Abstract
Plasmodium falciparum causes a spectrum of malarial disease from asymptomatic to uncomplicated through to severe. Investigations of parasite virulence have associated the expression of distinct variants of the major surface antigen of the blood stages known as Pf EMP1 encoded by up to 60 var genes per genome. Looking at the population genomics of var genes in cases of uncomplicated malaria, we set out to determine if there was any evidence of a selective sweep of specific var genes or clonal epidemic structure related to the incidence of uncomplicated disease in children. By sequencing the conserved DBLα domain of var genes from six sentinel sites in Uganda we found that the parasites causing uncomplicated P. falciparum disease in children were highly diverse and that every child had a unique var DBLα repertoire. Despite extensive var DBLα diversity and minimal overlap between repertoires, specific DBLα types and groups were conserved at the population level across Uganda. This pattern was the same regardless of the geographic distance or malaria transmission intensity. These data lead us to propose that any parasite can cause uncomplicated malarial disease and that these diverse parasite repertoires are composed of both upsA and non-upsA var gene groups.
Subject terms: Genetic variation, Malaria
Introduction
Children living in many regions of Africa are at risk of febrile episodes of malaria until they eventually acquire immunity that protects them against the development of clinical disease. This immunity is non-sterilizing and occurs after repeated exposure to Plasmodium spp1.
Plasmodium falciparum Erythrocyte Membrane Protein 1 (Pf EMP1) has been proposed as the major variant surface antigen (VSA) of the most virulent malaria parasite known as P. falciparum. This highly polymorphic antigen is encoded by up to 60 diverse var genes present in each parasite genome2–4. Differential expression of these genes leads to switching of Pf EMP1 variants in a single cell5–7. This mechanism of clonal antigenic variation occurring in a hierarchical manner allows the parasite to evade the host immune response to Pf EMP1 thereby establishing a chronic infection. High levels of diversity have been observed in the var gene repertoire in a single parasite8–11 as well as in populations12–15. Importantly, Pf EMP1 is also a virulence factor as it mediates adhesion of infected erythrocytes to host endothelial cells and expression of specific variants is associated with the pathogenesis of uncomplicated, severe and placental malaria (reviewed in e.g. ref.16).
Characterization of the structure of var genes has revealed a conserved domain architecture consisting of an N-terminal segment (NTS), followed by multiple Duffy-binding like (DBL) and cysteine-rich interdomain region (CIDR). This is despite the sequences being highly polymorphic8,11. Var genes can be further classified into four main sub-groups based on semi-conserved upstream promoter sequences (ups): groups A (upsA), B (upsB), C (upsC), and E (upsE) with group E consisting of only var2csa, a gene associated with placental malaria9,10,17,18. Recombination events occur mostly between var genes from the same group, generating high sequence diversity while still preserving the var domain architecture19. Despite this considerable diversity, comparisons to distantly related primate parasites (e.g. P. reichenowi) have shown that balancing selection has maintained ancient var sequence fragments over millions of years20. Where malaria transmission is high, multiple-clone P. falciparum infections are common21,22 leading to increased rates of meiotic recombination resulting in extensive repertoire diversity23,24. Moreover, high levels of mitotic recombination have been observed within cloned laboratory lines25. Whether this mitotic diversification process occurs as frequently in nature remains an open question.
Evidence from case-control studies have shown that group A var genes are preferentially expressed in children with severe and/or cerebral malaria, group B var genes are associated with clinical malaria (uncomplicated and severe) and cerebral malaria, and group C transcripts are present in larger proportions in children with chronic asymptomatic malaria infections26–34. Sequencing of seven P. falciparum genomes has shown that all parasites have the group A and B var genes11 so expression of these genes rather than the presence of these genes per se is associated with disease severity.
Population genetic studies of var genes have focused on sequencing the highly conserved DBLα domain and demonstrated by limited sampling that extensive diversity in DBLα sequences and repertoires exist in endemic areas of South America, Africa and Papua New Guinea12–14,35,36. Recently, we investigated the population structure of var DBLα sequences in parasites from asymptomatic children from an area of high malaria transmission in Gabon, West Africa15. Strikingly, in this first study of deep sampling of var genes in the asymptomatic reservoir in Africa we found that every parasite isolate had a distinct var DBLα repertoire with minimal sharing of DBLα types among 200 isolates. The absence of parasite genomes with high sharing of types was consistent with immune selection structuring a large effective parasite population size, as defined by DBLα diversity, into repertoires as different as possible in a transmission system favoring outcrossing. The case was made that these data provide evidence for a novel type of “strain structure” in a system where recombination is the key driver of diversification of the major genes under immune selection. These important observations regarding the var population genomics of asymptomatic infections in children lead us to ask the question: what is the population structure of var genes in uncomplicated malaria cases of P. falciparum in an area of high transmission in Africa? We wanted to explore whether they exhibit the same population structure or whether there is epidemic transmission of parasites with related repertoires of var genes causing uncomplicated malaria. Moreover, we wanted to explore the population structure of different groups of var genes (i.e., upsA and upsB/upsC) to see if they had distinct patterns of genome evolution under variable conditions of transmission.
Specifically, our experiments describe the var DBLα diversity of the parasite population causing uncomplicated P. falciparum cases in children under five. We present results for six sentinel health sites in Uganda and demonstrate that P. falciparum parasites causing uncomplicated cases in children were highly diverse with distinct var DBLα repertoires despite varying levels of transmission intensity across Uganda. In our study we found no evidence of epidemic expansion or clonal propagation of P. falciparum parasites with highly related var DBLα repertoires in the population.
These data led us to propose that any parasite can cause uncomplicated disease. Furthermore, these highly diverse parasite repertoires contain both upsA and non-upsA var gene groups in proportions expected from whole genome sequencing11.
Results
Diversity and frequency distribution of DBLα types
DBLα amplicons were pooled and sequenced from 517 of the 600 isolates from the six sentinel sites.
A total of 51,401 DBLα sequence reads were obtained from the 517 isolates and clustered based on average linkage using a 96% sequence identity threshold resulting in 21,156 unique DBLα sequences, or DBLα types. Sixteen (3%) isolates with less than 20 DBLα types were excluded from analyses due to limited DBLα type counts. From the 501 isolates utilized for this analysis, a total of 50,624 DBLα sequence reads were obtained (mean = 8,437; range = 4,347–10,950 per site) and we observed a total of 21,134 DBLα types (mean = 5,766; range = 3,012–7,366 DBLα types per sentinel site) (Table 1).
Table 1.
Var DBLα sampling by sentinel study site.
| Study Sites | No. isolates sampled | Total var DBLα sequences sampled | Mean no. DBLα types per isolatea (range) | Total no. DBLα types | No. DBLα types seen onceb (%) | No. upsA DBLα types (%)c |
|---|---|---|---|---|---|---|
| Kanungu | 64 | 4,374 | 68 (22–195) | 3,012 | 2,299 (76.3) | 369 (12.3) |
| Jinja | 89 | 8,713 | 98 (48–208) | 6,108 | 4,816 (78.9) | 697 (11.4) |
| Kyenjojo | 83 | 6,567 | 79 (40–165) | 4,421 | 3,339 (75.5) | 481 (10.9) |
| Arua | 97 | 10,950 | 113 (50–262) | 7,366 | 5,723 (77.7) | 838 (11.4) |
| Tororo | 91 | 10,726 | 118 (20–211) | 7,266 | 5,565 (76.6) | 732 (10.1) |
| Apac | 77 | 9,294 | 121 (46–254) | 6,424 | 5,043 (78.5) | 771 (12.0) |
| All Sites | 501 | 50,624 | 101 (20–262) | 21,134 | 12,493 (59.1) | 1,624 (7.7) |
aCalculated by the following formula: total var DBLα sequences sampled/no. isolates sampled in that particular site. bRefers to the DBLα types that were seen only once in all isolates from a given sentinel site based on the frequency distribution of DBLα types in that particular site (see Fig. 1). cCalculated by the following formula: total upsA DBLα types/total no. DBLα types in that particular site.
Within and among the sentinel Ugandan sites sampled the distribution of the DBLα types showed a similar pattern of abundance (Fig. 1). The majority of the DBLα types were rare and only found in one isolate per site, whereas very few DBLα types were seen more than once per site (Fig. 1). A total of 12,493 DBLα types (59.1%) were seen only once in all sentinel sites where the minimum and maximum number of times a type was seen was 1 and 288, respectively (Table 1, Fig. 1). Within each sentinel site the proportion of DBLα types seen once ranged from 75.5% to 78.9% (Table 1, Fig. 1). Interestingly, among all sites 8,641 DBLα types (40.9%) were seen more than once with 536 DBLα types (2.5%) seen in 10 or more isolates (Fig. 1). Of the 536 types seen in at least 10 isolates, 374 were observed in isolates from Gabon15 with 121 seen at a frequency above 2.5%. Note that due to the reoccurrence of some DBLα types among the sites, the total number of observed DBLα types in all sites is lower than the cumulative sum of DBLα types observed in each site (Table 1).
Figure 1.
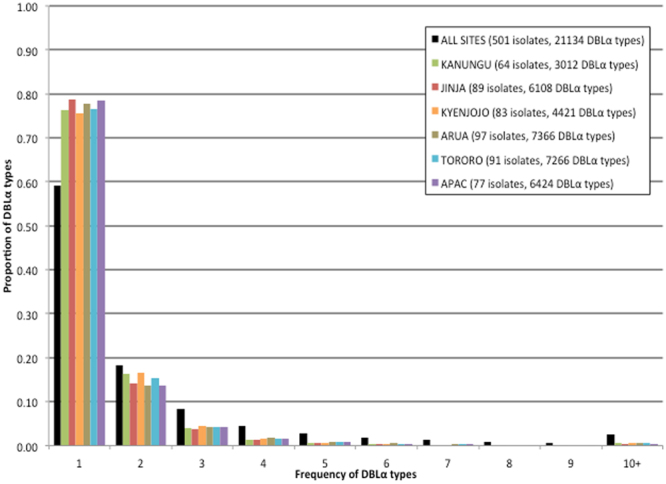
Proportion of DBLα types appearing 1 to 10 or more times within and among all sentinel sites.
We further classified the 21,134 DBLα types into upsA or upsB/upsC by translating the DBLα types and then characterizing them as upsA or upsB/upsC according to their domain class (see Methods and Supplementary Text 1). The DBLα type sequence groups were distributed as expected from whole genome sequencing (e.g. ref.11) with upsA representing 7.7% (1,624 DBLα types) and upsB/upsC representing 92.3% (19,510 DBLα types) of the total number of DBLα types sequenced in Uganda (Table 1). The distribution of the DBLα types showed type-specific patterns of abundance when the upsA and upsB/upsC DBLα types were considered separately (Supplementary Figure S1). When comparing the proportion of upsA to upsB/upsC DBLα types across all sentinel sites, although less abundant in the total population, specific upsA DBLα types were significantly more likely to be observed in 10 or more isolates (i.e. more conserved in the population) than the upsB/upsC DBLα types (p < 0.001). This pattern of increased abundance of the upsA DBLα types being more conserved was only statistically significant within the high transmission sites of Arua (p = 0.026), Tororo (p < 0.001), and Apac (p < 0.001). For the low to moderate transmission sites (Kanungu, Jinja, and Kyenjojo) there was no significant repeat occurrence of specific upsA DBLα types. It is important to note that the conserved upsA DBLα types represented a minority (13.1%) of the total upsA DBLα types observed.
Depth of DBLα sampling
Using previously published methods, the cumulative diversity curves were utilized to measure the depth of DBLα sampling in each of the six Ugandan sentinel sites and among all sites12,13. A curve that approaches an asymptote and levels off indicates a well-sampled population. Even though a large dataset of DBLα types was obtained for clinical cases, the cumulative diversity curves did not plateau for any of the sentinel sites, indicating the population was not sampled deeply enough to observe all types (Supplementary Figure S2)12,13. The cumulative diversity curve for upsA DBLα types appears to be leveling off (i.e., approaching saturation) in comparison to the curve for upsB/upsC DBLα types, indicating an approximation of the total upsA DBLα types in the total Ugandan population (Supplementary Figure S3). DBLα richness was estimated by calculating Chao2, ICE and Jackknife2 statistics, which are designed to capture the extent of missing data37–42. We estimated a total of 41,345 DBLα types in all sentinel sites (95% CI = 40,376–42,363) by Chao2 statistics, 44,092 types by ICE statistics and 42,207 types by Jackknife2 statistics (Supplementary Table S1). Using the Chao2 richness estimates, the proportion of DBLα types sampled was estimated to be relatively high in all the Ugandan sentinel sites (51.1%) (Supplementary Table S1). Within each sentinel site, the proportion of total DBLα types sampled by Chao2 richness estimates ranged from a minimum of 30.9% to a maximum of 37.1%, with a mean of 33.7% (Supplementary Table S1).
Relatedness of DBLα repertoires within isolates
For the 501 isolates analyzed for the six sentinel sites the size of DBLα repertoires ranged from the minimum DBLα type cut off of 20 (described above) to a maximum of 262 DBLα types per isolate (Table 1). The mean DBLα repertoire size was 101 DBLα types per isolate in all sentinel sites. In the low to moderate transmission sentinel sites (Kanungu, Jinja, Kyenjojo) the mean number of DBLα types in each isolate ranged from 68 to 98. In the high transmission sites (Arua, Tororo, Apac) the mean number of DBLα types in each isolate ranged from 113 to 121.We observed more than 60 DBLα types per isolate in 376 isolates (75.0%) consistent with the presence of multiple-clone infections in the majority of the population (Fig. 2).
Figure 2.
Schematic presentation of the major sequence group (upsA, and upsB/upsC) composition of the DBLα repertoires and the number of DBLα types per isolate for each of the six sentinel sites in Uganda. Left hand y-axis: For each sentinel site in Uganda the bar graph depicts each isolate’s DBLα repertoire major sequence group (upsA, and upsB/upsC) composition. The upsA proportion is represented by the use of lighter color tones in the upper portion of the column, whereas the upsB/upsC proportion is indicated by darker color tones in the lower portion. Right hand y-axis: Line graph depicting the number of DBLα types identified in each isolate. The line graphs have been ordered such that the minimum DBLα repertoire size (smallest number of DBLα types per isolate) is on the left and the maximum DBLα repertoire size (largest number of DBLα types per isolate) is on the right for each sentinel site.
When further considering the upsA and upsB/upsC composition for each DBLα repertoire, the proportions were similar within each sentinel site despite (i) the varying transmission intensities across Uganda and (ii) the number of DBLα types per isolate (i.e. DBLα repertoire size) (Fig. 2). The mean upsA and upsB/upsC DBLα type repertoire proportion for all sites was 0.16 upsA DBLα types (0.84 upsB/upsC). In each site the proportions ranged from 0.13 upsA DBLα types (0.87 upsB/upsC) in Kyenjojo to 0.18 upsA DBLα types (0.82 upsB/upsC) in Apac.
To determine whether epidemics of genomes with highly related DBLα repertoires were causing uncomplicated disease, we used pairwise type sharing (PTS) comparisons to examine the overlap of DBLα repertoires between isolates within and among the sentinel sites. Briefly, the DBLα repertoire in one isolate was compared to the DBLα repertoire in all other isolates within a site and among all sites. As mentioned above, 75.0% of isolates contained more than one parasite genome. Previous studies have shown that even when a parasite isolate contains more than one P. falciparum genome, PTS is a robust estimator of relatedness between isolates12. The presence of highly related genomes that share many DBLα types would be evidenced by high PTS scores between isolates. PTS among the 501 isolates resulted in 125,250 pairwise comparisons, of which 13,245 (10.6%) shared no DBLα types (Fig. 3). For 93,649 (74.8%) pairwise comparisons, the PTS score ranged from >0 to ≤0.05, indicating there was minimal sharing between all isolates (≤5% sharing) (Fig. 3). The median and mean PTS scores were 0.026 and 0.029 respectively among all sites (median PTS range across all sites = 0.025–0.033; mean PTS range across all sites = 0.028–0.037), with a maximum PTS score of 0.93 among all sites (maximum PTS range across all sites = 0.25–0.93). Only 17 (0.014%) pairwise comparisons had a PTS score >0.2 indicating that a strikingly limited number of isolates shared greater than 20% of their DBLα types. The overlap of DBLα repertoires within each sentinel site followed a similar pattern with an average of 81.4% of within site comparisons having a PTS score greater than 0 but less than or equal to 0.05, indicating that there was ≤5% sharing between DBLα repertoires in the majority of isolates within each sentinel site. This observed type sharing (~5%) is an upper limit since it is possible that type sharing can occur between different parasite genotypes within isolates with more than one parasite genome. Overall these results show that in this Ugandan parasite population uncomplicated malaria cases in children were caused by genetically distinct parasites with weakly overlapping DBLα repertoires (as defined by a PTS score ≤0.20) both on a local and national scale despite varying levels of transmission intensity across Uganda.
Figure 3.

(a) Heat map representation of the PTS of DBLα types among isolates within and among sentinel sites in Uganda. The DBLα repertoire in one isolate was compared to the DBLα repertoire in all other isolates within a site and among all sites. Different shading colors (Green = Kanungu; Red = Jinja; Orange = Kyenjojo; Taupe = Arua; Blue = Tororo; Purple = Apac; Grey = sharing between sites) are used to indicate the PTS values by site and among all sites. The darker the color of the box, the greater the total number of shared DBLα types in the DBLα repertoires between two isolates: no shading represents a PTS score of zero (i.e., no sharing), the darkest signifies a PTS score >0.2 (indicates >20% sharing of DBLα types in the DBLα repertoires between two isolates) and gradation in-between represent PTS scores as indicated in the color key provided. (b) The frequency distribution of the proportion of pairwise comparisions within particular PTS score ranges within and among all sites. N refers to the total number of pairwise comparisons between isolates within each sentinel site and among all sites. The proportion estimates were calculated as follows: # of pairwise comparisons within a specific PTS score range/total # pairwise comparisions in each site or among all sites.
To investigate whether there were any selective sweeps of specific var DBLα types we examined repertoire overlap based on the upsA and upsB/upsC DBLα types separately, as upsA DBLα types are expected to be more conserved relative to upsB/upsC DBLα types24. It is important to note that as expected the repertoire sizes being compared were smaller for the upsA comparisons as the upsA DBLα types comprised, on average, only 16% of the repertoires. For the upsA DBLα types the median and mean PTS scores were 0.06 and 0.06 respectively among all sites (median PTS range across all sites = 0–0.07; mean PTS range across all sites = 0.05–0.07), with a maximum PTS score of 1 among all sites (maximum PTS range across all sites = 0.35–1) (Supplementary Figure S4). For the upsB/upsC DBLα types, the median and mean PTS scores were 0.02 and 0.03 respectively among all sites (median PTS range across all sites = 0.02–0.03; mean PTS range across all sites = 0.02–0.03), with a maximum PTS score of 0.93 among all sites (maximum PTS range across all sites = 0.23–0.93) (Supplementary Figure S4). These low PTS values show no evidence of epidemics of genomes with highly related DBLα repertoires or selective sweeps of specific var genes, not even among the more conserved upsA DBLα types.
Geographic population structure of DBLα types
To assess the structuring of DBLα types on a geographic scale, we investigated the number of shared DBLα types between the sites. There was substantial sharing of DBLα types between sites regardless of the distance between them (Fig. 4a). The PTS scores of mean DBLα type sharing between two sites were also calculated and compared to the distance between the sites using “birds flight” and road distance. The distances between the sites ranged from 112.41 km (Jinja to Tororo) to 521.29 km (Tororo to Kanungu) by bird’s flight and from 130 km (Jinja to Tororo) to 729 km (Kanungu to Arua) by road distance (Supplementary Table S2). The calculated Spearman’s rank correlation coefficients (ρ) were determined to be −0.74 (p = 0.002) by bird’s flight and −0.72 (p = 0.003) by road (Fig. 4). The results from the distance analysis between sites using bird’s flight and road distance were statistically significant (p < 0.05), and indicate a negative correlation between distance and DBLα type sharing. Increasing the distance between sentinel sites decreases the sharing of DBLα types. Despite these trends, there was conservation of a large number of DBLα types, as defined by 96% sequence identity, across Uganda with 1,995 DBLα types seen in at least three sentinel sites and 277 DBLα types seen in all sites.
Figure 4.
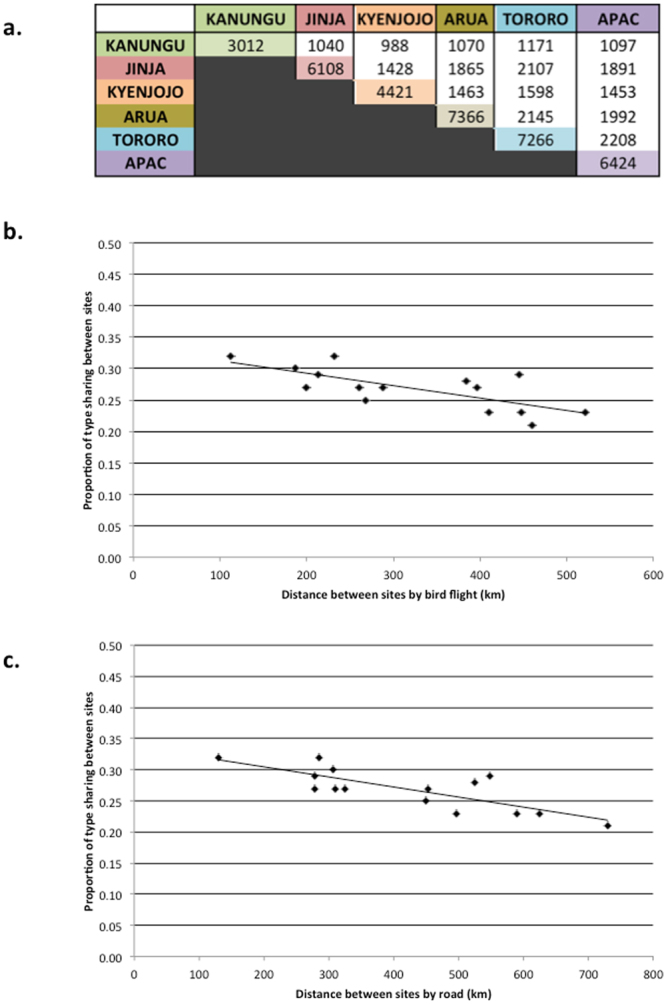
(a) Sharing of observed DBLα types (i.e., identical DBLα types) between the sentinel sites. (b) PTS (proportion) between each of the six Ugandan sentinel sites was compared to the distance between the sites using the “birds flight” and (c) road distance. The calculated Spearman’s rank correlation coefficients (ρ) were determined to be (b) −0.74 (p = 0.002) and (c) −0.72 (p = 0.003).
To take into account the patterns observed related to the upsA or upsB/upsC DBLα types, we investigated the number of shared DBLα types between sites independently for DBLα types classified as upsA and upsB/upsC. The same trend was observed for both upsA and upsB/upsC DBLα types with a negative correlation between distance and type sharing between sites, with sites farther apart sharing fewer DBLα types (Supplementary Figure S5). Interestingly, when the upsA and upsB/upsC DBLα types were analyzed independently, upsA DBLα type sharing between sites was significantly higher (range = 37–58% sharing, Supplementary Figure S5) compared to the upsB/upsC DBLα types (range = 18–29% sharing, Supplementary Figure S5) for all comparisons (p < 0.001). This is not altogether surprising, since upsA DBLα types have been reported to be more conserved, possibly due to balancing selection24.
Discussion
P. falciparum causes a spectrum of malarial disease from asymptomatic to uncomplicated through to severe. Approximately 1–2% of children who become ill with malaria develop severe disease and only 0.25% of cases result in death43,44. Investigations of parasite virulence have associated the expression of semi-conserved clades of various parts of var gene sequences (rather than the presence of specific var genes in a genome per se) with defined disease outcomes, especially severe disease, in case control studies26–34. Unlike previous studies we have asked a different question: looking at the population genomics of var genes in cases of uncomplicated malaria, we set out to determine if there was any evidence of a selective sweep of specific var genes or epidemic structure related to the incidence of uncomplicated disease in children.
We investigated the diversity and population structure of var gene sequences encoding the Pf EMP1 DBLα domain of P. falciparum causing uncomplicated malaria. This was examined in six sites across Uganda with varying malaria transmission intensities, from low (i.e., low recombination rates) to high (i.e., higher recombination rates). Surprisingly, even in low transmission sites we found no evidence of a selective sweep of any specific var DBLα types in the parasites causing uncomplicated malaria, even after stratifying by both the upsA and upsB/upsC DBLα types. In fact, a key genetic feature of these parasites was that they all showed minimal overlap with respect to their DBLα repertoires suggesting that there is selection for var diversity and heterogeneity at the DBLα repertoire level in this population. Indeed, there was no particular common genetic signature indicative of clonal propagation or epidemic expansion of related P. falciparum parasites causing uncomplicated malaria as has been reported in Senegal45 where, in comparison to our study sites, malaria control has been intense.
Our analysis of 21,134 unique DBLα types from 501 uncomplicated cases revealed extensive diversity of the DBLα domain of var genes, consistent with Chao estimates of DBLα diversity from previous limited sampling that we completed in African sites with high transmission13. Paradoxically, we found high diversity at the DBLα type level yet conservation of individual DBLα types with 40.9% of the DBLα types identified being conserved in Uganda (i.e., seen more than once when all the six surveyed populations were combined). These striking results would not be predicted by the extremely high rates of mitotic recombination that have been reported from in vitro experiments with culture adapted P. falciparum lines25. Based on the conservation of DBLα types between isolates and across sites, we propose that there are forces structuring the parasite population to maintain these DBLα types in the transmission system. This would be consistent with a mechanism such as balancing selection24.
Although we observed extensive diversity at the DBLα repertoire level, the proportional representation of upsA and upsB/upsC DBLα types within each repertoire was maintained as expected24. This is likely due to their chromosomal orientation and reduced levels of recombination with other var gene groups to maintain domain architecture46,47. Interestingly, when looking at the frequency of the DBLα types across all sites, the upsA were ~9x more likely to be seen in 10 or more isolates when compared to the upsB/upsC. This significant pattern was only observed in the high transmission sites (EIR >300) where exposure to malaria is intense. Thus, the assumed increased rates of meiotic recombination in high transmission sites have led to the dispersal of the same upsA DBLα types into a greater proportion of P. falciparum repertoires.
Every DBLα repertoire was unique in the 501 uncomplicated malaria cases with minimal overlap of the DBLα repertoires within, between and across all sites. The observed parasite population structure further supports the immune selection model described by Artzy-Randrup et al.23. They explored the population structuring of var genes by simulating the dynamics of all possible var gene combinations given a large and diverse pool of Pf EMP1 variants as well as patterns of immunity in the host population. They showed that despite high levels of outcrossing (i.e., high meiotic recombination rates) there is a defined parasite population structure that exhibits “strain” structuring23. This is consistent with our observations as the P. falciparum parasites causing uncomplicated malaria have distinct, minimally overlapping DBLα repertoires despite varying transmission intensities across Uganda. Furthermore we observed the same pattern locally (i.e., within a site) and nationally (i.e., across all sites) as predicted by the immune selection model where the same var repertoire structuring would be expected despite different host population sizes23. This is also the same pattern of “strain” structure that we observed in isolates from asymptomatic children in Bakoumba where a clear var population structure with limited overlap in the DBLα repertoires was described15. This structure would enhance the parasites’ ability to evade the host immune response under conditions of high transmission and high recombination rates.
A trade-off between maintaining parasite fitness in the host while still achieving successful transmission between hosts (i.e., immune evasion), has been described by Buckee and Recker24. They proposed an evolutionary model where parasite phenotypic plasticity could explain population structuring such that there is a trade-off of evolutionary strategies: conservation of var gene domain architecture (i.e., sequence length) serves to optimize parasite fitness and diversity at the var repertoire level maximizes immune evasion. Sequencing results from our study show for the first time both conservation of individual DBLα types (at 96% sequence identity) and yet high repertoire diversity in uncomplicated P. falciparum cases. The evolutionary forces maintaining this pattern of diversity need to be explored further with models that consider the large number of DBLα variants we have seen in nature. There is no question that the need to prolong infection so as to enable transmission requires evasion of the host immune system, which provides a strong selection force to diversify var genes as well as repertoires within a host.
Many studies have shown that individuals with uncomplicated malaria exhibit a broad range of antibody responses and harbor parasites that express diverse Pf EMP1 variants27,28,31,46,48,49. Although we define sequences of the same DBLα type as sharing 96% sequence identity, epitopes can be shared between sequences with a lower identity (e.g. the DBLβ3 domains of DC4 share 80% sequence identity but are antigenically cross-reactive50). Nonetheless, the prevalence of the host variant-specific anti-Pf EMP1 response will be dependent on the extent of Pf EMP1 variant diversity in the parasite population. Hence, acquisition of immunity to the less diverse upsA DBLα types would be expected to occur faster than the ~12x more diverse upsB/upsC DBLα types. Serological network studies by Buckee et al. (2009) propose that immune selection occurs at different levels within the upsA or upsB/upsC groups of the same var multigene family51 and this could influence the acquisition of immunity to specific var gene groups.
When considering geographic structuring of conserved DBLα types there was significantly higher sharing of upsA DBLα types (range 37–58%) between sites compared to the upsB/upsC DBLα types (range 18–29%) over the large geographic distances sampled in Uganda (~100 km to ~600 km). Paradoxically given this significant conservation of the upsA DBLα types across sites, we still report minimal PTS overlap of DBLα repertoires between isolates. This pattern of PTS was consistent no matter how we partitioned the repertoires per isolate: (i) the entire DBLα repertoire, (ii) only the upsB/upsC DBLα types, and perhaps most interestingly, (iii) even when exclusively examining the upsA DBLα types. This highlights the fact that there was no detectable evidence of linkage between the DBLα types at the isolate repertoire level.
We observed a paradoxical pattern of high diversity and minimal overlap at the parasite var DBLα repertoire level within a host, yet conservation of 40.9% of DBLα types. This overall pattern was observed repeatedly by our sampling at multiple sites in Uganda and was the same regardless of geographic distance or malaria transmission intensity. Whether demographic forces or immune selection are maintaining this pattern remains to be answered. Importantly, the low PTS among var DBLα repertoires show that any parasite with a diverse repertoire of var genes, rather than an epidemic of parasites with related var repertoires, can cause uncomplicated disease.
Methods
Ethical Statement
The study was reviewed and approved by the Uganda National Council of Science and Technology and by the institutional review boards/human ethics committees of Makerere University (Kampala, Uganda), University of California (San Francisco, US), New York University School of Medicine (New York, US), and The University of Melbourne (Melbourne, AU). Written informed consent was obtained in the local language from the parent/guardian(s) for the children enrolled in this study and for the future use of the biological specimens. Parent/guardian(s) of the children were asked to volunteer and were not coerced. The researchers in the study had no professional relationship with the individuals recruited for this study. Biological specimens collected consisted of dried blood spots on filter paper collected by trained personnel at the hospital. The consent process was consistent with the ethical expectations at the time of enrollment and the ethics committees approved these procedures.
Study sites and population
The study was performed in six independent sentinel health centers established by the Uganda Ministry of Health. Malaria endemicity is widely variable across Uganda and the six sentinel study sites were chosen to represent the geographic diversity of malaria transmission intensity: based on the annual entomological inoculation rates (EIR) Kanungu, Jinja and Kyenjojo experience a low to moderate transmission (EIR = 6, 6, 7, respectively) whereas the study sites Arua, Tororo and Apac experience relatively higher transmission intensities (EIR = 397, 563, 1586, respectively). A cross-sectional survey was used to consecutively enroll 1000 outpatients per sentinel site who had been referred to the laboratory for a diagnostic malaria blood smear in accordance with the standard of care for fever case management between May 2006 and February 2007. Details on the study population and data collection procedures have been published elsewhere52. Briefly, after obtaining informed consent from all individuals and/or their parents/guardians, the participants’ age and sex were recorded and a finger-prick blood isolate was obtained for a thick film blood smear, a rapid diagnostic test (RDT) and a dried blood spot (DBS) for molecular testing. From the 1000 outpatients who were enrolled at each of the six sentinel sites across Uganda (N = 6000), isolates from 100 children between 6 months and 5 years from each of the six sentinel sites (N = 600) with a microscopy confirmed P. falciparum infection were included in this analysis. For the purposes of this study all children who tested positive for P. falciparum by microscopy, were febrile (≥37.5 °C), and showed no additional symptoms to indicate severe disease when enrolled were defined as having an “uncomplicated P. falciparum infection”.
DNA extraction
Genomic DNA from each DBS isolate was extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Var DBLα PCR and sequencing
The P. falciparum var genes from genomic DNA were amplified using the DBLα domain as previously described with modifications12,13. From each isolate of genomic DNA, a ~550–700 bp region of the DBLα domain was amplified using a degenerate primer set (F: 5′-CMTGYGCDCCRTWYMGAMG, R: 5′-TCKGCCCATTCYTCRAACCA) designed against the semi-conserved blocks B and H of DBLα8. Each of the DBLα primers were designed by adding a GS FLX Titanium primer sequence 10 bp multiplex identifiers (MID) published by Roche53. These MID primers were used to ‘barcode’ and distinguish the DBLα sequences amplified from a unique isolate once all isolates were pooled and sequenced together54. The PCR conditions for the DBLα amplification were as follows: 2 μl of isolate genomic DNA, dNTPs at a final concentration of 0.07 mM, each primer (forward and reverse with same MID combination) at a final concentration of 0.375 μM, MgCl2 at a final concentration of 2 mM, 1x reaction buffer, and 3 units GoTaq Flexi polymerase (Promega) in a 40 μL total reaction volume. PCR cycling was carried out on an Eppendorf thermal cycler and involved an initial denaturation step of 95 °C for 2 min, 30 cycles of 95 °C × 40 sec, 49 °C × 90 sec, and 65 °C × 90 sec, followed by a final extension step of 65 °C for 10 min. Finally the isolate amplicons were pooled and sequenced using next generation 454 sequencing (Roche) performed at NYU School of Medicine at the Center for Health Informatics and Bioinformatics and the Memorial Sloan-Kettering Cancer Center Genomics Core Laboratory. The 454 sequencing provides average read lengths of 400 bp, therefore lending itself to the assembly of the individual var DBLα amplicons of 550–700 bp lengths using the forward and reverse sequence reads from each direction.
DBLα sequence analysis
A custom pipeline was developed to de-multiplex, de-noise and remove PCR and sequencing artefacts from the DBLα domain reads. The first part of the pipeline is available as the Multipass web server: http://www.cbs.dtu.dk/services/MultiPass-1.0, and the following cleaning steps described below are implemented in a python script available here: https://github.com/454data/postprocess. The sff-files obtained from each region on the 454-plate were divided into smaller isolate specific sff-files by identification of reads with exact matching MID sequences in both ends using BioPython v1.57. Ambiguous primer sites were then identified (exact match) and trimmed off the flowgrams, reverse reads were reverse complemented, and a dat-file (AmpliconNoise format) with the resulting flowgrams was created for each isolate, using BioPython v1.55. By combining the forward and reverse reads this method takes advantage of bi-directional amplicon sequencing, since the forward reads will have highest quality in the 5′-end of the target sequence, and the reverse reads will improve the 3′-end quality. Flowgram clustering was performed using PyroDist, FCluster and PyroNoiseM from the AmpliconNoise package v1.2556. The flowgram clusters produced by AmpliconNoise were base called using Multipass to obtain the most likely var DBLα sequences given the flowgrams and a high open reading frame likelihood, as described in ref.57. The nucleotide sequences generated by Multipass were clustered by 96% identity using Usearch v5.2.3258 with seeds (cluster member with support from highest number of reads after de-replication) as output. Chimeras were removed using Uchime implemented in Usearch v5.2.3258,59, first in de-novo mode where chimera detection is based on read abundance, all parents are expected to be present in the sequence set, and candidate parents must be at least 2x more abundant than the chimera candidate sequence; subsequently in database mode, where sequences are searched against self and chimeras are found irrespective of the abundance of the parents. To increase overall quality of the sequences remaining at this point, a minimal coverage threshold of three reads per sequence type was applied to remove the least supported sequences. Next, we screened for and removed non-target amplified human sequences by local alignment search against the BLAST human genomic databases (http://ftp.ncbi.nlm.nih.gov/blast/db/) using the blastn feature of BLAST+ 2.2.25 (NCBI), with expectation value criteria of 1e-50. Sequences were also searched using a DBLβ HMM with HMMer v3.1 (hmmer.org). After the human and non-target P. falciparum check, a small number of sequences remained that had no similarity to a DBLα-tag HMM and these were removed. The pipeline was validated and optimized on experimental sequence data generated on the laboratory clones (3D7, Dd2, and HB3) for which published genome sequence is available. More than 90% of the sequences obtained from the control samples had no errors when compared to the known reference, and the deviating sequences had maximally 5 errors. To subsequently determine DBLα types shared between isolates, the cleaned DBLα reads were clustered using a pipeline based on the USEARCH software suite version 8.1.183158. Initially duplicate reads were removed and the remaining reads were sorted by how many duplicates were present using the derep_prefix command. The remaining reads were then clustered at 96% pairwise identity using the usearch cluster_fast command. Finally, the original unfiltered reads were aligned back to the centroids of the clusters and an OTU table was generated using the usearch_global command before a binary version of the table was generated. The code for the pipeline is available on GitHub at https://github.com/UniMelb-Day-Lab/clusterDBLalpha.
Cumulative diversity curves
The cumulative diversity curves, analogous to species accumulation, were generated using EstimateS v9.160 to estimate var DBLα richness by sampling all DBLα types from each sentinel site and among all sites without replacement. All sampling depths between one and the number of DBLα types obtained from the study site were repeated 100-fold, whereupon the mean number of var DBLα types obtained for each sampling depth was calculated. The cumulative diversity curve was plotted using R v3.1361 to plot the number of var DBLα types as a function of the number of var DBLα sequences sampled.
Frequency plots
Frequency plots were tabulated using the data generated and plotted using Microsoft Excel.
Richness estimates
Using EstimateS v9.160 the diversity of DBLα types within and among all sites in Uganda were calculated by estimating the total number of DBLα types and the proportion of DBLα types shared between isolates. For each sentinel site two different statistical estimates of richness were used, non-parametric statistical Chao2 and incidence-based coverage estimator (ICE). Chao2 calculations were used to estimate the total number of DBLα types in a particular sentinel site based on singletons and doubletons62,63. ICE statistics were used to estimate the total number of DBLα types in a study site based on all the observed types by dividing them into two groups: rare and abundant types64. Jacknife2 calculations were used to estimate the total number of DBLα types in a particular sentinel site by resampling the unique and duplicate types, thereby reducing bias41.
Pairwise type sharing
For each sentinel site, pairwise type sharing (PTS) statistics were calculated to quantify the relatedness between the DBLα repertoires identified from two isolates. This methodology has been published elsewhere12,13 and is a useful statistic to analyze diversity and determine the number of DBLα types shared between isolates. Briefly, PTS is calculated as follows:
| 1 |
Thus, a PTS score is the ratio of the number of shared DBLα types between two isolates and the sum of DBLα types in both isolates. The ratio ranges between 0 and 1, where a PTS score of 0 signifies no DBLα repertoire similarity and 1 signifies identical DBLα repertoires.
Distance-based pairwise type sharing
PTS statistics were calculated to estimate the mean DBLα type sharing between two sentinel sites. PTS scores between sites were calculated as follows:
| 2 |
Thus, a PTS score is the ratio of the number of shared DBLα types between two sites and the sum of DBLα types in both sites. A comparison was performed to evaluate the effect of distance (“birds flight” distance and transit/road distance) on DBLα type sharing between sentinel sites. “Birds flight” distance between the sentinel sites was calculated using Google Maps (https://www.google.com/maps/preview) and transit/road distance was calculated using the Via Michelin Maps and Routes application (http://www.viamichelin.com). Spearman’s rank correlation coefficient was calculated to evaluate the trend and was performed using IBM SPSS (Version 22) software.
Classification of var gene A/non-A groups
Reads were translated into all six reading frames. Protein hidden Markov models of 150 VAR domains of Rask et al.11 were aligned to the translated sequences using HMMER v3.1b1 with an e-value of 0.01. Both the bias composition correction and composition filter were turned off as suggested in Rask et al. (–nonull2 —tobias)11. The most significant match to any of a reads six translated frames was then taken as the domain assignment for that read. Reads were further classified into upsA if their most significant match was with a DBLα1 domain and upsB/upsC (i.e., non-A) if that matched with either DBLα0 or DBLα2 domains. The domain models did not allow for an accurate distinction between var groups B and C. Cross validation was used to ascertain the success of this approach with the classification accuracy of upsA and upsB/upsC combined found to be 96.2% and 99.8%, respectively (see Supplementary Text 1).
Statistical analysis
Statistical analyses were carried out using IBM SPSS Statistics (Version 22) software and the open source EpiInfo v7 (https://www.cdc.gov/epiinfo/index.html) software. A test was deemed to be statistically significant if the p-value was less than 0.05. For all analyses the sentinel sites (Kanungu, Jinja, Kyenjojo, Arua, Tororo, and Apac) were categorized as defined in the study design. Chi-squared test (χ 2) was used for univariate analyses of discrete variables to compare proportions.
Data Availability Statement
The nucleotide sequences reported in this paper have been deposited in the NCBI database (Project no. PRJNA385208, Accession no. SAMN06833355-SAMN06833855).
Electronic supplementary material
Acknowledgements
We would like to thank the children and their families for their willingness to participate, as well as the Uganda Ministry of Health for their expertise and coordination at each of the sentinel sites across Uganda. We would like to thank all the field teams for the technical assistance and sample collection. Additionally we would like to recognize the laboratory personnel at New York University for their assistance with the laboratory experiments. Finally we thank everyone involved for his or her continued patience and understanding as this research was disrupted by Hurricane Sandy (New York, NY; October 29, 2012). This research was supported by the National Institutes of Allergy and Infectious Diseases, National Institutes of Health [grant number R01-AI084156] and Fogarty International Center, National Institutes of Health (Program on the Ecology and Evolution of Infectious Diseases (EEID)) [grant number R01-TW009670].
Author Contributions
S.R.-P., K.E.T., and K.P.D. designed research; S.R.-P. and K.E.T. performed research; G.T.-H. and T.R. performed bioinformatic analysis; M.R.K., B.G., and G.D. designed field study and collected samples; S.R.-P. and K.E.T. analyzed data; and S.R.-P., K.E.T., M.F.D., and K.P.D. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shazia Ruybal-Pesántez and Kathryn E. Tiedje contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-11814-9
References
- 1.Day, K. P. & Marsh, K. Naturally acquired immunity to Plasmodium falciparum. Immunol Today12, 68–71 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Baruch, D. I. et al. Cloning the P. falciparum gene encoding Pf EMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell82, 77–87 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Smith, J. D. et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell82, 101–110 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su, X. Z. et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell82, 89–100 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Scherf, A. et al. Antigenic variation in malaria: In situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J17, 5418–5426 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voss, T. S. et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature439, 1004–1008 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Dzikowski, R., Frank, M. & Deitsch, K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog2, 0184–0194 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith, J. D., Subramanian, G., Gamain, B., Baruch, D. I. & Miller, L. H. Classification of adhesive domains in the Plasmodium falciparum Erythrocyte Membrane Protein 1 family. Mol Biochem Parasitol110, 293–310 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature419, 498–511 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavstsen, T., Salanti, A., Jensen, A. T. R., Arnot, D. E. & Theander, T. G. Sequence Analysis of Coding and Non-Coding Regions.14, 1–14 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rask, T. S., Hansen, D. A., Theander, T. G., Pedersen, A. G. & Lavstsen, T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes - divide and conquer. PLoS Comput Biol6 (2010). [DOI] [PMC free article] [PubMed]
- 12.Barry, A. E. et al. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog3, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, D. S. et al. A molecular epidemiological study of var gene diversity to characterize the reservoir of Plasmodium falciparum in humans in Africa. PLoS One6, e16629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tessema, S. K. et al. Phylogeography of var gene repertoires reveals fine-scale geospatial clustering of Plasmodium falciparum populations in a highly endemic area. Mol Ecol24, 484–497 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Day, K. P. et al. Evidence of strain structure in Plasmodium falciparum var gene repertoires in children from Gabon, West Africa, doi:10.1073/pnas.1613018114 (2017). [DOI] [PMC free article] [PubMed]
- 16.Kraemer, S. M. & Smith, J. D. A family affair: var genes, Pf EMP1 binding, and malaria disease. Curr Opin Microbiol9, 374–80 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Kraemer, S. M. & Smith, J. D. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol50, 1527–1538 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Salanti, A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol49, 179–191 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Kraemer, S. M. et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics8, 45 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zilversmit, M. M. et al. Hypervariable antigen genes in malaria have ancient roots. BMC Evol Biol13, 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul, R. E. et al. Mating patterns in malaria parasite populations of Papua New Guinea [see comments]. Science (80-)269, 1709–1711 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Babiker, H. A. et al. Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology109(Pt 4), 413–421 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Artzy-Randrup, Y. et al. Population structuring of multi-copy, antigen-encoding genes in plasmodium falciparum. Elife2012, 1–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckee, C. O. & Recker, M. Evolution of the multi-domain structures of virulence genes in the human malaria parasite, Plasmodium falciparum. PLoS Comput Biol8 (2012). [DOI] [PMC free article] [PubMed]
- 25.Claessens, A. et al. Generation of Antigenic Diversity in Plasmodium falciparum by Structured Rearrangement of Var Genes During Mitosis. PLoS Genet10 (2014). [DOI] [PMC free article] [PubMed]
- 26.Jensen, A. T. R. et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses Pf EMP1 encoded by group A var genes. J Exp Med199, 1179–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaestli, M. et al. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis193, 1567–74 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rottmann, M. et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun74, 3904–3911 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyriacou, H. M. et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol150, 211–218 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Normark, J. et al. Pf EMP1-DBL1alpha amino acid motifs in severe disease states of Plasmodium falciparum malaria. Proc Natl Acad Sci USA104, 15835–40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warimwe, G. M. et al. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci USA106, 21801–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falk, N. et al. Analysis of Plasmodium falciparum var genes expressed in children from Papua New Guinea. J Infect Dis200, 347–356 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Kalmbach, Y. et al. Differential var gene expression in children with malaria and antidromic effects on host gene expression. J Infect Dis202, 313–317 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Warimwe, G. M. et al. Prognostic indicators of life-threatening malaria are associated with distinct parasite variant antigen profiles. Sci Transl Med4, 129ra45–129ra45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherf, A., Lopez-Rubio, J. J. & Riviere, L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol62, 445–470 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Dara, A. et al. New var reconstruction algorithm exposes high var sequence diversity in a single geographic location in Mali. Genome Med9, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao, A., Hwang, W. H., Chen, Y. C. & Kuo, C. Y. Estimating the number of shared species in two communities. Stat Sin10, 227–246 (2000). [Google Scholar]
- 38.Chazdon, R. L., Colwell, R. K., Denslow, J. S. & Guariguata, M. R. Statistical methods for estimating species richness of woody regeneration in primary and secondary rainforests of noetheastern Costa Rica. For Biodivers Res Monit Model20, 285–309 (1998). [Google Scholar]
- 39.Chao, A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics43, 783–791 (1987). [PubMed] [Google Scholar]
- 40.Burnham, K. P. & Overton, W. S. Robust Estimation of Population Size When Capture Probabilities Vary Among Animals. Ecology60, 927–936 (1979). [Google Scholar]
- 41.Smith, E. P. & van Belle, G. Nonparametric Estimation of Species Richness. Biometrics40, 119–129 (1984). [Google Scholar]
- 42.Palmer, M. W. Estimating Species Richness: The Second-Order Jackknife Reconsidered. Ecology72, 1512–1513 (1991). [Google Scholar]
- 43.Greenwood, B., Marsh, K. & Snow, R. Why do some African children develop severe malaria? Parasitol Today7, 277–281 (1991). [DOI] [PubMed] [Google Scholar]
- 44.Snow, R. W. & Marsh, K. New insights into the epidemiology of malaria relevant for disease control. 54, 293–309 (1998). [DOI] [PubMed]
- 45.Daniels, R. et al. Genetic surveillance detects both clonal and epidemic transmission of malaria following enhanced intervention in Senegal. PLoS One8, e60780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bull, P. C. et al. Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS Pathog1, 0202–0213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bull, P. C. et al. Plasmodium falciparum antigenic variation. Mapping mosaic var gene sequences onto a network of shared, highly polymorphic sequence blocks. Mol Microbiol68, 1519–1534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen, M. A. et al. Plasmodium falciparum Variant Surface Antigen Expression Varies Between Isolates Causing Severe and Nonsevere Malaria and Is Modified by Acquired Immunity. J Immunol168, 3444–3450 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Barry, A. E. et al. The Stability and Complexity of Antibody Responses to the Major Surface Antigen of Plasmodium falciparum Are Associated with Age in a Malaria Endemic Area. Mol Cell Proteomics10, M111.008326–M111.008326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bengtsson, A. et al. A Novel Domain Cassette Identifies Plasmodium falciparum Pf EMP1 Proteins Binding ICAM-1 and Is a Target of Cross-Reactive, Adhesion-Inhibitory Antibodies. J Immunol190, 240–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckee, C. O., Bull, P. C. & Gupta, S. Inferring malaria parasite population structure from serological networks. Proc Biol Sci276, 477–485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hopkins, H. et al. Rapid diagnostic tests for malaria at sites of varying transmission intensity in Uganda. J Infect Dis197, 510–8 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Roche. Using multiplex identifier (MID) adaptors for the GS FLX titanium chemistry–extended MID set. Tech Bull Genome Seq FLX Syst TCB No 005–2009 (2009).
- 54.Hamady, M., Walker, J. J., Harris, J. K., Gold, N. J. & Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods5, 235–237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cock, P. J. A. et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics25(11), 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quince, C., Lanzen, A., Davenport, R. J. & Turnbaugh, P. J. Removing Noise From Pyrosequenced Amplicons (2011). [DOI] [PMC free article] [PubMed]
- 57.Rask, T. S., Petersen, B., Chen, D. S., Day, K. P. & Pedersen, A. G. Using expected sequence features to improve basecalling accuracy of amplicon pyrosequencing data. BMC Bioinformatics17, 176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed]
- 60.Colwell, R. K. EstimateS: Statistical estimation of species richness and shared species from samples. Version 9.1. Available at: http://viceroy.eeb.uconn.edu/estimates/ (2006).
- 61.R Core Team. R: A language and environment for statistical computing (2015).
- 62.Chao, A. et al. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol Monogr84, 45–67 (2014). [Google Scholar]
- 63.Colwell, R. K. & Coddington, J. A. Estimating terrestrial biodiversity through extrapolation. Philos Trans Biol Sci345, 101–118 (1994). [DOI] [PubMed] [Google Scholar]
- 64.Chao, A. & Lee, S.-M. Estimating the Number of Classes via Sample Coverage. Journal of the American Statistical Association87, 210–217 (1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences reported in this paper have been deposited in the NCBI database (Project no. PRJNA385208, Accession no. SAMN06833355-SAMN06833855).



