Abstract
Measurement of biomarkers has been incorporated within clinical research of asthma to characterize the population and to associate the disease with environmental and therapeutic effects. Regrettably, at present, there are no specific biomarkers, none is validated or qualified, and endotype-driven choices overlap. Biomarkers have not yet reached clinical practice and are not included in current asthma guidelines. Last but not least, the choice of the outcome upholding the value of the biomarkers is extremely difficult, since it has to reflect the mechanistic intervention while being relevant to both the disease and the particular person. On the verge of a new age of asthma healthcare standard, we must embrace and adapt to the key drivers of change. Disease endotypes, biomarkers, and precision medicine represent an emerging model of patient care building on large-scale biologic databases, omics and diverse cellular assays, health information technology, and computational tools for analyzing sizable sets of data. A profound transformation of clinical and research pattern from population to individual risk and from investigator-imposed subjective disease clustering (hypothesis driven) to unbiased, data-driven models is facilitated by the endotype/biomarker-driven approach.
Keywords: Asthma, biomarkers, precision medicine
ASTHMA MANAGEMENT IN THE ADVENT OF PRECISION MEDICINE
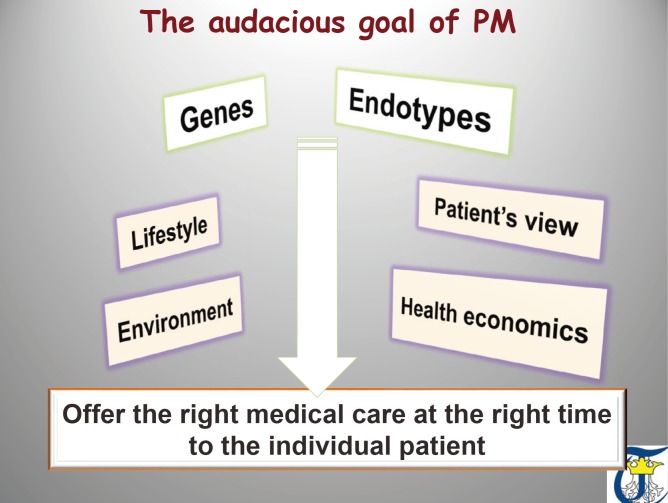
Precision medicine (PM) is an emerging approach for disease prevention and treatment that takes into account individual variability in environment, lifestyle, and genes for each person. This is in contrast to a “one-size-fits-all” approach, in which disease treatment and prevention strategies are developed for the average person, with less consideration for the differences between individuals. The concept embraces 4 key features: personalized care based on molecular, immunologic and functional endotypes of the disease, with participation of the patient in the decision making process of therapeutic actions, and taking into account predictive and preventive aspects of the treatment.1,2 PM is different from personalized medicine, and the terms should not be used interchangeably: in PM the focus is on identifying which approaches will be effective for which patients based on genetic, environmental, and lifestyle factors (individual risk) and on integrating research disciplines and clinical practice to guide individualized patient care (Fig. 1).
Fig. 1. The audacious goal of precision medicine. Understanding the complex networks of molecular, genetic and environmental in combination with strong health economics data and in alignment with patients participation will open the door for prevention strategies and curative therapies for asthma.
The concept of PM that takes individual variability into account is not new: starting with Archibald Garrod's pioneering research3 in 1902 advancing the hypothesis of “chemical individuality” in alkaptonuria, to transfusion selected by blood typing, anemia treatment guided by its mechanism, and allergen immunotherapy driven by the relevant sensitizing allergen to genetically targeted therapies in cancer or cystic fibrosis. The prospect of applying this concept broadly has been dramatically improved by the recent development of large-scale biologic databases, powerful methods for characterizing patients (such as proteomics, metabolomics, genomics, diverse cellular assays, and health information technology [HIT]), and computational tools for analyzing large sets of data.1,2,4,5
The emerging framework of PM brings together a variety of stakeholders: Academia, patients, ethics committees, regulators/policy makers, and diagnostic/pharmaceutical industry in a precision-medicine ‘ecosystem’ using a variety of tools such as omics, biobanks/registries, bioinformatics, and HIT, which all need to be accounted for. What is needed now is harmonization between stakeholders with agreement on a broad research program encouraging creative approaches and testing them rigorously both for robustness and for applicability for real life personalized care (Fig. 2).
Fig. 2. What we need to achieve the audacious goal of precision medicine. Harmonization between stakeholders and tools with agreement on a broad research program encouraging creative approaches and testing them rigorously both for robustness and for applicability for real life personalized care is needed to bring precision medicine to the clinic.
We are coming to understand the complex networks of molecular pathways and characteristics of the asthma endotypes that interact to drive inflammation and remodeling that need to be targeted, in combination, to develop prevention strategies and curative therapies for asthma.5,6,7,8,9 This subtle and highly complex architecture of disease-specific pathways (endotypes) steadily provides new targets for more efficient and tailored interventions.5,10,11 However, although the progress in understanding the mechanisms of asthma has been so significant during the last decade, it does not yet mirror the number of submission dossiers for new medicinal products to regulatory agencies. Instead, it seems likely that the pipeline in successful drug development is rather slowing down.12,13 The reasons for this declining trend could be the sluggish translational implementation of these discoveries and the uncertainty experienced by pharmaceutical companies due to the lack of guidance, which should be provided by the regulators. Expanding PM clinical trials in asthma is a must for moving the field forward, together with other initiatives, such as developing new laboratory models for research and national/regional and international asthma knowledge systems.2,5
The multilevel (clinical, functional, structural, and biological) and dynamic (i.e., subjected to variability with time) heterogeneity of asthma challenges a personalized approach for treating these patients. Thus, asthma is ideally suited for PM requiring an individualized approach based on biological mechanisms (endotypes) for a better selection of treatment responders, risk prediction, and design of disease-modifying strategies.2,4,5 A major critical step for this tailored, mechanistic approach will be the identification, validation, and qualification of pathways-specific biomarkers as companion diagnostics, which will ultimately enable the clinician to select ‘the right treatment for the right patient at the right time.’4,14,15 Therefore, the ultimate goal of the endotype/biomarker-driven approach in asthma is to develop the “magic bullet” linking drug response with the biological profile and to provide a safe and efficacious drug for a selected patient population. Besides the therapeutic purpose, the endotype/biomarker-driven approach should be able to open new avenues for prevention in high-risk individuals, early diagnosis and intervention, and disease modifying strategies.4,5,7
ASTHMA BIOMARKERS: A CRITICAL APPRAISAL
The ideal asthma biomarker links the disease endotype with the phenotype, predicts disease behaviors: exacerbation, severity, response to treatment, is stable over time or has a predictable variation pattern, and is easily replicated across populations with different genetic backgrounds.7,10 Biomarkers can be associated with the mechanistic pathway and thus guide treatment (for example sputum or blood eosinophils or serum periostin) or is the key molecule of the pathway and thus the biomarker itself is the target of the intervention. The challenge with asthma biomarkers is their variability across age and asthma severity and in time, thus incorporating time trends as a hallmark of asthma as a variable disease is a must in biomarkers research and validation.16 Another significant challenge is the complexity of a given endotype. We recently described the type 2 asthma as an example of a complex endotype with several major pathogenic pathways driven by interleukin (IL)-5, IL-4/IL-13, or immunoglobulin E (IgE) and with a multitude of modulators: genetic and epigenetic influences, barrier dysfunction, the exposome, nutrition, and metabolic pathways, etc.5,11,16 Last but not least, all biomarkers should be validated and qualified. Validation is the process of assessing the biomarker and its measurement performance characteristics, and determining the range of conditions under which the biomarker will give reproducible and accurate data. According to the Food and Drug Administration (FDA), a valid biomarker is defined as “measured in an analytical test system with well-established performance characteristics and for which there is an established scientific framework or body of evidence that elucidates the physiologic, toxicologic, pharmacologic, or clinical significance of the test results.”17 Qualification is the evidentiary process of linking a biomarker with biological processes and clinical end points.14,15 The European Medicines Agency (EMA) offers scientific advice to support the qualification of innovative development methods for a specific intended use in the context of research and development into pharmaceuticals. The Committee for Medicinal Products for Human Use (CHMP) gives the advice on the basis of recommendations by the Scientific Advice Working Party. This qualification process leads to a CHMP qualification opinion or CHMP qualification advice. The opinion is based on the assessment of data submitted to the Agency. Before final adoption of qualification opinion, the CHMP makes its evaluation open for public consultation by the scientific community. The advice is based on the evaluation of the scientific rationale and on the preliminary data submitted to EMA and is meant to help develop future protocols and methods toward qualification. To facilitate parallel submissions of applications for biomarker qualification to EMA and to FDA, the 2 agencies launched in December 2014, a joint letter of intent allows the 2 agencies to share scientific perspectives and advice. The agencies are also able to provide the same response to submitters.18
The most scrutinized biomarkers in asthma are related to eosinophilic inflammation and/or type 2 immune response (Table 1). However, the available biomarkers cannot distinguish between the innate and adaptive immune responses generating the type 2 milieu, very few have been evaluated in targeted interventions and none is validated and qualified. In addition, there are few studies linking type 2 biomarkers to disease subendotypes. High-throughput profiling studies of well-characterized patients, including gene expression (microarrays) and omics, can help identify combined signatures for type 2 asthma as per system medicine.19,20,21 One notable example of this approach using gene expression microarrays of bronchial epithelial cells obtained from patients with asthma has been the identification of periostin, an IL-13-responsive biomarker.19,22,23 Additional studies have aimed to determine disease endotypes by exploring the transcriptome of the airway compartment, including the airway epithelium24 and sputum.25 Unfortunately, even gene signatures are not highly specific for type 2 asthma: a recent study showed airway gene expression alterations specific for type 2 asthma present in a subset of patients with chronic obstructive lung disease (COPD).26 We have recently proposed a combination of biomarkers, such as IL-5 and IL-13, as the best predictor for blood eosinophilia in adult asthmatics.27
Table 1. Biomarkers linked to eosinophilic asthma and/or type 2 asthma.
| Biomarker | Experimental | Association | Intervention |
|---|---|---|---|
| IL-5 (serum, saliva)27–35 | ✔ | ✔ | ✔ |
| IL-13 (serum, sputum)27,36-38 | ✔ | ✔ | ✔ |
| IgE (serum)39–41 | ✔ | ✔ | ✔ |
| IL-4 (serum, sputum)38,42 | ✔ | ✔ | ✔ |
| Periostin (serum, lung biopsies, BAL, tears)43–45 | ✔ | ✔ | ✔ |
| Type 2 gene expression (periostin, serpin B2, CLCA-1) in bronchial biopsies/sputum cells19,22,46 | ✔ | ✔ | - |
| DPP-4 (serum)47 | ✔ | - | ✔ |
| Eotaxin, RANTES, GM-CSF (serum, saliva)48,49 | ✔ | ✔ | - |
| IL-9 (serum)50 | ✔ | ✔ | ? |
| IL-25 (bronchial epithelium, serum)51,52 | ✔ | ✔ | ? |
| TSLP; CRTH2 and DP1 receptors52–57 | ✔ | ✔ | Under investigation |
| CCR8; TARC; IL-31; IL-32 and T1/ST2; IL-19; NKT cells58–60 | ✔ | ✔ | - |
| IL-33, proangiogenic BM precursors, osteopontin, galectin 961–63 | ✔ | ✔ | - |
| CD48, leptin, lactoferin, IL-2364–66 | ✔ | - | - |
| IL-7 (serum, PBMCs)67 | ✔ | ✔ | - |
| ICOS/ICOS-L; IL-22; H4 receptors68–70 | ✔ | - | - |
| Il-5 and IL-13 producing Innate lymphoid cells in serum and sputum71 | ✔ | ✔ | - |
| DNA methylation profile72 | - | ✔ | - |
Most of the biomarkers are only for research purpose and none of them is validated or qualified.
BAL, broncho-alveolar lavage; BM, bone marrow; CCR, C-C chemokine receptor; CLCA-1, chloride channel accessory 1; CRTH2, G-protein-coupled chemokine receptor homologous molecule expressed on Th2 lymphocytes; DNA, deoxyribonucleic acid; DP1, the prostaglandin D2 receptor 1; DPP4, dipeptidyl peptidase-4; GM-CSF, granulocyte-macrophage colony-stimulating factor; H, histamine; ICOS, inducible co-stimulator; ICOS-L, inducible co-stimulator ligand; Ig, immunoglobulin; IL, interleukin; NKT, natural killer T cell; PBMC, peripheral blood mononuclear cells; RANTES, regulated on activation, normal T cell expressed and secreted; TARC, thymus- and activation-regulated chemokine; T1/ST2, immunoregulatory protein of the IL-1 receptor family; TSLP, thymic stromal lymphopoietin.
Less is known about the biomarkers of non-type 2 asthma, a distinct asthma endotype with several relevant clinical features, such as increased asthma severity, increased remodeling, and lower response to bronchodilator and anti-inflammatory treatment. Based on data from Th2 high/low molecular signature studies and induced sputum evaluation, the incidence of adult non-type 2 asthma reaches 30%–50%.19,20,73 The endotyping of non-type 2 is far behind the type 2 asthma, and until now, no endotype-driven interventions have been proved to be effective. Two major mechanisms leading to neutrophilic inflammation were postulated: the dysregulated innate immune response, including neutrophil intrinsic abnormalities, and the activation of the IL-17-dependent pathway.4,74 Several factors, such as age, metabolic or epigenetic factors, or the activation of the epithelial-mesenchimal trophic unit, have been identified as modulators. Non-type 2 asthma has a distinct DNA methylation profile in peripheral blood mononuclear cells involving secreted frizzled related protein 1 (sFRP1) as a key node, over representing the Wnt signaling pathway.72 Another example is the consumption of a high fat meal proven to increase neutrophilic airway inflammation in asthma subjects. This occurs through changes in expression of genes regulating airway inflammation and may provide useful therapeutic targets for immunomodulation, particularly relevant to obese asthmatics, who are habitually consuming diets with a high fat content.75 Corresponding biomarkers can be described for each non-type 2 asthma subendotypes (Tables 2 and 3); however, very few were tested as a potential therapeutic target with equivocal results (IL-17, tumor necrosis factor [TNF-α], interferon [IFN]-β) and none of them is validated or qualified. Some of these biomarkers, such as IL-17, have prognostic value related to asthma severity.76
Table 2. Biomarkers linked to neutrophilic inflammation in non-type-asthma.
| Biomarker | Experimental | Association | Intervention |
|---|---|---|---|
| IL-8, LTB477–79 | ✔ | ✔ | - |
| IL-12, IL-1880 | ✔ | ✔ | - |
| IL-17/TRIF-176,81–84 | ✔ | ✔ | ✔ |
| BDNF, MIP-3a/CCL-20, IL-1 β85–87 | ✔ | ✔ | - |
| IL-32; PAMPS, DAMPS, SDF87–89 | ✔ | ? (IL-32 in smokers) | - |
| Galectin-3 binding protein90 | - | ✔ | - |
| DNA methylation profile72 | - | ✔ | - |
Less is known about the biomarkers of non-type 2 asthma. Neutrophil intrinsic abnormalities, and the activation of the IL-17-dependent pathway have been postulated as disease subendotypes.
BDNF, brain-derived neurotrophic factor; CCL, C-C chemokine ligand; IL, interleukin; LT, leukotriene; DAMPS, damage-associated molecular patterns; MIP, macrophage inhibitory protein; PAMPS, pathogen-associated molecular patterns; SDF, stromal cell-derived factor; TRIF, toll/IL-1 receptor (TIR)-domain-containing adapter-inducing interferon-β.
Table 3. Biomarkers linked to dysregulation of innate immune response in non-type-2 asthma.
| Biomarker | Experimental | Association | Intervention |
|---|---|---|---|
| TNF-α91–93 | ✔ | ✔ | ✔ |
| IFNs91,94,95 | ✔ | ✔ | ✔ |
| NK cells, TLRs96–101 | ✔ | ✔ | - |
| Purinergic inflammation102,103 | ✔ | ✔ | - |
| Chitinase-like proteins104 | ✔ | ✔ | - |
| MBL, defensins, collectins, cathelidicin, granzyme, complement/C5L291,105-107 | ✔ | ✔ | - |
| IRAK-M; APRIL108 | ✔ | - | - |
| TREM1109 | ✔ | ✔ | - |
| Surfactant protein D110 | - | ✔ | - |
APRIL, a proliferation-inducing ligand; C5L2, C5a like receptor 2; IFN, interferon; IRAK-M, interleukin-1 receptor-associated kinase 3; MBL, mannan binding lectin; NK cell, Natural Killer cell; TLR, Toll-like receptor; TNF, tumor necrosis factor; TREM1, triggering receptor expressed on myeloid cells 1.
Biomarkers of defective resolution/repair (Table 4) are almost neglected in asthma research, although there are solid proofs that they can be linked to asthma severity and/or steroid responsiveness.111,112,113,114 For example, children with severe asthma have decreased lipoxin A4 concentrations in induced sputum, and this coupled with increased levels of leukotriene B4 (LTB4) promoting neutrophilic inflammation might be involved in the reduced ability of inhaled corticosteroids to control airway inflammation.115 Feeble attempts have been done to evaluate these biomarkers as therapeutic targets.116
Table 4. Biomarkers of defective resolution/repair.
Although airway remodeling is a prominent phenotypic trait of asthma and vast amounts of data accumulated from basic research, no attempt has been made to link a biomarker (Table 5) to a potential therapeutic intervention. Bronchial termoplasty (BT) trials, for example, were conducted in a population of severe asthmatic patients selected only based on positive methacholine challenge test as a surrogate for airway smooth muscle (ASM) abnormalities.127,128 Few clinics perform bronchoscopy with bronchial biopsies to substantiate ASM thickening. The intent of performing BT is to ablate ASM, but this response is variable—as was seen in the feasibility study—and there is a report of a patient who exhibited persistent smooth muscle hyperplasia following treatment.129 Therefore, there are likely other mechanisms by which BT results in improved asthma symptoms, and better understanding of the precise role of ASM in the pathogenesis of asthma is thus required. Remodeling is of particular importance both in severe asthma where it can be demonstrated within the proximal airway wall despite suppressed tissue inflammation130 and in steroid naïve asthma.131 There have been significant advancements in imaging techniques (computed tomography, magnetic resonance imaging, and positron emission tomography) for the evaluation of asthmatic patients, both from clinical and research perspectives. Imaging biomarkers can be linked to specific asthmatic phenotypes and provide a more detailed understanding of endotypes. Airway wall thickening, as measured through high-resolution computed tomography (HRCT), was associated with asthma severity, airflow obstruction, airway reactivity, and lung volume.132,133 Acinar ventilation heterogeneity (Sacin) was also associated with severe asthma.134 Both HRCT and magnetic resonance imaging can identify small airway disease in asthma, a special phenotype and potential new endotype in asthma.135,136
Table 5. Biomarkers of airway remodeling.
| Biomarker | Experimental | Association | Intervention |
|---|---|---|---|
| MMP/TIMP, TGF-β, IL-13, ADAMTS, ADAM-8, ADAM-7137–141 | ✔ | ✔ | - |
| VEGF/ADAM-33141–146 | ✔ | ✔ | ? (vitamin D) |
| Claudin, fibulin-1, endothelin-1, retinoid receptors147–150 | ✔ | ✔ | - |
| COX-2, TMEFF2, TRPM-1130 | - | ✔ | - |
| RELM-β, ICOS-L, relaxin, oncostatin M, decorin, amphiregulin, LIGHT, airway basal stem cells, CC/CXC chemokines131,137,151 | ✔ | - | - |
| Serum chitotriosidase activity and chitinase-3-like protein 1 levels152 | - | ✔ | - |
| Activin-A153 | - | ✔ | - |
| Serum periostin, osteopontin131,154 | ✔ | ✔ | - |
| Imaging biomarkers: HRCT airway wall thickening/wall area, MRI Sacin132–136 | - | ✔ | - |
ADAM, a disintegrin and metalloproteinase domain-containing protein; ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; COX-2, cyclo-oxygenase-2; HRCT, high-resolution computed tomography; ICOS-L, inducible co-stimulator ligand; LIGHT, homologous to lymphotoxin, exhibits inducible expression, and competes with HSV glycoprotein D for binding to herpesvirus entry mediator, a receptor expressed on T lymphocytes; MMP, metalloproteinase; MRI, magnetic resonance imaging; RELM-β, resistin-like molecule-β; Sacin, acinar ventilation heterogeneity; TGF, transforming growth factor; TIMP, tissue inhibitor of metalloproteinase; TMEFF2, transmembrane protein with epidermal growth factor like and two follistatin like domains 2; TRPM-1, transient receptor potential cationic channel subfamily M member 1; VEGF, vascular endothelial growth factor.
Future perspectives
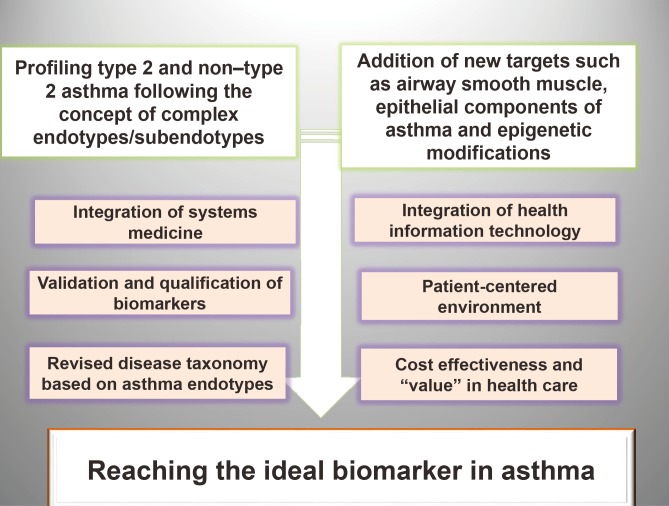
Key steps for moving the biomarkers field in asthma forward (Fig. 3) involve profiling asthma following the concept of complex endotypes and subendotypes linked to validated and qualified biomarkers resulting from the unbiased approach facilitated by the big data driven-models. Integrating HIT with systems medicine and with a patient centered environment is essential for coordination and alignment in translating the research data into functional clinical decision algorithms. A revised endotypic taxonomy of asthma can stimulate targeted research and interventions to identify biomarkers predicting the implication of distinct endotypes in disease pathogenesis. Policy makers feel threatened by analytical outputs and find reasons to reject them, unless they develop high levels of trust in their pedigree and provenance, thus healthcare systems need to adapt based on cost-effective delivering value grounds.
Fig. 3. Advancing the asthma biomarkers field. Profiling type 2 and non-type 2 asthma should follow the concept of complex endotypes/subendotypes in parallel with addition of new targets such as ASM, epithelial components of asthma and epigenetic modifications together with integration of systems medicine and advances in HIT. Validation and qualification of asthma biomarkers is an essential step for facilitating regulatory approval and acceptance into the health system. Improved understanding and common usage of disease phenotypes, endotypes, biomarkers, and precision therapies at the point of care is key for bringing the precision medicine into asthma clinic. Both full patient monitoring using novel digital technology and the concept of endotypes/novel biomarkers/patient centered care need to be reinforced as part of the healthcare system transformation. Development and implementation of a new asthma taxonomy including disease endotypes is highly needed. ASM, airway smooth muscle; HIT, health information technology.
Emerging technologies have generated a new wealth of knowledge that needs to be analyzed and translated into clinical decision algorithms, personalized asthma prevention, and management. Big data analytics, computer vision, image processing, sensors, and robotics with medical science are expected to benefit personalized asthma diagnosis and monitoring. Data mining as well as profiling and techniques for big data analytics are key tools used to discover and communicate meaningful patterns in personalized health data to promote early and instant disease diagnosis and personalized management. These advances are starting to improve the quality of health care and reduce costs. However, we should proceed on this pathway with care, and data mining should always be subjected to meaningful clinical interpretation and external validation.155,156,157
Rapid advances in HIT have created unprecedented opportunities to collect, analyze, and learn from vast amounts of “real-world” data that currently are locked away in unconnected servers and file cabinets. While clinical trials will likely remain the gold standard of evidence, crowdsourcing backed up by HIT advances promises to overcome the current limitations of observational data. By analyzing an immense body of observational data in real time, physicians and researchers can identify trends and associations between myriad variables and generate new hypotheses, and draw immediate practice-changing conclusions.158,159,160,161
In the near future, asthma practices will participate in HIT-based systems that securely compile and analyze information from individual electronic health records (major clinical endpoints, comorbidities, symptom scores, objective measurements, treatments, side effects, molecular profiles, and quality of life, etc.). The data collected will not be biased by any preselection criteria. Advanced HIT tools, such as rapid learning systems, will structure the huge body of unbiased data by normalizing similar information, even if provided in different formats, correcting for the wide variation in data standards. Then, data will be run through correlation and trend analysis tools, revealing connections that can be used to draw statistically valid conclusions and develop robust hypotheses.162,163,164,165
Global HIT systems will allow physicians anywhere in the world to benefit from the latest, best available knowledge.166 Asthma biomarkers research goes global, as HIT systems link researchers, patients, biobanks, registries, and research procedures, even in the most remote locations, so endotyping—a central element of PM—becomes affordable and universal.
CONCLUSION
There are several arguments strongly supporting the asthma biomarkers as valuable tools to bring precision medicine closer to the asthma clinic: the evidence for endotype/biomarkers-driven treatment of type 2 asthma is accumulating, the unbiased data-driven models allow for a shift from investigator-driven hypothesis research to hypothesis-generating research, health information technology is quickly advancing and there is significant pressure and support from patients and society to implement precision medicine for asthma. On the other hand, several challenges need to be overcome in the near future, including biomarkers validation and qualification as pathway specific, translation of big data into clinical decision algorithms, variation between countries in IT resources, and limits of patient expertise in population subgroups. In addition, regulators and policy makers need strong economic arguments to implement the changes. Failure to balance data volume and quality to support material and measurable decisions in asthma care will undermine the success of big data initiatives.
ACKNOWLEDGMENTS
Ioana Agache wrote the review as part of the preparation for the research project PN-II-RU-TE-2014-4-2303.
Footnotes
There are no financial or other issues that might lead to conflict of interest.
References
- 1.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 2.Muraro A, Fokkens WJ, Pietikainen S, Borrelli D, Agache I, Bousquet J, et al. European symposium on precision medicine in allergy and airways diseases: report of the European Union Parliament Symposium (October 14, 2015) Allergy. 2016;71:583–587. doi: 10.1111/all.12819. [DOI] [PubMed] [Google Scholar]
- 3.Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. Lancet. 1902;160:1616–1620. [Google Scholar]
- 4.Muraro A, Lemanske RF, Jr, Hellings PW, Akdis CA, Bieber T, Casale TB, et al. Precision medicine in patients with allergic diseases: airway diseases and atopic dermatitis-PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2016;137:1347–1358. doi: 10.1016/j.jaci.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Agache I, Akdis CA. Endotypes of allergic diseases and asthma: an important step in building blocks for the future of precision medicine. Allergol Int. 2016;65:243–252. doi: 10.1016/j.alit.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 7.Agache I, Akdis C, Jutel M, Virchow JC. Untangling asthma phenotypes and endotypes. Allergy. 2012;67:835–846. doi: 10.1111/j.1398-9995.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- 8.Lötvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 9.Ray A, Oriss TB, Wenzel SE. Emerging molecular phenotypes of asthma. Am J Physiol Lung Cell Mol Physiol. 2015;308:L130–L140. doi: 10.1152/ajplung.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agache IO. From phenotypes to endotypes to asthma treatment. Curr Opin Allergy Clin Immunol. 2013;13:249–256. doi: 10.1097/ACI.0b013e32836093dd. [DOI] [PubMed] [Google Scholar]
- 11.Agache I, Sugita K, Morita H, Akdis M, Akdis CA. The complex type 2 endotype in allergy and asthma: from laboratory to bedside. Curr Allergy Asthma Rep. 2015;15:29. doi: 10.1007/s11882-015-0529-x. [DOI] [PubMed] [Google Scholar]
- 12.European Medicines Agency, Committee for Medicinal Products for Human Use. Zelboraf: summary of opinion. London: European Medicines Agency; 2011. [Google Scholar]
- 13.Maggon K. New drug approvals FDA/EMA in 2010: declining R&D productivity? Knol Publishing Guild (KPG); 2011. [Google Scholar]
- 14.Wagner JA. Overview of biomarkers and surrogate endpoints in drug development. Dis Markers. 2002;18:41–46. doi: 10.1155/2002/929274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodsaid FM, Frueh FW, Mattes W. Strategic paths for biomarker qualification. Toxicology. 2008;245:219–223. doi: 10.1016/j.tox.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 16.Agache IO. Endotype driven treatment of asthma. Curr Treat Options Allergy. 2014;1:198–212. [Google Scholar]
- 17.U.S. Food and Drug Administration. Guidance for industry: pharmacogenomic data submissions [Internet] Silver Spring (MD): U.S. Food and Drug Administration; 2005. Available from: https://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm079849.pdf. [Google Scholar]
- 18.European Medicines Agency. Qualification of novel methodologies for drug development: guidance to applicants [Internet] London: European Medicines Agency; 2014. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2009/10/WC500004201.pdf. [Google Scholar]
- 19.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–160. 160.e1–160.e9. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Kuo CS, Pavlidis S, Loza M, Baribaud F, Rowe A, Pandis I, et al. A transcriptome-driven analysis of epithelial brushings and bronchial biopsies to define asthma phenotypes in U-BIOPRED. Am J Respir Crit Care Med. 2017;195:443–455. doi: 10.1164/rccm.201512-2452OC. [DOI] [PubMed] [Google Scholar]
- 22.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicodemus-Johnson J, Naughton KA, Sudi J, Hogarth K, Naurekas ET, Nicolae DL, et al. Genome-wide methylation study identifies an IL-13-induced epigenetic signature in asthmatic airways. Am J Respir Crit Care Med. 2016;193:376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190:1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan X, Chu JH, Gomez J, Koenigs M, Holm C, He X, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med. 2015;191:1116–1125. doi: 10.1164/rccm.201408-1440OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, et al. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191:758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agache I, Strasser DS, Klenk A, Agache C, Farine H, Ciobanu C, et al. Serum IL-5 and IL-13 consistently serve as the best predictors for the blood eosinophilia phenotype in adult asthmatics. Allergy. 2016;71:1192–1202. doi: 10.1111/all.12906. [DOI] [PubMed] [Google Scholar]
- 28.Brasier AR, Victor S, Boetticher G, Ju H, Lee C, Bleecker ER, et al. Molecular phenotyping of severe asthma using pattern recognition of bronchoalveolar lavage-derived cytokines. J Allergy Clin Immunol. 2008;121:30–37.e6. doi: 10.1016/j.jaci.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little FF, Delgado DM, Wexler PJ, Oppenheim FG, Mitchell P, Feldman JA, et al. Salivary inflammatory mediator profiling and correlation to clinical disease markers in asthma. PLoS One. 2014;9:e84449. doi: 10.1371/journal.pone.0084449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjermer L, Lemiere C, Maspero J, Weiss S, Zangrilli J, Germinaro M. Reslizumab for inadequately controlled asthma with elevated blood eosinophil levels: a randomized phase 3 study. Chest. 2016;150:789–798. doi: 10.1016/j.chest.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 31.Bleecker ER, FitzGerald JM, Chanez P, Papi A, Weinstein SF, Barker P, et al. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high-dosage inhaled corticosteroids and long-acting β2-agonists (SIROCCO): a randomised, multicentre, placebo-controlled phase 3 trial. Lancet. 2016;388:2115–2127. doi: 10.1016/S0140-6736(16)31324-1. [DOI] [PubMed] [Google Scholar]
- 32.Pavord ID, Korn S, Howarth P, Bleecker ER, Buhl R, Keene ON, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–659. doi: 10.1016/S0140-6736(12)60988-X. [DOI] [PubMed] [Google Scholar]
- 33.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–993. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 35.Cabon Y, Molinari N, Marin G, Vachier I, Gamez AS, Chanez P, et al. Comparison of anti-interleukin-5 therapies in patients with severe asthma: global and indirect meta-analyses of randomized placebo-controlled trials. Clin Exp Allergy. 2017;47:129–138. doi: 10.1111/cea.12853. [DOI] [PubMed] [Google Scholar]
- 36.Brightling CE, Chanez P, Leigh R, O'Byrne PM, Korn S, She D, et al. Efficacy and safety of tralokinumab in patients with severe uncontrolled asthma: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir Med. 2015;3:692–701. doi: 10.1016/S2213-2600(15)00197-6. [DOI] [PubMed] [Google Scholar]
- 37.Corren J, Lemanske RF, Jr, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 38.Wenzel S, Castro M, Corren J, Maspero J, Wang L, Zhang B, et al. Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: a randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet. 2016;388:31–44. doi: 10.1016/S0140-6736(16)30307-5. [DOI] [PubMed] [Google Scholar]
- 39.Ledford D, Busse W, Trzaskoma B, Omachi TA, Rosén K, Chipps BE, et al. A randomized multicenter study evaluating Xolair persistence of response after long-term therapy. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.08.054. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 40.Djukanovic R, Hanania N, Busse W, Price D. IgE-mediated asthma: new revelations and future insights. Respir Med. 2016;112:128–129. doi: 10.1016/j.rmed.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 41.Hanania NA, Wenzel S, Rosén K, Hsieh HJ, Mosesova S, Choy DF, et al. Exploring the effects of omalizumab in allergic asthma: an analysis of biomarkers in the EXTRA study. Am J Respir Crit Care Med. 2013;187:804–811. doi: 10.1164/rccm.201208-1414OC. [DOI] [PubMed] [Google Scholar]
- 42.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370:1422–1431. doi: 10.1016/S0140-6736(07)61600-6. [DOI] [PubMed] [Google Scholar]
- 43.Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. 2012;130:647–654.e10. doi: 10.1016/j.jaci.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanemitsu Y, Matsumoto H, Mishima M KiHAC Respiratory Medicine Group. Factors contributing to an accelerated decline in pulmonary function in asthma. Allergol Int. 2014;63:181–188. doi: 10.2332/allergolint.13-RA-0670. [DOI] [PubMed] [Google Scholar]
- 45.Matsusaka M, Kabata H, Fukunaga K, Suzuki Y, Masaki K, Mochimaru T, et al. Phenotype of asthma related with high serum periostin levels. Allergol Int. 2015;64:175–180. doi: 10.1016/j.alit.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–394. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephan M, Suhling H, Schade J, Wittlake M, Tasic T, Klemann C, et al. Effects of dipeptidyl peptidase-4 inhibition in an animal model of experimental asthma: a matter of dose, route, and time. Physiol Rep. 2013;1:e00095. doi: 10.1002/phy2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravensberg AJ, Ricciardolo FL, van Schadewijk A, Rabe KF, Sterk PJ, Hiemstra PS, et al. Eotaxin-2 and eotaxin-3 expression is associated with persistent eosinophilic bronchial inflammation in patients with asthma after allergen challenge. J Allergy Clin Immunol. 2005;115:779–785. doi: 10.1016/j.jaci.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 49.Zietkowski Z, Tomasiak MM, Skiepko R, Bodzenta-Lukaszyk A. RANTES in exhaled breath condensate of stable and unstable asthma patients. Respir Med. 2008;102:1198–1202. doi: 10.1016/j.rmed.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Parker JM, Oh CK, LaForce C, Miller SD, Pearlman DS, Le C, et al. Safety profile and clinical activity of multiple subcutaneous doses of MEDI-528, a humanized anti-interleukin-9 monoclonal antibody, in two randomized phase 2a studies in subjects with asthma. BMC Pulm Med. 2011;11:14. doi: 10.1186/1471-2466-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang W, Smith SG, Salter B, Oliveria JP, Mitchell P, Nusca GM, et al. Allergen-induced increases in interleukin-25 and interleukin-25 receptor expression in mature eosinophils from atopic asthmatics. Int Arch Allergy Immunol. 2016;170:234–242. doi: 10.1159/000449248. [DOI] [PubMed] [Google Scholar]
- 52.Yi L, Cheng D, Zhang K, Huo X, Mo Y, Shi H, et al. Intelectin contributes to allergen-induced IL-25, IL-33, and TSLP expression and type 2 response in asthma and atopic dermatitis. Mucosal Immunol. 2017 doi: 10.1038/mi.2017.10. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allakhverdi Z, Comeau MR, Smith DE, Toy D, Endam LM, Desrosiers M, et al. CD34+ hemopoietic progenitor cells are potent effectors of allergic inflammation. J Allergy Clin Immunol. 2009;123:472–478. doi: 10.1016/j.jaci.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Mutalithas K, Guillen C, Day C, Brightling CE, Pavord ID, Wardlaw AJ. CRTH2 expression on T cells in asthma. Clin Exp Immunol. 2010;161:34–40. doi: 10.1111/j.1365-2249.2010.04161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong CK, Hu S, Cheung PF, Lam CW. Thymic stromal lymphopoietin induces chemotactic and prosurvival effects in eosinophils: implications in allergic inflammation. Am J Respir Cell Mol Biol. 2010;43:305–315. doi: 10.1165/rcmb.2009-0168OC. [DOI] [PubMed] [Google Scholar]
- 56.Ying S, O'Connor B, Ratoff J, Meng Q, Mallett K, Cousins D, et al. Thymic stromal lymphopoietin expression is increased in asthmatic airways and correlates with expression of Th2-attracting chemokines and disease severity. J Immunol. 2005;174:8183–8190. doi: 10.4049/jimmunol.174.12.8183. [DOI] [PubMed] [Google Scholar]
- 57.Hammad H, Kool M, Soullié T, Narumiya S, Trottein F, Hoogsteden HC, et al. Activation of the D prostanoid 1 receptor suppresses asthma by modulation of lung dendritic cell function and induction of regulatory T cells. J Exp Med. 2007;204:357–367. doi: 10.1084/jem.20061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoshino M, Nakagawa T, Sano Y, Hirai K. Effect of inhaled corticosteroid on an immunoreactive thymus and activation-regulated chemokine expression in the bronchial biopsies from asthmatics. Allergy. 2005;60:317–322. doi: 10.1111/j.1398-9995.2005.00694.x. [DOI] [PubMed] [Google Scholar]
- 59.Mutalithas K, Guillen C, Raport C, Kolbeck R, Soler D, Brightling CE, et al. Expression of CCR8 is increased in asthma. Clin Exp Allergy. 2010;40:1175–1185. doi: 10.1111/j.1365-2222.2010.03504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao SC, Cheng YC, Wang YC, Wang CW, Yang SM, Yu CK, et al. IL-19 induced Th2 cytokines and was up-regulated in asthma patients. J Immunol. 2004;173:6712–6718. doi: 10.4049/jimmunol.173.11.6712. [DOI] [PubMed] [Google Scholar]
- 61.Koh YI, Shim JU. Association between sputum natural killer T cells and eosinophilic airway inflammation in human asthma. Int Arch Allergy Immunol. 2010;153:239–248. doi: 10.1159/000314364. [DOI] [PubMed] [Google Scholar]
- 62.Préfontaine D, Lajoie-Kadoch S, Foley S, Audusseau S, Olivenstein R, Halayko AJ, et al. Increased expression of IL-33 in severe asthma: evidence of expression by airway smooth muscle cells. J Immunol. 2009;183:5094–5103. doi: 10.4049/jimmunol.0802387. [DOI] [PubMed] [Google Scholar]
- 63.Ueno T, Miyazaki E, Ando M, Nureki S, Kumamoto T. Osteopontin levels are elevated in patients with eosinophilic pneumonia. Respirology. 2010;15:1111–1121. doi: 10.1111/j.1440-1843.2010.01825.x. [DOI] [PubMed] [Google Scholar]
- 64.Gangwar RS, Minai-Fleminger Y, Seaf M, Gutgold A, Shikotra A, Barber C, et al. CD48 on blood leukocytes and in serum of asthma patients varies with severity. Allergy. 2016 doi: 10.1111/all.13082. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 65.Grotta MB, Squebola-Cola DM, Toro AA, Ribeiro MA, Mazon SB, Ribeiro JD, et al. Obesity increases eosinophil activity in asthmatic children and adolescents. BMC Pulm Med. 2013;13:39. doi: 10.1186/1471-2466-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curran CS, Bertics PJ. Lactoferrin regulates an axis involving CD11b and CD49d integrins and the chemokines MIP-1α and MCP-1 in GM-CSF-treated human primary eosinophils. J Interferon Cytokine Res. 2012;32:450–461. doi: 10.1089/jir.2011.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kelly EA, Koziol-White CJ, Clay KJ, Liu LY, Bates ME, Bertics PJ, et al. Potential contribution of IL-7 to allergen-induced eosinophilic airway inflammation in asthma. J Immunol. 2009;182:1404–1410. doi: 10.4049/jimmunol.182.3.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maazi H, Patel N, Sankaranarayanan I, Suzuki Y, Rigas D, Soroosh P, et al. ICOS: ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42:538–551. doi: 10.1016/j.immuni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fang P, Zhou L, Zhou Y, Kolls JK, Zheng T, Zhu Z. Immune modulatory effects of IL-22 on allergen-induced pulmonary inflammation. PLoS One. 2014;9:e107454. doi: 10.1371/journal.pone.0107454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartwig C, Munder A, Glage S, Wedekind D, Schenk H, Seifert R, et al. The histamine H4-receptor (H4 R) regulates eosinophilic inflammation in ovalbumin-induced experimental allergic asthma in mice. Eur J Immunol. 2015;45:1129–1140. doi: 10.1002/eji.201445179. [DOI] [PubMed] [Google Scholar]
- 71.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134:671–678.e4. doi: 10.1016/j.jaci.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gunawardhana LP, Gibson PG, Simpson JL, Benton MC, Lea RA, Baines KJ. Characteristic DNA methylation profiles in peripheral blood monocytes are associated with inflammatory phenotypes of asthma. Epigenetics. 2014;9:1302–1316. doi: 10.4161/epi.33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology. 2006;11:54–61. doi: 10.1111/j.1440-1843.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 74.Agache I. Non-eosinophilic asthma endotypes. Curr Treat Options Allergy. 2015;2:257–267. [Google Scholar]
- 75.Li Q, Baines KJ, Gibson PG, Wood LG. Changes in expression of genes regulating airway inflammation following a high-fat mixed meal in asthmatics. Nutrients. 2016;8:E30. doi: 10.3390/nu8010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agache I, Ciobanu C, Agache C, Anghel M. Increased serum IL-17 is an independent risk factor for severe asthma. Respir Med. 2010;104:1131–1137. doi: 10.1016/j.rmed.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 77.Gibson PG, Simpson JL, Saltos N. Heterogeneity of airway inflammation in persistent asthma: evidence of neutrophilic inflammation and increased sputum interleukin-8. Chest. 2001;119:1329–1336. doi: 10.1378/chest.119.5.1329. [DOI] [PubMed] [Google Scholar]
- 78.Hicks A, Goodnow R, Jr, Cavallo G, Tannu SA, Ventre JD, Lavelle D, et al. Effects of LTB4 receptor antagonism on pulmonary inflammation in rodents and non-human primates. Prostaglandins Other Lipid Mediat. 2010;92:33–43. doi: 10.1016/j.prostaglandins.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 79.Wood LG, Simpson JL, Hansbro PM, Gibson PG. Potentially pathogenic bacteria cultured from the sputum of stable asthmatics are associated with increased 8-isoprostane and airway neutrophilia. Free Radic Res. 2010;44:146–154. doi: 10.3109/10715760903362576. [DOI] [PubMed] [Google Scholar]
- 80.Bogaert P, Naessens T, De Koker S, Hennuy B, Hacha J, Smet M, et al. Inflammatory signatures for eosinophilic vs. neutrophilic allergic pulmonary inflammation reveal critical regulatory checkpoints. Am J Physiol Lung Cell Mol Physiol. 2011;300:L679–L690. doi: 10.1152/ajplung.00202.2010. [DOI] [PubMed] [Google Scholar]
- 81.Roussel L, Houle F, Chan C, Yao Y, Bérubé J, Olivenstein R, et al. IL-17 promotes p38 MAPK-dependent endothelial activation enhancing neutrophil recruitment to sites of inflammation. J Immunol. 2010;184:4531–4537. doi: 10.4049/jimmunol.0903162. [DOI] [PubMed] [Google Scholar]
- 82.Hsia BJ, Whitehead GS, Thomas SY, Nakano K, Gowdy KM, Aloor JJ, et al. Trif-dependent induction of Th17 immunity by lung dendritic cells. Mucosal Immunol. 2015;8:186–197. doi: 10.1038/mi.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bullens DM, Truyen E, Coteur L, Dilissen E, Hellings PW, Dupont LJ, et al. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Respir Res. 2006;7:135. doi: 10.1186/1465-9921-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busse WW, Holgate S, Kerwin E, Chon Y, Feng J, Lin J, et al. Randomized, double-blind, placebo-controlled study of brodalumab, a human anti-IL-17 receptor monoclonal antibody, in moderate to severe asthma. Am J Respir Crit Care Med. 2013;188:1294–1302. doi: 10.1164/rccm.201212-2318OC. [DOI] [PubMed] [Google Scholar]
- 85.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125:1028–1036.e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kikuchi S, Kikuchi I, Takaku Y, Kobayashi T, Hagiwara K, Kanazawa M, et al. Neutrophilic inflammation and CXC chemokines in patients with refractory asthma. Int Arch Allergy Immunol. 2009;149(Suppl 1):87–93. doi: 10.1159/000211379. [DOI] [PubMed] [Google Scholar]
- 87.Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. 2010;31:318–324. doi: 10.1016/j.it.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seys SF, Hox V, Van Gerven L, Dilissen E, Marijsse G, Peeters E, et al. Damage-associated molecular pattern and innate cytokine release in the airways of competitive swimmers. Allergy. 2015;70:187–194. doi: 10.1111/all.12540. [DOI] [PubMed] [Google Scholar]
- 89.Petty JM, Sueblinvong V, Lenox CC, Jones CC, Cosgrove GP, Cool CD, et al. Pulmonary stromal-derived factor-1 expression and effect on neutrophil recruitment during acute lung injury. J Immunol. 2007;178:8148–8157. doi: 10.4049/jimmunol.178.12.8148. [DOI] [PubMed] [Google Scholar]
- 90.Gao P, Gibson PG, Baines KJ, Yang IA, Upham JW, Reynolds PN, et al. Anti-inflammatory deficiencies in neutrophilic asthma: reduced galectin-3 and IL-1RA/IL-1β. Respir Res. 2015;16:5. doi: 10.1186/s12931-014-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in amish and Hutterite farm children. N Engl J Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wenzel SE, Barnes PJ, Bleecker ER, Bousquet J, Busse W, Dahlén SE, et al. A randomized, double-blind, placebo-controlled study of tumor necrosis factor-alpha blockade in severe persistent asthma. Am J Respir Crit Care Med. 2009;179:549–558. doi: 10.1164/rccm.200809-1512OC. [DOI] [PubMed] [Google Scholar]
- 93.Holgate ST, Noonan M, Chanez P, Busse W, Dupont L, Pavord I, et al. Efficacy and safety of etanercept in moderate-to-severe asthma: a randomised, controlled trial. Eur Respir J. 2011;37:1352–1359. doi: 10.1183/09031936.00063510. [DOI] [PubMed] [Google Scholar]
- 94.Bullens DM, Decraene A, Dilissen E, Meyts I, De Boeck K, Dupont LJ, et al. Type III IFN-lambda mRNA expression in sputum of adult and school-aged asthmatics. Clin Exp Allergy. 2008;38:1459–1467. doi: 10.1111/j.1365-2222.2008.03045.x. [DOI] [PubMed] [Google Scholar]
- 95.Djukanović R, Harrison T, Johnston SL, Gabbay F, Wark P, Thomson NC, et al. The effect of inhaled IFN-β on worsening of asthma symptoms caused by viral infections. A randomized trial. Am J Respir Crit Care Med. 2014;190:145–154. doi: 10.1164/rccm.201312-2235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matangkasombut P, Marigowda G, Ervine A, Idris L, Pichavant M, Kim HY, et al. Natural killer T cells in the lungs of patients with asthma. J Allergy Clin Immunol. 2009;123:1181–1185. doi: 10.1016/j.jaci.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chun E, Lee SH, Lee SY, Shim EJ, Cho SH, Min KU, et al. Toll-like receptor expression on peripheral blood mononuclear cells in asthmatics; implications for asthma management. J Clin Immunol. 2010;30:459–464. doi: 10.1007/s10875-009-9363-z. [DOI] [PubMed] [Google Scholar]
- 98.Månsson Kvarnhammar A, Tengroth L, Adner M, Cardell LO. Innate immune receptors in human airway smooth muscle cells: activation by TLR1/2, TLR3, TLR4, TLR7 and NOD1 agonists. PLoS One. 2013;8:e68701. doi: 10.1371/journal.pone.0068701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Møller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Børglum AD. Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008;63:1064–1069. doi: 10.1136/thx.2007.094128. [DOI] [PubMed] [Google Scholar]
- 100.Roponen M, Yerkovich ST, Hollams E, Sly PD, Holt PG, Upham JW. Toll-like receptor 7 function is reduced in adolescents with asthma. Eur Respir J. 2010;35:64–71. doi: 10.1183/09031936.00172008. [DOI] [PubMed] [Google Scholar]
- 101.Wood LG, Simpson JL, Wark PA, Powell H, Gibson PG. Characterization of innate immune signalling receptors in virus-induced acute asthma. Clin Exp Allergy. 2011;41:640–648. doi: 10.1111/j.1365-2222.2010.03669.x. [DOI] [PubMed] [Google Scholar]
- 102.Denlinger LC, Shi L, Guadarrama A, Schell K, Green D, Morrin A, et al. Attenuated P2X7 pore function as a risk factor for virus-induced loss of asthma control. Am J Respir Crit Care Med. 2009;179:265–270. doi: 10.1164/rccm.200802-293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 104.Tang H, Fang Z, Sun Y, Li B, Shi Z, Chen J, et al. YKL-40 in asthmatic patients, and its correlations with exacerbation, eosinophils and immunoglobulin E. Eur Respir J. 2010;35:757–760. doi: 10.1183/09031936.00034409. [DOI] [PubMed] [Google Scholar]
- 105.Koponen P, He Q, Helminen M, Nuolivirta K, Korppi M. Association of MBL2 polymorphism with asthma after bronchiolitis in infancy. Pediatr Int. 2012;54:619–622. doi: 10.1111/j.1442-200X.2012.03651.x. [DOI] [PubMed] [Google Scholar]
- 106.Baines KJ, Wright TK, Simpson JL, McDonald VM, Wood LG, Parsons KS, et al. Airway β-defensin-1 protein is elevated in COPD and severe asthma. Mediators Inflamm. 2015;2015:407271. doi: 10.1155/2015/407271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bratke K, Klug A, Julius P, Kuepper M, Lommatzsch M, Sparmann G, et al. Granzyme K: a novel mediator in acute airway inflammation. Thorax. 2008;63:1006–1011. doi: 10.1136/thx.2007.091215. [DOI] [PubMed] [Google Scholar]
- 108.Xiao Y, Motomura S, Deyev V, Podack ER. TNF superfamily member 13, APRIL, inhibits allergic lung inflammation. Eur J Immunol. 2011;41:164–171. doi: 10.1002/eji.201040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Croteau-Chonka DC, Qiu W, Martinez FD, Strunk RC, Lemanske RF, Jr, Liu AH, et al. Gene expression profiling in blood provides reproducible molecular insights into asthma control. Am J Respir Crit Care Med. 2017;195:179–188. doi: 10.1164/rccm.201601-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mackay RM, Grainge CL, Lau LC, Barber C, Clark HW, Howarth PH. Airway surfactant protein d deficiency in adults with severe asthma. Chest. 2016;149:1165–1172. doi: 10.1016/j.chest.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kazani S, Planaguma A, Ono E, Bonini M, Zahid M, Marigowda G, et al. Exhaled breath condensate eicosanoid levels associate with asthma and its severity. J Allergy Clin Immunol. 2013;132:547–553. doi: 10.1016/j.jaci.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Planagumà A, Kazani S, Marigowda G, Haworth O, Mariani TJ, Israel E, et al. Airway lipoxin A4 generation and lipoxin A4 receptor expression are decreased in severe asthma. Am J Respir Crit Care Med. 2008;178:574–582. doi: 10.1164/rccm.200801-061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vachier I, Bonnans C, Chavis C, Farce M, Godard P, Bousquet J, et al. Severe asthma is associated with a loss of LX4, an endogenous anti-inflammatory compound. J Allergy Clin Immunol. 2005;115:55–60. doi: 10.1016/j.jaci.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 114.Dauletbaev N, Lands LC. Could relative abundance of airway lipoxins be the clue to restore corticosteroid sensitivity in severe asthma? J Allergy Clin Immunol. 2016;137:1807–1808. doi: 10.1016/j.jaci.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 115.Gagliardo R, Gras D, La Grutta S, Chanez P, Di Sano C, Albano GD, et al. Airway lipoxin A4/formyl peptide receptor 2-lipoxin receptor levels in pediatric patients with severe asthma. J Allergy Clin Immunol. 2016;137:1796–1806. doi: 10.1016/j.jaci.2015.11.045. [DOI] [PubMed] [Google Scholar]
- 116.Chung KF. Lipoxins and epoxyeicosatrienoic acids. Potential for inhibitors of soluble epoxide hydrolase in severe asthma? Am J Respir Crit Care Med. 2014;190:848–850. doi: 10.1164/rccm.201409-1659ED. [DOI] [PubMed] [Google Scholar]
- 117.Levy BD, Kohli P, Gotlinger K, Haworth O, Hong S, Kazani S, et al. Protectin D1 is generated in asthma and dampens airway inflammation and hyperresponsiveness. J Immunol. 2007;178:496–502. doi: 10.4049/jimmunol.178.1.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gu Z, Lamont GJ, Lamont RJ, Uriarte SM, Wang H, Scott DA. Resolvin D1, resolvin D2 and maresin 1 activate the GSK3β anti-inflammatory axis in TLR4-engaged human monocytes. Innate Immun. 2016;22:186–195. doi: 10.1177/1753425916628618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aoki H, Hisada T, Ishizuka T, Utsugi M, Kawata T, Shimizu Y, et al. Resolvin E1 dampens airway inflammation and hyperresponsiveness in a murine model of asthma. Biochem Biophys Res Commun. 2008;367:509–515. doi: 10.1016/j.bbrc.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 120.Aoki H, Hisada T, Ishizuka T, Utsugi M, Ono A, Koga Y, et al. Protective effect of resolvin E1 on the development of asthmatic airway inflammation. Biochem Biophys Res Commun. 2010;400:128–133. doi: 10.1016/j.bbrc.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 121.Haworth O, Cernadas M, Yang R, Serhan CN, Levy BD. Resolvin E1 regulates interleukin 23, interferon-gamma and lipoxin A4 to promote the resolution of allergic airway inflammation. Nat Immunol. 2008;9:873–879. doi: 10.1038/ni.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Haworth O, Cernadas M, Levy BD. NK cells are effectors for resolvin E1 in the timely resolution of allergic airway inflammation. J Immunol. 2011;186:6129–6135. doi: 10.4049/jimmunol.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, Porter TF, et al. Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. J Exp Med. 2009;206:15–23. doi: 10.1084/jem.20081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiao C, Puddicombe SM, Field S, Haywood J, Broughton-Head V, Puxeddu I, et al. Defective epithelial barrier function in asthma. J Allergy Clin Immunol. 2011;128:549–556. 556.e1–556.e12. doi: 10.1016/j.jaci.2011.05.038. [DOI] [PubMed] [Google Scholar]
- 125.de Boer WI, Sharma HS, Baelemans SM, Hoogsteden HC, Lambrecht BN, Braunstahl GJ. Altered expression of epithelial junctional proteins in atopic asthma: possible role in inflammation. Can J Physiol Pharmacol. 2008;86:105–112. doi: 10.1139/y08-004. [DOI] [PubMed] [Google Scholar]
- 126.Wawrzyniak P, Wawrzyniak M, Wanke K, Sokolowska M, Bendelja K, Rückert B, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. 2017;139:93–103. doi: 10.1016/j.jaci.2016.03.050. [DOI] [PubMed] [Google Scholar]
- 127.Cox G, Thomson NC, Rubin AS, Niven RM, Corris PA, Siersted HC, et al. Asthma control during the year after bronchial thermoplasty. N Engl J Med. 2007;356:1327–1337. doi: 10.1056/NEJMoa064707. [DOI] [PubMed] [Google Scholar]
- 128.Castro M, Rubin AS, Laviolette M, Fiterman J, De Andrade Lima M, Shah PL, et al. Effectiveness and safety of bronchial thermoplasty in the treatment of severe asthma: a multicenter, randomized, double-blind, sham-controlled clinical trial. Am J Respir Crit Care Med. 2010;181:116–124. doi: 10.1164/rccm.200903-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doeing DC, Husain AN, Naureckas ET, White SR, Hogarth DK. Bronchial thermoplasty failure in severe persistent asthma: a case report. J Asthma. 2013;50:799–801. doi: 10.3109/02770903.2013.796974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilson SJ, Ward JA, Sousa AR, Corfield J, Bansal AT, De Meulder B, et al. Severe asthma exists despite suppressed tissue inflammation: findings of the U-BIOPRED study. Eur Respir J. 2016;48:1307–1319. doi: 10.1183/13993003.01129-2016. [DOI] [PubMed] [Google Scholar]
- 131.Hoshino M, Ohtawa J, Akitsu K. Association of airway wall thickness with serum periostin in steroid-naive asthma. Allergy Asthma Proc. 2016;37:225–230. doi: 10.2500/aap.2016.37.3945. [DOI] [PubMed] [Google Scholar]
- 132.Lee YM, Park JS, Hwang JH, Park SW, Uh ST, Kim YH, et al. High-resolution CT findings in patients with near-fatal asthma: comparison of patients with mild-to-severe asthma and normal control subjects and changes in airway abnormalities following steroid treatment. Chest. 2004;126:1840–1848. doi: 10.1378/chest.126.6.1840. [DOI] [PubMed] [Google Scholar]
- 133.Aysola RS, Hoffman EA, Gierada D, Wenzel S, Cook-Granroth J, Tarsi J, et al. Airway remodeling measured by multidetector CT is increased in severe asthma and correlates with pathology. Chest. 2008;134:1183–1191. doi: 10.1378/chest.07-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gonem S, Hardy S, Buhl N, Hartley R, Soares M, Kay R, et al. Characterization of acinar airspace involvement in asthmatic patients by using inert gas washout and hyperpolarized (3)helium magnetic resonance. J Allergy Clin Immunol. 2016;137:417–425. doi: 10.1016/j.jaci.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 135.Ueda T, Niimi A, Matsumoto H, Takemura M, Hirai T, Yamaguchi M, et al. Role of small airways in asthma: investigation using high-resolution computed tomography. J Allergy Clin Immunol. 2006;118:1019–1025. doi: 10.1016/j.jaci.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 136.Hogg JC, McDonough JE, Suzuki M. Small airway obstruction in COPD: new insights based on micro-CT imaging and MRI imaging. Chest. 2013;143:1436–1443. doi: 10.1378/chest.12-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gagliardo R, La Grutta S, Chanez P, Profita M, Paternò A, Cibella F, et al. Non-invasive markers of airway inflammation and remodeling in childhood asthma. Pediatr Allergy Immunol. 2009;20:780–790. doi: 10.1111/j.1399-3038.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- 138.Dolhnikoff M, da Silva LF, de Araujo BB, Gomes HA, Fernezlian S, Mulder A, et al. The outer wall of small airways is a major site of remodeling in fatal asthma. J Allergy Clin Immunol. 2009;123:1090–1097.e1. doi: 10.1016/j.jaci.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 139.Obase Y, Rytilä P, Metso T, Pelkonen AS, Tervahartiala T, Turpeinen M, et al. Effects of inhaled corticosteroids on metalloproteinase-8 and tissue inhibitor of metalloproteinase-1 in the airways of asthmatic children. Int Arch Allergy Immunol. 2010;151:247–254. doi: 10.1159/000242362. [DOI] [PubMed] [Google Scholar]
- 140.Mukhopadhyay S, Sypek J, Tavendale R, Gartner U, Winter J, Li W, et al. Matrix metalloproteinase-12 is a therapeutic target for asthma in children and young adults. J Allergy Clin Immunol. 2010;126:70–76.e16. doi: 10.1016/j.jaci.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 141.Chen J, Deng L, Dreymüller D, Jiang X, Long J, Duan Y, et al. A novel peptide ADAM8 inhibitor attenuates bronchial hyperresponsiveness and Th2 cytokine mediated inflammation of murine asthmatic models. Sci Rep. 2016;6:30451. doi: 10.1038/srep30451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–871. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 143.Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34:757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- 144.Kim SH, Pei QM, Jiang P, Yang M, Qian XJ, Liu JB. Effect of active vitamin D3 on VEGF-induced ADAM33 expression and proliferation in human airway smooth muscle cells: implications for asthma treatment. Respir Res. 2017;18:7. doi: 10.1186/s12931-016-0490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res. 2006;7:91. doi: 10.1186/1465-9921-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Detoraki A, Granata F, Staibano S, Rossi FW, Marone G, Genovese A. Angiogenesis and lymphangiogenesis in bronchial asthma. Allergy. 2010;65:946–958. doi: 10.1111/j.1398-9995.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- 147.Fujita H, Chalubinski M, Rhyner C, Indermitte P, Meyer N, Ferstl R, et al. Claudin-1 expression in airway smooth muscle exacerbates airway remodeling in asthmatic subjects. J Allergy Clin Immunol. 2011;127:1612–1621.e8. doi: 10.1016/j.jaci.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 148.Liu G, Cooley MA, Jarnicki AG, Hsu AC, Nair PM, Haw TJ, et al. Fibulin-1 regulates the pathogenesis of tissue remodeling in respiratory diseases. JCI Insight. 2016;1:e86380. doi: 10.1172/jci.insight.86380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lau JY, Oliver BG, Baraket M, Beckett EL, Hansbro NG, Moir LM, et al. Fibulin-1 is increased in asthma--a novel mediator of airway remodeling? PLoS One. 2010;5:e13360. doi: 10.1371/journal.pone.0013360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Druilhe A, Zahm JM, Benayoun L, El Mehdi D, Grandsaigne M, Dombret MC, et al. Epithelium expression and function of retinoid receptors in asthma. Am J Respir Cell Mol Biol. 2008;38:276–282. doi: 10.1165/rcmb.2006-0453OC. [DOI] [PubMed] [Google Scholar]
- 151.Grainge C, Dulay V, Ward J, Sammut D, Davies E, Green B, et al. Resistin-like molecule-β is induced following bronchoconstriction of asthmatic airways. Respirology. 2012;17:1094–1100. doi: 10.1111/j.1440-1843.2012.02215.x. [DOI] [PubMed] [Google Scholar]
- 152.James AJ, Reinius LE, Verhoek M, Gomes A, Kupczyk M, Hammar U, et al. Increased YKL-40 and chitotriosidase in asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;193:131–142. doi: 10.1164/rccm.201504-0760OC. [DOI] [PubMed] [Google Scholar]
- 153.Samitas K, Poulos N, Semitekolou M, Morianos I, Tousa S, Economidou E, et al. Activin-A is overexpressed in severe asthma and is implicated in angiogenic processes. Eur Respir J. 2016;47:769–782. doi: 10.1183/13993003.00437-2015. [DOI] [PubMed] [Google Scholar]
- 154.Simoes DC, Xanthou G, Petrochilou K, Panoutsakopoulou V, Roussos C, Gratziou C. Osteopontin deficiency protects against airway remodeling and hyperresponsiveness in chronic asthma. Am J Respir Crit Care Med. 2009;179:894–902. doi: 10.1164/rccm.200807-1081OC. [DOI] [PubMed] [Google Scholar]
- 155.van Staa TP, Goldacre B, Buchan I, Smeeth L. Big health data: the need to earn public trust. BMJ. 2016;354:i3636. doi: 10.1136/bmj.i3636. [DOI] [PubMed] [Google Scholar]
- 156.Schneeweiss S. Learning from big health care data. N Engl J Med. 2014;370:2161–2163. doi: 10.1056/NEJMp1401111. [DOI] [PubMed] [Google Scholar]
- 157.Williams SM, Moore JH. Big data analysis on autopilot? BioData Min. 2013;6:22. doi: 10.1186/1756-0381-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chaudhry B, Wang J, Wu S, Maglione M, Mojica W, Roth E, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med. 2006;144:742–752. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- 159.Blumenthal D. Stimulating the adoption of health information technology. N Engl J Med. 2009;360:1477–1479. doi: 10.1056/NEJMp0901592. [DOI] [PubMed] [Google Scholar]
- 160.Gibson CJ, Dixon BE, Abrams K. Convergent evolution of health information management and health informatics: a perspective on the future of information professionals in health care. Appl Clin Inform. 2015;6:163–184. doi: 10.4338/ACI-2014-09-RA-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Milgrom H, Tran ZV. The rise of health information technology. Curr Opin Allergy Clin Immunol. 2010;10:178–180. doi: 10.1097/ACI.0b013e32833954ac. [DOI] [PubMed] [Google Scholar]
- 162.Howard R, Rattray M, Prosperi M, Custovic A. Distinguishing asthma phenotypes using machine learning approaches. Curr Allergy Asthma Rep. 2015;15:38. doi: 10.1007/s11882-015-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Custovic A, Ainsworth J, Arshad H, Bishop C, Buchan I, Cullinan P, et al. The Study Team for Early Life Asthma Research (STELAR) consortium ‘Asthma e-lab’: team science bringing data, methods and investigators together. Thorax. 2015;70:799–801. doi: 10.1136/thoraxjnl-2015-206781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matui P, Wyatt JC, Pinnock H, Sheikh A, McLean S. Computer decision support systems for asthma: a systematic review. NPJ Prim Care Respir Med. 2014;24:14005. doi: 10.1038/npjpcrm.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Morrison D, Wyke S, Agur K, Cameron EJ, Docking RI, Mackenzie AM, et al. Digital asthma self-management interventions: a systematic review. J Med Internet Res. 2014;16:e51. doi: 10.2196/jmir.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haux R. Health information systems - past, present, future. Int J Med Inform. 2006;75:268–281. doi: 10.1016/j.ijmedinf.2005.08.002. [DOI] [PubMed] [Google Scholar]