Abstract
Ewing Sarcoma (ES) is a highly aggressive bone tumor with peak incidence in the adolescent population. It has a high propensity to metastasize, which is associated with dismal survival rates of approximately 25%. To further understand mechanisms of metastasis we investigated microRNA regulatory networks in ES. Our studies focused on miR-130b due to our analysis that enhanced expression of this microRNA has clinical relevance in multiple sarcomas, including ES. Our studies provide insights into a novel positive feedback network involving the direct regulation of miR-130b and activation of downstream signaling events contributing towards sarcoma metastasis. Specifically, we demonstrated miR-130b induces proliferation, invasion, and migration in vitro and increased metastatic potential in vivo. Using microarray analysis of ES cells with differential miR-130b expression we identified alterations in downstream signaling cascades including activation of the CDC42 pathway. We identified Arhgap1, which is a negative regulator of CDC42, as a novel, direct target of miR-130b. In turn, downstream activation of PAK1 activated the JNK and AP-1 cascades and downstream transcriptional targets including IL-8, MMP1 and CCND1. Furthermore, chromatin immunoprecipitation of endogenous AP-1 in ES cells demonstrated direct binding to an upstream consensus binding site within the miR-130b promoter. Finally, small molecule inhibition of PAK1 blocked miR-130b activation of JNK and downstream AP-1 target genes, including primary miR-130b transcripts, and mir-130b oncogenic properties, thus identifying PAK1 as a novel therapeutic target for ES. Taken together, our findings identify and characterize a novel, targetable miR-130b regulatory network that promotes ES metastasis.
Keywords: metastasis, microRNA, PAK1, Ewing sarcoma, CDC42
Introduction
Ewing sarcoma (ES) is the second most common bone tumor in children and adolescents. Tumors often arise in the central axis and are identified by the presence of the oncogenic EWS-FLI1 fusion transcript 1. Treatment consists of surgery and/or radiation therapy along with high dose cytotoxic systemic chemotherapy that has improved survival rates for patients with localized disease to approximately 70–75%. However, ES has a high propensity for metastasis to the lung, liver and other bones, which are associated with extremely poor survival rates of approximately 25% that have remained stagnant over the past 30 years 2. Therefore, understanding the biology of metastatic disease is of utmost clinical importance due to high mortality and morbidity Ewing sarcoma patients.
MicroRNAs (miRNAs) are 18–22 nucleotide non-coding RNAs that have the ability to regulate protein translation and degradation through complementary binding to target mRNAs 3. Recently, miRNAs have emerged as key regulators of numerous normal cellular processes such as proliferation, growth, differentiation, and apoptosis. Although miRNAs are critical for normal cellular homeostasis, a vast amount of miRNA research demonstrates that miRNAs are often aberrantly expressed in various neoplasms and can promote aggressive, metastatic disease 4–6. Accumulating evidence suggests miR-130b is overexpressed in a number of malignancies and therefore acts as an oncomiR in these tumors 7–9. It has been shown that overexpression of miR-130b increases stemness and chemoresistance in glioblastoma and increases cell migration and invasion in glioma 10,11. Furthermore, miR-130b was identified as oncogenic miR and is part of pan-cancer oncogenic superfamily that targets key pathways often deregulated in cancer including p53 and TGFβ12. The majority of miR-130b studies have been in epithelial based tumors; but several sarcomas, including leiomyosarcoma, osteosarcoma, and ES have shown enhanced expression of miR-130b 13–15. Overexpression of miR-130b correlated with poor event free survival and overall survival in ES; however, its function and role in dictating sarcoma biology and metastasis is not understood 14. In addition, the transcriptional regulation and downstream gene regulatory networks of miR-130b have not been well elucidated in sarcomas.
There are relatively few studies dissecting the molecular alterations of metastasis in Ewing sarcoma. Prior studies have highlighted that alterations in the cytoskeleton and RhoGTPases may be important for the promotion of metastasis in ES16,17. The RhoGTPases encompass the Rho, Rac, and Cdc42 subfamilies which act as molecular switches activated in response to cellular stimuli such as, growth factors18. Upon activation, RhoGTPases signal through several effector proteins including, p21 activated kinases (PAKs) to mediate changes in cytoskeleton architecture, cell cycle, and gene transcription18. PAKs are serine/threonine kinases that function as downstream effectors for several oncogenic signaling networks and alterations in PAKs have been attributed to carcinogenesis and metastasis in several tumors 19,20. Upon activation PAKs can phosphorylate various downstream effectors including c-JUN N-terminal kinase (JNK), with the subsequent phosphorylation and nuclear localization of key oncogenic transcription factors such as Activator Protein 1 (AP-1), which is composed of the members of the JUN, FOS, MAF, and ATF protein families.21,22. Active AP-1 in the nucleus binds preferentially, to 12-O-tetradecanoylphorbol-13-acetate (TPA) responsive elements in canonical target gene promoters such as CCND1, MMP3, and IL-8 23–26. The role of PAK1 has been studied in many neoplasms, however; the function and contribution of PAK1 in ES tumorigenesis and metastasis is yet to be explored
Our studies investigated the oncogenic role of miR-130b in Ewing sarcoma, where we identified a novel miR-130b-AP-1 signaling axis that promotes transcription and activation of miR-130b and subsequent contributions to the invasive and metastatic properties of Ewing sarcoma.
Materials and Methods
Cell culture and transfection
Human ES cell lines TC71, MHH-ES1, and A4573 were grown in RPMI 1640 Medium supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Fisher Scientific, Austria), 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 at 37°C. TC71 cells were derived from the humerus of male Ewing sarcoma patient with progressive disease. MHH-ES-1 cells were derived from the ascites fluid of male Ewing sarcoma patient with a primary tumor in the left pelvis. All cell lines were authenticated at the MD Anderson Characterized Cell Line Core Facility prior to studies being initiated. Cell lines were validated by STR DNA fingerprinting using the Promega 16 High Sensitivity STR Kit (Catalog # DC2100). The STR profiles were compared to online search databases (DSMZ/ATCC/JCRB/RIKEN) of 2455 known profiles; along with the MD Anderson Characterized Cell Line Core (CCLC) database of 2556 know profiles. The STR profiles matched known DNA fingerprints or were unique. TC71 and MHH-ES1, A4573 cells were kindly provided by Dr. David Loeb (Johns Hopkins Children’s Center, Baltimore, Maryland, 2008).
RNA oligoribonucleotides were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). A final concentration of 30 nM RNA miRNA mimics, 100 nM miRNA inhibitors, or 10nM siRNA pool was used for each transfection, unless otherwise indicated. Co-transfection of the RNA duplex and plasmid DNA were conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). All transfections were performed in accordance with the manufacturer’s protocol.
RNA oligoribonucleotides, plasmids, and inhibitors
All RNA oligoribonucleotides and negative controls were purchased from Ambion (Austin, TX). mirVana miRNA mimics and inhibitors corresponded to the mature miR-130b. On-Target Plus siRNA pool to Arhgap1 was purchased from Dharmacon (Lafayette, CO). FRAX597 and IPA3 were purchased from Tocris (Minneapolis, MN). JNK-IN-8 was purchased from Sigma-Aldrich (St. Louis, MO).
The miR-130b expression plasmid, pEZX-MR03, was purchased from GeneCopoeia (GeneCopoeia Inc., Rockville, MD, USA). For generation of stable cell lines, cells were transfected with miR-130b using Lipofectamine 2000. 48 hours post transfection media was replaced with selection media containing puromycin to generate TC71 control and TC71/130b cells. Cells were selected with puromycin and sorted for GFP expression. The miR-130b knockdown plasmid, pEZX-AMO3, was purchased from GeneCopoeia. MHH-ES-1 cells were transfected with anti-miR-130b plasmid to generate MHH control and MHH/anti-miR-130b cells. Cells were selected with hygromycin and sorted for mCherry expression. Cells were selected with 2ug/ml Puromycin and 200ug/ml Hygromycin, 72 hours post transfection. Bioinformatics analysis predicted one conservative miR-130b binding site in ARHGAP1 3′-UTR. The ARHGAP1 3′UTR construct was a generous gift from the Kurie Lab (M.D. Anderson, Houston, TX) The mutant 3′-UTR, which contained the mutated sequence in the complementary site for the seed region of miR-130b, was generated using the Stratagene QuickChangeII Kit (Agilent Technologies, Santa Clara, CA).
Luciferase assay
For miR-130b targeted 3′-UTR assays, TC71 cells in a 6-well plate were co-transfected with 250 ng of the indicated hRL(Renilla luciferase)-ARHGAP1 wild-type or mutant reporter plasmids and 30 nM of either miR-130b or scrambled control mimics. After 48 hours, Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Renilla luciferase activity of each sample was normalized by Firefly luciferase activity.
CDC42 Activity Assay
CDC42 activation was measured with a CDC42 G-LISA Activation Assay (Colorimetric format) purchased from Cytoskeleton Inc. (Denver, CO). Briefly, TC71 cells were transfected with 30nM miR-130b mimics or scrambled control. 48 hours post transfection cells were harvested and processed according to the manufacturer’s protocol. Activity of CDC42 was measured as colorimetric change in absorbance at 490nM.
Microarray Profiling
Briefly, RNA samples were isolated from TC71 cells transiently expressing miR-130b mimic or a scrambled control for target preparation. High quality total RNA was amplified and purified using the Illumina TotalPrep RNA Amplification Kit (Ambion, Cat. no. IL1791) following the manufacturer’s instructions. An aliquot of 750 ng of amplified products was loaded on Illumina HumanHT-12 v4 Expression BeadChip arrays and hybridized at 58 °C in an Illumina Hybridization Oven (Illumina, San Diego, CA, USA; Cat. no. BD-103-0204) for 17 h, washed and incubated with straptavidin-Cy3 to detect biotin-labeled cRNA on the arrays. Arrays were dried and scanned with the BeadArray Reader (Illumina). Data were analyzed using the Genome Studio software (Illumina).
The microarray data were generated by the University of Texas Medical School Genomics Core Facility. Pathway Analysis was performed using MetaCore™ Pathway Analysis v 6.15 (Thomson Reuters). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus27 and are accessible through GEO Series accession number GSE83548 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83548). Venn diagrams were generated using the program BioVenn28 (http://www.cmbi.ru.nl/cdd/biovenn).
Analysis of gene expression
Total RNA was harvested was harvested using the miRNeasy Mini Kit (Qiagen, Germantown, MD) according to manufacturer protocol. For miRNA cDNA synthesis, total RNA (1 μg) were used for cDNA synthesis using the High-Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) and a miRNA specific primer from TaqMan MicroRNA Assay kit (Applied Biosystems). Otherwise, 1μg total RNA was used synthesize cDNA using the qScript cDNA SuperMix (QuantaBio, Beverly, MA, USA). Quantitative RT-PCR (qPCR) was used to evaluate gene expression. AP-1 (c-JUN and c-FOS) and AP-1 target genes, including MMP1, MMP3, CCND1 and IL-8, were quantified by Fast SYBR® Green Master Mix and normalized to GAPDH expression. Primary miR-130b (pri-miR-130b) and Mature microRNA expression was detected using a TaqMan MicroRNA Assays kit and TaqMan® Fast Universal PCR Master Mix (2X), no AmpErase® UNG. All reactions were performed on a StepOnePlus™ System. All reactions were run in triplicate. The mature miR-130b level was normalized to the level of RNU44 and pri-miR0130b was normalized to GAPDH to yield a 2−ΔΔCt value. All genes were normalized to GAPDH to yield a 2−ΔΔCt value. MMP1 (Cat. No. HQP055159), MMP3 (Cat. No, HQP011270), AND IL-8 (Cat. No. HQP009678) primers were purchased from GeneCopoeia (Rockville, MD). Primary and mature miR-130b primers were purchased from Applied Biosystems (Cat. No. 4427012 and A25576). Primer sequences are included in Supplementary table S3.
Ewing Sarcoma Patient Samples
The research, H-32129, was approved by the Institutional Review Boards for Baylor College of Medicine and Affiliated Institutions (IRB00002649). The BCM IRB has approved a waiver of consent/HIPAA authorization and has determined that all requirements are met by this protocol in order to grant the waiver. Patient clinical annotations are outlined in Supplemental Table S2.
Gene Ontology Analysis
Differentially expressed genes that were differences of 1.5 fold change and P-values, as evaluated by two tailed t-test, P<0.05 were subjected to gene ontology analysis using Thomson Reuters Metacore™ program.
RPPA
Reverse phase protein array assays were carried out as described previously with minor modifications29. Additional details are described in supplemental methods.
Western blot analysis
Briefly, total cell lysates were isolated in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 lysis buffer supplemented with protease inhibitors (Complete mini, Roche, Indianapolis, IN, USA) and phosphatase inhibitors (Sigma, P0044 and P5726). Nuclear lysates were isolated utilizing the NE-PER nuclear and cytoplasmic extraction reagents (ThermoScientific, Grand Island, NY, USA, 78833) following the manufacturer’s instructions. Lysates were boiled in NuPAGE LDS sample buffer (Invitrogen), separated on NuPAGE Novex 4%–12% Bis-Tris Gels (Invitrogen), and transferred to polyvinylidene difluoride (PVDF) membranes. Blots were incubated sequentially overnight with primary and secondary antibodies. Immunoreactive signals were developed with ECL kit (Thermo Scientific, Waltham, MA, USA). The antibodies used for immunoblotting were as follows: rabbit monoclonal antibodies (Ab) Phospho-PAK1 (Thr423)/PAK2 (Thr402) Antibody (2601), PAK1 Antibody (2602), CDC42 (2466), HDAC1 (2062), Phospho-c-Jun (Ser63) (D47G9) XP® (3270S), c-JUN (60A8), JNK (56G8), phospho-JNK (Thr183/Tyr185) (81E11) were purchased from Cell Signaling Technology (Beverly, MA, USA), anti-ARHGAP1 (Thermo Scientific, Grand Island, NY, USA, PA5-27426), GAPDH (AB2302), Anti-β-Tubulin, clone AA2, (05-661) (EMD Millipore, Billerica, MA, USA).
Chromatin Immunoprecipitation
ChIP assay was performed using EZ-Magna ChIP™ A Chromatin Immunoprecipitation Kit (Cat# 17-408, Millipore) according to the manufacturer’s instructions. Briefly, MHH-ES1 cells were then crosslinked using formaldehyde to a final concentration of 1%, and genomic DNA sonicated using an EpiShear™ Probe Sonicator to generate 500bp fragments. Cells were sonicated at 35% amplitude for 10s pulses and 30s rest for a total of 10 cycles. Soluble chromatin was then immunoprecipitated with a c-JUN antibody and amplified using specific PCR primers for the AP-1 binding site.
Cell Proliferation Assay
Growth assays were assessed by plating 1000 cells per well in triplicate per day for 4 days in a 96-well plate unless otherwise indicated. Cell proliferation was detected through the addition of the colorimetric reagent Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan), according to the manufacturer’s instructions (Cell counting kit-8 (CCK-8) assay kit; Dojindo Laboratories). Each value represents the triplicate average for that day.
Migration and invasion assay
Migration/Invasion assays were analyzed in a 24-well Boyden chamber with an 8-μm pore size polycarbonate membrane (Corning Glass Works, Corning, NY, USA). For invasion assays, to mimic the extracellular matrix, chambers were coated with 30μg/ml collagen and incubated at 37°C overnight. Cells were suspended in 100 μl serum-free RPMI and added to the upper chamber, whereas the lower compartment was filled with 300 μl RPMI containing 10% FBS. After incubation at 37°C for the 24 hours, cells were fixed and stained with 0.1% crystal violet. The cells on the top layer were removed with a cotton swab. All the migrated cells on the lower membrane were quantified using ImageJ. Invasion assay was done by the same procedure, except that the membrane was coated with collagen (3432-005-01, R&D Systems, MN, USA) before adding cells to the upper chamber.
In vivo xenograft assay
1 × 106 TC71 cells stably overexpressing miR-130b or scrambled control were injected into the tail vein of 10 (5 per group) Rag2−/− mice. Six weeks post injection all animals were sacrificed and tissues were isolated. For the MHH-ES1 studies, a total of 10 (5 per group) NOD-SCID-gamma (NSG) mice were injected with 1 × 106 cells stably expressing anti-miR-130b or a scrambled control. All mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Six weeks post injection all mice were sacrificed and tissues were isolated. All animal experiments were conducted according to institutional animal care and use committee protocols after approval was obtained from the BCM Institutional Review Board (BCM Animal Protocol AN-5225).
Immunohistochemistry
At the time of sacrifice, lung tissue was inflated with formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. Slides were scanned in the Department of Veterinary Medicine and Surgery at the MD Anderson Cancer Center. Stained tumor sections were scanned, and the number of lung nodules per specimen was quantified utilizing the Aperio ImageScope Viewer (Leica Biosystems, Buffalo Grove, IL, USA).
Clinical significance assessment of miRNA expression in Sarcoma patients
We evaluated first the prognostic significance of miRNA expression in the TCGA Sarcoma patient cohort (https://tcga-data.nci.nih.gov/tcga/) using a methodology previously described. For each miRNA and for each Sarcoma specimen, we computed the z-score for its expression within the TCGA cohort. Specimens were then ranked according to their individual miRNA z-score, and association with survival was evaluated by comparing the bottom half of the ranked specimens with the rest of the specimens (top half) using the log-rank test. Survival significance was evaluated by using the package survival in the R statistical system; significance was achieved for P-value<0.05. Next, we evaluated the association of miRNA expression with categorical clinical variables compiled for the TCGA Sarcoma patient cohort. We scored the specimens as described above, then collected the miRNA expression scores for each value of the categorical clinical variable. We evaluated association using the ANOVA method, as implemented in the R statistical system; significance was achieved for P-value<0.05.
Statistical Analysis
The significant differences of control and miR-130b mimic or control versus anti-miR-130b under different conditions were determined by Student’s t-test. A P-value <0.05 was considered significant. A two-sided Student’s t-test (paired) was used to calculate the level of statistical significance for metastatic lesions in control/miR-130b and control/anti-miR-130b mice. All tests of data were performed using GraphPad Prism.
Results
miR-130b is overexpressed in Ewing sarcoma and promotes invasion, migration, proliferation in vitro
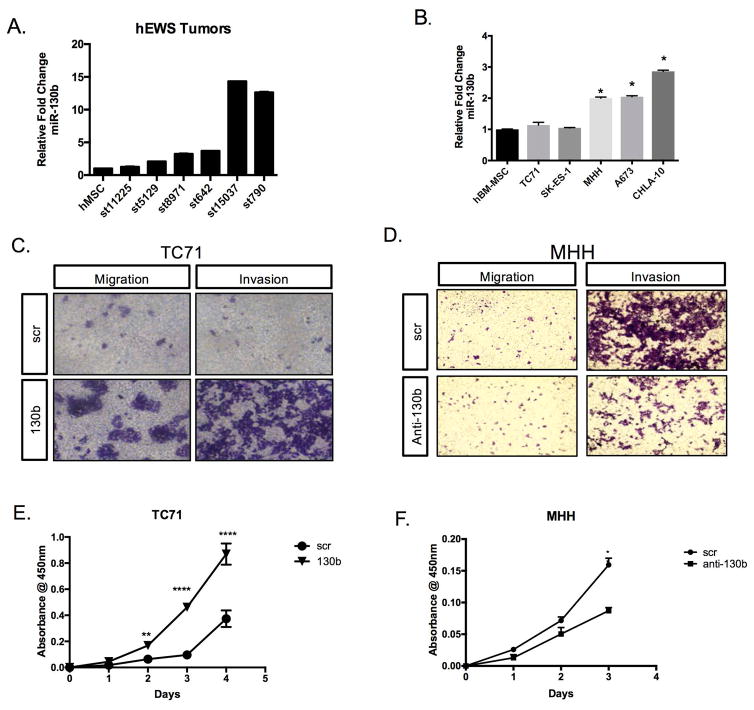
Ewing sarcoma is a rare but aggressive pediatric tumor, therefore access to adequate numbers of patient samples is limited. However, using institutional patient tumor samples, we analyzed the relative abundance of miR-130b and determined relative expression levels in ES compared to normal human mesenchymal stem cells (hMSCs). We noted enhanced expression of miR-130b in the tumor specimens compared to MSCs (Figure 1A). Furthermore, analysis of The Cancer Genome Atlas (TCGA) sarcoma cohort suggests there is clinical significance of high miR-130b in not only pediatric, but also adult sarcomas. Specifically, we analyzed the expression and clinical outcomes of miR-130b in (TCGA) sarcoma cohort and noted high miR-130b expression correlates with poor survival (Supplemental Figure 1), therefore suggesting the importance of this microRNA, and its downstream regulation across the various sarcoma subtypes.
Figure 1. miR-130b is overexpressed in Ewing sarcoma.
(A) Relative expression of miR-130b by qPCR from patient sample obtained from Texas Children’s Cancer Center compared to human mesenchymal stem cells (hMSC). (B) Relative expression of miR-130b by qPCR compared to MSCs. (C&D) Boyden chamber invasion and migration assays for TC71 and MHH-ES1 cells stained with 0.05% crystal violet and imaged using EVOS XL core imaging system. (E&F) CCK-8 colorimetric cell proliferation assay for TC71 and MHH-ES1 cells. Values represent the average of triplicate samples, *P<0.05, **P<0.01, ***P<0.001. scr=TC71 scrambled control ES cells, 130b =TC71 miR-130b mimic ES cells, anti-130b= anti-miR-130b oligo ES cells.
We then examined the expression of miR-130b in ES cell lines relative to MSCs and decided to overexpress miR-130b in TC71 cells and inhibit miR1-30b in MHH cells for functional studies (Figure 1B). To determine the functional significance of miR-130b in Ewing sarcoma, we performed complementary gain and loss of function studies. After determining relative expression of miR-130b in various ES cell lines (Figure 1B), TC71 and A4573 cells were transiently transfected with miR-130b mimics or a negative control and subjected to transwell assay. Overexpression of miR-130b increased the migratory and invasive capacity of TC71 and A4573 cells (Figure 1C, Supplemental Figure 2A, 2B). Furthermore, overexpression of miR-130b increased the proliferation of TC71 cells (Figure 1E). Conversely, loss-of-function studies demonstrated that inhibition of miR-130b levels in the MHH-ES1 cell line decreases tumor cell proliferation and invasive potential (Figure 1D, 1F). Taken together these results suggest miR-130b can contribute to the more aggressive, disseminated properties of Ewing sarcoma.
miR-130b expression alters in vivo metastatic potential for Ewing sarcoma
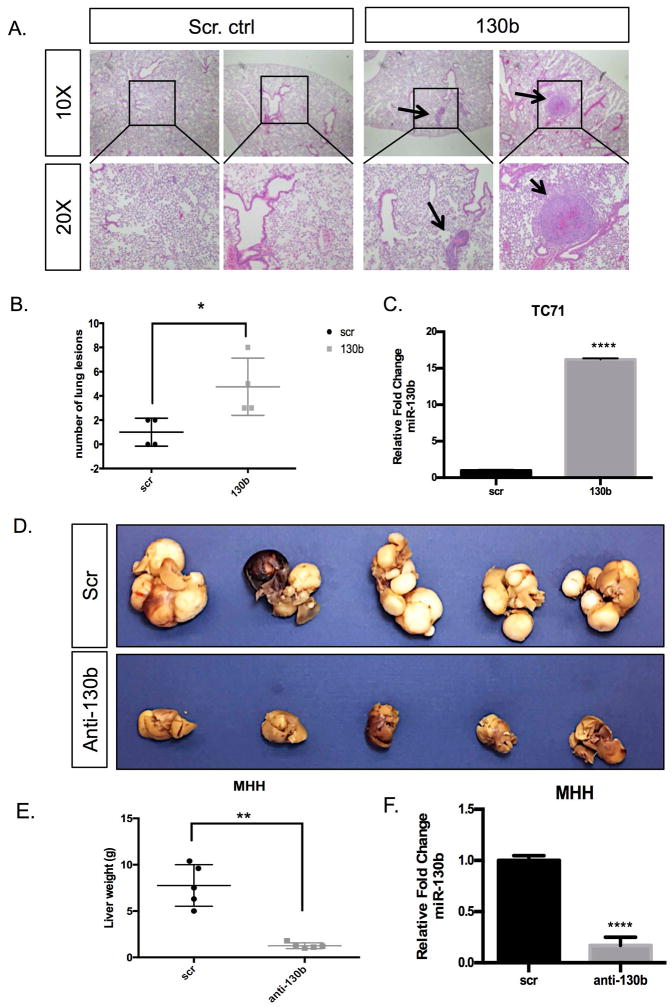
After determining miR-130b contributes to ES in vitro metastatic properties, we investigated the in vivo significance of miR-130b on ES metastatic potential. We performed both gain and loss-of-function studies using TC71 and MHH-ES1 ES cells, respectively, through tail vein injection of immunodeficient mice as described in methods section. Specifically, we injected stable TC71 control and TC71/130b overexpressing cells via tail vein into mice and six weeks post-injection all mice were sacrificed and necropsies performed. TC71/130b injected mice showed significantly increased number of metastatic lung nodules compared to the control mice at the time of sacrifice (Figure 2A, 2B). Subsequently, we injected stable MHH-ES1 control and MHH-ES1/anti-miR-130b ES cells via tail vein and observed significantly decreased metastatic liver lesions in the miR-130b knockdown mice compared to the control mice (Figure 2C). Thus, we demonstrate that in vivo miR-130b is able to significantly alter ES dissemination, with organ tropism most likely dependent on inherent properties of the cell line. TC71 has been shown to have the ability to metastasize to the lungs in prior studies, while our experience with MHH-ES-1 cells have demonstrated propensity for metastasis to the liver, which might be attributed to their derivation from ascites fluid of the abdomen30,31. Overall our in vitro and in vivo studies provide convincing evidence that miR-130b contributes to metastatic properties and phenotype seen in ES tumors.
Figure 2. miR-130b alters metastatic potential in vivo.
(A) H&E-stained lung sections from the control and miR-130b expressing groups following tail vein injections of 1 × 106 TC71 cells with and without stable overexpression of miR-130b. Black arrows highlight evident microscopic lesions. (B) Quantification of pulmonary nodules in each group. Error bars were generated from control (n=5) and miR-130b (n=4) in each group. *P<0.05 (C) qPCR expression of miR-130b in TC71/scr and TC71/130b stable cell lines. (D) Images of metastatic livers of scrambled control and anti-miR-130b stable expressing groups following tail vein injections of 1 × 106 MHH cells with and without knockdown of miR-130b. (E) Quantification of miR-130b expression in MHH/scr and MHH/anti-130b stable cell lines. Error bars were generated from control (n=5) and anti-miR-130b (n=5) in each group. **P<0.05. Black boxes indicate areas magnified and arrows represent metastatic lesion observed.
miR-130b promotes the activation of CDC42 through negative regulation of previously unknown target, ARHGAP1, in Ewing sarcoma
After determining the contributions of miR-130b towards the metastatic potential for Ewing sarcoma, we were interested in determining miR-130b functional activities that could contribute towards driving ES metastasis. miRNAs are estimated to regulate approximately 30% of the genome through inverse relationships with target mRNAs 32. To understand how miR-130b could regulate the transcriptome of ES, we performed gene expression profiling and subsequent validation of differentially expressed genes (Supplemental Figure 3A). Pathway analysis using MetaCore™ software (Thomson Reuters) was performed on significant (p<0.05) differentially expressed genes. We analyzed genes downregulated with transient miR-130b overexpression in TC71 cells and then overlapped these candidate genes with predicted targets of miR-130b by TargetScan32. The overlap resulted in approximately 70 genes (Supplemental Figure 3B). Supplemental table 1 highlights the significant pathways altered, which included genes not only involved in cell morphology, but also interestingly significant alterations of genes involved in regulating migratory pathways, including the CDC42 pathway (Supplemental Table 1).
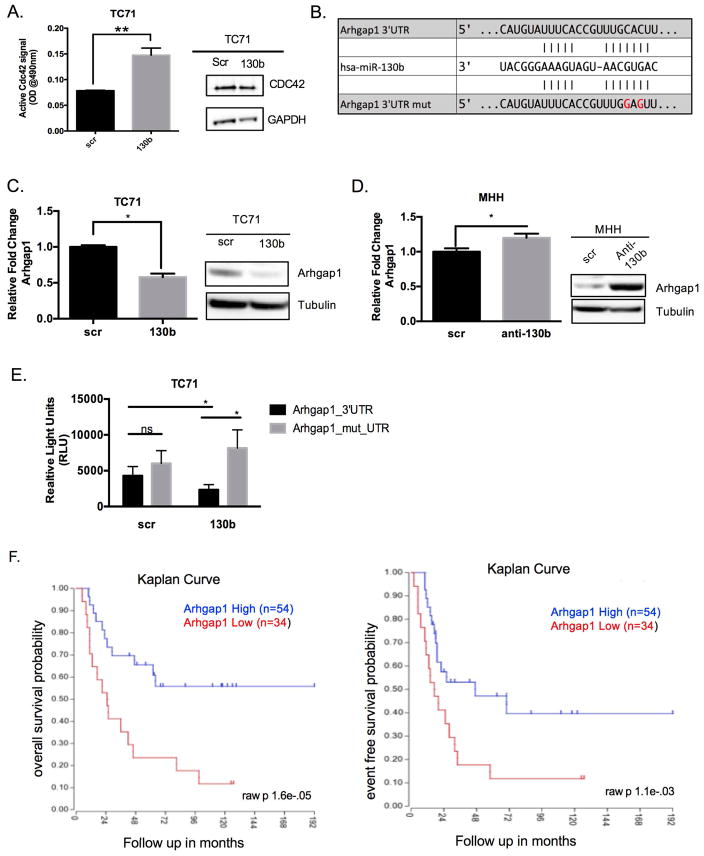
Subsequently, to investigate the functional significance of CDC42, we performed an ELISA assay to measure the activity of CDC42 in the TC71 miR-130b overexpressing cells. CDC42 is a member the RHO, RAC, and CDC42 subfamilies that make up the RhoGTPases33. In response to various cellular stimuli such as integrin engagement, growth factors, and stress, RhoGTPases are guanine exchange factors (GEFs) that act on numerous cellular processes such as proliferation, cytoskeletal remodeling, and gene transcription. Adding an extra layer of regulation, GTPase-Activating Proteins (GAP) hydrolyze GTP inactivating the GTPase34. We observed miR-130b activated CDC42, as measured by colorimetric ELISA, whereas total CDC42 protein levels remained unchanged (Figure 3A).
Figure 3. miR-130b activates CDC42 through direct target, ARHGAP,1 in Ewing sarcoma.
(A) G-LISA assay of TC71 cells overexpressing miR-130b. Values represent the average of triplicate samples. P-values were determined comparing scramble control lysates to miR-130b mimic lysates. *P<0.01, CDC42 western blot of TC71 scrambled control and miR-130b mimic transfected cells. (B) Representation of the seed pairing between miR-130b and Arhgap1 3′UTR. Bottom represents mutated nucleotides for luciferase assay. (C) qPCR and western blot of ARHGAP1 expression in TC71 cells ectopically overexpressing miR-130b mimic. P-values were determined comparing scramble control to miR-130b overexpression using Student’s t-test *P<0.05 (D) qPCR and western blot of ARHGAP1 expression in MHH-ES1 cells expressing anti-mir to miR-130b. P-values were determined comparing scramble control to anti-miR-130b using Student’s t-test *P<0.05 (E) Luciferase assay of TC71 cells co-transfected with either miR-130b mimic or scrambled control and wildtype 3′UTR ARHGAP1 plasmid or seed mutant plasmid. P-values were determined comparing scramble control to miR-130b overexpression using Student’s t-test *P<0.05. (F) Kaplan-Meier scan for ARHGAP1 derived from clinically annotated human Ewing Sarcoma gene expression database (dataset Mixed Ewing Sarcoma - Savola - 117 - MAS5.0 - u133p2 “http://hgserver1.amc.nl/cgi-bin/r2/main.cgi”. High expression (blue line) and low expression (red line) are compared. Scr = TC71 scrambled control ES cells, 130b = TC71 miR-130b mimic ES cells, anti-130b= anti-miR-130b oligo ES cells.
Using the pathway analysis and TargetScan miRNA prediction algorithm to identify mRNAs that had seed sequence for miR-130b and functioned in CDC42 pathway, we identified ARHGAP1 as a putative target of miR-130b (Figure 3B), which was downregulated in our gene microarray. ARHGAP1 is a high-affinity negative regulator of CDC42 that hydrolyzes active CDC42 into the inactive GDP state35,36. Previous studies investigating ARHGAP1 demonstrate that loss of ARHGAP1 leads to constitutive activation of CDC42 and subsequent downstream effector signaling37. We therefore hypothesized that downregulation of ARHGAP1 by miR-130b contributed to the observed activation of CDC42 in our cells. TC71 cells overexpressing miR-130b demonstrated downregulation of ARHGAP1 mRNA and protein by qPCR and western blot (Figure 3C). Conversely, knockdown of miR-130b in MHH cells resulted in modest mRNA, but significant protein level increases for ARHGAP1 (Figure 3D). To determine whether or not this interaction was by direct binding, we used a luciferase reporter assay containing the ARHGAP1 3′UTR to verify the interaction. miR-130b was able to significantly repress luciferase activity while the seed mutant restored luciferase expression (Figure 3E). Finally, using the publically available R2 Microarray visualization platform “http://hgserver1.amc.nl/cgi-bin/r2/main.cgi” we noted that ARHGAP1 expression levels are adversely associated with event-free and overall survival rates for patients with Ewing sarcoma (Figure 3F). These results suggest that interaction between miR-130b and ARHGAP1 is specific and demonstrate ARHGAP1 is a novel direct target of miR-130b that has clinical ramifications in patients with Ewing sarcoma.
miR-130b promotes the activation of an oncogenic CDC42/PAK1/JNK gene regulatory network
Upon activation, CDC42 signals downstream to PAK1, which autophosphorylates in response to CDC42 binding 38,39. In response to miR-130b overexpression, PAK1 has enhanced phosphorylation by immunoblotting analyses (Figure 4A). Conversely, knockdown of miR-130b resulted in decreased phospho-PAK1 levels (Figure 4B). Furthermore, we demonstrated that direct transient knockdown of ARHGAP1 expression mimicked the overexpression of miR-130b with enhanced downstream activation of PAK1 and c-Jun (Supplemental Figure 4A).
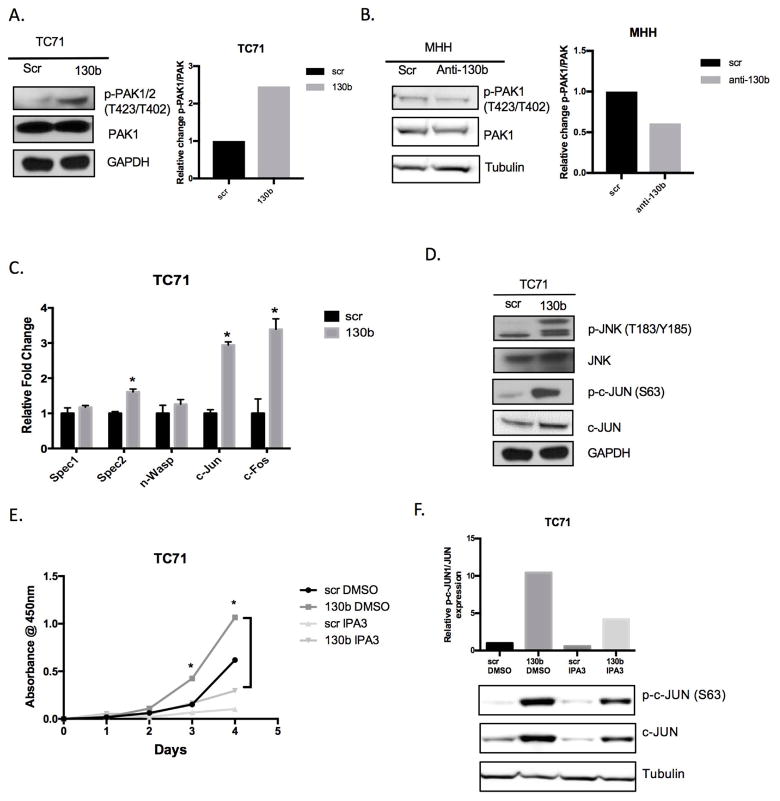
Figure 4. miR-130b modulates downstream activation of PAK1 and JNK.
(A, B) Phospho-PAK1/2 (Thr423/Thr402) and total PAK1 western blot of TC71 and MHH-ES1 miR-130b altered cells. GAPDH and Tubulin were used as loading controls. Western blots were quantified using ImageJ software. (C) qPCR analysis of Cdc42/Pak1 downstream effector after transfection of miR-130b mimic or scrambled control. P-values were calculated using Student’s t-test comparing scramble control to miR-130b mimic cells. (D) Western Blot of increased expression of the JNK cascade after miR-130b transfection. GAPDH was used as loading control. (E) Colorimetric cell proliferation assay of TC71 cells with and without IPA3 (10μM) treatment. Values represent the average of triplicates for each day. P-values were determined comparing miR-130b DMSO and miR-130b IPA3. *P<0.05 (F) Western blot analysis of phosphorylated and total c-JUN in total cell lysates isolated from TC71 scrambled control or miR-130b mimic cells treated with IPA3 (10 μM) for 24 h. Loading controls are identical in 4A and 4D since western blots were reprobed with different antibodies. scr=TC71 scrambled control ES cells, 130b =TC71 miR-130b mimic ES cells, anti-130b= anti-miR-130b oligo ES cells. Tubulin was used for loading control. (+) and (−) indicated with and without treatment of IPA3. p-c-JUN and p-PAK1= phosphorylated
PAK1 overexpression has been demonstrated in many tumors contributing to oncogenic signaling cascades, however the role and regulation of PAK1 in ES remains unknown. We observed an increase in the mRNA expression of PAK1 in ES patient samples and a negative correlation with ES patient survival (Supplemental Figure 4B, 4C). PAK1 links CDC42 to nuclear signals and cytoskeletal rearrangements. Expression levels of several cytoskeletal reorganization genes, n-WASP, SPEC1, and SPEC 2 remained relatively unchanged when miR-130b is overexpressed suggesting miR-130b may be stimulating downstream PAK1 effectors (Figure 4C). Evidence suggests that PAK1 can signal through JNKs resulting in the activation of c-JUN21. Further investigation of our gene expression array revealed a dramatic upregulation of c-JUN and c-FOS (Figure 4C). Coinciding with the enhanced gene expression seen in the microarray, we performed Reverse Phase Protein Array (RPPA) analysis that showed upregulation of phosphorylated JNK, phosphorylated c-JUN and c-JUN protein levels, which were subsequently validated by western blot analysis (Figure 4D, Supplemental Figure 5A, 5B). Taken together, these data suggest that miR-130b regulation of CDC42 and PAK1 results in downstream activation of the JNK pathway.
In order to determine if the effects of miR-130b could be pharmacologically mitigated, we used an inhibitor of PAK1, IPA3. IPA3 acts as an allosteric inhibitor of PAK1 resulting in an unfavorable conformation of the PAK1 active site40. Using IPA3, the increased proliferation due to miR-130b in TC71 cells was attenuated (Figure 4E). Furthermore, IPA3 also reduced the c-JUN and JNK levels induced by miR-130b (Figure 4F). To provide confirmation that PAK1 inhibition can attenuate the effects of miR-130b, we used the PAK1 inhibitor FRAX59741. Treatment with FRAX597 rescued the proliferative effects or miR-130b (Supplemental Figure 4D). Collectively, these data confirm a targetable CDC42/PAK1/JNK regulatory network promoted by miR-130b.
Downstream activation of AP-1 and target genes by miR-130b
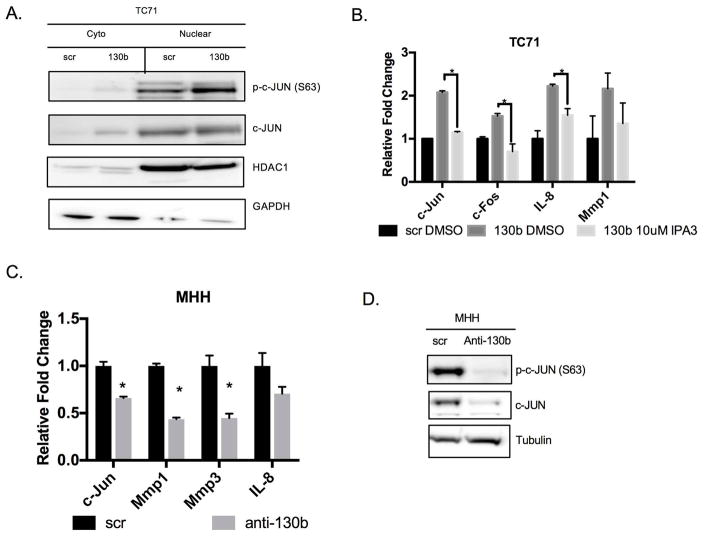
The microarray data and RPPA provide evidence that miR-130b induces the activation of JNK/SAPK and subsequent phosphorylation of its downstream target, c-JUN. The AP-1 transcription factor translocates to the nucleus upon activation and binds to the promoter region of target genes. To further confirm AP-1 is activated, we fractionated protein lysates from cells overexpressing miR-130b. Western blot analysis demonstrates that phosphorylated c-JUN and total c-JUN are increased in response to miR-130b overexpression but phospho-c-JUN is enriched in the nuclear fraction further confirming AP-1 is being activated (Figure 5A), while treatment with IPA3 and FRAX597 rescued the increased expression of c-JUN and AP-1 target genes (Figure 5B, Supplemental Figure 4E, 4F). In addition, use of the AP-1 inhibitor, SR11302, and JNK1 inhibitor, JNK-IN-8 also suppressed miR-130b induced c-JUN and AP-1 target gene expression by qPCR, which further confirms miR-130b activation of this pathway (Supplemental Figure 4G). Finally, knockdown of miR-130b also decreased the expression of c-JUN and AP-1 targets by qPCR and western blot analysis, suggesting miR-130b is specifically mediating this oncogenic transcriptional program (Figure 5C, 5D). Taken together our results highlight that miR-130b and CDC42, PAK1, and AP-1 form a critical oncogenic network within Ewing sarcoma.
Figure 5. miR-130b activates AP-1 signaling.
(A) Western blot of TC71 cells expressing miR-130b or scrambled control (right) western blot of fractionated lysates (left). GAPDH and HDAC1 were used as loading controls. (B) qPCR was performed to assess the expression levels of c-Jun and AP-1 targets. Total RNA was isolated 24 hours after IPA3 (10uM) treatment. P-values were determined using Student’s t-test comparing miR-130b mimic cells treated with IPA3 (10μM) to DMSO treated cells **P<0.01, ***P<0.001. (C) Assessment of c-Jun and AP-1 targets with miR-130b knockdown in MHH cells. Expression was measured by qPCR. P-values were calculated using Student’s t-test comparing scramble control to anti-miR-130b cells. *P<0.05. (D) Western blot of phosphorylated and total c-JUN after miR-130b knockdown in lysates isolated from MHH cells 48 hours post-transfection. Tubulin was used as loading control. Loading controls are identical in 4B and 5D since western blots were reprobed with different antibodies. Scr = TC71 scrambled control ES cells, 130b = TC71 miR-130b mimic ES cells, anti-130b= anti-miR-130b oligo ES cells.
AP-1 binds to the miR-130b promoter and regulates miR-130b transcription
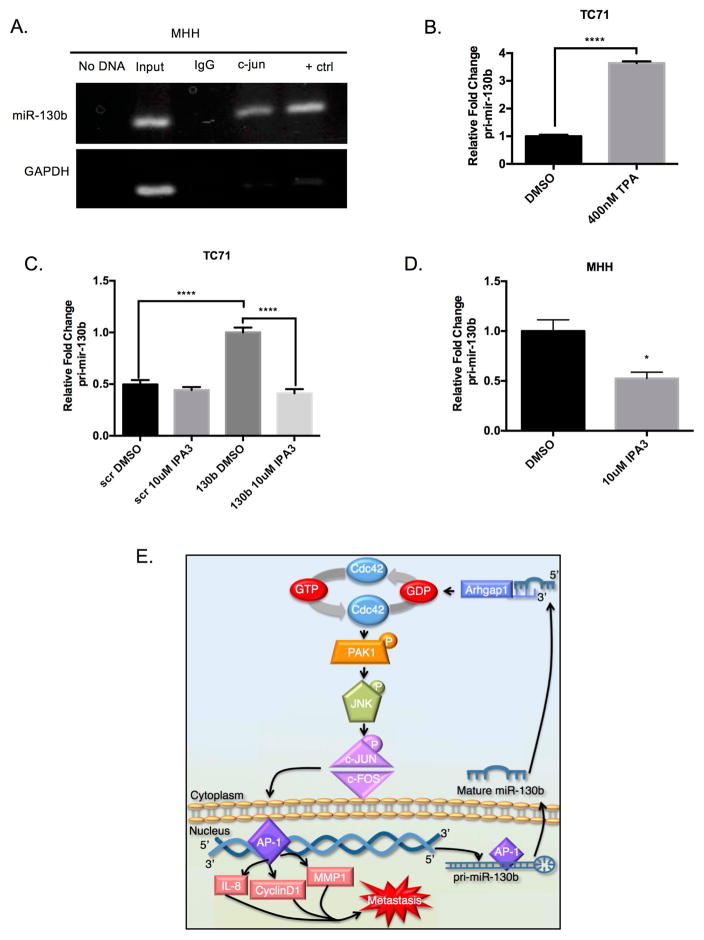
The causes of miRNA dysregulation have been linked to numerous processes including chromosomal aberrations, miRNA processing defects, and transcription factor binding42. After demonstrating the functional significance of miR-130b in Ewing sarcoma, we decided to look at the regulation of miR-130b. We analyzed an approximately 4kb region upstream of the miR-130b precursor using the UCSC ENCODE DNA browser (http://genome.ucsc.edu/) 43,44. Utilizing the publically available ChIP-SEQ data in this region we observed an enriched H3K27ac region, which is associated with active enhancers. We next examined predicted transcription factor binding sites within this region. Interestingly, an AP-1 binding site was predicted approximately 1kb upstream from the miR-130b precursor (Supplemental Figure 6). In order to examine whether AP-1 was binding to the miR-130b promoter, we performed Chromatin Immunoprecipitation (ChIP) assays on endogenous levels of AP-1. Genomic DNA was isolated from MHH cells and a site-specific band was amplified using primers covering the AP-1 region from DNA immunoprecipitated with a c-JUN antibody indicating the occupancy of the transcription factor at the promoter of miR-130b (Figure 6A).
Figure 6. AP-1 binds to miR-130b and regulates expression.
(A) Chromatin immunoprecipitation of MHH-ES1 cells using primers specific to AP-1 in the miR-130b promoter. (B) qPCR of pri-miR-130b expression after 4hour treatment with 400nM TPA. P-value was determined using Student’s t-test comparing TC71 treated with DMSO to TPA (400nM) treated cells. (C) qPCR of primary miR-130b transcript levels in TC71 after transfection with miR-130b and 24hour treatment with 10uM IPA3. P-value was determined using Student’s t-test comparing miR-130b mimic cells treated with IPA3 (10μM) to DMSO treated cells. *P<0.001. (D) qPCR of primary miR-130b transcript levels in MHH-ES1 cells after 24 hour treatment with 10μM IPA3. P-value was determined using Student’s t-test comparing MHH-ES1 cells treated with IPA3 (10μM) to DMSO treated cells. *P<0.05. (E) Summary diagram of miR-130b/CDC42/PAK1/AP-1 oncogenic, positive feedback loop in Ewing sarcoma. Scr = TC71 scrambled control ES cells, 130b = TC71 miR-130b mimic ES cells.
To understand if AP-1 is promoting transcription of miR-130b we treated TC71 cells with 12-O-tetradecanoylphorbol-13-acetate (TPA), which is an activator of c-JUN and AP-1. TPA stimulation led to an induction of the pri-miR-130b transcript after just 4 hours suggesting AP-1 can quickly induce miR-130b expression (Figure 6B). The induction of the pri-miR-130b transcript by TPA and the induction of JNK, c-JUN, and AP-1 target genes by overexpression of miR-130b suggest that a positive feedback loop is functioning to drive an oncogenic, metastatic gene regulatory network in Ewing sarcoma. To further investigate the presence of a positive feedback loop, we overexpressed miR-130b in TC71 cells and treated the cells with IPA3. After treatment with IPA3, we observed a decrease in the pri-miR-130b transcript (Figure 6C). Furthermore, treatment of MHH cells with IPA3 also decreased the levels of the pri-miR-130b transcript (Figure 6D). In sum, these results suggest a positive feedback loop exists between miR-130b expression and AP-1 activation. Specifically, miR-130b overexpression leads to the stimulation of CDC42 through downregulation of CDC42 negative regulator, ARHGAP1, which in turn leads to the autophosphorlyation of PAK1 and the activation of AP-1 and its target genes. Subsequently, AP-1 promotes the expression of miR-130b, thus completing an oncogenic positive feedback loop in Ewing sarcoma (Figure 6E).
Discussion
miR-130b has been reported to have oncogenic properties in numerous cancers, but its direct targets and their function, along with the regulation of miR-130b, remain largely unknown. Here we demonstrate a previously unknown gene regulatory network regulated by miR-130b in Ewing sarcoma that contributes metastatic phenotypes both in vitro and in vivo. We have further elucidated a mechanism in which miR-130b promotes sustained CDC42 activation through downregulation of ARHGAP1, a negative regulator of CDC42 and novel direct target of miR-130b. Prolonged activation of CDC42 by miR-130b leads to downstream AP-1 activation and target gene expression that promotes sarcoma invasion, migration, and proliferation. A positive feedback network was demonstrated by the ability of AP-1 to bind to the miR-130b promoter and induce transcription of the primary miR-130b. Finally, we could abrogate the effects induced by miR-130b and AP-1 using a PAK1 inhibitor, IPA3.
p21 activated kinases (PAKs) are a group of serine threonine kinases that are key signaling nodes for the activation of pro-tumorigenic pathways including the activation of pathways involved in proliferation, evasion of apoptosis, and invasion45. Numerous studies have also demonstrated that PAKs are mutated, overexpressed, or activated in cancer, making them exciting therapeutic targets for anti-cancer therapies46,47. Interestingly, the contribution of PAK1 in Ewing sarcoma development and metastasis has been severely understudied, and inhibition of this signaling node, in combination with approved standard treatment protocols, may offer new therapeutic options for patients with metastatic disease. We have shown novel insights into the effects of PAK1 in ES. We demonstrate that PAK1 is upregulated in ES patient tumors, enhanced expression correlates with poor patient outcome, and the use of the PAK1 inhibitors IPA3 and Frax597 attenuate the pro-proliferative effects of miR-130b. We did observe an increase in phospho-c-JUN levels with IPA3 which is most likely due to stress caused by treatment with IPA3. Furthermore, because c-JUN is downstream of stress response pathways, we did not observe a complete rescue in phosphorylation of c-JUN. However, these proof of principle studies demonstrate IPA3 and next generation PAK inhibitors such as, FRAX597 may prove promising to further test in an in vivo model for Ewing sarcoma.
It is well known that miRNAs can have numerous targets, and therefore simultaneous effects within the cell. As mentioned above, we prioritized ARHGAP1-mediated signaling based on analysis of our microarray data demonstrating ARHGAP1 downregulation and components of the CDC42-PAK pathway were significantly altered. In addition, Gene Ontology analysis further suggested deregulation of other pathways, such as the ROBO pathway, but we were unable to validate these findings. PAK1 activation intersects the regulation of several oncogenic pathways and is known to have various downstream effectors involved in cytoskeleton remodeling. Overall our studies, including the analysis and integration of microarray and RPPA data, suggest that miR-130b-mediated PAK1 activation of JNK signaling in contrast to traditional cytoskeleton remodeling genes. However, we cannot exclude other significant miR-130b mediated biological phenotypes in Ewing sarcoma that might be worth exploring.
While we provide evidence of a novel regulatory mechanism in Ewing sarcoma, these pathways can have various modes of activation. Several studies have demonstrated that EWS-FLI1 is capable of modulating miRNA expression; however, miR-130b expression was not shown to be altered in these studies48,49. While miR-130b has been independently shown to contribute to metastatic properties in other malignancies, we cannot exclude the contribution of the fusion transcript to aid in metastatic potential dictated by miR-130b7,50. Overall, we highlight this as a potential mechanism to promote metastasis in Ewing sarcoma, but cannot exclude other mechanisms of action such as input from other GTPases or components of the MAPK pathway. Future and ongoing studies include testing of relevant inhibitors of the MAPK and JNK as well as PAK1 in vitro and in vivo.
In summary, our functional genomic studies demonstrate that miR-130b has significant impact on the phenotypes that contribute to metastatic foci formation. Our detailed molecular mechanism highlights the importance of miR-130b in sustaining an oncogenic signaling loop in Ewing sarcoma while also elucidating novel druggable targets for the treatment of metastatic Ewing sarcoma.
Supplementary Material
Novelty and Impact.
We have defined a novel metastatic-promoting pathway directly regulated by the oncomiR-130b. We demonstrate that enhanced expression of miR-130b directly regulates Arhgap1, a negative regulator of CDC42 and stimulates a CDC42/PAK1/AP-1 axis. This leads to direct binding and regulation of miR-130b, thus providing a positive feedback loop in Ewing sarcoma. We have delineated a novel mechanism involved in sarcoma metastasis and revealed new therapeutic targets for the treatment of metastatic Ewing sarcoma.
Acknowledgments
This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom. This work was partially supported by the Cancer Prevention and Research Institute of Texas (RP170005, RP150648) and the NCI Cancer Center Support Grant (P30CA125123)
Cancer Prevention & Research Institute of Texas Proteomics & Metabolomics Core Facility Support Award (RP120092) (DPE and SH) and NCI Cancer Center Support Grant to Antibody-based Proteomics Core/Shared Resource (P30CA125123) (DPE and SH).
STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core, NCI # CA016672.
Research was supported through funding by The Faris D. Virani Ewing Sarcoma Center (JTY), the Gillson Longenbaugh Foundation (JTY) and St. Baldrick’s Foundation (JTY). LS was supported by a pre-doctoral fellowship award from the Cancer Prevention Research Institute of Texas (CPRIT) (RP140102).
References
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–5. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Deley M-CL, Kovar H, Grimer R, Whelan J, Claude L, Delattre O, Paulussen M, et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J Clin Oncol. 2015;33:3036–46. doi: 10.1200/JCO.2014.59.5256. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Budd WT, Seashols-Williams SJ, Clark GC, Weaver D, Calvert V, Petricoin E, Dragoescu EA, O’Hanlon K, Zehner ZE. Dual Action of miR-125b As a Tumor Suppressor and OncomiR-22 Promotes Prostate Cancer Tumorigenesis. PloS One. 2015;10:e0142373. doi: 10.1371/journal.pone.0142373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drebber U, Lay M, Wedemeyer I, Vallböhmer D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409–15. doi: 10.3892/ijo.2011.1036. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi C, Zhou X, Bian X, Ping Y, Wen Y, Zhao S, et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget. 2015;6:29129–42. doi: 10.18632/oncotarget.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang R-M, Xu J-F, Fang F, Yang H, Yang L-Y. MicroRNA-130b promotes proliferation and EMT-induced metastasis via PTEN/p-AKT/HIF-1α signaling. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016 doi: 10.1007/s13277-016-4919-z. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Zhao X, Zhang H, Yuan H, Zhu M, Sun Q, Lai X, Wang Y, Huang J, Yan J, Yu J. MiR-130b suppresses prostate cancer metastasis through down-regulation of MMP2. Mol Carcinog. 2015;54:1292–300. doi: 10.1002/mc.22204. [DOI] [PubMed] [Google Scholar]
- 9.Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To K-F, Zheng B-J, Lai PBS, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Sheng X, Chen H, Wang H, Ding Z, Xu G, Zhang J, Lu W, Wu T, Zhao L. MicroRNA-130b promotes cell migration and invasion by targeting peroxisome proliferator-activated receptor gamma in human glioma. Biomed Pharmacother Bioméd Pharmacothérapie. 2015;76:121–6. doi: 10.1016/j.biopha.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhu G, Wang Y, Mijiti M, Wang Z, Wu P-F, Jiafu D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem Biophys Res Commun. 2015;465:194–9. doi: 10.1016/j.bbrc.2015.07.149. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton MP, Rajapakshe K, Hartig SM, Reva B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler DA, Coarfa C, McGuire SE. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielson LS, Menendez S, Attolini CS-O, Guijarro MV, Bisogna M, Wei J, Socci ND, Levine DA, Michor F, Hernando E. A Differentiation-Based MicroRNA Signature Identifies Leiomyosarcoma as a Mesenchymal Stem Cell-Related Malignancy. Am J Pathol. 2010;177:908–17. doi: 10.2353/ajpath.2010.091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, Alberghini M, Grilli A, Knuutila S, Schaefer K-L, Mattia G, Negrini M, et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- 15.Yu L-D, Jin R-L, Gu P-C, Ling Z-H, Lin X-J, Du J-Y. Clinical significance of microRNA-130b in osteosarcoma and in cell growth and invasion. Asian Pac J Trop Med. 2015;8:752–6. doi: 10.1016/j.apjtm.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Krook MA, Hawkins AG, Patel RM, Lucas DR, Van Noord R, Chugh R, Lawlor ER. A bivalent promoter contributes to stress-induced plasticity of CXCR4 in Ewing sarcoma. Oncotarget. 2016 doi: 10.18632/oncotarget.11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza-Naranjo A, El-Naggar A, Wai DH, Mistry P, Lazic N, Ayala FRR, da Cunha IW, Rodriguez-Viciana P, Cheng H, Tavares Guerreiro Fregnani JH, Reynolds P, Arceci RJ, et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol Med. 2013;5:1087–102. doi: 10.1002/emmm.201202343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadok A, Marshall CJ. Rho GTPases: masters of cell migration. Small GTPases. 2014;5:e29710. doi: 10.4161/sgtp.29710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasenthil S, Sahin AA, Barnes CJ, Wang R-A, Pestell RG, Vadlamudi RK, Kumar R. p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J Biol Chem. 2004;279:1422–8. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 20.Carter JH, Douglass LE, Deddens JA, Colligan BM, Bhatt TR, Pemberton JO, Konicek S, Hom J, Marshall M, Graff JR. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res Off J Am Assoc Cancer Res. 2004;10:3448–56. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 21.Bagrodia S, Dérijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–8. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 22.Brown JL, Stowers L, Baer M, Trejo J, Coughlin S, Chant J. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol CB. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 23.Murayama T, Ohara Y, Obuchi M, Khabar KS, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF-kappaB-binding sites of the interleukin-8 gene. J Virol. 1997;71:5692. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res Ther. 2001;4:157. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras Mutants and c-Ets-2 Activate the Cyclin D1 Promoter through Distinguishable Regions. J Biol Chem. 1995;270:23589–97. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 26.Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim H-T, Brown PH. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2007;27:366–77. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- 27.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C-H, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S, Rosen JM. Mammary Stem Cells and Tumor-Initiating Cells Are More Resistant to Apoptosis and Exhibit Increased DNA Repair Activity in Response to DNA Damage. Stem Cell Rep. 2015;5:378–91. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S-H, Tilan JU, Galli S, Izycka-Swieszewska E, Polk T, Horton M, Mahajan A, Christian D, Jenkins S, Acree R, Connors K, Ledo P, et al. High neuropeptide Y release associates with Ewing sarcoma bone dissemination - in vivo model of site-specific metastases. Oncotarget. 2015;6:7151–65. doi: 10.18632/oncotarget.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Heyking K, Calzada-Wack J, Göllner S, Neff F, Schmidt O, Hensel T, Schirmer D, Fasan A, Esposito I, Müller-Tidow C, Sorensen PH, Burdach S, et al. The endochondral bone protein CHM1 sustains an undifferentiated, invasive phenotype, promoting lung metastasis in Ewing sarcoma. Mol Oncol. 2017 doi: 10.1002/1878-0261.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–42. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 34.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Wang Z-X, Zheng Y. Characterization of the Interactions between the Small GTPase Cdc42 and Its GTPase-activating Proteins and Putative Effectors COMPARISON OF KINETIC PROPERTIES OF Cdc42 BINDING TO THE Cdc42-INTERACTIVE DOMAINS. J Biol Chem. 1997;272:21999–2007. doi: 10.1074/jbc.272.35.21999. [DOI] [PubMed] [Google Scholar]
- 36.Low BC, Seow KT, Guy GR. The BNIP-2 and Cdc42GAP Homology Domain of BNIP-2 Mediates Its Homophilic Association and Heterophilic Interaction with Cdc42GAP. J Biol Chem. 2000;275:37742–51. doi: 10.1074/jbc.M004897200. [DOI] [PubMed] [Google Scholar]
- 37.Yang L, Wang L, Zheng Y. Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia induction, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol Biol Cell. 2006;17:4675–85. doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenke FT, King CC, Bohl BP, Bokoch GM. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–73. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- 39.Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–63. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deacon SW, Beeser A, Fukui JA, Rennefahrt UEE, Myers C, Chernoff J, Peterson JR. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Licciulli S, Maksimoska J, Zhou C, Troutman S, Kota S, Liu Q, Duron S, Campbell D, Chernoff J, Field J, Marmorstein R, Kissil JL. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem. 2013;288:29105–14. doi: 10.1074/jbc.M113.510933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manser E, Leung T, Salihuddin H, Zhao Z, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–6. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 46.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 47.Radu M, Semenova G, Kosoff R, Chernoff J. Pak Signaling in the Development and Progression of Cancer. Nat Rev Cancer. 2014;14:13. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franzetti G-A, Laud-Duval K, Bellanger D, Stern M-H, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2013;32:3915–21. doi: 10.1038/onc.2012.403. [DOI] [PubMed] [Google Scholar]
- 49.Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DNT, Schaefer K-L, Nakatani F, Scotlandi K, Reiter M, Strunk D, Speleman F, et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene. 2011;30:2173–80. doi: 10.1038/onc.2010.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian J, Hu L, Li X, Geng J, Dai M, Bai X. MicroRNA-130b promotes lung cancer progression via PPARγ/VEGF-A/BCL-2-mediated suppression of apoptosis. J Exp Clin Cancer Res CR. 2016;35:105. doi: 10.1186/s13046-016-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.