIP-qPCR experiments on ES cells to published ChIP-seq data (Chen et al., 2008; Marson et al., 2008). We found that Oct4 (Figure 5B,E,I,L, blue bars) and Sox2 (Figure 5C,F,J,M, blue bars) binding were enriched at locations previously detected in ChIP-Seq experiments (blue hash marks, Figure 5A,D,H,K) near the genomic loci of Nanog (Figure 5A,B,C blue bars), Sox2 (Figure 5D,E,F, blue bars), Oct4 (Figure H,I,J blue bars), and Brachyury (Figure 5K,L,M, blue bars). The correspondence between the peaks in our ChIP-qPCR measurement and peak calls in published ChIP-Seq data suggested that the ChIP-qPCR accurately reflects Oct4 and Sox2 binding.
Figure 5. Oct4 and Sox2 bind asymmetrically in NE and ME regulatory regions during differentiation.
ChIP-qPCR assays for Oct4 and Sox2 binding at Nanog (A,B,C), Sox2 (D,E,F,G), Oct4 (H,I,J), and Brachyury (K,L,M,N) loci in ES (blue bars), ME (red bars), and NE (green bars) cells. Each genomic region is depicted with qPCR primer coordinates (orange hashes) and previously reported Oct4 and Sox2 ES cell-specific binding sites (blue hashes, hash heights proportional to published ChIP-seq peak heights). Mean fold enrichment is normalized to input, and error bars represent ±SEM of technical triplicates. X-axis represents positions relative to the transcriptional start site. (G) ME lineage specific binding of Oct4 at the Sox2 +3700bp neural enhancer probed with multiple primer pairs. Each point and error bars represent mean enrichment values ±SEM for 3 biological replicates (p=0.0014). (N) Lineage specific binding of Sox2 at the Brachyury −4250bp region was probed with multiple primer pairs. Points show mean enrichment values for Sox2 in the NE lineage (green circles) and ME lineage (red circles) over 3 biological replicates. Error bars (black for NE lineage, gray ME lineage) represent ±SEM of biological replicates. NE peak at −4250bp has p = 0.0171 while ME enrichment has p= 0.37.
As ES cells differentiate, Oct4 and Sox2 protein levels first fall and then are differentially regulated during ME and NE lineage selection (Figure 2, 4). Consistent with the fall, during both ME and NE differentiation, Oct4 and Sox2 enrichment decreased at the Nanog promoter (Figure 5B,C, red bars for ME and green bars for NE lineage) and also at the −3.5kb ES specific regulatory region of Sox2 (Figure 5E,F red and green bars).
However, a class of regulatory sites became differentially occupied by Oct4 or Sox2 during lineage choice. In ME progenitor cells, Oct4 is induced and Sox2 down regulated (Figure 4). In these cells, Oct4 binding was enriched at the +3.7kB regulatory region of the Sox2 locus (red bars in Figure 5E, Figure 5G, p=0.0014), while Sox2 enrichment was uniformly depleted across the entire Sox2 locus in ME cells (Figure 5F red bars). The +3.7kb (downstream) site is known to regulate Sox2 expression in neural progenitor cells (Miyagi et al., 2006; Sikorska et al., 2008; Tomioka et al., 2002).
In NE progenitor cells, Sox2 is induced while Oct4 is down regulated (Figure 4). In these cells, Sox2 binding was enriched in the +3.7kb NE regulatory region of its own enhancer, consistent with the role of this region in controlling Sox2 expression in NE cells (Figure 5F green bar at +3700 from Sox2 TSS, p=0.0019). Sox2 enrichment also increased at the −4.3kB region of the Brachyury promoter (Figure 5M green bar at −4250bp from Brachyury TSS, Figure 5N, green line, p=0.0171) where Sox2 binding was enriched in all 3 NE biological replicates. In ME cells, Sox2 enrichment at this locus was detected in only one of three biological replicates (Figure 5M red bar at −4250bp from Brachyury TSS, Figure 5N, red line), likely due to contaminating subpopulations of NE progenitors, and was not statistically significant (p= 0.37).
In this way, while many ES cell-specific binding events are depleted of Oct4 and Sox2 during differentiation, a fraction of regions become differentially occupied by either Oct4 or Sox2. As Oct4 protein levels increase relative to Sox2, Oct4 binding increases in regions of the genome associated with NE differentiation. As Sox2 protein levels increase, Sox2 binding increases in the Brachyury promoter. Thus, these two regulatory regions become enriched for either Oct4 or Sox2 during lineage choice. These results together with our imaging data suggest that Oct4 and Sox2 might perform lineage specific repression during differentiation. In such a model, Oct4 would repress the NE lineage and Sox2 the ME lineage.
Perturbation of Oct4 or Sox2 protein level modulates cell fate selection
To test the functional roles of Oct4 and Sox2 in lineage selection, we interfered with the levels of these proteins by transfecting differentiating cells with over-expression plasmids and siRNA constructs (Figure S5). In these experiments, we drove differentiation into NE or ME progenitor cells at high efficiency by adding the induction signals retinoic acid or CHIR to ES cells after 48 hours in N2B27 (see the second results section). We performed the transfection experiments without selection markers so that cells transfected with a given plasmid or siRNA occurred next to untransfected cells, and in a single field of view we could examine perturbed and unperturbed cells (Figure S5).
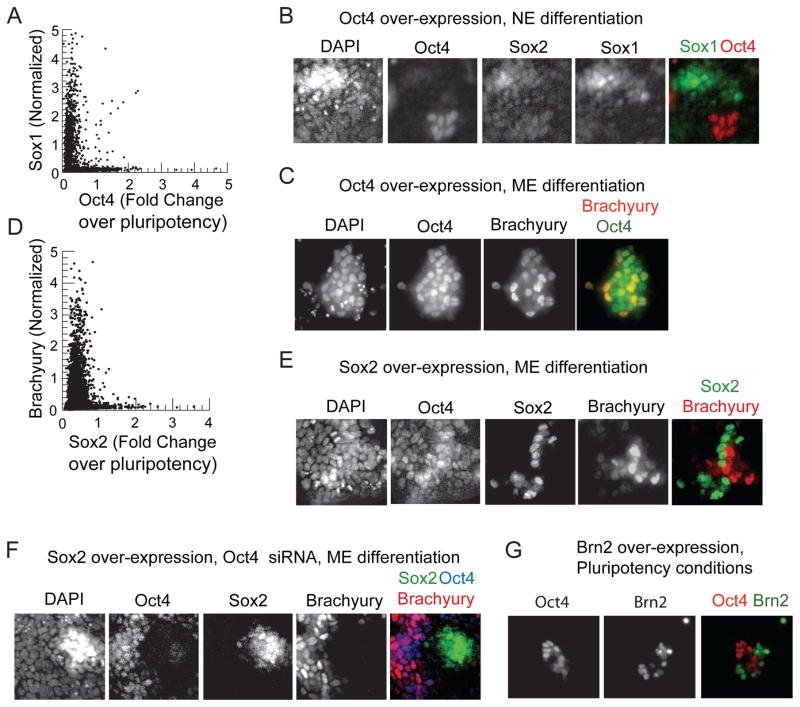
We confirmed that Oct4 inhibits NE differentiation by transfecting cells with a constitutively expressing Oct4 plasmid while inducing NE differentiation with retinoic acid. Cells with high Oct4 did not express Sox1, and cells expressing Sox1 had low Oct4 levels (Figure 6A,B):, as illustrated by the “L” shape of the scatter plot (Figure 6A). In contrast, Oct4 over expression during mesendodermal differentiation did not block Brachyury expression (Figure 6C)..
Figure 6. Oct4 specifically represses the NE lineage and Sox2 the ME lineage.
(A) Scatter plot of Oct4 vs. Sox1 in 2500 cells from a population in which constitutive Oct4 plasmid has been transfected and neural differentiation inducing signal retinoic acid (1μM) was added. Oct4 levels are measured as fold change over levels in ES cells under pluripotency conditions (B) Images of cells from A co-stained for Oct4, Sox2, Sox1, and DAPI. (C) Images of cells transfected with a constitutive Oct4 plasmid for 24 hours prior to CHIR induced ME differentiation co-stained for Oct4, Brachyury, and DAPI. (D) Scatter plot of Brachyury vs. Sox2 in over 5000 cells transfected with a Sox2 plasmid 24 hours prior to the induction of mesendodermal differentiation with 200ng/ml Wnt3a (identical results obtained with CHIR, data not shown) co-stained for Sox2, Brachyury and DAPI (E) Images of cells from D (F) Images of cells transfected with siRNA against Oct4 and a constitutive Sox2 plasmid co-stained for Oct4, Sox2, Brachyury and DAPI. (G) Images of ES cells transfected with a Brn2 plasmid and stained for Brn2 and Oct4 under pluripotency promoting conditions. See also Figure S5.
We confirmed that Sox2 inhibits ME differentiation by driving constitutive expression of Sox2 during mesendodermal differentiation driven by CHIR. Cells with high Sox2 expression did not express Brachyury (Figure 6D,E), again highlighted by the “L” shaped scatter plot (Figure 6D). Further, cells in which Sox2 was driven constitutively and Oct4 expression was abrogated using siRNA did not express Brachyury (Figure 6F) showing that ME inhibition by Sox2 was independent of Oct4. Furthermore, Brachyury expression was confined to cells that retained Oct4 expression suggesting that Oct4 is necessary for mesendodermal differentiation.
Further, over expressing Sox2 while abrogating Oct4 expression under conditions supporting pluripotency led to a fraction of cells activating the neural ectodermal regulator Sox1 (Figure S5) suggesting that Sox2 drives NE differentiation.
As another test of relevance, we asked whether forced expression of terminal lineage markers induces the observed pattern of Oct4 or Sox2 protein expression. Brn2 is a core regulator of neural identity that is induced in NE cells in our experimental system at a similar time as Sox1 (Figure S1). Forced Brn2 expression can convert fibroblasts directly into neurons (Vierbuchen et al., 2010). We found that constitutive expression of Brn2 in ES cells growing in pluripotency promoting conditions reproducibly down regulated Oct4 in these cells, as shown by images in Figure 6G.
These experiments show that Oct4 specifically represses Sox1 and the NE lineage, and Sox2 specifically represses Brachyury and the mesendodermal lineage. Thus, the differential activation of these genes critically regulates cell fate choice.
Differential regulation of Oct4 and Sox2 enables signal integration
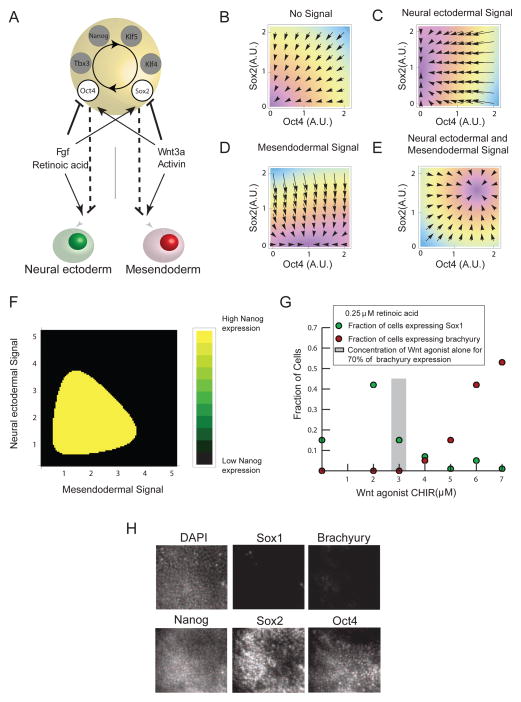
Together, the data presented above lead us to a model of pluripotency circuit regulation during differentiation and lineage choice. In the pluripotent state, positive regulatory interactions between Oct4, Sox2, Nanog, Klf4, Klf5, and Tbx3 maintain these proteins at high and correlated levels, preventing differentiation. We verified positive regulatory interactions between Oct4, Sox2, and Nanog using siRNA (Figure S5,S6). Upon withdrawal of pluripotency promoting factors, LIF and BMP, or with the direct abrogation of Nanog expression with siRNA, the levels of pluripotency factors fall. This fall allows the differentiation signals, Wnt and retinoic acid or FGF, to differentially regulate elements of the circuit. The differential regulation competes with the intrinsic positive interactions between the pluripotency factors. The ME inducing signals drive Sox2 levels down and Oct4 levels up, while the NE inducing signals drive Oct4 down and Sox2 up. Because Oct4 specifically inhibits the NE lineage and Sox2 specifically inhibits the ME lineage, the cells make a lineage choice.
We modeled the competition between the intrinsic positive regulation of the pluripotency factors and the differential regulation by signals. Model analysis (Supplemental Experimental Procedures, Figures 7B,C,D,E,F, S6C,D) revealed that this competition allows the pluripotency circuit to perform signal integration, so that one differentiation signal can change the cell’s interpretation of the other. Specifically, the model predicted that exposure of the cell to Wnt and retinoic acid simultaneously can jam the pluripotency circuit in a high Oct4 and high Sox2 state, leading to the expression of Nanog (Figure 7F) and, thus, preventing the activation of either Brachyury or Sox1
Figure 7. The architecture of the pluripotency circuit enables signal integration and lineage choice.
(A) Model for the coupling between the pluripotency circuit, differentiation signals, and cell fates based upon data from Figures 3, 4 and 5. The asymmetric inhibition of Oct4 and Sox2 by differentiation signals competes with positive feedback between circuit elements (circular arrow), Oct4, Sox2, Nanog, Klf5, Klf4, Tbx3. (B–E) Plots of the Oct4 and Sox2 phase space showing the stable fixed points in the presence of different combinations of differentiation signals as defined by the mathematical model (Supplemental Experimental Procedures). In each panel, arrows depict the magnitude and direction of the time rate of change of Oct4 and Sox2 concentration in Oct4 and Sox2 space. Conditions are (B) in the absence of differentiation signals (C) In the presence of NE inducing signals (D) In the presence of ME inducing signals (E) In the presence of inducers of both lineages. (F) Plot of steady state Nanog level for combinations of ME and NE inducing signals. Nanog is near zero in black regions but is present at ES cell levels in yellow regions. (G) A titration of the Wnt agonist CHIR in cells also treated with 250nM retinoic acid. Green dots show fraction of cells activating Sox1 and red dots indicate fraction of cells activating Brachyury. (H) Images of Sox1-GFP cells treated with 5uM CHIR and 250nM retinoic acid immunostained for Brachyury, Oct4, Sox2, and Nanog. See also Figure S6.
To test the prediction of our model, we added combinations of RA and CHIR to differentiating ES cells and quantified the fraction of cells that activated the germ layer markers Sox1-GFP or Brachyury by microscopy. Experimentally, we found that RA changes the cell’s interpretation of the CHIR signal. We added 250nM RA to cells and titrated the concentration of CHIR. In cells treated with CHIR between 3–5uM and 250nM retinoic acid, we did not detect either Sox1-GFP or Brachyury staining by microscopy (Figure 7G), while cells treated with these concentrations of CHIR alone would have activated Brachyury at 70% efficiency. To ascertain the fate of these cells, we stained the same cells for Oct4, Sox2, and Nanog, and found that many cells contained levels of these proteins similar to levels observed under pluripotency promoting conditions (Figure 7H). This implies that the cells do not enter either the ME or NE fate within the duration of the experiment. This could be because of delayed lineage choice or because the cells have entered a distinct state from which they cannot reach the ME and NE fate. A detailed analysis of the gene expression pattern, epigenetic state, and potency of these cells is the subject of future work and will give additional insight into the state of these jammed cells.
Discussion
In this work we asked how embryonic stem cells leave the pluripotent state and select between the ME and NE cell fate. As a cell transitions from ES cell to germ layer progenitor, several hundred genes are down regulated. By reconstituting the lineage branch in vitro, we could classify DNA binding proteins based on their expression pattern during lineage selection. We found that very few proteins that are present in ES cells are regulated in a lineage specific fashion during differentiation providing a substantial simplification of the complex, underlying transcriptional circuit. By using the underlying lineage branch as a guide, we can identify small, crucial sets of factors that control lineage choice within the complex regulatory circuits described by genomic methods.
Through this approach we found that pluripotency maintenance and lineage choice are intricately linked. The pluripotency circuit is known to act as a unit that strongly represses lineage specific gene expression in ES cells (Figure 1B). However, rather than being a monolithic entity, the pluripotency circuit components have lineage specific roles, so that the same proteins can also be used for lineage selection. While the intact pluripotency circuit inhibits all germ layer differentiation, Oct4 specifically represses only the neural ectodermal fate, while Sox2 specifically represses only the mesendodermal fate. Together, Oct4 and Sox2 repress differentiation into either germ layer fate. When these two proteins are differentially regulated leading to high Oct4 and low Sox2 levels, or low Oct4 and high Sox2 levels, either the mesendodermal fate or the neural ectodermal fate becomes available to the cell. Thus, by modulating the levels of the same proteins that together repress differentiation, a cell can select a germ layer fate.
Terminal markers of lineage choice, like Sox1 and Brachyury, have been identified in many progenitor cell populations. However, such terminal markers only report on the outcome of differentiation and do not allow examination of the events leading to cell fate choice. Oct4 and Sox2 protein levels provide a way to track the state of the cell continuously from ES cell to NE or ME progenitor cell. By studying proteins that are present and modulated continuously during lineage selection, we gain access to the detailed dynamics of the process. For example, sorting cells by their Oct4 or Sox2 levels during the cell fate transition would enable examination of epigenetic regulation and transcription factor binding patterns as a function of the cell’s differentiation status. Having continuous markers of lineage selection can thus illuminate the temporal coordination of a broad array of cellular processes and their impact on a cell’s developmental decision.
In many systems, a progenitor cell’s response to differentiation signals depends on the cell’s history, its context, and the presence of other signals in the environment. For example, in the developing limb bud, FGF and Wnt signals together retain progenitor cells in a multipotent state but individually promote differentiation and cell lineage specification (Hu et al., 2009; ten Berge et al., 2008a). Our study demonstrates that a context dependent interpretation of the signals can arise in part from the architecture of the transcriptional circuit that governs the multipotent state. External signals differentially activate Oct4 and Sox2, so that conflicting signals alter the cell’s interpretation of one another through their combined influence on the pluripotency circuit dynamics as seen in Figure 7.
Core circuits of transcription factors also operate in neural progenitor cells (Briscoe et al., 2000) and hematopoietic progenitor cells (Cantor and Orkin, 2001). These circuits might also play a role in signal integration and cell fate choice. While each type of progenitor cell might use a different combination of transcription factors to make lineage choices, circuits that make developmental decisions might use similar principles to integrate information from external signals and select a fate (Anderson, 2001). Our study provides a general approach for isolating decision-making circuits and for relating broad scale circuit structure to the dynamic behavior of single cells.
Experimental Procedures
ES Cell Culture and Differentiation Assays
Cells were propagated in feeder-free, serum-free N2B27 media supplemented with LIF and BMP as described previously (Ying and Smith, 2003; Ying et al., 2008). ES cell media was supplemented with the FGF receptor inhibitor PD173074 (Sigma, StemGent) at 100 nM to suppress background differentiation. For differentiation toward the neural ectoderm or mesendoderm lineages, ES cells were plated into N2B27 for 48 hr, followed by addition of Wnt3a (200 ng/ml) or CHIR99021 (3 μM) for ME differentiation (ten Berge et al., 2008b), or RA (500 nM) for NE differentiation (Ying et al., 2003b). For differentiation experiments, cells were immunostained 36 hr after addition of the differentiation signal.
Immunofluorescence and FACS
Immunofluorescence and flow cytometry were performed using standard techniques. (Supplementary Experimental Procedures).
Live Cell Fluorescence Microscopy
Cells were seeded on glass bottom dishes (MatTek) and imaged on an environmentally enclosed Zeiss Axiovision.
Analysis of Published Microarray and ChIP-seq Data
Transcription factors and DNA binding proteins in ES, ME, and NE cells were identified from GEO GSE12982 (Shen et al., 2008). Published ChIP-seq data (Chen et al., 2008; Marson et al., 2008) were analyzed using custom code in Mathematica (Wolfram) (Supplementary Experimental Procedures).
Gene Expression Microarray for ES vs. 48 hr state
Microarrays were conducted in triplicates on Illumina Mouse-Ref8 BeadChips, according to the manufacturer’s protocol. Quantile normalization was used in Figure 2B. Microarray data were deposited at GSE 29005.
Overexpression and siRNA Knockdown
Overexpression experiments used CAG promoter-driven Oct4 and Sox2 plasmids (Mitsui et al., 2003). siRNA constructs were validated in previous studies (Hu et al., 2009). Plasmid and/or siRNA transfections were performed 24 hours before addition of differentiation signals.
Generation of Oct4-mCitrine Transgenic ES Cell Line
Oct4-mCitrine fusion construct was generated using Red/ET Recombination (Gene Bridges). The construct consisted of the distal and proximal enhancers of Oct4, the Oct4 ORF, the linker, mCitrine cDNA, the native 3′ UTR of Oct4, and the loxP-PGK-gb2-neo-loxP selection cassette (Gene Bridges).
Chromatin Immunoprecipitation and qPCR
Chromatin Immunoprecipitation (ChIP) was performed essentially according to (Mikkelsen et al., 2007). qPCR was performed on an ABI 7900HT using primers from ChIP-qPCR tiling arrays (SABiosciences) and custom primer sets (Operon). Fold enrichment was calculated relative to input.
Mathematical Modeling of Pluripotency Circuit
Simulation of mathematical models was performed using custom written code in MATLAB (MathWorks) (Supplementary Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Doug Melton, Sean Eddy, Areez Mody, and Alexander Schier for scientific discussions and Rene Maehr and Masa Yamagata for discussions and technical assistance. We thank Manfred Baetscher and the Harvard Genome Modification Facility for assistance with the cell line generation. We thank Bodo Stern, Nicole Francis, and in particular Sean Eddy and three anonymous referees for extensive comments on the manuscript. We thank the Harvard Stem Cell Institute Seed Grant for support (SR). Microarray hybridization and measurements were performed by the Molecular Genetics Core Facility at Children’s Hospital Boston supported by NIHP50-NS40828, and NIH-P30-HD18655.
Footnotes
Author Contributions MT and SR conceived the project. MT established the experimental system and performed the immunofluorescence and perturbation experiments. SJL and MT built the cell lines and performed microarray experiments. LNZ performed the FACS experiments and established techniques for long term microscopy of ES cells on glass. MT and SJL performed the time-lapse microscopy experiments. ZS and AM did the ChIP for Oct4 and Sox2. SJL, MT and SR performed the tiling qPCR experiments. MT, SR, and SJL performed the data and mathematical analyses and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS One. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron. 2001;30:19–35. doi: 10.1016/s0896-6273(01)00260-4. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jackson SA, Schiesser J, Stanley EG, Elefanty AG. Differentiating embryonic stem cells pass through ‘temporal windows’ that mark responsiveness to exogenous and paracrine mesendoderm inducing signals. PLoS One. 2010;5:e10706. doi: 10.1371/journal.pone.0010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Masui S, Ohtsuka S, Yagi R, Takahashi K, Ko MS, Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyagi S, Nishimoto M, Saito T, Ninomiya M, Sawamoto K, Okano H, Muramatsu M, Oguro H, Iwama A, Okuda A. The Sox2 regulatory region 2 functions as a neural stem cell-specific enhancer in the telencephalon. J Biol Chem. 2006;281:13374–13381. doi: 10.1074/jbc.M512669200. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Jakt LM, Era T. Embryonic stem-cell culture as a tool for developmental cell biology. Nat Rev Mol Cell Biol. 2007;8:502–507. doi: 10.1038/nrm2189. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H. Mouse ES cell culture system as a model of development. Dev Growth Differ. 2010;52:275–283. doi: 10.1111/j.1440-169X.2009.01166.x. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, Ribecco-Lutkiewicz M, Bani-Yaghoub M, Walker PR. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. J Neurosci Res. 2008;86:1680–1693. doi: 10.1002/jnr.21635. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008a;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008b;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003b:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.