Abstract
Biological systems have evolved to harness non-equilibrium processes from the molecular to the macro scale. It is currently a grand challenge of chemistry, materials science, and engineering to understand and mimic biological systems that have the ability to autonomously sense stimuli, process these inputs, and respond by performing mechanical work. New chemical systems are responding to the challenge and form the basis for future responsive, adaptive, and active materials. In this article, we describe a particular biochemical-biomechanical network based on the microtubule cytoskeletal filament – itself a non-equilibrium chemical system. We trace the non-equilibrium aspects of the system from molecules to networks and describe how the cell uses this system to perform active work in essential processes. Finally, we discuss how microtubule-based engineered systems can serve as testbeds for autonomous chemical robots composed of biological and synthetic components.
Introduction
Microtubules, tubular structures with a diameter of 25 nm and a length of several micrometers formed from the protein tubulin, are a unique system to study non-equilibrium assembly processes.1, 2 Starting with the near-equilibrium polymerization of tubulin into microtubules, over the complex behaviors and functions enabled by dynamic instability, to the close-to-macroscopic behavior of microtubule assemblies, recent research is illustrating the transition from chemistry to robotics. A special emphasis of current research are situations where biomolecular motors, such as kinesin, can couple the system to stores of chemical energy while thermal fluctuations still remain critical for the system behavior. In biological systems, microtubules act as “robotic arms” grasping chromosomes or waving cilia, and the study of these systems generates valuable insights into non-equilibrium thermodynamics as well as molecular robotics. Our discussion of recent contributions to the field is loosely organized around the “distance from equilibrium,” starting with microtubule assembly and ending with molecular robotics.
Microtubules are polymers of alpha-beta tubulin dimers that assemble non-covalently using hydrophobic and electrostatic interactions (Fig. 1a). They are important biological structures that form the structural support girders of the cell (Fig. 1b). They mechanically support long extensions in axons, dendrites, cilia, and flagella. Microtubules act as load-bearing compression rods in muscle that enable contracting muscle fibers to spring back fully3. They set up the cell division machinery, the mitotic spindle, and their disassembly acts to pull apart chromosomes in the dividing cell (Fig. 1b).
Figure 1.
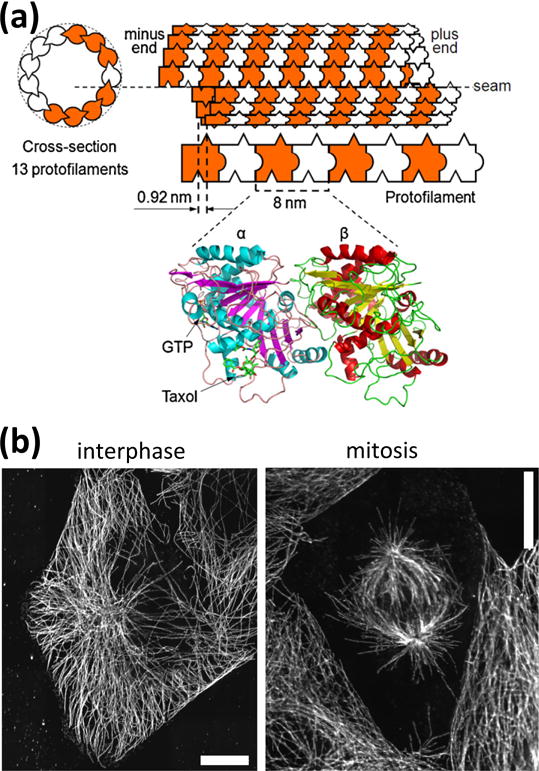
(a) Schematic representation of a microtubule with 13 protofilaments. Protofilaments are made from tubulin dimers which bind to each other in a head-to-tail fashion. Protofilaments bind to each other with an offset which results in a seam in the microtubule structure. The ribbon diagram (bottom) is derived from electron crystallography4 and shows the GTP and taxol binding sites. (b) During interphase, the microtubule cytoskeleton is radially organized, with microtubules emerging from a microtubule-organizing center located in the vicinity of the nucleus (visible as the microtubule free central region). During mitosis, two spindles of oriented microtubules form, then the microtubules attach to the chromosomes initially located along the centerline of the dividing cell and separate them by exerting forces generated from depolymerization. Scale bars: 10 μm.
Structurally, microtubules are built to withstand mechanical stresses. As hollow cylinders, 25 nm in outer diameter and 17 nm in inner diameter, they are built to withstand compressive loads and are significantly stiffer (persistence length of 1 mm) compared to other filaments that are built like twisted strings, such as actin or DNA. The microtubule tube is composed of a lattice of alpha-beta tubulin dimers that bind in a rhombic lattice that closes into a tube with 13 longitudinal lattices, called protofilaments (Fig. 1). The individual tubulin dimers are actually enzymes that bind and hydrolyze GTP as an energy source causing the filament length to change in time. Thus, microtubules are inherently non-equilibrium and dynamic. Further, microtubules couple to a host of associated proteins and enzymes that all act in concert to create the essential cellular structures described above. These individual structures may appear static, but all are dynamical steady-states. In this review, we will describe the non-equilibrium world of the microtubule from their individual dynamics to the coupling of many filaments with motors in simplified in vitro systems to describing how the complex biological machines of the cell use them to sense, decide, and react to cause motion and work. We highlight recent efforts to utilize microtubules and kinesin motors as components in molecular systems, which may evolve into autonomous molecular robots. Finally, we discuss studies pointing to the fundamental limits in the performance of molecular robots.
Equilibrium Aspects of the Assembly of Tubulin into Microtubules
The polymerization of filamentous microtubules is an entropically-driven process and the polymeric form is actually the minimum-energy state for the system at 37°C. Experimentally, we know this because when we assemble tubulin dimers using a slowly-hydrolyzable analog of GTP, GMPCPP, the filaments assemble normally. Although it may be surprising that assembly could be entropically driven, it is possible because the water molecules, which are far more numerous composing the bulk of the system, have increased entropy when microtubules polymerize. It is the hydrophobic, greasy patches between dimers that restrict the accessible states of the nearby water molecules. When the dimers bind, these hydrophobic patches exclude the water.
Once polymerized with GMPCPP, microtubules are fairly stable structures until the GMPCPP is hydrolyzed. Stability in this case means they stay polymerized in different conditions including cold temperatures (4–10°C) and dilution of the filaments below the typical critical concentration for polymerization. In addition to GMPCPP, there are other stabilizing effectors, such as the chemotherapeutic drug, paclitaxel (Taxol). Like GMPCPP, Taxol stabilizes the microtubule against cold depolymerization and lowers the critical concentration, no matter which nucleotide is in the binding pocket (GTP, GDP, GMPCPP). Taxol is a hydrophobic molecule that binds to the beta tubulin monomer near the GTP-binding site.
The length distribution of rapidly grown and subsequently stabilized microtubules has been found to follow a Schulz distribution,5 which is frequently used to describe molecular weight distributions of synthetic polymers.6 Schulz distributions have a linear increase at low molecular weights, a peak at intermediate molecular weights, and an exponential decrease towards higher molecular weights. They arise from the kinetics of polymerization at more than one end of the polymer chain.7 Growth by tubulin addition terminates when a local equilibrium between the growing microtubule end and the local free tubulin concentration has been established. In conditions typically used to polymerize microtubules in vitro, this occurs on a timescale of less than an hour.8
The observation that microtubules join end-to-end on a timescale of days, most recently illustrated by the work of Bachand et al.9, demonstrates that the microtubule length distribution continues to evolve towards a global minimum in free energy. However, the slow translational and particularly rotational diffusion of these polymers with molecular weights in the GigaDalton range leads to extremely slow reaction kinetics.
The example of tubulin polymerization illustrates nicely that our notion of equilibrium is inevitably associated with a specific timescale. Depending on the timescale (here minutes vs. days), the system is capable of overcoming kinetic barriers of different sizes and thereby explores different regions of the phase space.
Assembled microtubules are known for their superior mechanical properties, which play essential roles in their cellular abilities. Stabilized microtubules are important to the stability and length of the axon, for instance. In the axon, microtubules are stabilized by the neuronal microtubule-associated protein (MAP) tau, which also stiffens the filaments.11–13 Interestingly, Taxol has generally been shown to make microtubules more flexible both by direct experiments and from molecular dynamics simulations,13–18 whereas GMPCPP microtubules are measured to be stiffer.13, 16, 19 Microtubule stabilization also appears to coincide with certain post-translational modification states, such as acetylation, but it remains unclear if acetylation causes stiffer filaments, or if acetylation recruits stabilizing and stiffening MAPs.
Non-Equilibrium Aspects of the Assembly of Tubulin into Microtubules (Dynamic Instability)
Although microtubule polymerization is a process moving towards equilibrium, the individual tubulin dimers themselves are enzymes that bind and hydrolyze GTP to GDP. That hydrolysis causes conformational changes within the tubulin dimer subunit that tilts and shifts the interactions between the dimers. These conformational changes were once thought to be a bending within the dimer because the equilibrium structures of GDP tubulin are GDP-rings composed of microtubule protofilaments that are bent back and around20. Recent high resolution structural work shows that the conformational changes are much more complex but resemble a compaction that results in a tilting of the interdimer locations.21, 22 The end result is still a dimer that changes from a straight conformation to a tilted back conformation upon hydrolysis of GTP (Fig. 3).
Figure 3.
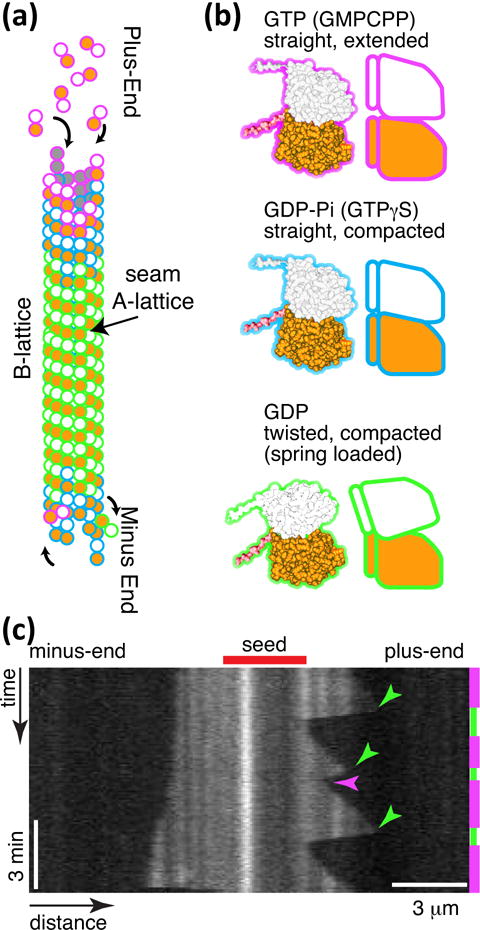
Microtubule dynamics. (a) Microtubule lattice with GTP cap at plus and minus ends, and GDP tubulin in the body. The lattice is a B-lattice, with alpha-alpha lateral interactions. (b) Tubulin conformations GTP, straight and extended conformation, GDP-Pi straight and compacted conformation, and GDP in the compacted and twisted conformation. (c) Space-time plot (kymograph) of microtubule length dynamics over time. Images of microtubules are laid adjacent to show the length changes over time. Magenta time segments on the right denote growth, and green segments denote depolymerisation. Catastrophes are denoted with green arrow heads, and one rescue is denoted with a magenta arrowhead. This experiment was performed at 20 μM free tubulin. The plus-end is on the right, and the minus-end is on the left. Vertical scale bar: 3 min. Horizontal scale bar: 3 μm.
GTP hydrolysis within a tubulin dimer occurs only at the beta-tubulin at the exchangeable site (E-site), and the binding of the next dimer longitudinally on the same protofilament catalyzes hydrolysis and the subsequent conformational change. Thus, the dimers at the end of the filament have a high probability of having GTP at the E-site and being in the straight conformation (Fig. 3). Once the next dimer binds, the probability of hydrolysis increases, so the dimers within the body of the microtubule filament are most likely in the GDP, tilted conformation. Despite their desire to alter their internal conformation to the compacted form, GDP-dimers in the body are constrained to stay bound to their neighbors - forced into the straight conformation. This causes the dimers to be at a higher potential, conformationally spring loaded.
The stored strain energy favors the disassembly of the microtubule, but the presence of GTP-dimers at the microtubule end (a “GTP cap”) creates a kinetic barrier, which maintains the microtubule in a metastable state. Slowing addition of GTP-dimers, either as a result of low dimer concentrations or as a stochastic event, may cause the loss of the GTP cap by conversion of the GTP-dimers into GDP-dimers. Without a GTP cap, there is no barrier to disassembly and the microtubule depolymerizes rapidly (an event referred to as “catastrophe”). When the depolymerization process is arrested and reversed to growth (termed “rescue”) it is typically either due to GTP-dimers remaining in the lattice or the binding of stabilizing associated proteins.
While the entire solution (buffer, tubulin, microtubules, GTP, GDP, phosphate) evolves towards equilibrium, the GTPase character of the tubulin couples microtubule assembly to the exothermic conversion of GTP into GDP and maintains the microtubule population in a non-equilibrium state. In a cyclic process, binding of GTP primes tubulin for assembly, assembled tubulin converts GTP to GDP, the presence GDP tubulin enables disassembly followed by GDP unbinding and restoration of free tubulin. While free in solution, tubulin dissociates GDP and rebinds GTP, making it competent to polymerize once again. Most assays are done in a GTP-rich environment, where GTP is not the limiting factor. While the coupling of an exothermic to an endothermic reaction is of course present in many biochemical systems, the intrinsic microtubule mechanics gives the microtubule polymerization in the presence of GTP an unusual kinetics.
The long-time equilibrium of tubulin is not that interesting. When GTP runs out, tubulin stays in the dimer state. Other polymerization species can be formed, such as GDP rings20. Ultimately, tubulin will unfold and aggregate non-specifically. Tubulin requires a chaperone to fold, so once it is unfolded, it cannot return to the properly folded state outside of the cell. Misfolding of tubulin is often triggered by reactive oxygen species (ROS). One recent work has shown enhanced and elongated microtubule dynamics when the ROS are removed from the solutions.23
For most in vitro assays, the total concentration of tubulin is fairly small (1 μM – 45 μM) compared to GTP (1 mM), allowing multiple rounds of GTP hydrolysis and GTP rebinding. Although the exchange of GDP to GTP in free tubulin is fast, it can become rate limiting and lead to coupled oscillations in the microtubule phase. Studies performed at an order of magnitude higher tubulin concentrations than typical (50 – 400 μM), show that the entire microtubule population can oscillate in autonomous polymerization-depolymerization cycles.24, 25 Theoretical modeling shows that the oscillations emerge from the replacement of GTP in the released tubulin limiting the reaction and the return to polymer.24 These results are strikingly different from those at low concentration where every individual microtubule is performing dynamic instability such that the total background tubulin concentration stays constant.
The original measurements demonstrating dynamic instability required there to be two states (GTP and GDP) in order to recapitulate the long growth and fast shrinkage times observed.26 Thus, since the system was observed to have dynamic instability, it was known to be a non-equilibrium process. Mitchison and Kirshner stated, “The observed length fluctuations are much too large to be explained by random fluctuations of an equilibrium polymer. Using reasonable rate constants, a polymer 15 μm long would take a year to fluctuate to zero by fluctuations at equilibrium, and the addition of treadmilling would not give the observed rates.”26 Thus, the first realization of how fast microtubule dynamics were at equilibrium lead immediately to the need for non-equilibrium processes (energy-utilizing conformational state changes) to reconcile the dynamics. No equilibrium process, even one with cooperativity, would enable such fast kinetics.
As stated above, the microtubule dynamics are inexorably linked to the mechanical nature of the filaments. The mechanical properties of dynamic microtubules have been measured and, interestingly, are dependent on the growth rate of the tubulin.27, 28 This is due to the fact that fast polymerization results in microtubule lattices with defects. As we know from other crystalline growth processes, fast, stochastic growth can lead to lattice defects, such as empty lattice sites and dislocations. Additionally, the fact that microtubules can end-to-end anneal (Fig. 2c) can lead to lattice defects at the annealing site.
Figure 2.
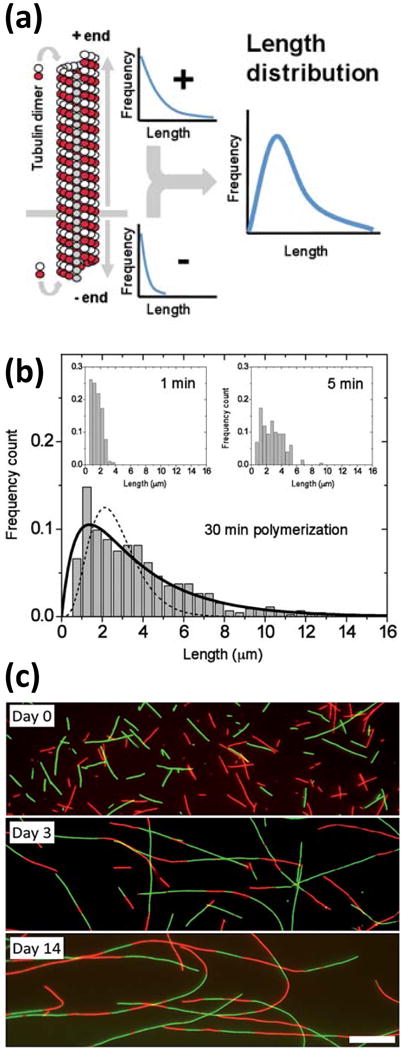
(a) Microtubule assembly proceeds at both ends with unequal rates, giving rise to a length distribution with a peak at intermediate lengths. (b) An equilibrium between tubulin addition and removal is established within minutes. (c) Mixing two microtubule populations of different fluorescent labels (red and green) reveals end-to-end joining of microtubules observed on a timescale of days. Scale bar: 10 μm. (a) & (b) reproduced from10 and (c) reproduced from9 with permission from the Royal Society of Chemistry.
Recent work has shown that when free tubulin is present, defects in the microtubule wall lattice can heal by the addition of new dimers to open sites.28, 29 When the free tubulin associates within the lattice defect, it is in the GTP form, in the straight conformation.29 The healed site becomes mechanically and chemically more stable.28, 29 A recent cellular study has shown that the healing of microtubules enhances the mechanical stability in cells.29 The enhanced stability of the microtubule cytoskeleton in one particular region of the cell can cause the cytoskeleton to increase growth in that direction.29 Since the microtubules are the steering wheel of motile cells,30, 31 increased growth in one region of the cell edge can turn a motile cell.29 Thus, the local mechanical nature of microtubules within a cell can direct the cell’s motion.
Microtubules’ ability to self-heal by the addition of new tubulin dimers is important for a number of cellular activities. For instance, it has been previously shown that katanin, a microtubule severing enzyme, targets microtubule defects.32, 33 That implies that a defect, if left unhealed, could potentially lead to the severing and collapse of the filament. Further, recent work has shown that microtubule defects can inhibit the transport of motor proteins needed to traffic intracellular cargos.34, 35 Another recent study demonstrated that intracellular transport can be enhanced when microtubules are stretched in live neurons.36 These studies imply that mechanical stresses and even damage to microtubules can affect the biological functions and activities of microtubule filaments.
Feedback and Control in the Assembly of Tubulin into Microtubules
As described above, tubulin at very high concentrations can form a negative feedback loop due to the lag in re-binding of GTP.24, 25 This feedback causes the polymerization and depolymerization to coordinate into oscillating waves.24 In cells, tubulin concentration is not that high, yet microtubule growth and shrinkage is coordinated through microtubule-associated proteins (MAPs) that bind to the filaments and alter the polymerization, depolymerization, catastrophe, and rescue rates. There have been a number of studies that have catalogued the effects of individual types of MAPs on microtubule dynamics.37–40
Some MAPs individually alter the microtubule dynamics. For instance, tau protein, a MAP found exclusively in the axons of neurons that is misfolded and aggregated in patient brains with Alzheimer’s disease and dementia is known to stabilize filaments, promote polymerization, and mechanically rigidify microtubules (Fig. 4a).12, 13, 41, 42 Another positive microtubule regulator is doublecortin, which binds preferentially to and creates 13-protofilament microtubules.43 Doublecortin binds to the junction between dimers, which could help it to control the position of the next dimer into a perfect 13-protofilament lattice.44
Figure 4.
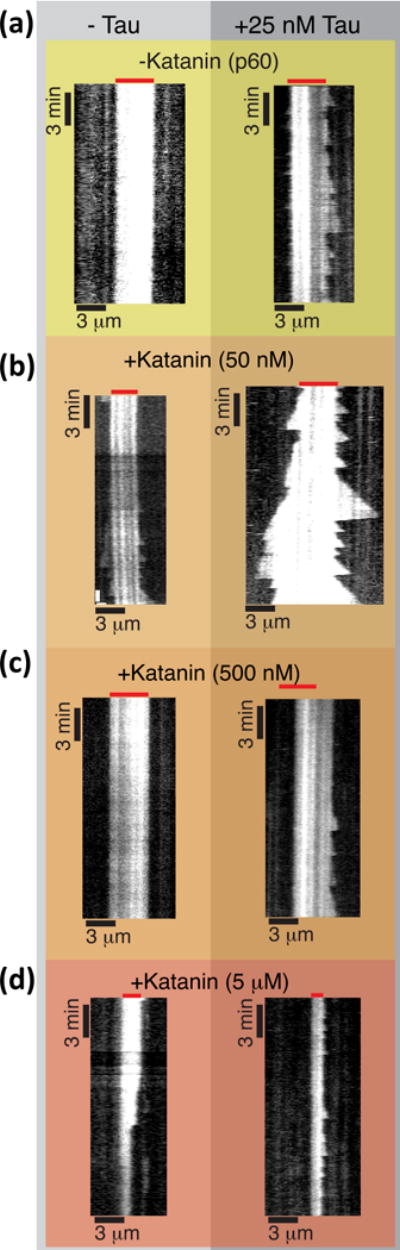
Dynamic instability of 7 μM tubulin in the presence of stabilizers (25 nM tau) and destabilizers (katanin) at 23°C. (a) Microtubules are not dynamic in the presence of 7 μM tubulin without tau (left), but grows with 25 nM tau (right). (b) The addition of 50 nM katanin increases the dynamics of microtubules without tau (left) and with tau (right). (c) Adding 500 nM katanin removes all dynamics without tau (left), and inhibits growth with tau (right). (d) Adding 5 μM katanin causes the GMPCPP seeds to be depolymerized without tau (left), but minimal growth with tau (right).
There are several MAPs that have been shown to have altered affinity for different types of tubulin enzymatic states. As described above, tubulin has different conformations depending on the nucleotide in the E-site of the beta tubulin (GTP, GDP-Pi, or GDP). Both end-binding proteins (EB1, 2, 3) and TOG-domain proteins (such as XMAP215 and CLASPs) appear to have weaker affinity for the GDP state.22, 45–47 Because only the GTP cap of the microtubule has GTP or GDP-Pi (Fig. 3), these proteins preferentially bind to and “track” the growing microtubule ends. Interestingly, most of these proteins act to decrease the critical concentration – enabling growth at lower tubulin concentrations.48 But, these proteins have differential effects on microtubule dynamics including: enhancing polymerization rates (XMAP215),46, 49–54 enhancing rescues and inhibiting catastrophes (CLASP)46, 50, 55 or enhancing catastrophe (EB proteins)48, 52 of dynamic microtubules. This could be due to limiting the ability of tubulin to bind just from steric effects.
There are two main types of destabilizing MAPs, all of which are ATPase enzymes. One type is a variant of kinesin motor proteins, which typically walk along microtubules and carry cargos to the microtubule plus-ends. The kinesin-family of destabilizers use the ATPase to destabilize the microtubule protofilaments from their ends and peel them back. The first discovered microtubule destabilizer was MCAK, discovered from Xenopus egg extracts.56 Since MCAK’s discovery, several branches of the kinesin family tree have been shown to have a penchant for destabilizing microtubules including the kinesin-8s and kinesin-13s.37 Depolymerizing kinesins play a major role in mitosis to control microtubule dynamics, length, and stability throughout the process. The depolymerizing activity of these kinesins has been recapitulated using purified proteins in vitro. In the presence of dynamic microtubules with 15 μM free tubulin, MCAK has been shown to be a catastrophe factor57 or completely inhibit growth.48 Interestingly, they can also disassemble stabilized microtubules, but not “doubly stabilized” microtubules that have both GMPCPP and Taxol present.58
The second family of microtubule destabilizing enzymes are the microtubule severing proteins. These proteins are evolutionarily distinct from kinesin enzymes.40, 59 They are from the ATPases Associated with various cellular activities (AAA+) enzyme family – an ancient enzyme family with members in all cell types and organisms. The AAA+ enzymes form hexameric rings with an active pore in the middle. Severing enzymes come from three families, katanin, spastin, and fidgetin. They all appear to form rings where the active pore recognizes the carboxy-terminal tail of tubulin dimers, but the specific mechanism for how severing enzymes work to remove dimers from the lattice is still a mystery. Interestingly, katanin can sever stabilized and doubly stabilized microtubules,33 but not dynamic microtubules (Fig. 4b). We have found that this is because free tubulin dimers in solution can inhibit the binding of katanin.60 Spastin and fidgetin have not been tested on dynamic microtubules, but may face the same challenge.
The microtubule depolymerizing and severing enzymes can break microtubules down into dimers, but can also create other tubulin assembly byproducts. The breaking pathways and subsequent tubulin products of destruction can be informative about how the microtubule is assembled and the energetics that hold it together and allow it to heal. Studies on microtubule structural failure have informed on the ability of depolymerizers and severing enzymes to exert forces to destroy microtubules. In particular, molecular dynamics simulations can apply pulling or pushing forces at particular locations along the microtubule lattice to determine the work required to destroy the lattice.61–64 In these works, it is clear that the intramolecular bonds between dimers are the weak points in the lattice, and they come apart before dimers are unraveled.64 These studies are complimented by several experimental studies of the structure of filaments during and after forced destruction by depolymerizing enzymes or severing enzymes.65–67 Other studies have used atomic force microscopy (AFM) techniques to specifically push on microtubules.68–70 The microtubule cylinder can buckle under such pushing, and individual dimers can also be removed, if the AFM cantilever tip is small enough to apply a contact pressure at one point.70
In the cell, there are multiple microtubule regulators that can all act simultaneously. In order to dissect the synergistic, cooperative, or antagonistic effects of these regulators in concert, researchers are systematically combining these regulators to determine if they stymie or enhance microtubule dynamics. Some of the first studies focused on depolymerizing kinesins (MCAK) with plus-end stabilizers (EB3).48 MCAK has an EB-binding domain, an SxIP motif, which caused the MCAK to localize to the growing plus-ends. The MCAK and EB3 were able to overpower one another at extreme ratios, but when balanced, they increased the dynamicity of the microtubules giving the microtubules a longer average length with frequent catastrophes and rescues.
We have combined the stabilizer, tau, with the microtubule severing enzyme, katanin (Fig 4), and find that together, microtubules are more dynamic. This is similar to the effect of EB3 with MCAK. Microtubules are protected from severing by free dimers alone until katanin is at very high concentrations (Fig. 4). Tau protects, stabilizes, and promotes growth, even in the presence of katanin, inhibiting the loss of polymer (Fig. 4).
An exciting study was published recently using five different microtubule regulating MAPs to determine a complete biochemical network of microtubule dynamic activity.55 The proteins used included EB1, TOG proteins XMAP215 (msps) and CLASP (Mast/Orbit), SLAIN2 (Sentin), and depolymerizing kinesin kinesin-13 (Klp10A). Interestingly, the Drosophila CLASP, called Mast/Orbit, suppresses microtubule growth alone, which is different from prior work with CLASPs.46 In combination, these five factors occupy a phase space made up of growing, shrinking, and paused microtubules. Interestingly, the paused state was truly pausing – not growth and shrinkage with rapid dynamics, as observed with MCAK and EB3. Future studies with severing enzymes acting as negative regulators would shed even more light onto the control of microtubule dynamics.
Stabilizers and destabilizers in combination and competition provide regulatory feedback loops to control microtubule dynamics in cells. For instance, depolymerizing motors work to detach incorrect or ineffective microtubule-kinetochore attachments in the mitotic spindle. Many of the motors and associated proteins are controlled by kinases that alter binding through specific phosphorylation sites Together, this regulation constitutes the spindle checkpoint – a feedback system used to ensure correct chromosome separation during mitosis.71 We discuss these biological examples of microtubule network organization under feedback control and producing work in live cells below.
Equilibrium Self-Assembly of Microtubules in vitro
The transition from equilibrium to non-equilibrium dynamics described above for the assembly of microtubules from tubulin proteins can also be found in the assembly of higher-order structures made from microtubule filaments. Microtubules are negatively charged rigid rods in typical buffers, and therefore will assemble into a nematic phase at sufficiently high concentrations (Fig. 5a).72 Manipulation of the microtubule-microtubule interactions by addition of counterions can induce attraction and the formation of microtubule bundles.73 Similarly, depletion agents can induce attractive interactions between microtubules and bundle formation.73, 74
Figure 5.
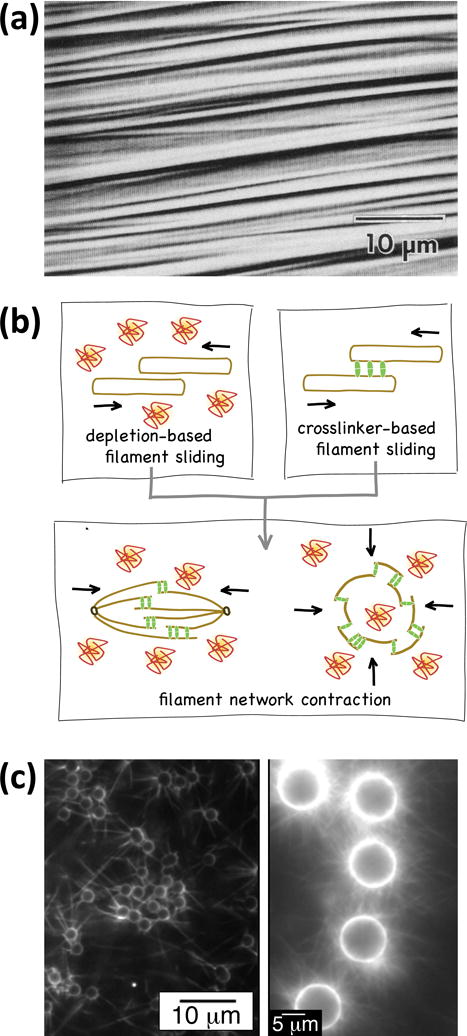
Equilibrium self-assembly of microtubules. (a) Microtubules at high concentration self-assemble into ordered arrays (a nematic crystalline phase). This research was originally published in72. © the American Society for Biochemistry and Molecular Biology. (b) The combination of microtubule aggregation due to depletion forces and forces exerted by cross-linkers capable of passively diffusing along the microtubule can give rise to contractile networks. Reproduced with permission from76. (c) Microtubule asters can be assembled by end-specific attachment of microtubules to microspheres (left), or by polymerization of microtubules from the surface of microspheres (right). Adapted with permission from84, 88. Copyright 2008, 2013 American Chemical Society.
A particularly interesting system are microtubules interacting via Ase1 proteins.75 Ase1, also known as MAP65 in plants and PRC1 in mammals, is capable of cross-linking microtubules while being able to laterally diffuse along the tubulin lattice. This entropically favors overlap between the cross-linked microtubules and generates a thermodynamic driving force towards contraction of a microtubule network, which can be well-understood with equilibrium thermodynamics (Fig. 5b).76, 77 Despite being transient, weak and thermodynamically controlled, many of these interactions coupled together can overpower active motor proteins.78
In contrast, cross-linking of microtubules by strong cross linkers, such as streptavidin proteins cross-linking biotinylated microtubules, leads to highly disordered networks of microtubules.79 Although parallel bundles of microtubules would maximize the number of cross-linkers and minimize the elastic energy stored in bent microtubules, the system is quickly trapped in a local energy minimum where the number of cross-links is low and the stored elastic energy is high.80, 81 Since the lifetime of each biotin-streptavidin bond is on the timescale of hours,82 the evolution towards the equilibrium configuration is extremely slow.83
Binding of microtubules to a predetermined attachment site by specific interactions can be used to construct microtubule structures with desired geometries. For example, streptavidin-coated microspheres or surface patches can organize biotinylated microtubules into aster shapes (Fig. 5c).84, 85{Armstrong2014} However, the low translational and rotational diffusion coefficients of long microtubules can increase the assembly time under equilibrium conditions so much that this approach becomes impractical for larger structures.86 The preferred approach is therefore to let short microtubules self-assemble onto the attachment site and then extend the microtubules by further polymerization with added tubulin.87, 88
“Passive” self-assembly, where the building blocks move by diffusion and mismatched connections are broken by thermal forces, can yield beautiful equilibrium structures (such as DNA origami89 and of course microtubules themselves). However, passive self-assembly is opposed by strong bonds and slow diffusion, and both factors start to act in the assembly of microtubules-based structures.
Non-Equilibrium Self-Assembly and Self-Organization of Microtubules in vitro
The limitations of slow diffusive transport can of course be overcome by manipulating microtubules with external fields. In particular, dielectrophoresis has proven to be a useful tool to attract microtubules to specific sites and form structures resembling e.g. the mitotic spindles described below.90, 91
However, rather than the organization of microtubules with long range fields, the emergent phenomena induced by the local interactions of microtubules with motor proteins have captured the imagination of researchers.92, 93 Kinesin and dynein motor proteins interacting with microtubules permit the coupling of microtubule manipulation to a large reservoir of chemical energy in the form of ATP.94, 95 This chemical energy can be either used to speed up assembly processes (overcoming either kinetic barriers or friction and viscosity) or to reach high energy metastable states.86 A third and maybe the most exciting possibility is to use the energy to maintain dissipative non-equilibrium states, which will revert to equilibrium states when the ATP is exhausted.74, 77, 96, 97
The acceleration of assembly and the creation of stable but strained structures due to the mechanical work performed by molecular motors is best characterized as “active self-assembly” because the system does not quickly revert to equilibrium after the flow of chemical energy is stopped. In contrast, the emergence of ordered states which depend on the constant flow of energy is a classic example of “self-organization”. Active self-assembly appeals to the engineer as a bridge between self-assembly utilizing diffusive transport and robotic assembly where the sequence of assembly steps is programmed98. However, biological systems demonstrate that self-organized structures enable a desirable mix of adaptability, robustness, and longevity, which researchers try to replicate in the creation and study of “active matter”.99
The state of the art in the active self-assembly of microtubules is described in a recent review.100 The initial descriptions of the nanowire and nanospool structures formed when microtubules gliding on a surface coated with kinesin motors can interact with each other via biotin-streptavidin linkages80, 81 have been complemented by detailed studies elucidating the assembly process.101–116 Under suitable conditions, microtubule bundles with close to millimeter dimensions have been obtained, and a mathematical description of the process captured the fundamental differences between diffusive and active transport of building blocks in self-assembly (Fig. 6a).117 Equally important is the ability to utilize some of the mechanical work by the motors to create highly stressed structures, that is microtubule spools with a diameter of a few micrometers. Finally, the forces produced by the molecular motors enable the breaking of unwanted connections and thereby let the system overcome kinetic barriers on its path to assembly.
Figure 6.
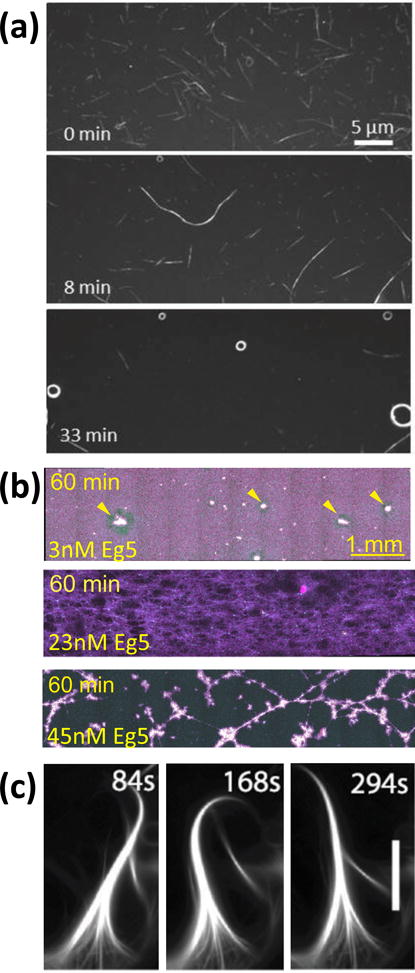
Self-assembly and self-organization in kinesin-microtubule systems. (a) Biotinylated microtubules partially covered with streptavidin assemble into wires and spools as they are transported by surface-adhered kinesins. Adapted with permission from81. Copyright 2005 American Chemical Society. (b) Different microtubule networks form depending on the concentration of motile cross-linkers (here the kinesin Eg5). Adapted from118. (c) Kinesin cross-linkers and depletion forces lead to the self-organization of microtubules into beating cilia. Scale bar: 5 μm. From77. Reprinted with permission from AAAS.
The study of microtubule self-organization processes mediated by kinesin motors in vitro was pioneered by Nedelec et al.92 who demonstrated that microtubules interacting with motile cross linkers formed by kinesin motor assemblies evolve into ordered structures reminiscent of mitotic spindles. Several recent high profile studies expanded upon this theme and demonstrated filament/motor systems exhibiting self-organization, ranging from networks formed by actin filaments and myosin motors,119, 120 over vortex lattices emerging from microtubules moving on surface-adhered motors,94, 97, 121 to microtubule bundle networks exhibiting motion and internal flows driven by kinesin motors.74, 77,118, 122
For both self-assembly and self-organization processes we can consider a notion of efficiency.123 If a highly strained structure, such as a “microtubule spool”, is formed, how much chemical energy has been expended relative to the stored strain energy? If the assembly of a structure, such as a “microtubule wire”, is accelerated by motor-driven transport, how much chemical energy has been utilized to produce a given amount of heat as result of viscous friction? If the maintenance of a self-organized structure requires a certain flow of energy, how much larger is the consumption of chemical energy?
The principal appeal of motor proteins is their stunning efficiency in converting chemical energy into mechanical work (frequently larger than 40%).124–126 However, this potential for high efficiency is frequently not utilized by the system. For example, the assembly of extended microtubule wires relies on the active transport of microtubules by surface-adhered kinesins, but the viscous force resisting the microtubule gliding is on the order of tens of femtoNewtons while the hundreds of kinesin motors propelling the microtubule can supply hundreds of picoNewtons as each one of them consumes one ATP molecule for every 8 nm step it takes. Thus the system efficiency is below 0.001%.
Well-designed macroscale robotic systems exhibit much higher efficiencies. Further, the efficiency of self-assembly by diffusive transport – as encountered in chemical reactions – is high in processes near equilibrium where little of the free energy of the system is dissipated during the assembly process in excess of what is needed to perform the assembly. Biological self-organized systems tend to achieve high efficiency not only at the level of the employed motor, but often also at the level of the overall system, such as a muscle or in neuronal transport. We believe that low efficiency will be a major roadblock to the utilization of non-equilibrium processes,127 and that progress towards the thermodynamic limits will be essential.
Another design feature of using microtubule-based systems is the mechanical properties. As described above, microtubules are incredibly stiff and capable of withstanding compressive loads. Further, the ability of microtubules to self-repair after degradation by the addition of new dimers to the ends or sides of the filaments allows them to withstand molecular wear. A number of recent papers have demonstrated that simple kinesin-microtubule interactions can cause wear and even tearing apart of stabilized microtubules.128–130 These reports, combined with those mentioned above, detail the limitations of stable microtubules with respect to mechanical failure. The good news is that dynamic microtubules give a solution to these limitations. The ability of dynamic microtubules to regrow and self-heal in the presence of free tubulin dimers enables microtubules to be essential parts of cellular machines (see next section). Further, understanding and using this property will allow us to design and use microtubules in engineering and practical applications to be apart of molecular robot systems of the future (see below).
Complex, Controlled Non-Equilibrium Feedback in Biology
As just described, microtubules are the ideal structural basis on which to build biological machines of the cell. Such essential cellular organizations are dynamically arranged and maintained through non-equilibrium processes. Two specific examples include the cilium and the mitotic spindle. For instance, the cilium structure appears quiescent, yet the organization of microtubules, associated proteins, and axonemal dynamics required for cilia length maintenance and motility is truly in a dynamical steady state maintaining the structure.131–134 With genetic manipulation, the steady state can be altered to have shorter flagella or different beating patterns.131, 133, 134 When mechanical manipulation is performed, such as removal of one flagella from a Chlamydomonas cell, the system is out of steady state but recovers by shrinking one cilium while growing the other until the two catch up and grow together.133, 135 Interestingly, models to describe such systems are reminiscent of “control theory,” an interdisciplinary branch of engineering and mathematics that deals with the behavior of dynamical systems with inputs, and how their behavior is modified by feedback.133
The mitotic spindle is another microtubule-based organization that appears quiescent, but observations point to a dynamical steady state of tubulin proteins turning over along the length of the football-shaped structure. The mitotic spindle turn over comes from both microtubule nucleation, dynamic instability, and the activity of individual motors and crosslinking proteins acting along the length of the spindle.136
There are also known gradients of kinases and phosphatases that control the state and interaction of a host of microtubule-associated proteins and motors.137 The underlying structure of the mitotic spindle is not that complex, but the non-equilibrium maintenance, information processing, feedback control, and resulting work that results in the splitting of the genetic material into two new cells is still being elucidated. A recent review by Trivedi and Stukenberg postulate a double feedback loop control system to control the mechanical and chemical signaling in the kinetochore/centromere “checkpoint” system that could control when the spindle decides to divide,71 so we will not go through it completely here. Instead, we will highlight a few examples involving microtubules and motors relevant to the active, non-equilibrium systems described above.
Such microtubule systems are not simply chemical, but utilize the mechanical nature of the filaments to create forces to perform mechanical work to move objects in the cell. This is perhaps more obvious in the mitotic spindle (Fig. 7). At early stages, microtubule polymerization and association of proteins act to completely rearrange the microtubules and genetic chromosomal materials into the center of the cell. The astral microtubules that extend to the edges of the cell from the microtubule organizing centers act to keep the spindle in the exact center of the cell longitudinally (Fig. 7(b)). Work in reconstituted systems show that microtubule-organizing centers can center themselves in a microchamber coated with dynein motors.138 Interestingly, the microtubules are not being pushed in this system, but rather pulled. The centering comes when the number of microtubules on each side is equal, making the sum of the forces equal to zero. Lateral spindle centering has also been recently investigated in live C. elegans egg cells using magnetic tweezers (Fig. 7(d)). The resultant force measurements show that there is a combination of viscous and elastic elements keeping the spindle in the center and not drifting side-to-side.140
Figure 7.
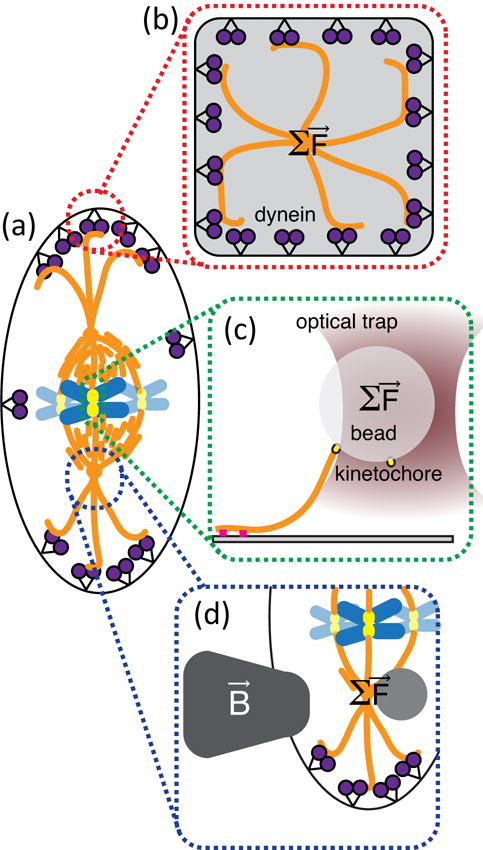
The microtubule network in dividing cells in a mechanochemical network that has chemical and force-dependent feedback loops. (a) A dividing cell in metaphase can be tested by recapitulating various parts in vitro using force measurements.138 (b) Optical trapping combined with nanolithography was used to recapitulate centrosome centering within dynein-lined microchambers. (c) Optical trapping was used to show that kinethochores use tension-sensitive slip-bonds to hold onto depolymerizing microtubules during anaphase.139 (d) Magnetic tweezers were used in live cells to measure the restoring force controlling midplane centering.140
Another location in the metaphase mitotic spindle where force-feedback plays an essential role is in the link between the microtubules of the spindle and the kinetochores (Fig. 7(c)). Over the past several years, sensitive force measurements have shown that kinetochores can bind to microtubules with a catch-bond type of interaction that strengthens under tension – much like a finger-trap toy.139, 141 These tension sensors hold tight when under tension between the two sister chromatids, turning off the spindle check-point when ready to split into two. This alignment is an essential organization to count and catalogue the genetic material. Once aligned and processed, the system changes to pulling apart the chromosomes to the two far ends of the spindle to create the centers of the two, new daughter cells.
These examples are just a few of many exciting and clean quantitative measurements now accessible to understand biological processes involving microtubules. The resultant new information is allowing specific and accurate modeling to feed predictions into more experiments. The feedback of these systems are beginning to be understood, and once they are understood, we may begin to use the same ideas to create autonomously self-organizing and responsive “robot” systems of the same microtubule materials in vitro.
Engineering Autonomous Molecular Robots
In our efforts to engineer robots at the molecular and nanoscale it may be helpful to achieve clarity about language and goals. What is an autonomous molecular robot?
In traditional usage, a robot is a programmable machine. It is characterized by both information processing and performance of mechanical work. Without mechanical movement, we may have a computer or sensor, but not a robot. Without programmability, we have a machine. The design and synthesis of molecular machines is worthy of a Nobel Prize in Chemistry, as the award to Feringa, Sauvage, and Stoddart in 2016 demonstrates, but they are not robots. Many machines can sense, compute and act, but they do so in a “hard-wired” fashion. In other words, a mousetrap is not a robot.
An objection is that a molecular robot is programmed at the moment of its creation, that a molecular entity - by necessity – follows a set of instructions encoded into it and a different set of instructions change the molecular entity. For example, a program can be encoded in a DNA strand, but the information is inseparable from the molecular structure. However, some molecular structures can clearly be programmed to change their behavior, e.g. by changes in redox states, conformational changes, or post-translational modifications. For example, the Bryant group has developed kinesin motor proteins which alter their direction of movement along the microtubule in response to optical stimuli.142 Thus it seems fair to require programmability even for molecular robots.
Biological systems however illustrate the challenge to conceptual clarity. The ribosome performs a sequence of assembly steps defined by the binding mRNA, but is the mRNA-ribosome complex a robot executing a program, or is the ribosome a machine resembling a printing press which inalterably performs the sequence of functions dictated by the mRNA? Are the proteins binding to microtubules and modifying their dynamic behavior, instruction being stored on the surface of a microtubule robot, or parts modifying a machine?
Similar confusion can be induced by the macroscopic “foals” created by artist Douglas Repetto.143 Despite having barely more parts than a mousetrap, these wooden structures powered by a constantly rotating electric motor exhibit seemingly awareness of their physical environment and complex movements which can be altered by minor modifications to their structure. In robotics, the concept of passive-dynamic walking machines challenges our ideas what a robot has to be.144
Progress in robotics as well as our increasing familiarity with macroscale autonomous robots, such as the “Roomba” or the “Aibo”, has somewhat shifted the concept of a robot away from a programmable machine to an autonomous device capable of sensing, computing and actuating even if the responses are hard-wired. Walter’s tortoises,145 Braitenberg’s vehicles,146 and an electronic mousetrap would be considered by most as a “robot”. Thus Winfree suggested at one point that “robot” is not a useful technical term.147
Murata et al. define a molecular robot as “a set of molecular devices such as sensors, logic gates, and actuators integrated into a consistent system”,148 which appears to gloss over the distinction between a machine and a robot. But the research described by Murata et al. reveals an emphasis on mobility and thereby suggests that a robot can be thought of as a mobile machine. From this point of view, a ribosome (stationary) is a machine, RNA polymerase (moving along the DNA strand) is a robot. Thus a slight revision of Murata’s definition to “a set of molecular devices such as sensors, logic gates, and actuators integrated into a mobile system” may capture the spirit of molecular robotics.
What constitutes autonomy at the molecular scale is similarly worth asking.149 At the macroscale, autonomy is conferred by energy storage and the ability to sense, compute and act without communication with a controller. In biology, cells are clearly autonomous units capable of storing energy, processing inputs, and responding with complex actions. Which of these characteristics can be obtained in molecular robots?
Molecular shuttles assembled from microtubules and kinesin motor proteins are model systems for autonomous molecular robots, and they are well-suited to illustrate the meaning of “a set of molecular devices integrated into a mobile system” as well as to examine the meaning of autonomy at the molecular scale.150, 151
In the most frequently employed design, kinesin motor proteins are adhered to a surface and propel microtubules stabilized against depolymerization with velocities up to 1 μm/s (Fig. 8a).151 Various passive and active means to control the velocity and direction of microtubule gliding have been developed.151–155 For example, ATP concentration or temperature can control microtubule speed, and guiding channels and external fields can control the direction of movement. Cargo can be attached to and removed from gliding microtubules via specific cross linkers, including antibodies, DNA strands, and biotin streptavidin bonds.
Figure 8.
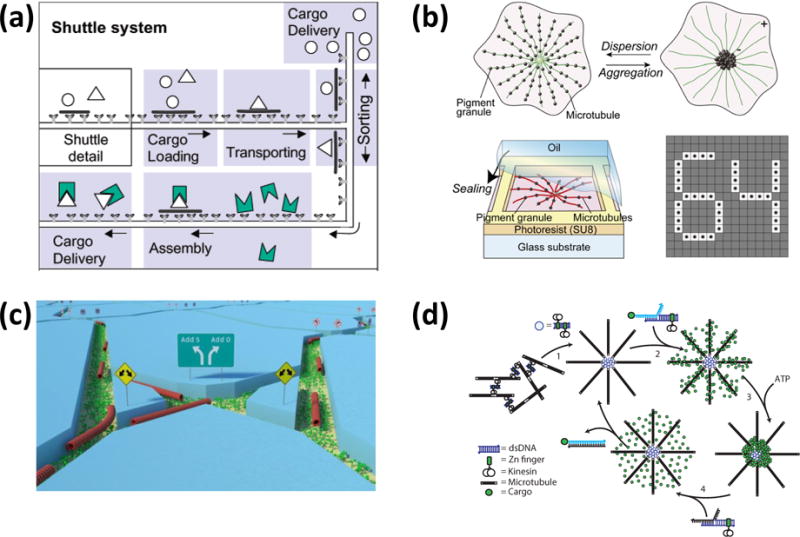
Molecular robots based on the kinesin-microtubule system. (a) Molecular shuttles as conceived by Vogel and Howard utilize kinesins adhered in tracks and controllably activated with defined concentrations of ATP to propel cargo-carrying microtubules. Adapted with permission from177. Copyright 2003 Americal Chemical Society. (b) In “artificial melanophores”, kinesin-driven aggregation of fluorescent material along microtubule asters creates a switchable pixel. From87. (c) Microtubules traversing a network of channels can be used for energy-efficient biocomputation. From160. (d) DNA connections introduce programmability into the self-organization process. From164. Reprinted by permission from Macmillan Publishers Ltd: Nature Nanotechnol. 9, 44-47, copyright 2014.
An individual microtubule and its associated motors can be considered a molecular robot, although the identity of kinesin motors constantly changes as the microtubule is propelled across the surface. The kinesin motors in turn obtain the chemical energy for propulsion from ATP in the solution. Therefore, this molecular robot harvests not only energy from the environment without any ability to store it, but also continuously exchanges the molecular devices composing it with the environment. The “set of molecular devices” referred to by Murata et al.148 can therefore be in a dynamic state.
An alternative point of view is to consider the entire experiment with millions of microtubules, trillions of kinesin motors and nanomoles of ATP molecules as a molecular robot. This “molecular robot” is certainly a system of molecular devices but now only with internal mobility and macroscopic dimensions. However, enclosure of microtubules and motors into membranes could achieve autonomous microscopic systems resembling cells capable of mobility and energy storage, as shown by the Dogic group.74
For a chemist, the first view with a focus on individual supramolecular structures exchanging energy and building blocks with the environment may seem more natural and better able to capture the molecular character of the system. For example, the variability in microtubule length originates in the chemistry of its assembly process (Fig. 2), the elevation of gliding microtubules above the surface has been shown to be determined by the free energy of the kinesin tails,156, 157 and the direction of movement is influenced by thermal fluctuations. Nevertheless, some molecular fluctuations are already smoothed out at the scale of an individual gliding microtubule. For example, while the stepping of an individual kinesin motor exhibits a randomness related to its chemomechanical cycle, the fluctuations in microtubule gliding velocity arise from the randomness in the motor positions on the surface relative to the microtubule.158 In some applications, each microtubule performs individually the required task and their large number merely amplifies the effect. For example in “smart dust” biosensors based on kinesin-powered molecular shuttles, each microtubule captures analytes, picks up fluorescent tags, and delivers the tags to a collection site.159 Inspired by the camouflage mechanism of fish, Aoyama et al.’s artificial melanophores utilize the transport of fluorescent material by kinesin motors along a radial array of microtubules to switch the optical properties of a “pixel” in a large array of such devices (Fig. 8b).87
However, in other applications the “set of molecular devices” encompasses more logically many microtubules and motors. Nicolau’s biocomputation concept160 (Fig. 8c) relies on a large number of kinesin-propelled microtubules (or myosin-propelled actin filaments) to traverse a network of channels encoding a mathematical problem. The solution to the problem is given by the distribution of microtubules along different exit channels of the network. Thus, the solution to the problem emerges from the observation of a “swarm” of microtubules, similar to demonstrations of surface imaging by molecular shuttles to reveal topography161, 162 or surface deformation163.
While the kinesin-microtubule system allows rapid and efficient conversion of chemical energy into mechanical work, it is not naturally suited for sensing and computing functions. However, state-of-the-art protein engineering has been shown to imbue kinesins with controllability142, and the integration of DNA building blocks has enabled programmability164 (Fig. 8d).
The issues of feedback and control have arisen in several studies of the kinesin-microtubule system in different guises. Van den Heuvel et al. sorted gliding microtubules by creating a feedback mechanism which resorts to macroscopic components (a camera images the fluorescently labeled microtubules, a computer evaluates the image and an electric power supply adjusts the voltages of electrodes creating an electric field acting on the microtubules).165 Similar systems with macroscopic components in the control loop have been repeatedly studied, but they clearly remove the autonomy of the molecular robot and turn the kinesin-microtubule system into a machine with remote control.166–172 However, a gliding microtubule navigating through channels and around obstacles on a surface exhibits autonomous behavior similar to Walter’s tortoises. The gliding microtubule “chooses” a path which minimizes the bending of the microtubule. Thermal fluctuations introduce noise to the microtubule bending and thereby the direction of movement.173, 174 Constraining the direction of movement of the microtubule in a general direction (e.g. in a guiding channel) requires the addition of mechanical work at every encounter between the microtubule and the wall of the guiding channel to counteract the loss of directionality in the direction of movement between wall contacts.175, 176
Autonomous feedback and control in molecular robots can often exploit the tendency of the system to approach thermodynamic equilibrium. The theory of molecular level control is therefore in principle closely connected to non-equilibrium thermodynamics. However, the results of modern non-equilibrium thermodynamics yield insights primarily when thermal fluctuations are dominant.178 This is the case e.g. for the directionality of smoothly gliding microtubules over distances exceeding the persistence length,169, 173, 176, 179 but frequently the deviations from the intended direction or desired velocity of a gliding microtubule are not thermal in origin (originating e.g. from the encounter with a defective kinesin motor).109, 174, 180 In these situations, feedback mechanisms can be created which arise from state-dependent rates in a non-equilibrium reaction network (inspired by signal transduction cascades). Such feedback is reminiscent of the formation of an ant trail, where a trail is stabilized by the pheromones deposited by ants traveling on it.181
The general robustness of kinesin-driven transport of microtubules has also enabled first investigations into engineering challenges such as extending the lifetime of the system. The first measure is of course to remove reactive oxygen species (ROS).182, 183 However microtubule shrinking, breaking and stopping can be observed unrelated to reactive oxygen species, and recent studies have aimed to characterize and minimize these degradation processes.129, 184, 185 The example of biological systems suggests that a constant turnover of molecular building blocks is required to construct molecular robots with lifetimes exceeding a few hours. Schaedel et al. have demonstrated that microtubules possess the capability to self-heal after mechanically induced damage, again utilizing microtubules as model systems to illustrate molecular engineering concepts.28
In summary, microtubules and their associated motors are an ideal model system to explore the engineering principles and potential applications of autonomous chemical robots, because they bridge the molecular and mesoscopic scales and permit the application of a broad range of techniques from biochemistry, biophysics, and nanoscale engineering. While several of the characteristics of an autonomous molecular robot (here defined as a mobile system of sensors, logic, and actuators), are so far only present in rudimentary form, it is our hope that future researchers will use the term “embryonic” to describe today’s molecular systems.
Fundamental limits for molecular robots
An exciting aspect of molecular robots is that they may be designed to operate at the physical limits for sensing, computing, and force generation, for example those imposed by the second law of thermodynamics. These limits have been investigated in many biophysical studies aiming to achieve an understanding of biological systems. The classic paper by Adam and Delbrueck describing how a reduction in the dimensionality of diffusion processes can facilitate intracellular transport186 has inspired investigations into how molecular shuttles can assist in the capture of analytes by nanoscale sensors.187, 188 The principles governing information flow in biochemical networks189–191 also apply to computations performed by molecular robots.192 Universal bounds apply to the energy efficiency of molecular motors,126, 193 and it does not matter if their origin is biological or synthetic. Schneider extensively discussed the applications of information theory to molecular machine design.194 In fact, a molecular robot may be buffeted by thermal fluctuations to such a degree that it is not obviously recognizable as an energy-consuming machine, and a recent advance is a procedure to identify non-equilibrium processes from violations of detailed balance,195 the organizing principle of molecular machines.196 A corollary to the Bell equation defines the optimal loading of a molecular bond,197 providing a guidepost to the design of molecular structures in the way yield strength guides structure design at the macroscale.
Due to their small energy consumption, molecular robots may even be powered by information reservoirs capable of supplying a few bits (or kTs).198 Recent experimental demonstrations supplied a stream of information by modulating external fields,199 but it is conceivable that more elegant ways to store and supply information at the molecular scale will be found.
Many interesting questions with respect to the fundamental limits of molecular robots and their subsystems however cannot be readily answered using thermodynamics or information theory. A major practical limitation of the kinesin-microtubule system is the relatively short lifetime, however it is not clear if that is a fundamental limitation of molecular motors or specific to protein-based motors.200 Marden and Allen discovered an empirical relationship between motor force output and motor mass which holds from the macroscale to the molecular scale and whose origin is completely unclear (Fig. 9a).201 Armstrong and Hess127 discussed a requirement for molecular robots rather than a limitation, based on an analogy with relationships discovered in ecology: In order to permit their deployment in large numbers, the molecular motors employed in the robotic systems have to reach energy efficiencies comparable to macroscale engines (Fig. 9b).
Figure 9.
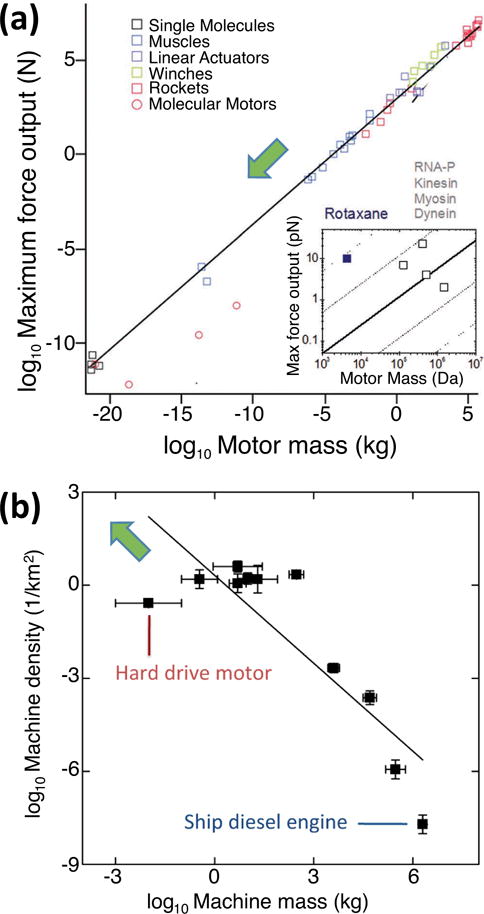
Empirical relationships discovered for motors raise questions for molecular motor and robot development. (a) Relation between maximum force of motors as function of their mass as discovered by Marden and Allen. Adapted with permission from201. Copyright 2002, National Academy of Science USA. The inset extrapolates the trend to the molecular scale. The rotaxane employed for artificial muscles is significantly stronger than what would be expected from this universal scaling law, which potentially accounts for its small lifetime.202 (b) Motors appear to replicate the inverse relationship between species size and abundance observed in ecosystems, and technological development drives the emergence of small and ubiquitous motors. However, motor efficiency has to be maintained as it is across species in biology. Adapted from127. Copyright 2014 American Chemical Society.
Maybe the biggest question is if future molecular robots populating the world will resemble biological systems like computers physically resemble brains (meaning not at all), or if they will conform to their biological role models by necessity and be soft, fragile, and short-lived.
Conclusions
The future of engineering clearly lies in technologies incorporating soft, active, and autonomous systems that can sense, process information, and respond to their environment to create forces and work to perform specific tasks to solve currently difficult problems. Indeed, many of our most challenging problems involve soft, granular, and biological systems that require knowledge and manipulation of such systems. For instance, could we design a chemical system to seek out underground or underwater oil spills from pipelines to cap and seal before environmental disaster strikes? Can we create systems that will seek out survivors from beneath the rubble of collapsed buildings to not only find them, but to also reorganize or reorder to create escape pathways for the survivors? Can we build medical nanorobots which assist the immune system and reverse aging at the cellular level?
Our deeper fundamental understanding of such chemically autonomous systems can be gleaned from further study and understanding of the biological systems we describe in this review. Biological systems have evolved to autonomously organize, sense, compute, respond, move, create force, and perform work in integrated systems ranging from molecules over tissues to animals. We still have a lot to learn from these systems in order to engineer our own autonomous materials. At the same time chemically autonomous systems begin to inform our understanding of the designs which have evolved in biological systems, an approach pursued by the emerging field of synthetic biology.203
Supplementary Material
Acknowledgments
Images of microtubules in LLPCK2 pig epithelial cells for Figure 1 provided by Patricia Wadsworth, University of Massachusetts Amherst. Kymograph images of dynamic microtubules taken by Dr. Megan Bailey under the direction of JLR.
HH is supported by the U.S. Army Research Office grant W911NF-13–1–0390 and thanks S. Murata, M. Stojanovic, E. Winfree and H. Palacci for fruitful discussions.
JLR is supported by a research grant from the Mathers Foundation, Scialog grant 4308.1 from the Moore Foundation and Research Corporation for Science Advancement, Army Research Office grant DoD ARO MURI 67455-CH-MUR (lead by Thayamanavan), NIH grant R01-GM109909 (lead by D. Sharp), and NSF INSPIRE grant MCB-1344203.
Notes and references
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. Fourth. Garland; New York: 2002. 4 edn. [Google Scholar]
- 2.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 3.Robison P, Caporizzo MA, Ahmadzadeh H, Bogush AI, Chen CYX, Margulies KB, Shenoy VB, Prosser BL. Science. 2016;352:428. doi: 10.1126/science.aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nogales E, Wolf SG, Downing KH. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 5.Jeune-Smith Y, Hess H. Soft Matter. 2010;6:1778–1784. [Google Scholar]
- 6.Sperling LH. Introduction to Physical Polymer Science. 4th. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2006. [Google Scholar]
- 7.Schulz GV. Zeitschrift Fur Physikalische Chemie-Abteilung B-Chemie Der Elementarprozesse Aufbau Der Materie. 1939;43:25–46. [Google Scholar]
- 8.Howard J, Hyman AA. Meth Cell Biology. 1993;39:105–113. doi: 10.1016/s0091-679x(08)60164-8. [DOI] [PubMed] [Google Scholar]
- 9.Bachand M, Bouxsein NF, Cheng S, von Hoyningen-Huene SJ, Stevens MJ, Bachand GD. Rsc Adv. 2014;4:54641–54649. [Google Scholar]
- 10.Jeune-Smith Y, Agarwal A, Hess H. Journal of Visualized Experiments. 2010 doi: 10.3791/2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mickey B, Howard J. J Cell Biol. 1995;130:909–917. doi: 10.1083/jcb.130.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felgner H, Frank R, Biernat J, Mandelkow EM, Mandelkow E, Ludin B, Matus A, Schliwa M. J Cell Biol. 1997;138:1067–1075. doi: 10.1083/jcb.138.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins TL, Sept D, Mogessie B, Straube A, Ross JL. Biophys J. 2013;104:1517–1528. doi: 10.1016/j.bpj.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gittes F, Mickey B, Nettleton J, Howard J. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felgner H, Frank R, Schliwa M. J Cell Sci. 1996;109:509–516. doi: 10.1242/jcs.109.2.509. [DOI] [PubMed] [Google Scholar]
- 16.Donhauser ZJ, Jobs WB, Binka EC. Biophys J. 2010;99:1668–1675. doi: 10.1016/j.bpj.2010.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sept D, MacKintosh FC. Phys Rev Lett. 2010;104:018101. doi: 10.1103/PhysRevLett.104.018101. [DOI] [PubMed] [Google Scholar]
- 18.Valdman D, Atzberger PJ, Yu D, Kuei S, Valentine MT. Biophys J. 2012;102:1144–1153. doi: 10.1016/j.bpj.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez BJ, Valentine MT. Cytoskeleton. 2014;71:530–541. doi: 10.1002/cm.21190. [DOI] [PubMed] [Google Scholar]
- 20.Carlier MF, Didry D, Pantaloni D. Biophys J. 1997;73:418–427. doi: 10.1016/S0006-3495(97)78081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Alushin GM, Brown A, Nogales E. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Islam MS, Kabir AMR, Inoue D, Sada K, Kakugo A. Biophys Chem. 2016;211:1–8. doi: 10.1016/j.bpc.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Carlier MF, Melki R, Pantaloni D, Hill TL, Chen Y. P Natl Acad Sci USA. 1987;84:5257–5261. doi: 10.1073/pnas.84.15.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandelkow EM, Lange G, Jagla A, Spann U, Mandelkow E. Embo J. 1988;7:357–365. doi: 10.1002/j.1460-2075.1988.tb02821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchison T, Kirschner M. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 27.Janson ME, Dogterom M. Biophys J. 2004;87:2723–2736. doi: 10.1529/biophysj.103.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaedel L, John K, Gaillard J, Nachury MV, Blanchoin L, Thery M. Nat Mater. 2015;14:1156–1163. doi: 10.1038/nmat4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aumeier C, Schaedel L, Gaillard J, John K, Blanchoin L, Théry M. Nat Cell Biol. 2016;18:1054–1064. doi: 10.1038/ncb3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao D, Su Z, Wang W, Wu H, Liu X, Akram S, Qin B, Zhou J, Zhuang X, Adams G. J Biol Chem. 2015;290:23766–23780. doi: 10.1074/jbc.M115.673517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams G, Zhou J, Wang W, Wu H, Quan J, Liu Y, Xia P, Wang Z, Zhou S, Jiang J. J Biol Chem. 2016;291:20692–20706. doi: 10.1074/jbc.M116.732719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis LJ, Odde DJ, Block SM, Gross SP. Biophys J. 2002;82:2916–2927. doi: 10.1016/S0006-3495(02)75632-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz-Valencia JD, Morelli MM, Bailey M, Zhang D, Sharp DJ, Ross JL. Biophys J. 2011;100:2440–2449. doi: 10.1016/j.bpj.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang WH, Li Q, Faysal KR, King SJ, Gopinathan A, Xu J. Biophys J. 2016;110:2229–2240. doi: 10.1016/j.bpj.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gramlich MW, Conway L, Liang WH, Labastide JA, King SJ, Xu J, Ross JL. Sci Rep-Uk. 2017 doi: 10.1038/srep44290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed WW, Saif TA. Sci Rep-Uk. 2014;4:4481. doi: 10.1038/srep04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walczak CE, Gayek S, Ohi R. Annu Rev Cell Dev Bi. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- 38.Ohi R, Zanic M. F1000Research. 2016;5:314. doi: 10.12688/f1000research.7439.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nogales E, Zhang R. Curr Opin Struc Biol. 2016;37:90–96. doi: 10.1016/j.sbi.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey ME, Jiang N, Dima RI, Ross JL. Biopolymers. 2016;105:547–556. doi: 10.1002/bip.22842. [DOI] [PubMed] [Google Scholar]
- 41.Schaap IAT, Hoffmann B, Carrasco C, Merkel R, Schmidt CF. J Struct Biol. 2007;158:282–292. doi: 10.1016/j.jsb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Levy SF, LeBoeuf AC, Massie MR, Jordan MA, Wilson L, Feinstein SC. J Biol Chem. 2005;280:13520–13528. doi: 10.1074/jbc.M413490200. [DOI] [PubMed] [Google Scholar]
- 43.Bechstedt S, Lu K, Brouhard GJ. Curr Biol. 2014;24:2366–2375. doi: 10.1016/j.cub.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 44.Fourniol FJ, Sindelar CV, Amigues B, Clare DK, Thomas G, Perderiset M, Francis F, Houdusse A, Moores CA. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur ELF. P Natl Acad Sci USA. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouveia SM, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, Wordeman L, Steinmetz MO, Akhmanova A. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 49.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Bassam J, Chang F. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanic M, Widlund PO, Hyman AA, Howard J. Nat Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
- 53.Podolski M, Mahamdeh M, Howard J. J Biol Chem. 2014;289:28087–28093. doi: 10.1074/jbc.M114.584300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussmann F, Drummond DR, Peet DR, Martin DS, Cross RA. Sci Rep-Uk. 2016;6:20653. doi: 10.1038/srep20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriwaki T, Goshima G. J Cell Biol. 2016;215:357–368. doi: 10.1083/jcb.201604118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wordeman L, Mitchison TJ. J Cell Biol. 1995;128:95–105. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 58.Varga V, Leduc C, Bormuth V, Diez S, Howard J. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 59.Sharp DJ, Ross JL. J Cell Sci. 2012;125:2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey ME, Sackett DL, Ross JL. Biophys J. 2015;109:2546–2561. doi: 10.1016/j.bpj.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theisen KE, Zhmurov A, Newberry ME, Barsegov V, Dima RI. The Journal of Physical Chemistry B. 2012;116:8545–8555. doi: 10.1021/jp212608f. [DOI] [PubMed] [Google Scholar]
- 62.Theisen KE, Desai NJ, Volski AM, Dima RI. The Journal of Chemical Physics. 2013;139:09B629_621. doi: 10.1063/1.4819817. [DOI] [PubMed] [Google Scholar]
- 63.Kononova O, Kholodov Y, Theisen KE, Marx KA, Dima RI, Ataullakhanov FI, Grishchuk EL, Barsegov V. J Am Chem Soc. 2014;136:17036–17045. doi: 10.1021/ja506385p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang N, Bailey ME, Burke J, Ross JL, Dima RI. Cytoskeleton. 2016 doi: 10.1002/cm.21346. [DOI] [PubMed] [Google Scholar]
- 65.Tan D, Rice WJ, Sosa H. Structure. 2008;16:1732–1739. doi: 10.1016/j.str.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang D, Grode KD, Stewman SF, Diaz-Valencia JD, Liebling E, Rath U, Riera T, Currie JD, Buster DW, Asenjo AB. Nat Cell Biol. 2011;13:361–369. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H. Cell reports. 2013;3:759–768. doi: 10.1016/j.celrep.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 68.de Pablo PJ, Schaap IA, MacKintosh FC, Schmidt CF. Phys Rev Lett. 2003;91:098101. doi: 10.1103/PhysRevLett.91.098101. [DOI] [PubMed] [Google Scholar]
- 69.Schaap IA, De Pablo PJ, Schmidt CF. European Biophysics Journal. 2004;33:462–467. doi: 10.1007/s00249-003-0386-8. [DOI] [PubMed] [Google Scholar]
- 70.Schaap IA, Carrasco C, de Pablo PJ, MacKintosh FC, Schmidt CF. Biophys J. 2006;91:1521–1531. doi: 10.1529/biophysj.105.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trivedi P, Stukenberg PT. Trends in biochemical sciences. 2016;41:160–174. doi: 10.1016/j.tibs.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hitt AL, Cross AR, Williams RC. J Biol Chem. 1990;265:1639–1647. [PubMed] [Google Scholar]
- 73.Needleman DJ, Ojeda-Lopez MA, Raviv U, Miller HP, Wilson L, Safinya CR. P Natl Acad Sci USA. 2004;101:16099–16103. doi: 10.1073/pnas.0406076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanchez T, Chen DTN, DeCamp SJ, Heymann M, Dogic Z. Nature. 2012;491:431–434. doi: 10.1038/nature11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lansky Z, Braun M, Ludecke A, Schlierf M, ten Wolde PR, Janson ME, Diez S. Cell. 2015;160:1159–1168. doi: 10.1016/j.cell.2015.01.051. [DOI] [PubMed] [Google Scholar]
- 76.Braun M, Lansky Z, Hilitski F, Dogic Z, Diez S. Bioessays. 2016;38:474–481. doi: 10.1002/bies.201500183. [DOI] [PubMed] [Google Scholar]
- 77.Sanchez T, Welch D, Nicastro D, Dogic Z. Science. 2011;333:456–459. doi: 10.1126/science.1203963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pringle J, Muthukumar A, Tan A, Crankshaw L, Conway L, Ross JL. J Phys-Condens Mat. 2013;25:374103. doi: 10.1088/0953-8984/25/37/374103. [DOI] [PubMed] [Google Scholar]
- 79.Idan O, Lam A, Kamcev J, Gonzales J, Agarwal A, Hess H. Nano Letters. 2012;12:240–245. doi: 10.1021/nl203450h. [DOI] [PubMed] [Google Scholar]
- 80.Bachand M, Trent AM, Bunker BC, Bachand GD. Journal of Nanoscience and Nanotechnology. 2005;5:718–722. doi: 10.1166/jnn.2005.112. [DOI] [PubMed] [Google Scholar]
- 81.Hess H, Clemmens J, Brunner C, Doot R, Luna S, Ernst KH, Vogel V. Nano Letters. 2005;5:629–633. doi: 10.1021/nl0478427. [DOI] [PubMed] [Google Scholar]
- 82.He SH, Lam AT, Jeune-Smith Y, Hess H. Langmuir. 2012;28:10635–10639. doi: 10.1021/la302034h. [DOI] [PubMed] [Google Scholar]
- 83.Yang YL, Bai M, Klug WS, Levine AJ, Valentine MT. Soft Matter. 2013;9:383–393. doi: 10.1039/C2SM26934A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spoerke ED, Bachand GD, Liu J, Sasaki D, Bunker BC. Langmuir. 2008;24:7039–7043. doi: 10.1021/la800500c. [DOI] [PubMed] [Google Scholar]
- 85.Portran D, Gaillard J, Vantard M, Thery M. Cytoskeleton. 2013;70:12–23. doi: 10.1002/cm.21081. [DOI] [PubMed] [Google Scholar]
- 86.Hess H. Soft Matter. 2006;2:669–677. doi: 10.1039/b518281f. [DOI] [PubMed] [Google Scholar]
- 87.Aoyama S, Shimoike M, Hiratsuka Y. Proceedings of the National Academy of Sciences. 2013;110:16408–16413. doi: 10.1073/pnas.1306281110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spoerke ED, Boal AK, Bachand GD, Bunker BC. Acs Nano. 2013;7:2012–2019. doi: 10.1021/nn303998k. [DOI] [PubMed] [Google Scholar]
- 89.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 90.Uppalapati M, Huang YM, Jackson TN, Hancock WO. Biophys J. 2007:308a–309a. [Google Scholar]
- 91.Noel JA, Teizer W, Hwang W. Acs Nano. 2009;3:1938–1946. doi: 10.1021/nn900325m. [DOI] [PubMed] [Google Scholar]
- 92.Nedelec FJ, Surrey T, Maggs AC, Leibler S. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- 93.Bachand GD, Spoerke ED, Stevens MJ. Biotechnol Bioeng. 2015;112:1065–1073. doi: 10.1002/bit.25569. [DOI] [PubMed] [Google Scholar]
- 94.Sumino Y, Nagai KH, Shitaka Y, Tanaka D, Yoshikawa K, Chate H, Oiwa K. Nature. 2012;483:448–452. doi: 10.1038/nature10874. [DOI] [PubMed] [Google Scholar]
- 95.Ito M, Kabir AMR, Inoue D, Torisawa T, Toyoshima Y, Sada K, Kakugo A. Polym J. 2014;46:220–225. [Google Scholar]
- 96.Keber FC, Loiseau E, Sanchez T, DeCamp SJ, Giomi L, Bowick MJ, Marchetti MC, Dogic Z, Bausch AR. Science. 2014;345:1135–1139. doi: 10.1126/science.1254784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue D, Mahmot B, Kabir AMR, Farhana TI, Tokuraku K, Sada K, Konagaya A, Kakugo A. Nanoscale. 2015;7:18054–18061. doi: 10.1039/c5nr02213d. [DOI] [PubMed] [Google Scholar]
- 98.Morris CJ, Stauth SA, Parviz BA. IEEE Transactions on Advanced Packaging. 2005;28:600–611. [Google Scholar]
- 99.Helbing D, Deutsch A, Diez S, Peters K, Kalaidzidis Y, Padberg-Gehle K, Lammer S, Johansson A, Breier G, Schulze F, Zerial M. Advances in Complex Systems. 2009;12:533–548. [Google Scholar]
- 100.Lam AT, VanDelinder V, Kabir AMR, Hess H, Bachand GD, Kakugo A. Soft Matter. 2016;12:988–997. doi: 10.1039/c5sm02042e. [DOI] [PubMed] [Google Scholar]
- 101.Liu HQ, Spoerke ED, Bachand M, Koch SJ, Bunker BC, Bachand GD. Advanced Materials. 2008;20:4476–4481. [Google Scholar]
- 102.Kawamura R, Kakugo A, Shikinaka K, Osada Y, Gong JP. Biomacromolecules. 2008;9:2277–2282. doi: 10.1021/bm800639w. [DOI] [PubMed] [Google Scholar]
- 103.Kakugo A, Tamura Y, Shikinaka K, Yoshida M, Kawamura R, Furukawa H, Osada Y, Gong JP. J Am Chem Soc. 2009;131:18089–18095. doi: 10.1021/ja901538n. [DOI] [PubMed] [Google Scholar]
- 104.Kawamura R, Kakugo A, Osada Y, Gong JP. Langmuir. 2010;26:533–537. doi: 10.1021/la902197f. [DOI] [PubMed] [Google Scholar]
- 105.Kawamura R, Kakugo A, Osada Y, Gong JP. Nanotechnology. 2010;21:145603. doi: 10.1088/0957-4484/21/14/145603. [DOI] [PubMed] [Google Scholar]
- 106.Liu H, Bachand GD. Soft Matter. 2011;7:3087–3091. [Google Scholar]
- 107.Kakugo A, Kabir AMR, Hosoda N, Shikinaka K, Gong JP. Biomacromolecules. 2011;12:3394–3399. doi: 10.1021/bm200829t. [DOI] [PubMed] [Google Scholar]
- 108.Tamura Y, Kawamura R, Shikinaka K, Kakugo A, Osada Y, Gong JP, Mayama H. Soft Matter. 2011;7:5654–5659. [Google Scholar]
- 109.Luria I, Crenshaw J, Downs M, Agarwal A, Seshadri SB, Gonzales J, Idan O, Kamcev J, Katira P, Pandey S, Nitta T, Phillpot SR, Hess H. Soft Matter. 2011;7:3108–3115. [Google Scholar]
- 110.Kabir AMR, Wada S, Inoue D, Tamura Y, Kajihara T, Mayama H, Sada K, Kakugo A, Gong JP. Soft Matter. 2012;8:10863–10867. [Google Scholar]
- 111.Inoue D, Kabir AMR, Mayama H, Gong JP, Sada K, Kakugo A. Soft Matter. 2013;9:7061–7068. [Google Scholar]
- 112.Gutmann G, Inoue D, Kakugo A, Konagaya A. Comm Com Inf Sc. 2014;461:13–22. [Google Scholar]
- 113.Wada S, Kabir AMR, Ito M, Inoue D, Sada K, Kakugo A. Soft Matter. 2015;11:1151–1157. doi: 10.1039/c4sm02292k. [DOI] [PubMed] [Google Scholar]
- 114.Wada S, Kabir AMR, Kawamura R, Ito M, Inoue D, Sada K, Kakugo A. Biomacromolecules. 2015;16:374–378. doi: 10.1021/bm501573v. [DOI] [PubMed] [Google Scholar]
- 115.VanDelinder V, Brener S, Bachand GD. Biomacromolecules. 2016;17:1048–1056. doi: 10.1021/acs.biomac.5b01684. [DOI] [PubMed] [Google Scholar]
- 116.Ito M, Kabir AMR, Islam MS, Inoue D, Wada S, Sada K, Konagaya A, Kakugo A. Rsc Adv. 2016;6:69149–69155. [Google Scholar]
- 117.Idan O, Lam A, Kamcev J, Gonzales J, Agarwal A, Hess H. Nano Letters. 2011;12:240–245. doi: 10.1021/nl203450h. [DOI] [PubMed] [Google Scholar]
- 118.Torisawa T, Taniguchi D, Ishihara S, Oiwa K. Biophys J. 2016;111:373–385. doi: 10.1016/j.bpj.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koenderink GH, Dogic Z, Nakamura F, Bendix PM, MacKintosh FC, Hartwig JH, Stossel TP, Weitz DA. P Natl Acad Sci USA. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kohler S, Schaller V, Bausch AR. Nat Mater. 2011;10:462–468. doi: 10.1038/nmat3009. [DOI] [PubMed] [Google Scholar]
- 121.Liu L, Tuzel E, Ross JL. J Phys-Condens Mat. 2011;23:374104. doi: 10.1088/0953-8984/23/37/374104. [DOI] [PubMed] [Google Scholar]
- 122.DeCamp SJ, Redner GS, Baskaran A, Hagan MF, Dogic Z. Nat Mater. 2015;14:1110–1115. doi: 10.1038/nmat4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koper GJM, Boekhoven J, Hendriksen WE, van Esch JH, Eelkema R, Pagonabarraga I, Rubi JM, Bedeaux D. Int J Thermophys. 2013;34:1229–1238. [Google Scholar]
- 124.Schmiedl T, Seifert U. Epl. 2008;81 [Google Scholar]
- 125.Wang W, Chiang TY, Velegol D, Mallouk TE. J Am Chem Soc. 2013;135:10557–10565. doi: 10.1021/ja405135f. [DOI] [PubMed] [Google Scholar]
- 126.Pietzonka P, Barato A, Seifert U. Journal of Statistical Mechanics. 2016:124004. [Google Scholar]
- 127.Armstrong MJ, Hess H. Acs Nano. 2014;8:4070–4073. doi: 10.1021/nn502431c. [DOI] [PubMed] [Google Scholar]
- 128.Kabir AMR, Inoue D, Hamano Y, Mayama H, Sada K, Kakugo A. Biomacromolecules. 2014;15:1797–1805. doi: 10.1021/bm5001789. [DOI] [PubMed] [Google Scholar]
- 129.Dumont E, Do LPC, Hess H. Nat Nano. 2015;10:166–169. doi: 10.1038/nnano.2014.334. [DOI] [PubMed] [Google Scholar]
- 130.VanDelinder V, Adams PG, Bachand GD. Sci Rep-Uk. 2016;6:39408. doi: 10.1038/srep39408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marshall WF, Rosenbaum JL. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marshall WF, Qin HM, Brenni MR, Rosenbaum JL. Mol Biol Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chan YHM, Marshall WF. Science. 2012;337:1186–1189. doi: 10.1126/science.1223539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Broekhuis JR, Leong WY, Jansen G. Int Rev Cel Mol Bio. 2013;303:101–138. doi: 10.1016/B978-0-12-407697-6.00003-9. [DOI] [PubMed] [Google Scholar]
- 135.Rosenbaum JL, Moulder JE, Ringo DL. The Journal of Cell Biology. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brugués J, Nuzzo V, Mazur E, Needleman DJ. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 137.Etemad B, Kops GJ. Current Opinion in Cell Biology. 2016;39:101–108. doi: 10.1016/j.ceb.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 138.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Nature. 2010;468:576–U255. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garzon-Coral C, Fantana HA, Howard J. Science. 2016;352:1124–1127. doi: 10.1126/science.aad9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miller MP, Asbury CL, Biggins S. Cell. 2016;165:1428–1439. doi: 10.1016/j.cell.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nakamura M, Chen L, Howes SC, Schindler TD, Nogales E, Bryant Z. Nat Nanotechnol. 2014;9:693–697. doi: 10.1038/nnano.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Repetto D. foal. http://sites.music.columbia.edu/douglas/portfolio/foal/
- 144.Collins SH, Wisse M, Ruina A. Int J Robot Res. 2001;20:607–615. [Google Scholar]
- 145.Walter WG. Sci Am. 1950;182:42–45. [Google Scholar]
- 146.Braitenberg V. Vehicles: Experiments in synthetic psychology. MIT Press; Cambridge, MA: 1984. [Google Scholar]
- 147.Winfree E. personal communication [Google Scholar]
- 148.Hagiya M, Konagaya A, Kobayashi S, Saito H, Murata S. Accounts Chem Res. 2014;47:1681–1690. doi: 10.1021/ar400318d. [DOI] [PubMed] [Google Scholar]
- 149.Hess H. The Bridge. 2010;40:51–56. [Google Scholar]
- 150.Hess H, Vogel V. Reviews in Molecular Biotechnology. 2001;82:67–85. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- 151.Agarwal A, Hess H. Prog Polym Sci. 2010;35:252–277. [Google Scholar]
- 152.Hiratsuka Y, Tada T, Oiwa K, Kanayama T, Uyeda TQ. Biophys J. 2001;81:1555–1561. doi: 10.1016/S0006-3495(01)75809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kamei T, Fukaminato T, Tamaoki N. Chemical Communications. 2012;48:7625–7627. doi: 10.1039/c2cc33552b. [DOI] [PubMed] [Google Scholar]
- 154.Sikora A, Ramón-Azcón J, Kim K, Reaves K, Nakazawa H, Umetsu M, Kumagai I, Adschiri T, Shiku H, Matsue T. Nano letters. 2014;14:876–881. doi: 10.1021/nl4042388. [DOI] [PubMed] [Google Scholar]
- 155.Jia Y, Dong W, Feng X, Li J, Li J. Nanoscale. 2015;7:82–85. doi: 10.1039/c4nr04454a. [DOI] [PubMed] [Google Scholar]
- 156.Kerssemakers J, Howard J, Hess H, Diez S. PNAS. 2006;103:15812–15817. doi: 10.1073/pnas.0510400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dumont ELP, Belmas H, Hess H. Langmuir. 2013;29:15142–15145. doi: 10.1021/la4030712. [DOI] [PubMed] [Google Scholar]
- 158.Palacci H, Idan O, Armstrong MJ, Agarwal A, Nitta T, Hess H. Langmuir. 2016;32:7943–7950. doi: 10.1021/acs.langmuir.6b02369. [DOI] [PubMed] [Google Scholar]
- 159.Fischer T, Agarwal A, Hess H. Nat Nanotechnol. 2009;4:162–166. doi: 10.1038/nnano.2008.393. [DOI] [PubMed] [Google Scholar]
- 160.Nicolau DV, Lard M, Korten T, van Delftf FCMJM, Persson M, Bengtsson E, Mansson A, Diez S, Linke H, Nicolau DV. P Natl Acad Sci USA. 2016;113:2591–2596. doi: 10.1073/pnas.1510825113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hess H, Clemmens J, Howard J, Vogel V. Nano Letters. 2002;2:113–116. [Google Scholar]
- 162.Kerssemakers J, Ionov L, Queitsch U, Luna S, Hess H, Diez S. Small. 2009;5:1732–1737. doi: 10.1002/smll.200801388. [DOI] [PubMed] [Google Scholar]
- 163.Inoue D, Nitta T, Kabir AMR, Sada K, Gong JP, Konagaya A, Kakugo A. Nat Commun. 2016;7 doi: 10.1038/ncomms12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wollman AJ, Sanchez-Cano C, Carstairs HM, Cross RA, Turberfield AJ. Nat Nanotechnol. 2014;9:44–47. doi: 10.1038/nnano.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van den Heuvel MGL, De Graaff MP, Dekker C. Science. 2006;312:910–914. doi: 10.1126/science.1124258. [DOI] [PubMed] [Google Scholar]
- 166.Ionov L, Stamm M, Diez S. Nano Letters. 2006;6:1982–1987. doi: 10.1021/nl0611539. [DOI] [PubMed] [Google Scholar]
- 167.Tucker R, Katira P, Hess H. Nano Letters. 2008;8:221–226. doi: 10.1021/nl072516n. [DOI] [PubMed] [Google Scholar]
- 168.Korten T, Birnbaum W, Kuckling D, Diez S. Nano Letters. 2012;12:348–353. doi: 10.1021/nl203632y. [DOI] [PubMed] [Google Scholar]
- 169.Nitta T, Tanahashi A, Hirano M. Lab on a Chip. 2010;10:1447–1453. doi: 10.1039/b926210e. [DOI] [PubMed] [Google Scholar]
- 170.Isozaki N, Shintaku H, Kotera H, Meyhofer E, Yokokawa R. Proc IEEE Micr Elect. 2015:238–241. [Google Scholar]
- 171.Isozaki N, Ando S, Nakahara T, Shintaku H, Kotera H, Meyhofer E, Yokokawa R. Sci Rep-Uk. 2015;5 doi: 10.1038/srep07669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kumar KS, Amrutha AS, Tamaoki N. Lab on a Chip. 2016;16:4702–4709. doi: 10.1039/c6lc01098a. [DOI] [PubMed] [Google Scholar]
- 173.Duke T, Holy TE, Leibler S. Phys Rev Lett. 1995;74:330–333. doi: 10.1103/PhysRevLett.74.330. [DOI] [PubMed] [Google Scholar]
- 174.Nitta T, Hess H. Nano Letters. 2005;5:1337–1342. doi: 10.1021/nl050586t. [DOI] [PubMed] [Google Scholar]
- 175.Clemmens J, Hess H, Lipscomb R, Hanein Y, Boehringer KF, Matzke CM, Bachand GD, Bunker BC, Vogel V. Langmuir. 2003;19:10967–10974. [Google Scholar]
- 176.Nitta T, Tanahashi A, Hirano M, Hess H. Lab on a Chip. 2006;6:881–885. doi: 10.1039/b601754a. [DOI] [PubMed] [Google Scholar]
- 177.Hess H, Matzke CM, Doot RK, Clemmens J, Bachand GD, Bunker BC, Vogel V. Nano Letters. 2003;3:1651–1655. [Google Scholar]
- 178.England JL. Nat Nanotechnol. 2015;10:919–923. doi: 10.1038/nnano.2015.250. [DOI] [PubMed] [Google Scholar]
- 179.Vikhorev PG, Vikhoreva NN, Sundberg M, Balaz M, Albet-Torres N, Bunk R, Kvennefors A, Liljesson K, Nicholls IA, Nilsson L, Omling P, Tagerud S, Montelius L, Mansson A. Langmuir. 2008;24:13509–13517. doi: 10.1021/la8016112. [DOI] [PubMed] [Google Scholar]
- 180.Bourdieu L, Duke T, Elowitz MB, Winkelmann DA, Leibler S, Libchaber A. Phys Rev Lett. 1995;75:176–179. doi: 10.1103/PhysRevLett.75.176. [DOI] [PubMed] [Google Scholar]
- 181.Bonabeau E, Théraulaz G. Sci Am. 2000;282:72–79. doi: 10.1038/scientificamerican0300-72. [DOI] [PubMed] [Google Scholar]
- 182.Brunner C, Hess H, Ernst KH, Vogel V. Nanotechnology. 2004;15:S540–S548. [Google Scholar]
- 183.Kabir AMR, Inoue D, Kakugo A, Kamei A, Gong JP. Langmuir. 2011;27:13659–13668. doi: 10.1021/la202467f. [DOI] [PubMed] [Google Scholar]
- 184.Korten T, Chaudhuri S, Tavkin E, Braun M, Diez S. IEEE T Nanobiosci. 2016;15:62–69. doi: 10.1109/TNB.2016.2520832. [DOI] [PubMed] [Google Scholar]
- 185.Reuther C, Diego AL, Diez S. Nat Nanotechnol. 2016;11:914–915. doi: 10.1038/nnano.2016.231. [DOI] [PubMed] [Google Scholar]
- 186.Adam G, Delbrueck M. In: Structural Chemistry and Molecular Biology. Rich A, Davidson N, editors. W H Freeman and Co.; New York: 1968. pp. 198–215. [Google Scholar]
- 187.Katira P, Hess H. Nano Letters. 2010;10:567–572. doi: 10.1021/nl903468p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nitta T, Hess H. Cel Mol Bioeng. 2013;6:109–115. [Google Scholar]
- 189.Detwiler PB, Ramanathan S, Sengupta A, Shraiman BI. Biophys J. 2000;79:2801–2817. doi: 10.1016/S0006-3495(00)76519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tostevin F, Ten Wolde PR, Howard M. PLoS Comput Biol. 2007;3:e78. doi: 10.1371/journal.pcbi.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mehta P, Lang AH, Schwab DJ. Journal of Statistical Physics. 2016;162:1153–1166. [Google Scholar]
- 192.Parrondo JM, Horowitz JM, Sagawa T. Nat Phys. 2015;11:131–139. [Google Scholar]
- 193.Seifert U. Rep Prog Phys. 2012;75:126001. doi: 10.1088/0034-4885/75/12/126001. [DOI] [PubMed] [Google Scholar]
- 194.Schneider TD. J Theor Biol. 1991;148:125–137. doi: 10.1016/s0022-5193(05)80467-9. [DOI] [PubMed] [Google Scholar]
- 195.Battle C, Broedersz CP, Fakhri N, Geyer VF, Howard J, Schmidt CF, MacKintosh FC. Science. 2016;352:604–607. doi: 10.1126/science.aac8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Astumian RD. Nat Nano. 2012;7:684–688. doi: 10.1038/nnano.2012.188. [DOI] [PubMed] [Google Scholar]
- 197.Hess H. Nano Letters. 2012;12:5813–5814. doi: 10.1021/nl303157n. [DOI] [PubMed] [Google Scholar]
- 198.Astumian RD. Nat Nanotechnol. 2016;11:582–583. doi: 10.1038/nnano.2016.98. [DOI] [PubMed] [Google Scholar]
- 199.Toyabe S, Sagawa T, Ueda M, Muneyuki E, Sano M. Nat Phys. 2010;6:988–992. [Google Scholar]
- 200.Hess H, Dumont ELP. Small. 2011;7:1619–1623. doi: 10.1002/smll.201100240. [DOI] [PubMed] [Google Scholar]
- 201.Marden JH, Allen LR. Proc Natl Acad Sci U S A. 2002;99:4161–4166. doi: 10.1073/pnas.022052899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu Y, Flood AH, Bonvallett PA, Vignon SA, Northrop BH, Tseng HR, Jeppesen JO, Huang TJ, Brough B, Baller M, Magonov S, Solares SD, Goddard WA, Ho CM, Stoddart JF. J Am Chem Soc. 2005;127:9745–9759. doi: 10.1021/ja051088p. [DOI] [PubMed] [Google Scholar]
- 203.Schwille P, Diez S. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:223–242. doi: 10.1080/10409230903074549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


