Abstract
AIM
To investigate the potential effect of curcumin on hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) and the underlying mechanism.
METHODS
A HepG2.2.15 cell line stably transfected with HBV was treated with curcumin, and HBV surface antigen (HBsAg) and e antigen (HBeAg) expression levels were assessed by ELISA. Intracellular HBV DNA replication intermediates and cccDNA were detected by Southern blot and real-time PCR, respectively. The acetylation levels of histones H3 and H4 were measured by Western blot. H3/H4-bound cccDNA was detected by chromatin immunoprecipitation (ChIP) assays. The deacetylase inhibitors trichostatin A and sodium butyrate were used to study the mechanism of action for curcumin. Additionally, short interfering RNAs (siRNAs) targeting HBV were tested along with curcumin.
RESULTS
Curcumin treatment led to time- and dose-dependent reductions in HBsAg and HBeAg expression and significant reductions in intracellular HBV DNA replication intermediates and HBV cccDNA. After treatment with 20 μmol/L curcumin for 2 d, HBsAg and cccDNA levels in HepG2.2.15 cells were reduced by up to 57.7% (P < 0.01) and 75.5% (P < 0.01), respectively, compared with levels in non-treated cells. Meanwhile, time- and dose-dependent reductions in the histone H3 acetylation levels were also detected upon treatment with curcumin, accompanied by reductions in H3- and H4-bound cccDNA. Furthermore, the deacetylase inhibitors trichostatin A and sodium butyrate could block the effects of curcumin. Additionally, transfection of siRNAs targeting HBV enhanced the inhibitory effects of curcumin.
CONCLUSION
Curcumin inhibits HBV gene replication via down-regulation of cccDNA-bound histone acetylation and has the potential to be developed as a cccDNA-targeting antiviral agent for hepatitis B.
Keywords: Curcumin, Hepatitis B virus, Covalently closed circular DNA, Histone deacetylation
Core tip: We showed that curcumin inhibited hepatitis B virus (HBV) replication and expression via reductions in covalently closed circular DNA-bound histone acetylation. Furthermore, siRNAs targeting HBV acted synergistically with curcumin, resulting in enhanced inhibition of HBV.
INTRODUCTION
Hepatitis B virus (HBV) is a species of the genus Orthohepadnavirus. HBV infection leads to severe diseases, including hepatitis, liver cirrhosis and hepatocellular carcinoma[1]. HBV has also been suggested to be involved in the development of pancreatic cancer[2]. Approximately 350 million individuals are infected with HBV, and more than 0.6 million people with HBV infection die every year as a result of end-stage liver disease and hepatocellular carcinoma worldwide[1,3]. The HBV genome is made of partially double-stranded relaxed circular DNA (rcDNA), 3020-3320 bp in size[4]. After the virus infects hepatocytes, rcDNA is released into the nucleus and converted to covalently closed circular DNA (cccDNA), which, along with histone binding, constitutes a minichromosome. The mean copies of cccDNA per HBV-infected hepatocyte range from 5 to 50[5,6]. Moreover, cccDNA serves as the critical template for viral replication and mRNA synthesis, which is the source of persistent and recurrent HBV infection. Nucleos(t)ide analogues (NAs) can efficiently inhibit HBV replication; however, they do not promote clearance of residual cccDNA[7].
Several treatment strategies are available to target cccDNA by inhibiting cccDNA minichromosome formation or inducing cccDNA degradation. Disubstituted sulfonamide (DSS) compounds have been reported to be specific inhibitors that prevent rcDNA conversion into cccDNA; however, DSS cannot promote cccDNA decay[8]. Lucifora et al[9] showed that inducing apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3A (APOBEC3A) and 3B (APOBEC3B) cytidine deaminase by interferon-α and lymphotoxin-β receptor activation induces cccDNA degradation via cytidine deamination and apurinic/apyrimidinic site formation. However, the absence of specificity of these cytidine deaminases results in genomic damage and cell-cycle arrest[10]. Recently, with the aid of DNA-cleaving enzymes, including zinc-finger nucleases (ZFN), TAL effector nucleases (TALENs), and CRISPR-associated system 9 (Cas9) proteins, specific targeting of HBV cccDNA was shown to cleave cccDNA[11-15]. Nevertheless, chronic expression of enzymes leads to off-target cleavage at homologous sequences in the human genome and represents a major limitation.
Furthermore, cccDNA-bound acetylated histones can modulate HBV replication and expression[16,17]. Hepatitis B virus X (HBx) protein can be recruited onto a cccDNA minichromosome to accelerate acetylation. Using a cccDNA chromatin immunoprecipitation (ChIP)-Seq assay, Tropberger et al[18] reported that low levels of histone posttranslational modifications (PTMs) were associated with transcriptional repression and promoter silencing.
Curcumin [1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione] was isolated from the rhizome of Curcuma longa L. (Zingiberaceae family), which exhibits antimicrobial activities against various bacteria, viruses, fungi, and parasites[19-23]. Curcumin can inhibit HBV via down-regulation of the gluconeogenesis gene coactivator PGC-1α[24] or trans-activation of transcription and increased stability of p53[25].
Based on findings that curcumin can inhibit p300 histone acetyltransferase activity[26,27], we hypothesized that deacetylation of cccDNA-bound histones may contribute to the inhibitory activities of curcumin on HBV. Therefore, the effects of curcumin on cccDNA-bound histones and on steady-state levels of HBV cccDNA were investigated in detail in the present study.
MATERIALS AND METHODS
Cell culture and transfection
HepG2.2.15 cells (an HBV stably transfected human hepatocarcinoma cell line) were maintained in DMEM medium (Gibco, Carlsbad, CA, United States) supplemented with 10% foetal bovine serum (Gibco), 1% GlutaMAX-I (Gibco) and 1% MEM Non-Essential Amino Acids Solution (Gibco). Transfection of siRNAs into HepG2.2.15 cells was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). The sequences were 5’-GAAUCCUCACAAUACCGCAtt and 5’-UGAGAGUCCAAGAGUCCUCtt for HBx-siRNA and 5’-GAAUCCUCACAAUACCGCAtt and 5’-UGCGGUAUU GUGAGGAUUCtt for hepatitis B virus S antigen (HBs)-siRNA[28].
Curcumin and deacetylase inhibitor treatments
Cells were seeded at approximately 60%-80% confluence 24 h prior to treatment with different concentrations of curcumin (Sigma, St. Louis, MO, United States) dissolved in dimethyl sulphoxide (DMSO). For histone acetylation blocking assays, cells were co-treated with 20 μmol/L curcumin and either 5 mmol/L sodium butyrate (Sigma) or 1 μmol/L trichostatin A (TSA, Sigma).
Nucleoprotein extraction and Western blot analysis
Nucleoproteins were extracted using a Nucleoprotein Extraction Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. For Western blot analysis, proteins were subjected to SDS-polyacrylamide gel electrophoresis on 12.5% gels and were then electrophoretically transferred to nitrocellulose membranes (Millipore, Billerica, MA, United States). The membranes were blocked with 5% non-fat milk in Tris-buffered saline (TBS; pH 7.5) with 0.05% Tween 20 for 2 h at RT and were then probed with rabbit polyclonal anti-acetyl-histone H3 (Abnova, diluted 1:1000) overnight at 4 °C. Mouse monoclonal anti-histone H3 (Beyotime Biotechnology, diluted 1:1000) served as an internal control protein. Horseradish peroxidase-conjugated goat anti-mouse antibody (Biosharp, 1:5000) was used as a secondary antibody. Protein brands were visualized by enhanced chemiluminescence (ECL) using an ECL kit (Millipore).
Extraction and quantification of HBV cccDNA
HepG2.2.15 cells were lysed in 800 μL of lysis buffer [50 mmol/L Tris-HCl (pH 8.0), 10 mmol/L EDTA, 150 mmol/L NaCl and 1% SDS] and incubated for 30 min at 4 °C. Cell lysates were adjusted to 0.5 mol/L KCl and centrifuged for 1 min at 10000 g to precipitate protein-bound DNA. Supernatants were digested with 0.5 mg/mL proteinase K for 2 h at 55 °C. The cccDNA was purified by phenol/chloroform (1:1) extraction and isopropanol precipitation in the presence of 15 μg of tRNA and 200 mM NaAc (pH 5.2).
Purified DNA was digested with Plasmid-Safe ATP-Dependent DNase (Epicenter, Madison, WI, United States) to degrade contaminating HBV inserted in cellular genomic DNA and OC (open circular) species and was then subjected to PCR amplification to select HBV cccDNA forms, as previously described[15]. The cccDNA was later subjected to real-time-PCR using SYBR Green Real-time PCR Master Mix (Roche, Mannheim, Germany) and cccDNA-specific primers: 5’-TGCACTTCGCTTCACCT (forward) and 5’-AGGGGC ATTTGGTGGTC (reverse). PCR was performed using an Applied Biosystems StepOne Real-Time PCR System. cccDNA copy numbers were quantified according to a standard curve generated from an HBV plasmid in a concentration range of 102-108 copies.
Extraction and quantification of HBV mRNA
Total RNA was extracted from HepG2.2.15 cells using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions and was then subjected to real-time-PCR using primers for cccDNA quantification; β-actin mRNA transcript levels were used to normalize the expression of each RNA sample.
cccDNA acetyl-histone H3 and acetyl-histone H4 ChIP assays
cccDNA ChIP assays were performed using EpiQuik Acetyl-Histone H3 ChIP and EpiQuik Acetyl-Histone H4 ChIP kits (EpiGentek, Farmingdale, NY, United States). Briefly, cells were collected and in vivo cross-linked in fresh culture medium containing 1% formaldehyde for 10 min at RT and were then lysed in 200 μL CP3A for 10 min at RT to isolate nuclear pellets. Chromatin solutions were sonicated for 4 pulses of 12 s each at level 5 using a Branson Microtip probe, followed by a 40-s rest on ice between each pulse to generate 200- to 1000-bp DNA fragments. Supernatants were diluted with CP4 at a 1:1 ratio, and 5 μL was removed as “input DNA”. Chromatin was then subjected to immunoprecipitation for 1 h at RT using strip wells pre-coated with antibodies specific to acetyl-histone H3, acetyl-histone H4 or normal mouse IgG. After six washes with CP1, immunoprecipitated chromatins and input DNA coated on the strip wells were digested with proteinase K and then purified using collection tubes. Purified DNA was subjected to Plasmid-Safe ATP-Dependent DNase digestion and real-time PCR amplification, as described above.
Quantification of HBV antigens
Culture supernatants of HepG2.2.15 cells were harvested and analysed for HBV surface antigen (HBsAg) and e antigen (HBeAg) levels using a Thermo Scientific Multiskan FC Microplate Photometer and ELISA kits (InTec, Xiamen, China).
Purification of HBV DNA from intracellular core particles and Southern blot analysis
HepG2.2.15 cells were washed twice with ice-cold PBS and lysed in 800 μL of lysis buffer [50 mmol/L Tris-HCl (pH 7.4), 1 mmol/L EDTA and 1% NP-40]. Cell lysates were centrifuged for 1 min at 10000 g to precipitate cell nuclei. Cellular genomic DNA and cccDNA were removed by the addition of 10 mM MgCl2 and 100 μg/mL DNase I, and the mixture was incubated for 30 min at 37 °C. Digestion was stopped by the addition of 25 mmol/L EDTA (pH 8.0). Proteins were digested with 0.5 mg/mL proteinase K and 1% SDS for 2 h at 55 °C. HBV DNA from intracellular core particles was purified by phenol/chloroform (1:1) extraction and isopropanol precipitation in the presence of 15 μg of tRNA and 200 mmol/L NaAc (pH 5.2)[28,29].
For Southern blot analysis, HBV DNA was subjected to agarose gel electrophoresis, followed by denaturation and transfer to nylon membranes using a Model 785 Vacuum Blotter (Bio-Rad, Hercules, CA, United States). DNA was fixed to membranes using an Ultraviolet Crosslinker (UVP, Upland, CA, United States). DNA hybridization and detection were performed using the DIG High Prime DNA Labelling and Detection Starter Kit II (Roche).
Statistical analysis
Statistical analyses were performed using unpaired t-tests. The results were evaluated with GraphPad Prism 5 and are expressed as the mean ± SD. P values < 0.05 (a) or < 0.01 (b) were set as the level of significance.
RESULTS
Curcumin inhibits HBV replication and expression
To confirm whether curcumin directly inhibits HBV expression, HepG2.2.15 cells were treated with various concentrations of curcumin for three consecutive days. Cell cytotoxicity was assessed using the CCK-8 assay, which revealed that there was no detectable toxic effect when cells were treated with less than 20 μmol/L curcumin (Figure 1). Cell culture supernatants from each day were collected and analysed for levels of HBsAg and HBeAg. Curcumin decreased HBsAg levels both dose- and time-dependently; HBsAg levels were reduced by up to 57.7%, 2 d after treatment with 20 μmol/L curcumin (Figure 2A). HBeAg was not reduced by 20 μmol/L of curcumin (Figure 2B). HBV cccDNA and HBV DNA from intracellular core particles were assayed 2 d after treatment with 20 μmol/L curcumin. RT-PCR and Southern blot experiments revealed strong reductions in cccDNA (75.5%; Figure 2C), mRNA (24.4%; Figure 2D) and HBV replication intermediates (Figure 2E).
Figure 1.
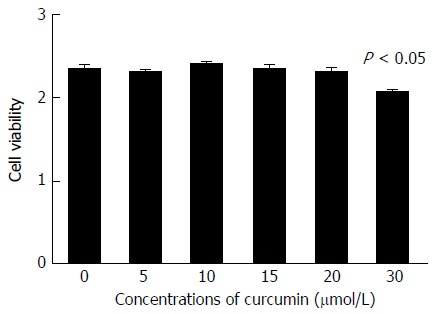
Cell cytotoxicity of curcumin. HepG2.2.15 cells were treated with 0, 5, 10, 15, 20 or 30 μmol/L curcumin for 2 d and then subjected to CCK-8 assay to detect toxic effect. The experiment was performed in duplicate and repeated at least three times.
Figure 2.
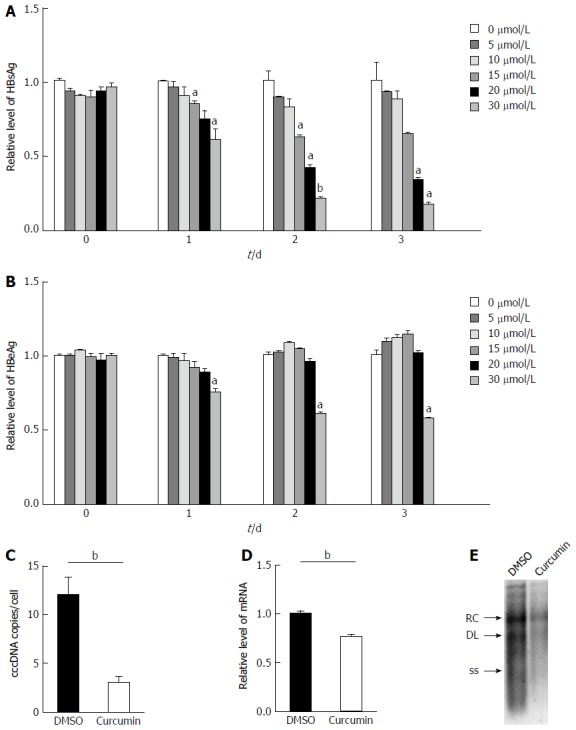
Curcumin inhibits hepatitis B virus replication and expression. A: HepG2.215 cells were treated with 0, 5, 10, 15, 20 or 30 μmol/L curcumin for three consecutive days. Culture medium from each day was collected and analysed for levels of HBsAg; B: Culture medium from each day was collected and analysed for levels of HBeAg; C: Hepatitis B virus (HBV) cccDNA accumulation in HepG2.215 cells treated with 20 μmol/L curcumin or DMSO for 2 d. HBV cccDNA was digested with Plasmid-Safe ATP-Dependent DNase to degrade contaminating HBV that had inserted in cellular genomic DNA and OC species and was then subjected to PCR amplification to amplify HBV cccDNA forms. Results are expressed as numbers of cccDNA copies per cell; D: Total RNA was extracted from HepG2.215 cells treated with 20 μmol/L curcumin or DMSO for 2 d and was subjected to real-time PCR to quantify HBV mRNA transcript levels; E: HBV DNA was extracted from intracellular core particles in HepG2.215 cells treated with 20 μmol/L curcumin or DMSO for 2 d. Southern blot analysis of HBV DNA replicative intermediates. RC, DL and SS represent relaxed circular, double linear and single-stranded forms of HBV DNA, respectively. All experiments were repeated at least three times; ELISA and RT-PCR were performed in duplicate. aP < 0.05; bP < 0.01. cccDNA: Covalently closed circular DNA; DMSO: Dimethyl sulphoxide.
Curcumin mediates the deacetylation of chromosomal and cccDNA-bound histones
To investigate the effects of curcumin on chromosomal histone acetylation, equivalent numbers of HepG2.2.15 cells were treated with various concentrations of curcumin for 2 d or with 20 μmol/L curcumin for different durations. As shown in Figure 3A and B, curcumin decreased histone acetylation levels both dose- and time-dependently. We found that 5 μmol/L curcumin was sufficient to decrease histone acetylation. When cells were treated with 20 μmol/L curcumin, acetylated histone H3 was reduced 30 min after treatment. Using a cccDNA ChIP assay, we found that the acetylation levels of cccDNA-bound histone H3 and histone H4 were significantly reduced when cells were treated with 20 μmol/L curcumin (Figure 3C). This finding is similar to our observed reduction of chromosomal histone acetylation.
Figure 3.
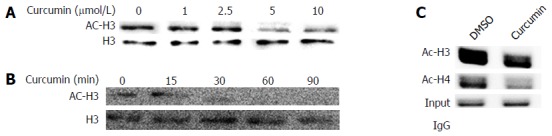
Curcumin mediates chromosomal and covalently closed circular DNA-bound histone deacetylation. A: HepG2.215 cells were treated with 0, 1, 2.5, 5 or 10 μmol/L curcumin and incubated at 37 °C for 2 d. The acetylation status of cellular H3 histones was analysed by Western blot; B: HepG2.215 cells were treated with 20 μmol/L curcumin for the indicated periods of time; C: ChIP on HepG2.215 cells treated with 20 μmol/L curcumin or DMSO for 2 d was performed using specific antibodies to acetyl-histone H3 (AcH3), acetyl-histone H4 (AcH4) or control IgG. Immunoprecipitated chromatins were digested with Plasmid-Safe ATP-Dependent DNase to degrade contaminating HBV that had inserted in cellular genomic DNA and OC species and then were subjected to PCR amplification to select HBV cccDNA forms. All experiments were repeated at least three times. cccDNA: Covalently closed circular DNA; HBV: Hepatitis B virus; DMSO: Dimethyl sulphoxide.
Histone deacetylase inhibitors block the inhibitory effect of curcumin
The histone deacetylase inhibitors sodium butyrate and TSA were used to test whether curcumin inhibits HBV by decreasing histone acetylation. HepG2.2.15 cells were treated with 20 μmol/L curcumin for 2 d, which resulted in significant reductions in the levels of HBsAg (Figure 4A) and cccDNA (Figure 4B). HBsAg and cccDNA levels increased by 31.3% and 2.4-fold, respectively, when cells were co-treated with curcumin and sodium butyrate compared with cells treated with curcumin alone. The histone deacetylase inhibitor TSA also partially blocked the inhibitory effect of curcumin on HBV, although the effect was much weaker than that of sodium butyrate. These findings suggest that curcumin inhibits HBV by inducing histone deacetylation.
Figure 4.
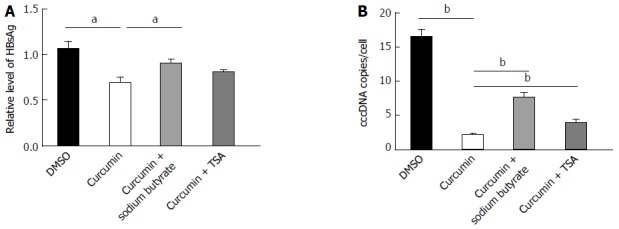
Histone deacetylase inhibitors block the inhibitory effect of curcumin. A: HepG2.215 cells were treated with 20 μmol/L curcumin alone or with either 5 mmol/L sodium butyrate or 1 μmol/L TSA for 2 d. Medium was collected and analysed for levels of HBsAg; B: HBV cccDNA was extracted and subjected to real-time qPCR. All experiments were performed in duplicate and repeated at least three times. aP < 0.05; bP < 0.01. cccDNA: Covalently closed circular DNA; DMSO: Dimethyl sulphoxide; TSA: Trichostatin A; HBV: Hepatitis B virus; HBsAg: HBV surface antigen.
siRNAs against HBV enhance the inhibitory effects of curcumin
Because HBx also regulates epigenetic modifications of cccDNA-bound histones[16], HBx-siRNA and HBs-siRNA were used to enhance the inhibitory effects of curcumin. HepG2.2.15 cells were transfected with 20 nmol/L siRNAs and treated with 20 μmol/L curcumin for 2 subsequent days. Transfection with siRNAs alone significantly reduced HBsAg expression to a level below the limit of detection (Figure 5A). Moreover, a further reduction was observed in HBV DNA levels from intracellular core particles extracted from cells that received combined treatment with siRNAs and curcumin compared with cells treated with curcumin alone (Figure 5B).
Figure 5.
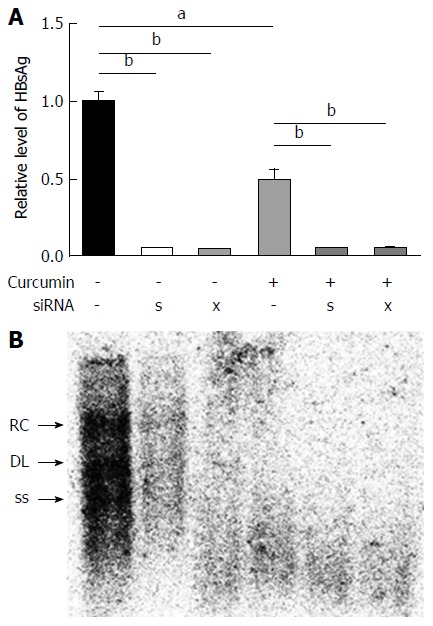
siRNAs against hepatitis B virus enhance the inhibitory effects of curcumin. HepG2.215 cells were transfected with 20 nmol/L siRNAs or negative control (HC) siRNA and were treated with 20 μmol/L curcumin or dimethyl sulphoxide for the next 2 d. HBsAg in cell culture supernatants and intracellular HBV replicative intermediates were detected by ELISA (A) and Southern blot analysis (B), respectively. All experiments were repeated at least three times; ELISA was performed in duplicate. aP < 0.05; bP < 0.01. HBV: Hepatitis B virus; HBsAg: HBV surface antigen.
DISCUSSION
The present study showed that curcumin not only inhibited intracellular HBV replication and HBsAg and HBeAg expression but also exhibited potent inhibitory effects on HBV cccDNA. Moreover, curcumin also reduced levels of both chromosomal and cccDNA-bound histones H3/H4, and addition of the histone deacetylase inhibitors sodium butyrate and TSA blocked the inhibitory effects of curcumin on HBV. These findings suggest that curcumin may induce the deacetylation of cccDNA-bound histones H3/H4, disrupt the steady state of HBV cccDNA and lead to the potent inhibition of HBV mRNA transcription and protein expression, along with a reduction in DNA replication. Furthermore, enhanced inhibition of intracellular HBV replication was revealed when curcumin was combined with siRNAs against HBV. Since HBx is pivotal for the steady state of cccDNA by regulating epigenetic modifiers of cccDNA-bound histones[16,30], the combination of curcumin with siRNAs, especially those targeting HBx, may lead to a synergistic effect in modulating the cccDNA steady state.
Several host-related factors, including histone proteins, regulate transcription and translation processes of cccDNA minichromosomes. Acetylation status changes of cccDNA-bound histones can regulate cccDNA transcription[16]. Previous studies have indicated that acetyltransferase inhibitors and deacetylase activators inhibit cccDNA transcription[16,31]. Curcumin has been reported to be an inhibitor of histone acetyltransferase (HAT), which specifically represses the activity of the p300/CREB-binding protein (CBP) HAT[26,27]. In the present study, curcumin mediated reductions in the levels of chromosomal and cccDNA-bound histone acetylation, which might result from the induction of histone deacetylation, because the deacetylase inhibitors TSA and sodium butyrate could block the inhibitory effects of curcumin on HBV.
HBV cccDNA is the primary template that allows for HBV gene expression and viral replication, and the steady state of cccDNA minichromosomes in the nuclei of hepatocytes contributes to persistent infection by HBV[32]. While there are no therapeutics currently available that target cccDNA, targeted therapeutics are attractive because the elimination of cccDNA results in the genomic cure of HBV infection[5]. Although genomic editing techniques, such as the CRISPR/CAS9 system, have shown promise in their capacity to edit HBV cccDNA, translating this finding into the clinic will be a challenge, mostly because of the need for the development of safe vectors for gene therapies.
Curcumin has been widely used as a dietary supplement for food colouring and flavouring, suggesting that it is safe for humans. However, poor bioavailability represents the biggest challenge for the clinical application of curcumin[33]. Poor absorption and rapid metabolism of curcumin in the body lead to low levels in the plasma and tissue. Fortunately, various efforts have been undertaken to promote its bioavailability. Removing the β-diketone moiety prevents curcumin from retro-aldol decomposition at physiological pH values[34]. Curcumin conjugated to two folic acid molecules (curcumin-2FA) increases water solubility and exhibits more efficient targeting of cancer cells that overexpress folic acid receptors[35]. Additionally, many other curcumin analogues can improve its bioavailability[36-40]. Moreover, curcumin-modified silver nanoparticles exhibited more efficient inhibition of respiratory syncytial virus[21]. Improving the bioavailability of curcumin will extend its clinical applications, especially in the treatment of HBV infection. Nevertheless, the long-term effects of curcumin on HBV cccDNA and its role in the elimination of cccDNA, along with combined effects of curcumin with other therapeutics, should be investigated in detail.
Taken together, the present study demonstrates that curcumin inhibits HBV by reducing cccDNA-bound histone acetylation and could potentially be developed as a cccDNA-targeting therapeutic for anti-HBV therapy.
COMMENTS
Background
Hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) is the critical template for viral replication and mRNA synthesis. The elimination of cccDNA is indicative of complete cure of hepatitis B. Nucleos(t)ide analogues (NAs) can efficiently inhibit HBV replication but do not result in clearance of residual cccDNA. Curcumin can inhibit HBV; however, its mechanism is unclear. The potential role of curcumin in the regulation of the steady state of cccDNA is thus of interest.
Research frontiers
The steady state of cccDNA minichromosomes can be regulated through deacetylation of cccDNA-bound histones. Hepatitis B virus X (HBx) protein is recruited onto cccDNA minichromosomes and plays a pivotal role in the steady state of cccDNA by regulating epigenetic modifiers of cccDNA-bound histones.
Innovations and breakthroughs
This is the first study indicating that curcumin inhibits HBV gene replication by reducing cccDNA-bound histone acetylation.
Applications
Curcumin has the potential to be developed as a cccDNA-targeting antiviral agent for hepatitis B.
Terminology
HBV covalently closed circular DNA (cccDNA): After the HBV infects hepatocytes, virus genome is released into the nucleus and converted to cccDNA, which, along with histone binding, constitutes a minichromosome and serves as the critical template for viral replication and mRNA synthesis.
Peer-review
The authors have evaluated the effects of curcumin on HBV DNA replication and gene expression in the HepG2.2.15 cell line. Interestingly, curcumin showed robust inhibitory effects on HBV cccDNA. In addition, the authors have described the mechanism underlying the effect of curcumin on HBV cccDNA, which might be through histone deacetylation or reducing histone acetylation.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: The authors have no conflicts of interest associated with this manuscript.
Data sharing statement: No additional data are available.
Peer-review started: June 8, 2017
First decision: July 13, 2017
Article in press: August 15, 2017
P- Reviewer: Farshadpour F, Inoue K, Jarcuska P, Kim K, Larrubia JR S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang FF
Contributor Information
Zhi-Qiang Wei, Institute of Biomedical Research, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China; Department of Infectious Diseases, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Yong-Hong Zhang, Institute of Wudang Chinese Medicine, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Chang-Zheng Ke, Department of Infectious Diseases, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Hong-Xia Chen, Institute of Biomedical Research, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Pan Ren, Hubei University of Chinese Medicine, Wuhan 430000, Hubei Province, China.
Yu-Lin He, Institute of Biomedical Research, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Pei Hu, Department of Pharmacy, Zhongnan Hospital of Wuhan University, Wuhan 430000, Hubei Province, China.
De-Qiang Ma, Department of Infectious Diseases, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Jie Luo, Center for Evidence-Based Medicine and Clinical Research, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China.
Zhong-Ji Meng, Institute of Biomedical Research, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China; Department of Infectious Diseases, Taihe Hospital, Hubei University of Medicine, Shiyan 442000, Hubei Province, China. zhongji.meng@taihehospital.com.
References
- 1.Fourati S, Pawlotsky JM. Recent advances in understanding and diagnosing hepatitis B virus infection. F1000Res. 2016;5:pii: F1000. doi: 10.12688/f1000research.8983.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassan MM, Li D, El-Deeb AS, Wolff RA, Bondy ML, Davila M, Abbruzzese JL. Association between hepatitis B virus and pancreatic cancer. J Clin Oncol. 2008;26:4557–4562. doi: 10.1200/JCO.2008.17.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Kang HS, Kim KH. Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J Gastroenterol. 2016;22:7017–7029. doi: 10.3748/wjg.v22.i31.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 6.Taranta A, Tien Sy B, Zacher BJ, Rogalska-Taranta M, Manns MP, Bock CT, Wursthorn K. Hepatitis B virus DNA quantification with the three-in-one (3io) method allows accurate single-step differentiation of total HBV DNA and cccDNA in biopsy-size liver samples. J Clin Virol. 2014;60:354–360. doi: 10.1016/j.jcv.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 7.You CR, Lee SW, Jang JW, Yoon SK. Update on hepatitis B virus infection. World J Gastroenterol. 2014;20:13293–13305. doi: 10.3748/wjg.v20.i37.13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, Mason WS, Xu X, Guo JT, Block TM, et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob Agents Chemother. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Zhang W, Lin J, Wang F, Wu M, Chen C, Zheng Y, Peng X, Li J, Yuan Z. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Mol Ther. 2014;22:303–311. doi: 10.1038/mt.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF-alpha promoter in monocytes and macrophages. J Leukoc Biol. 2003;73:862–871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 18.Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci USA. 2015;112:E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moghadamtousi SZ, Kadir HA, Hassandarvish P, Tajik H, Abubakar S, Zandi K. A review on antibacterial, antiviral, and antifungal activity of curcumin. Biomed Res Int. 2014;2014:186864. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali A, Banerjea AC. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Sci Rep. 2016;6:27539. doi: 10.1038/srep27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XX, Li CM, Huang CZ. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale. 2016;8:3040–3048. doi: 10.1039/c5nr07918g. [DOI] [PubMed] [Google Scholar]
- 22.Pécheur EI. Curcumin against hepatitis C virus infection: spicing up antiviral therapies with ‘nutraceuticals’? Gut. 2014;63:1035–1037. doi: 10.1136/gutjnl-2013-305646. [DOI] [PubMed] [Google Scholar]
- 23.Anggakusuma, Colpitts CC, Schang LM, Rachmawati H, Frentzen A, Pfaender S, Behrendt P, Brown RJ, Bankwitz D, Steinmann J, et al. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut. 2014;63:1137–1149. doi: 10.1136/gutjnl-2012-304299. [DOI] [PubMed] [Google Scholar]
- 24.Rechtman MM, Har-Noy O, Bar-Yishay I, Fishman S, Adamovich Y, Shaul Y, Halpern Z, Shlomai A. Curcumin inhibits hepatitis B virus via down-regulation of the metabolic coactivator PGC-1alpha. FEBS Lett. 2010;584:2485–2490. doi: 10.1016/j.febslet.2010.04.067. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Yoo HS, Kim JC, Park CS, Choi MS, Kim M, Choi H, Min JS, Kim YS, Yoon SW, et al. Antiviral effect of Curcuma longa Linn extract against hepatitis B virus replication. J Ethnopharmacol. 2009;124:189–196. doi: 10.1016/j.jep.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Li Q, Chang R, Yang D, Song Z, Guo Q, Huang C. Curcumin alleviates neuropathic pain by inhibiting p300/CBP histone acetyltransferase activity-regulated expression of BDNF and cox-2 in a rat model. PLoS One. 2014;9:e91303. doi: 10.1371/journal.pone.0091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 28.Meng Z, Xu Y, Wu J, Tian Y, Kemper T, Bleekmann B, Roggendorf M, Yang D, Lu M. Inhibition of hepatitis B virus gene expression and replication by endoribonuclease-prepared siRNA. J Virol Methods. 2008;150:27–33. doi: 10.1016/j.jviromet.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Z, Qiu S, Zhang X, Wu J, Schreiter T, Xu Y, Yang D, Roggendorf M, Schlaak J, Lu M. Inhibition of woodchuck hepatitis virus gene expression in primary hepatocytes by siRNA enhances the cellular gene expression. Virology. 2009;384:88–96. doi: 10.1016/j.virol.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Guidotti LG, Isogawa M, Chisari FV. Host-virus interactions in hepatitis B virus infection. Curr Opin Immunol. 2015;36:61–66. doi: 10.1016/j.coi.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 34.Kumari N, Kulkarni AA, Lin X, McLean C, Ammosova T, Ivanov A, Hipolito M, Nekhai S, Nwulia E. Inhibition of HIV-1 by curcumin A, a novel curcumin analog. Drug Des Devel Ther. 2015;9:5051–5060. doi: 10.2147/DDDT.S86558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra A, Das BC. Curcumin as an anti-human papillomavirus and anti-cancer compound. Future Oncol. 2015;11:2487–2490. doi: 10.2217/fon.15.166. [DOI] [PubMed] [Google Scholar]
- 36.Bharitkar YP, Das M, Kumari N, Kumari MP, Hazra A, Bhayye SS, Natarajan R, Shah S, Chatterjee S, Mondal NB. Synthesis of Bis-pyrrolizidine-Fused Dispiro-oxindole Analogues of Curcumin via One-Pot Azomethine Ylide Cycloaddition: Experimental and Computational Approach toward Regio- and Diastereoselection. Org Lett. 2015;17:4440–4443. doi: 10.1021/acs.orglett.5b02085. [DOI] [PubMed] [Google Scholar]
- 37.Ahsan N, Mishra S, Jain MK, Surolia A, Gupta S. Curcumin Pyrazole and its derivative (N-(3-Nitrophenylpyrazole) Curcumin inhibit aggregation, disrupt fibrils and modulate toxicity of Wild type and Mutant α-Synuclein. Sci Rep. 2015;5:9862. doi: 10.1038/srep09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gogoi B, Sen Sarma N. Curcumin-cysteine and curcumin-tryptophan conjugate as fluorescence turn on sensors for picric Acid in aqueous media. ACS Appl Mater Interfaces. 2015;7:11195–11202. doi: 10.1021/acsami.5b01102. [DOI] [PubMed] [Google Scholar]
- 39.Pan Y, Wang Y, Cai L, Cai Y, Hu J, Yu C, Li J, Feng Z, Yang S, Li X, et al. Inhibition of high glucose-induced inflammatory response and macrophage infiltration by a novel curcumin derivative prevents renal injury in diabetic rats. Br J Pharmacol. 2012;166:1169–1182. doi: 10.1111/j.1476-5381.2012.01854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Xiao J, Zhou H, Yang S, Wu X, Jiang C, Zhao Y, Liang D, Li X, Liang G. A novel monocarbonyl analogue of curcumin, (1E,4E)-1,5-bis(2,3-dimethoxyphenyl)penta-1,4-dien-3-one, induced cancer cell H460 apoptosis via activation of endoplasmic reticulum stress signaling pathway. J Med Chem. 2011;54:3768–3778. doi: 10.1021/jm200017g. [DOI] [PubMed] [Google Scholar]


