Abstract
Conditional gene knockout using the Cre/loxP system is instrumental in advancing our understanding of the function of genes in a wide range of disciplines. It is becoming increasingly apparent in the literature that some Cre transgenes mediated recombination occurs in unexpected tissues. Dermo1-Cre (Twist2-Cre) has been widely used to target skeletal lineage cells as well as other mesoderm-derived cells. Here we report that Dermo1-Cre exhibits spontaneous male germline recombination activity leading to a Cre-mediated recombination of a floxed Ptk2 (Protein tyrosine kinase 2, also known as Fak [Focal adhesion kinase]) allele but not a floxed Rb1cc1 (RB1 inducible coiled-coil 1, also known as Fip200 [FAK-family Interacting Protein of 200 kDa]) allele at high frequency. This ectopic germline activity of Dermo1-Cre occurred in all or none manner in a given litter. We demonstrated that the occurrence of germline recombination activity of Dermo1-Cre transgene can be avoided by using female mice as parental Dermo1-Cre carriers.
Keywords: Cre-loxP, conditional knockout, Fak, Fip200, Dermo1-Cre, Twist2-Cre, germline
Dermo1 (also named Twist2) is highly expressed in condensed mesenchyme during skeletal development and later in perichondrial and periosteal cells surrounding cartilage (Li et al., 1995). Similar to the Dermo1 expression pattern, Cre-recombinase activity in Dermo1-Cre mice was detected as early as E9.5 in mesodermal tissues. During endochondral ossification, Dermo1-Cre recombinase activity is detected in condensed mesenchyme from which both osteoblasts and chondrocytes are derived (Yu et al., 2003). Thus, Dermo1-Cre has been widely used as a tool to target skeletal lineage cells (Elefteriou and Yang, 2011). In addition, Dermo1-Cre has also been frequently used to target other mesenchymal lineage cells (Cornett et al., 2013; Geske et al., 2008; Lavine et al., 2008; Lin et al., 2008; Yin et al., 2008).
FAK (Focal adhesion kinase) is an intracellular non-receptor tyrosine kinase and a major mediator of signal transduction by integrins (Guan and Shalloway, 1992). Disruption of Fak gene in mice resulted in an early embryonic lethal phenotype, which precludes the thorough examination of tissue-specific phenotypes in postnatal life (Ilic et al., 1995). We have used the Cre-loxP recombination system to circumvent the early embryonic lethality by targeting the Fak gene disruption to the tissue of interest (Nagy et al., 2007; Peng et al., 2008; Shen et al., 2005; Sun et al., 2016). Recently, we reported that Fak deletion in osteoblast progenitor cells leads to osteopenia in mice (Sun et al., 2016). In order to elucidate the role of Fak in mesenchymal and osteochondrogenitor cells, we are employing the Dermo1-Cre transgenic mouse line (Yu et al., 2003). Fak floxed mice were bred with Dermo1-Cre mice to generate Fak conditional knockout mice. Cre-mediated recombination of the floxed allele inactivates the Fak function in Dermo1-Cre expressing cells and their descendants. The Cre transgene, wild type (WT or +), floxed (Flox), and Cre-recombined floxed Fak alleles (FAKR) were detected by PCR analysis of tail-tip genomic DNA with allele specific primers (Figure 1). To generate the conditional knockout mice, male FAKF/+;Dermo1-Cre/+ mice were bred with female FAKF/F mice. Tail-tip DNA was used to perform PCR to genotype the offspring. In this mating scheme, 4 genotypes (FAKF/F;Dermo1-Cre, FAKF/F, FAKF/+;Dermo1-Cre, and FAKF/+) were expected at 25% ratio for each (Figure 2A, top numbers). The Cre-mediated recombination should only occur in offspring carrying Dermo1-Cre transgene but not the offspring without Dermo1-Cre transgene including FAKF/F and FAKF/+. We obtained a total of 121 mice from 16 litters of the offspring. Surprisingly, thirteen percent (16/121) of offspring had unexpected genotype in which there was the absence of wild type allele and Dermo1-Cre transgene but the presence of Fak floxed allele and a Cre-recombined allele (designated as FAKF/R). There was 15% of offspring had the expected genotype of FAKF/F. Thus, 47% (16/34) of the offspring whose genotype showed the presence of Fak floxed allele and absence of both wild type allele and Dermo1-Cre transgene (thus genotypically “homozygous” for Fak floxed allele) showed the presence of the Cre-recombined allele (FAKR) (Figure 2a and 2b). Noticeably, whenever there was FAKF/F offspring in one litter, there was no FAKF/R mouse and vice versa (Table I), suggesting this unexpected recombination occurred in all or none manner.
Figure 1. Fak floxed locus and rearrangement.
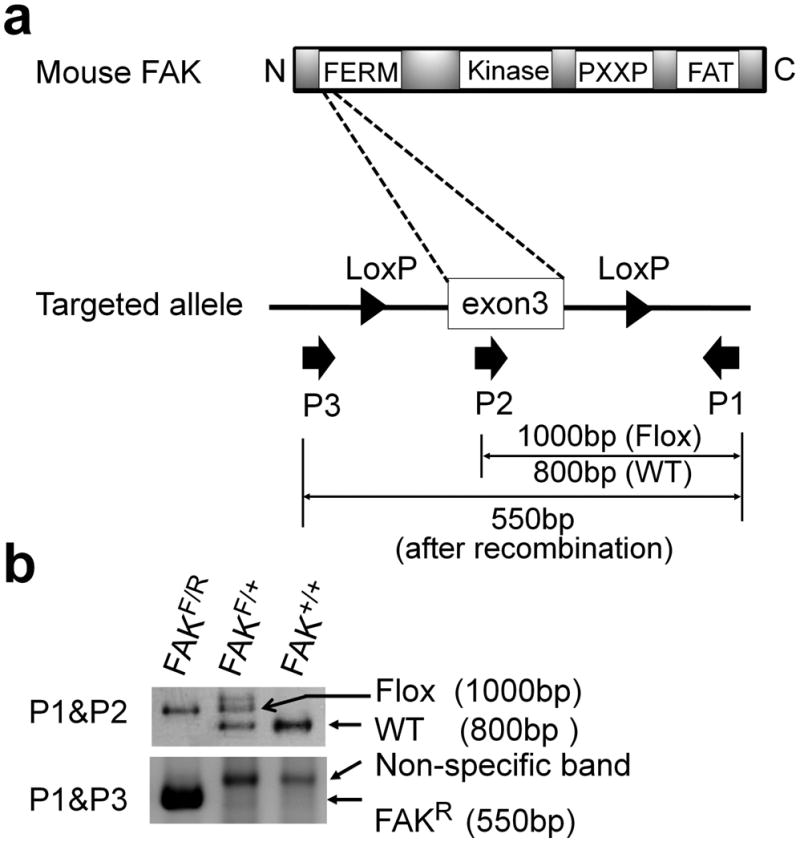
(a) Schematic of mouse Fak gene and targeted allele. Fak gene is composed of FERM, Kinase, PXXP, and FAT domains from N terminal to C terminal. Exon3 locating at FERM domain was flanked by LoxP sites. Primers used to identify the wild type (WT), floxed (Flox) and rearranged (FAKR) alleles are shown as solid arrows: primer pair P1/P2 amplifies a 800-bp WT and 1000-bp Flox bands; primer pair P1/P3 amplifies a 550-bp rearranged floxed Fak allele (FAKR). (b) Genomic DNA was extracted from mouse tail and analyzed by PCR using P1, P2, and P3 primers to distinguish different types of alleles with or without the rearrangement of Fak floxed allele.
Figure 2. Unexpected Fak allele recombination in the absence of Dermo1-Cre transgene.
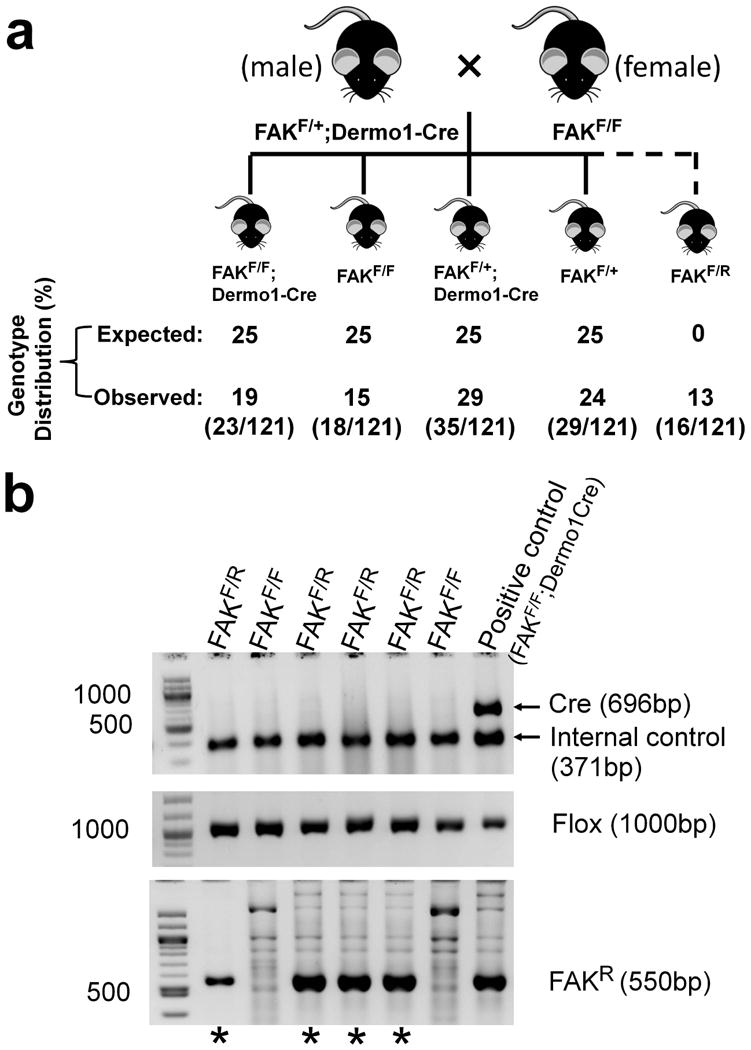
(a) Schematic showing breeding strategies and Fak gene deletion using male mice as paternal Dermo1-Cre carrier to generate Fak conditional knockout mice. Male mice heterozygous for both the Fak floxed allele and the Dermo1-Cre transgene were mated with female mice homozygous for Fak floxed allele. Offspring exhibiting Fak deletion without Dermo1-Cre transgene was observed (indicated by broken line). The table below the scheme shows the expected as well as the observed genotype distribution. The numbers of animals per total number of animals (n=121) is shown in parentheses. (b) Representative PCR reaction showing genotyping result of the mice whose genotype showed the presence of Fak floxed allele and absence of both wild type allele and Dermo1-Cre transgene. Primer pair Cre 1/Cre 2 was used to amplify Dermo1-Cre transgene as a 696-bp band and Primer pair Alk2-5/Alk2-3 was used to amplify Alk2 gene as an internal DNA control (upper panel). Primer pair P1/P2 was used to amplify the floxed Fak allele (Flox) as 1000-bp band (middle panel). Primer pair P1/P3 was used to amplify a 550-bp rearranged floxed Fak allele (FAKR) (lower panel). Asterisks at the bottom of the gel indicate progeny with unexpected rearrangement of Fak floxed allele (FAKR).
Table I. Distribution of FAKF/F and FAKF/R mice in different litters from three breeding pairs between male FAKF/+;Dermo1-Cre and female FAKF/F mice.
| Breeding Male | Litter Number | Litter Size | Number of FAKF/F mice | Number of FAKF/R mice |
|---|---|---|---|---|
| Male #1 | #1 | 13 | 4 | 0 |
| Male #1 | #2 | 7 | 2 | 0 |
| Male #1 | #3 | 9 | 2 | 0 |
| Male #1 | #4 | 8 | 1 | 0 |
| Male #1 | #5 | 9 | 3 | 0 |
| Male #1 | #6 | 12 | 3 | 0 |
| Male #2 | #7 | 5 | 1 | 0 |
| Male #3 | #8 | 3 | 1 | 0 |
| Male #3 | #9 | 7 | 1 | 0 |
| Male #1 | #10 | 2 | 0 | 1 |
| Male #2 | #11 | 9 | 0 | 3 |
| Male #2 | #12 | 8 | 0 | 2 |
| Male #2 | #13 | 7 | 0 | 2 |
| Male #2 | #14 | 8 | 0 | 3 |
| Male #2 | #15 | 8 | 0 | 3 |
| Male #2 | #16 | 6 | 0 | 2 |
The recombination of floxed allele in the absence of Cre transgene has been reported in using different promoters to drive Cre (Cochrane et al., 2007; Hayashi et al., 2003; Lallemand et al., 1998; Ramirez et al., 2004; Sakai and Miyazaki, 1997; Vincent and Robertson, 2003; Zhang et al., 2013). To determine whether this unexpected Dermo1-Cre transgene independent DNA recombination occurred at specific tissues or a more global manner, e.g. germline level, female FAKF/R mice were mated with male WT mice (Figure 3a). About 50% (22/45) of the offspring have FAKF/+ genotype, and the other 50% (23/45) of the offspring have FAKR/+ genotype (Fig 3). Thus, the recombined Fak allele can be transmitted to the offspring in Mendelian ratio, indicating a global floxed Fak allele recombination in FAKF/R mice. These results suggest that the unexpected recombination occurs either during spermatogenesis or soon after fertilization.
Figure 3. Schematic showing breeding strategies to determine whether the occurrence of Fak floxed allele recombination is at germline level.
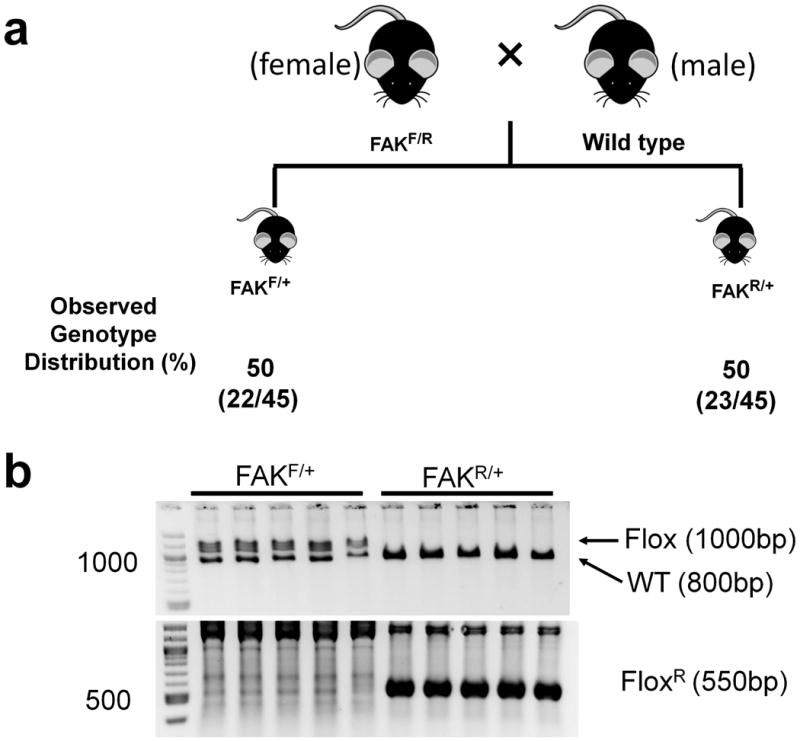
(a) Female FAKF/R mice were mated with male wild type mice. The table below the scheme shows the observed genotype distribution. The numbers of animals per total number of animals (n=45) is shown in parentheses. (b) Representative PCR reaction showing genotyping result of the offspring of female FAKF/R mice and male wild type mice. Primer pair P1/P2 was used to amplify WT and floxed Fak allele (Flox) as 800-bp and 1000-bp bands, respectively. Primer pair P1/P3 was used to amplify a 550-bp rearranged floxed Fak allele (FAKR).
To determine whether Cre mRNA and/or protein carried by sperms that are genotypically negative for Dermo1-Cre can cause the recombination at or after zygote stage, male Dermo1-Cre/+ mice were mated with female FAKF/F mice (Figure 4). In this experimental model, all offspring showed expected genotypes and there was no unexpected recombination in Cre-negative mice (Fig 4). Thus, we concluded that the unexpected Fak allele recombination only occurred before zygote stage. To further support this conclusion, there was no offspring with FAK+/R genotype shown in Fig 2. Because FAK+/R genotype can only occur when the maternally contributed floxed Fak allele is recombined, the absence of FAK+/R genotype suggests that the recombination did not occur at or after the fertilization, otherwise the maternally contributed floxed Fak allele has the equal opportunity as paternally contributed floxed Fak allele to be recombined in zygotes and consequently this should lead to the occurrence of FAK+/R genotype.
Figure 4. Schematic showing breeding strategies to determine whether the occurrence of Fak floxed allele recombination is at or after zygote stage.
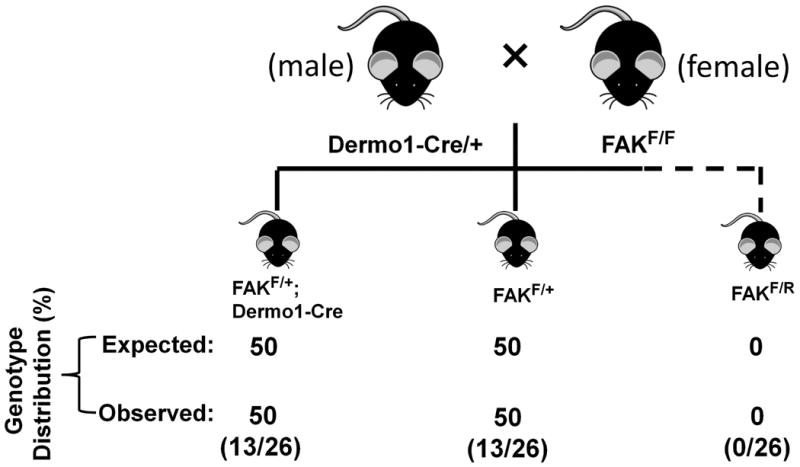
Male mice heterozygous for Dermo1-Cre transgene were mated with female mice homozygous for Fak floxed allele. The table below the scheme shows the expected as well as the observed genotype distribution. The numbers of animals per total number of animals (n=26) is shown in parentheses.
We identified the unexpected Fak floxed allele recombination occurs in male germline cells, next we investigated whether this unfavorable recombination can be avoided by using female mice as parental Cre carriers. Female FAKF/+;Dermo1-Cre/+ mice were mated with male FAKF/F mice and we evaluated F1 progeny for recombination of the inherited Flox allele in Cre-negative mice (Figure 5). Our data demonstrated that there was no recombination of the Fak allele occurred in Cre-negative progeny in this mating scheme (Fig 5).
Figure 5. Schematic showing breeding strategies and Fak gene deletion using female mice as Dermo1-Cre carrier.
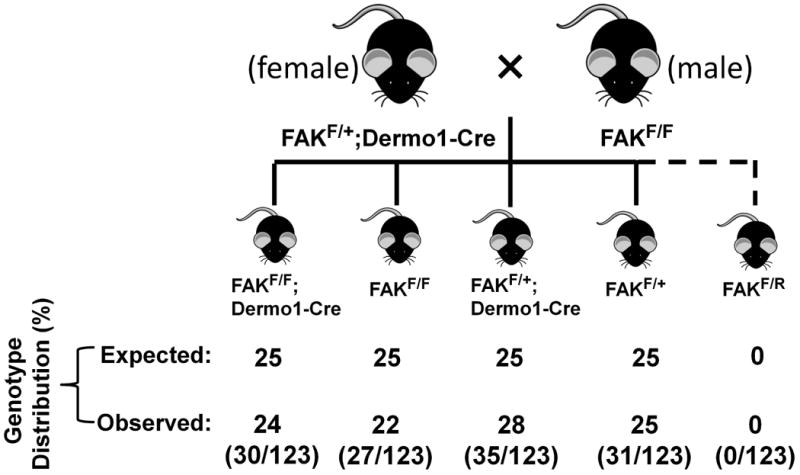
Female mice heterozygous for both the Fak floxed allele and the Dermo1-Cre transgene were mated with male mice homozygous for Fak floxed allele. The table below the scheme shows the expected as well as the observed genotype distribution. The numbers of animals per total number of animals (n=123) is shown in parentheses. The data were obtained from 9 different female breeders.
To determine whether Dermo1-Cre causes universal male germline recombination of floxed alleles, we examined the possible ectopic Dermo1-Cre recombination event in mouse with floxed allele of Fip200 (FAK-family Interacting Protein of 200 kDa) gene, whose product was identified as a FAK interacting protein (Ueda et al., 2000). In this experimental model, male FIP200F/+;Dermo1-Cre/+ mice were bred with female FIP200F/F mice using a similar breeding scheme shown in Fig. 2, and we evaluated F1 progeny for recombination of the inherited floxed allele in Cre-negative mice. In contrast to Fak floxed alleles, no recombination of the Fip200 allele occurred in Cre-negative progeny (0/48). This result indicates that the germline recombination of floxed alleles by Dermo1-Cre is not universal and only some genes may be susceptible to the amount of Cre recombinase produced. Dermo1-Cre mice were generated by inserting the Cre transgene within the first exon of Dermo1 gene (Yu et al., 2003). The germline Fak floxed allele recombination identified in the offspring of the breeding using male mice to carry Dermo1-Cre suggests that Dermo1 gene may be expressed in male germline cells. The absence of floxed Fip200 allele recombination suggests that the Fip200 floxed allele is not susceptible to the amount of Cre protein produced by Dermo1-Cre transgene in germline cells. Fak gene is located at mouse chromosome 15 (Fiedorek and Kay, 1995) and it is expressed during spermatogenesis (Gungor-Ordueri et al., 2014). This may indicate a more “open” chromatin structure where Fak gene locates, which may make Fak floxed alleles more susceptible to Cre recombinase at this developmental stage.
It is often reported that the frequency of Cre transgene-independent recombination occurs more often when female mice are used to carry Cre transgene (Cochrane et al., 2007; Zhang et al., 2013) or only “maternal inheritance” but not “paternal inheritance” occurs (Hayashi et al., 2003). Our data showed that female mice may be the preferred maternal Cre carrier when Dermo1-Cre is used. However, it takes more effort to maintain mating units using female as Cre carrier especially for embryonic studies. If germline recombination of floxed allele does occur when male mice are used as paternal Dermo1-Cre carrier, PCR analysis with tail tip DNA in progenies without Dermo1-Cre transgene can be employed to effectively identify the mice having germline recombination. However, since germline deletion may also occur in the presence of Dermo1-Cre, FAKF/F;Dermo1-Cre/+ mice could be indeed FAKF/R;Dermo1-Cre/+ mice but it is not possible to distinguish them by PCR analysis using the tail-tip DNA because rearranged Fak allele is expected in the tail tissue of both genotypes. Intriguingly, our data showed that the unexpected germline recombination of Fak floxed allele was an “all or none” event evidenced by the exclusive presence of either FAKF/F or FAKF/R genotype when male mice were used as paternal Dermo1-Cre carrier. This suggests that the Fak floxed allele recombination either happened or not happened to all the sperms used to produce one particular litter from the FAKF/+;Dermo1-Cre/+ mice.
Due to the limitation and scope of current report, neither the exact timing of male germline Fak allele recombination nor the mechanism of the “all or none” phenomenon is known. However, our data have two important implications. First, our data calls for the necessity to examine potential germline recombination when Dermo1-Cre is carried by male mice to target any other genes of interest. Second, we demonstrated that using female mice as Dermo1-Cre carriers can avoid the germline recombination of floxed alleles.
Materials and Methods
Animals
The floxed Fak (FAKF/F) mice and floxed Fip200 (FIP200F/F) mice were generated by us previously (Gan et al., 2006; Shen et al., 2005). Generation of transgenic mice was described previously (Yu et al., 2003) and they were obtained from Jackson laboratory (Bar Harbor, ME, strain 008712). All mice were backcrossed for at least 8 generations onto a C57BL/6NCrl background. Mice were housed under pathogen-free conditions at 22 ± 2 °C on a 12:12-h light/dark cycle, fed with 5001 or 5008 (for breeding pairs) rodent diet (LabDiet). All animal handling protocols were approved by IACUC at the University of Michigan.
Genotype Analysis by PCR
Genomic DNA from tail tip was prepared as described previously (Liu et al., 2013; Liu et al., 2010). DNA extracts were amplified by PCR using primer pairs to detect the Cre transgenes, wild type, floxed, and Cre-recombined Fak or Fip200 alleles as we previously described (Gan et al., 2006; Shen et al., 2005). PCR products were electrophoresed on agarose gels, stained with ethidium bromide, and imaged using UV light. Cre transgenes were amplified and identified as a 696-bp band using the Cre 1 (5′-GAGTGATGAGGTTCGCAAGA-3′) and Cre 2 (5′ CTACACCAGAGACGGAAATC 3′). As an internal DNA control, primers Alk2-5 (5′-ATGCTAGACCTGGGCAGCCATA-3′) and Alk2-3 (5′-CATGCTAGCAGCTCGGAGAAAC-3′) were applied simultaneously with Cre primers, generating a 371-bp amplicon. The reaction cycles for Cre and internal control are: 94°C, 1 min; 67 °C, 1 min; 72 °C, 1min; 25 cycles. Fak alleles were identified with primer set: P1 (5′-GCTGATGTCCCAAGCTATTCC-3′) and P2 (5′-TGGCCTGCTATGGATTTCGC-3′) using reaction cycles: 94°C, 1 min; 67 °C, 1 min; 72 °C, 2min; 32 cycles. The wild type and floxed Fak alleles were detected as 800-bp and 1000-bp products, respectively. To detect Cre-mediated recombination of the floxed Fak allele, primes P1 and P3 (5′-AGGGCTGGTCTGCGCTGACAGG-3′) were used under the same conditions. The rearranged Fak allele was detected as a 550-bp product. Fip200 alleles were identified with primer set: FP2 (5′-CAAAGAACAACGAGTGGCAGTAG -3′) and FP3 (5′-CATCAGATACACTAGAGCTGG-3′) using reaction cycles: 3 cycles at 94°C for 3 min, 60°C for 1 min, and 72°C for 2 min, followed by 33 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min, and 1 cycle at 94°C for 1 min, 60°C for 1 min, and 72°C for 10 min. The wild type and floxed Fip200 alleles were detected as 262-bp and 225-bp products, respectively. To detect Cre-mediated recombination of the floxed Fip200 allele, primes FP1 (5′-GGAACCACGCTGACATTTGACACTG-3′) and FP3 were used under the same conditions. The recombined Fip200 allele was detected as an 800-bp product.
Acknowledgments
This work was supported by National Institutes of Health grant (R01AR062030 to FL, R01DE020843 to YM, and R01CA163493, R01CA211066 and R01NS094144 to JLG), Scientific Research Fund of Sichuan Provincial Education Department (13ZB0337 to YH), Chengdu University School Fund (2012XJZ08 to YH). The authors declare that they have no conflict of interest.
References
- Cochrane RL, Clark SH, Harris A, Kream BE. Rearrangement of a conditional allele regardless of inheritance of a Cre recombinase transgene. Genesis. 2007;45:17–20. doi: 10.1002/dvg.20259. [DOI] [PubMed] [Google Scholar]
- Cornett B, Snowball J, Varisco BM, Lang R, Whitsett J, Sinner D. Wntless is required for peripheral lung differentiation and pulmonary vascular development. Dev Biol. 2013;379:38–52. doi: 10.1016/j.ydbio.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F, Yang X. Genetic mouse models for bone studies--strengths and limitations. Bone. 2011;49:1242–1254. doi: 10.1016/j.bone.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorek FT, Jr, Kay ES. Mapping of the focal adhesion kinase (Fadk) gene to mouse chromosome 15 and human chromosome 8. Mamm Genome. 1995;6:123–126. doi: 10.1007/BF00303256. [DOI] [PubMed] [Google Scholar]
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske MJ, Zhang X, Patel KK, Ornitz DM, Stappenbeck TS. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JL, Shalloway D. Regulation of focal adhesion-associated protein tyrosine kinase by both cellular adhesion and oncogenic transformation. Nature. 1992;358:690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- Gungor-Ordueri NE, Mruk DD, Wan HT, Wong EW, Celik-Ozenci C, Lie PP, Cheng CY. New insights into FAK function and regulation during spermatogenesis. Histol Histopathol. 2014;29:977–989. doi: 10.14670/hh-29.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development. 2008;135:3161–3171. doi: 10.1242/dev.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- Lin C, Yin Y, Long F, Ma L. Tissue-specific requirements of beta-catenin in external genitalia development. Development. 2008;135:2815–2825. doi: 10.1242/dev.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Fang F, Yuan H, Yang D, Chen Y, Williams L, Goldstein SA, Krebsbach PH, Guan JL. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J Bone Miner Res. 2013;28:2414–2430. doi: 10.1002/jbmr.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, Guan JL. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy T, Wei H, Shen TL, Peng X, Liang CC, Gan B, Guan JL. Mammary epithelial-specific deletion of the focal adhesion kinase gene leads to severe lobulo-alveolar hypoplasia and secretory immaturity of the murine mammary gland. J Biol Chem. 2007;282:31766–31776. doi: 10.1074/jbc.M705403200. [DOI] [PubMed] [Google Scholar]
- Peng X, Wu X, Druso JE, Wei H, Park AY, Kraus MS, Alcaraz A, Chen J, Chien S, Cerione RA, Guan JL. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proc Natl Acad Sci U S A. 2008;105:6638–6643. doi: 10.1073/pnas.0802319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, Tusell L, Genesca A, Whitaker DA, Melton DW, Jorcano JL. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Shen TL, Park AY, Alcaraz A, Peng X, Jang I, Koni P, Flavell RA, Gu H, Guan JL. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol. 2005;169:941–952. doi: 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Yuan H, Wang L, Wei X, Williams L, Krebsbach PH, Guan JL, Liu F. FAK Promotes Osteoblast Progenitor Cell Proliferation and Differentiation By Enhancing Wnt Signaling. J Bone Miner Res. 2016 doi: 10.1002/jbmr.2908. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000;149:423–430. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Robertson EJ. Highly efficient transgene-independent recombination directed by a maternally derived SOX2CRE transgene. Genesis. 2003;37:54–56. doi: 10.1002/gene.10226. [DOI] [PubMed] [Google Scholar]
- Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dublin P, Griemsmann S, Klein A, Brehm R, Bedner P, Fleischmann BK, Steinhauser C, Theis M. Germ-line recombination activity of the widely used hGFAP-Cre and nestin-Cre transgenes. PLoS One. 2013;8:e82818. doi: 10.1371/journal.pone.0082818. [DOI] [PMC free article] [PubMed] [Google Scholar]


