Abstract
Triple-negative breast cancer (TNBC) refers to one aggressive histological subtype of breast cancer with high heterogeneity and poor prognosis after standard therapy. Lack of clearly established molecular mechanism driving TNBC progression makes personalized therapy more difficult. Thus, identification of genetic variants associated with TNBC prognosis will show clinic significance for individualized treatments. Our study is aimed to evaluate the prognostic value of the genome wide association study (GWAS)-identified CHST9 rs1436904 and AQP4 rs527616 genetic variants in our established early-stage TNBC sample database. Cox regression was used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs). CHST9 rs1436904G allele was significantly associated with decreased disease-free survival time (DFS) (8.5 months shorter in GG genotype carriers compared to TT genotype carriers, HR = 1.70, 95% CI = 1.03–2.81, P = 0.038). Stratified analyses showed an increased risk of cancer progression in CHST9 rs1436904G allele carriers harboring larger tumor (tumor size > 2 cm), without lymph-node metastasis, being premenopausal at diagnosis or with vascular invasion (P = 0.032, 0.017, 0.008 or 0.003). Our findings demonstrate that the GWAS-identified 18q11.2 CHST9 rs1436904 polymorphism significantly contributes to prognosis of early-stage TNBC, suggesting its clinical potential in the screening of high-risk TNBC patients for recurrence and the possibility of patient-tailored therapeutic decisions.
Introduction
Breast cancer is one of the most common malignancies worldwide, with an estimated 255,180 new cases and 41,070 deaths in the United States in 20171. Notably, the incidence and mortality of breast cancer have increased tremendously in developing countries including China. Breast cancer is responsible for 15% of all new Chinese women cancer patients2. With similar carcinoma genetic mechanisms to other kinds of solid tumor, development of breast cancer is a chronic and multiple-step process involving accumulation of genetic and epigenetic alterations3,4. Risk factors for breast cancer include obesity, lack of physical exercise, alcohol abuse, hormone replacement therapy during menopause, ionizing radiation, early age at first menstruation, having children late or not at all, older age, and family history3–9. Triple-negative breast cancer (TNBC) refers to one type of breast cancer that does not express estrogen receptor (ER), progesterone receptor (PR) and Her2/neu10–14. TNBC patients usually have relatively poor outcomes due to its intrinsically aggressive behaviors and requires combination therapies instead of common chemotherapies because loss of target receptors. Thus, more effective and sensitive prognostic markers are instantly needed to guide clinical management of TNBC precisely.
Many genetic variants have been described to contribute to breast cancer risk since the discovery of BRCA1 and BRCA2 in 1990s15–22. These genetic inheritable variants are associated with familial breast cancer. Thus, identifying new genetic variants will show great significance in breast cancer prediction and treatments. Recent genome-wide analyses based on large consortia do avoid false positive identification of candidate genes23. Two 18q11.2 genetic variants (CHST9 rs1436904 and AQP4 rs527616) were identified as novel breast cancer susceptibility components based on GWAS24. One nested case-control study based on female Chinese patients within Singapore Chinese Health Study were also performed to verify the roles of these SNPs. It has been demonstrated that genetic variants on top of conventional risk factors did improve the risk prediction of breast cancer in Chinese women25, but not clear enough to declare whether CHST9 rs1436904 and AQP4 rs527616 affect prognosis of TNBC. To test this, we conducted a hospital-based cohort study of early-stage TNBC to further illustrate the role of these two genetic variants in breast cancer progress. We found that the CHST9 rs1436904 polymorphism might be a potential prognostic biomarker for early-stage TNBC, especially in the patients harboring larger tumor (tumor size > 2 cm), without lymph-node metastasis, being premenopausal at diagnosis or with vascular invasion.
Materials and Methods
Study subjects
A total of 381 TNBC patients were recruited between January 2008 and December 2015 at Cancer Hospital, Chinese Academy of Medical Sciences (Beijing, China). These patients were followed until May 6, 2016 in order to collect data on clinicopathological characteristics, treatments, and vital status, such as recurrence and death. Disease free survival (DFS) was defined as the time from the date of diagnosis until the date of the first locoregional recurrence, first distant metastasis, or death due to any cause. Patients known to be alive with no evidences of disease progression were censored at the last follow-up date or on May 6, 2016 (whichever came first). All subjects were ethnic Han Chinese. As we can see from histopathological data, most majority of recruited TNBC patients were at early-stage. At recruitment, the informed consent was obtained from each subject. This study was approved by the institutional Review Boards of Cancer Hospital, Chinese Academy of Medical Sciences and Shandong Cancer Hospital affiliated to Shandong University.
Immunohistochemistry (IHC) of formalin-fixed, paraffin-embedded breast cancer tissue samples obtained from the patients was used to evaluate ER or PR status with anti-ER and anti-PR antibodies. A positive ER or PR status was defined by nuclear staining of more than 1% based on guidelines of American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) in 2010. To determine the HER2 status, IHC or gene amplification was performed by fluorescence in situ hybridization (FISH). Tumors negative for ER, PR, and HER2 were defined as TNBCs.
Genotyping
A total of 1 mL blood sample was collected from each patient upon recruitment. Genomic DNA was extracted from the blood. The CHST9 rs1436904 and AQP4 rs527616 polymorphisms were analyzed by the MassArray system (Sequenom Inc., San Diego, California, USA) as described previously26–29. CHST9 rs1436904 PCR primers are 5′-ACGTTGGATGCTTCCCTGCAAGACTATGTG-3′ (Forward) and 5′-ACGTTGGATGGCAAGACAGGAGACAGATTC-3′ (Reverse). CHST9 rs1436904 UEP_SEQ primer is 5′-CCCCCTTGTGTCTCATTCCTCA-3′. CHST9 rs1436904 EXT1_SEQ primer is 5′-CCCCCTTGTGTCTCATTCCTCAG-3′. CHST9 rs1436904 EXT2_SEQ primer is 5′-CCCCCTTGTGTCTCATTCCTCAT-3′. AQP4 rs527616 PCR primers are 5′-ACGTTGGATGTTACACGAGACTGAGCCAAC-3′ (Forward) and 5′-ACGTTGGATGGAAATGCCCCTTAGGACAAG-3′ (Reverse). AQP4 rs527616 UEP_SEQ primer is 5′-GAGCTCCAGTGCTATTT-3′. AQP4 rs527616 EXT1_SEQ primer is 5′-GAGCTCCAGTGCTATTTC-3′. AQP4 rs527616 EXT2_SEQ primer is 5′-GAGCTCCAGTGCTATTTG-3′. A 20% blind, random sample of study subjects was genotyped in duplicates and the reproducibility was 100%.
Statistics
The differences of patient clinical characteristics were calculated using Student’s t test or χ2 test. DFS was calculated as the time to progression or death without progression from the date of diagnosis. Survival distributions were estimated with the Kaplan-Meier method and were compared using log-rank test. The multivariate Cox proportional hazards model was applied to estimate effects of prognostic factors on DFS, using proverbial clinical factors, including age of onset, body mass index (BMI), tumor size, lymph-node metastasis, histological type, histological grade, menopausal status, vascular invasion, breast or ovarian cancer history, surgical method, taxane/anthracycline-based chemotherapy and radiotherapy, where it was appropriate. References for multivariate analyses were without family history of breast cancer or ovarian cancer for breast cancer or ovarian cancer history, postmenopausal at diagnosis for menopausal status at diagnosis, modified radical mastectomy for operation method,histological grade I for histological grade, without vascular invasion for vascular invasion, tumor size ≤ 2 cm for tumor size, without lymph-node involvement for lymph-node involvement, without acceptance of chemotherapy for taxane/anthracycline-based chemotherapy, and without acceptance of radiotherapy for radiotherapy. P value of less than 0.05 was used as the criterion of statistical significance. All statistical procedures were conducted using SPSS software (version 16.0).
Results
TNBC patients’ characteristics and clinical outcomes
A total of 381 TNBC patients were enrolled in this study. All individuals were female ethnic Han Chinese. The distribution of demographic and clinical characteristics of patients were showed in Supplementary Table 1. By the time of the final analysis (May 2016), the median follow-up time of the patients was 45.5 months. One hundred and eighty-two patients (49.2%) had disease progression and the median DFS time was 36.0 months (range, 0–204 months).
Comparison of survival according to baseline characteristics of TNBC patients
To test whether various clinical characteristics contribute to DFS, patients were grouped according to age of onset, BMI, tumor size, lymph-node metastasis, histological type, histological grade, menopausal status, vascular invasion, breast or ovarian cancer history, surgical method, taxane/anthracycline-based chemotherapy and radiotherapy, respectively. DFS was compared between (or among) different sub-groups. As shown in Supplementary Table 2, BMI, histological grade, vascular invasion, lymph-node metastasis and radiotherapy can significantly influence DFS independently (P < 0.05). However, other baseline characteristics did not affect DFS (P > 0.05). After adjustments for other clinicopathologic factors, only BMI and vascular invasion showed statistically significant impacts on patient prognosis (Supplementary Table 2).
Effects of CHST9 rs1436904 and AQP4 rs527616 polymorphisms on TNBC DFS
It has been demonstrated that CHST9 rs1436904 and AQP4 rs527616 are breast cancer susceptibility single nucleotide polymorphisms (SNPs)24,25. However, the role of those two genetic variations in TNBC patients’ outcome has not been examined. Genotype frequencies of CHST9 rs1436904 and AQP4 rs527616 SNPs among patients were summarized in Table 1. Interestingly, only CHST9 rs1436904 polymorphism was significantly associated with DFS of TNBC patients. The mean DFS of TNBC patients with the CHST9 rs1436904 GG genotype (46.8 months) or the GT genotype (50.1 months) was significantly shorter than that of the TT group (55.3 months). Moreover, both univariate and multivariate Cox proportional hazards model indicated that the CHST9 rs1436904 genetic variation was significantly associated with disease progression of TNBC patients (Table 1 and Fig. 1). After adjustments of multiple clinical factors, the CHST9 rs1436904 GG genotype was still significantly associated with disease progression compared to subjects with the TT genotype (HR = 1.70, 95% CI = 1.03–2.81, P = 0.038). Similarly, the risk of early recurrence for TNBC patients carrying the CHST9 rs1436904 G allele (GT and GG genotype) increased about 1.51-folds (95% CI = 1.03–2.22) in comparison with TT genotype patients (P = 0.033).
Table 1.
Genotype frequencies of rs1436904 and rs527616 polymorphism among TNBC patients and their association with DFS.
| SNPs | Genotype | Patients No. (%) | DFS (month) Mean (25th, 75th) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||||
| rs1436904 | 378 | ||||||
| TT | 97(25.7) | 55.3(30, 68) | Reference | Reference | |||
| GT | 194(51.3) | 50.1(24, 65) | 1.31(0.91–1.88) | 0.151 | 1.42(0.95–2.13) | 0.086 | |
| GG | 87(23.0) | 46.8(24, 62) | 1.58(1.03–2.41) | 0.036 | 1.70(1.03–2.81) | 0.038 | |
| GT + GG | 281(74.3) | 49.1(24, 64) | 1.38(0.98–1.95) | 0.069 | 1.51(1.03–2.22) | 0.033 | |
| rs527616 | 375 | ||||||
| GG | 167(44.5) | 48.1(25, 62) | Reference | Reference | |||
| GC | 179(47.8) | 54.1(30, 71) | 0.84(0.62–1.14) | 0.256 | 0.78(0.56–1.09) | 0.143 | |
| CC | 29(7.7) | 48.9(24, 61) | 1.06(0.62–1.82) | 0.828 | 1.21(0.62–2.38) | 0.582 | |
| GC + CC | 208(55.5) | 53.4(27, 70) | 0.87(0.65–1.16) | 0.332 | 0.81(0.59–1.12) | 0.211 | |
Note: DFS, disease–free survival time; HR, hazard ratio; CI, confidence interval.
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNP and disease-free survival time (DFS) were estimated by Cox regression adjusted by age of onset, BMI, tumor size, lymph-node involvement, histological type, histological grade, menopausal status, vascular invasion, breast or ovarian cancer history, surgical method, taxane/anthracycline-based chemotherapy and radiotherapy.
Figure 1.
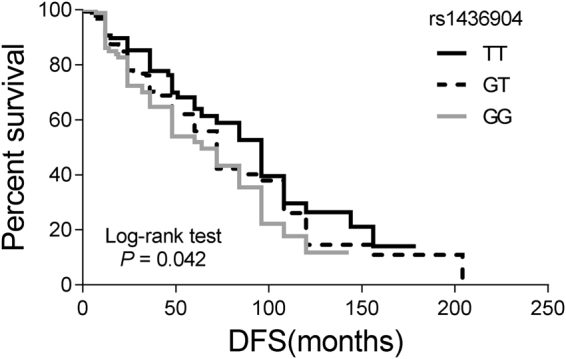
Kaplan-Meier curves of DFS for TNBC patients harboring different genotypes of CHST9 rs1436904 genetic variations. Survival curves of different genotypes were compared by Kaplan-Meier method, followed by long-rank test (P = 0.042).
Stratified analyses of the effects CHST9 rs1436904 on DFS of TNBC patients
The association between CHST9 rs1436904 polymorphism and DFS of TNBC patients was further examined by stratifying for age of onset, BMI, tumor size, lymph-node metastasis, histological type, histological grade, menopausal status, vascular invasion, breast or ovarian cancer history, surgical method, taxane/anthracycline-based chemotherapy and radiotherapy, respectively (Table 2 and Supplementary Table 3–7).
Table 2.
DFS of TNBC associated with CHST9 rs1436904 genotypes by tumor size, lymph-node involvement, menopausal status as well as vascular invasion
| Variable | CHST9 rs1436904 | |||||
|---|---|---|---|---|---|---|
| Genotypes | Patients No. (%) | DFS (month) Mean(25th, 75th) | HR (95% CI) | P | ||
| Tumor size | ≤ 2 cm | 179 | ||||
| TT | 46(25.7) | 54.9(32, 72) | Reference | |||
| GT | 90(50.3) | 53.2(29, 67) | 1.11(0.61–2.02) | 0.726 | ||
| GG | 43(24.0) | 47.7(24, 69) | 1.36(0.62–2.99) | 0.444 | ||
| GT + GG | 133(74.3) | 51.5(27, 69) | 1.19(0.69–2.06) | 0.534 | ||
| > 2 cm | 195 | |||||
| TT | 50(25.6) | 55.9(28, 64) | Reference | |||
| GT | 102(52.3) | 46.3(24, 62) | 1.61(0.88–2.95) | 0.122 | ||
| GG | 43(22.1) | 46.4(24, 60) | 2.16(0.97–4.81) | 0.060 | ||
| GT + GG | 145(74.4) | 46.4(24, 62) | 1.88(1.06–3.35) | 0.032 | ||
| Lymph-node involvement | No | 229 | ||||
| TT | 58(25.3) | 62.6(36, 72) | Reference | |||
| GT | 118(51.5) | 53.9(31, 72) | 1.85(1.01–3.37) | 0.046 | ||
| GG | 53(23.2) | 48.1(26, 70) | 2.27(1.07–4.83) | 0.033 | ||
| GT + GG | 171(74.7) | 52.1(30, 71) | 2.01(1.13–3.57) | 0.017 | ||
| Yes | 142 | |||||
| TT | 38(26.8) | 44.5(15, 55) | Reference | |||
| GT | 73(51.4) | 42.7(20, 57) | 1.15(0.61–2.16) | 0.675 | ||
| GG | 31(21.8) | 45.3(24, 60) | 1.39(0.60–3.23) | 0.450 | ||
| GT + GG | 104(73.2) | 43.5(20, 58) | 1.29(0.72–2.32) | 0.390 | ||
| Menopausal status at diagnosis | Premenopausal | 179 | ||||
| TT | 46(25.7) | 48.0(36, 83) | Reference | |||
| GT | 94(52.5) | 48.0(27, 62) | 2.07(1.18–3.65) | 0.011 | ||
| GG | 39(21.8) | 36.0(32, 64) | 2.18(1.12–4.22) | 0.021 | ||
| GT + GG | 133(74.3) | 45.0(27, 62) | 2.10(1.22–3.63) | 0.008 | ||
| Postmenopausal | 39 | |||||
| TT | 46(28.8) | 43.0(26, 61) | Reference | |||
| GT | 76(47.5) | 51.5(28, 71) | 0.96(0.51–1.82) | 0.904 | ||
| GG | 38(23.8) | 40.5(30, 63) | 1.41(0.68–2.93) | 0.360 | ||
| GT + GG | 114(71.2) | 48.0(29, 71) | 1.09(0.60–1.97) | 0.779 | ||
| Vascular invasion | No | 334 | ||||
| TT | 85(25.4) | 48.0(31, 64) | Reference | |||
| GT | 174(52.1) | 48.0(27, 65) | 1.29(0.85–1.95) | 0.227 | ||
| GG | 75(22.5) | 43.0(32, 67) | 1.19(0.73–1.95) | 0.484 | ||
| GT + GG | 249(74.6) | 48.0(27, 65) | 1.26(0.85–1.87) | 0.252 | ||
| Yes | 39 | |||||
| TT | 14(35.9) | 41.0(27, 92) | Reference | |||
| GT | 16(41.0) | 40.0(12, 60) | 3.71(0.99–13.94) | 0.052 | ||
| GG | 9(23.1) | 18.0(12, 24) | 39.37(7.27–213.22) | <0.001 | ||
| GT + GG | 25(64.1) | 24.0(12, 57) | 6.51(1.89–22.36) | 0.003 | ||
Note: DFS, disease–free survival time; HR, hazard ratio; CI, confidence interval.
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the association between SNP and disease-free survival time (DFS) were estimated by Cox regression adjusted by age of onset, BMI, tumor size, lymph-node involvement, histological type, histological grade, menopausal status, vascular invasion, breast or ovarian cancer history, surgical method, taxane/anthracycline-based chemotherapy and radiotherapy.
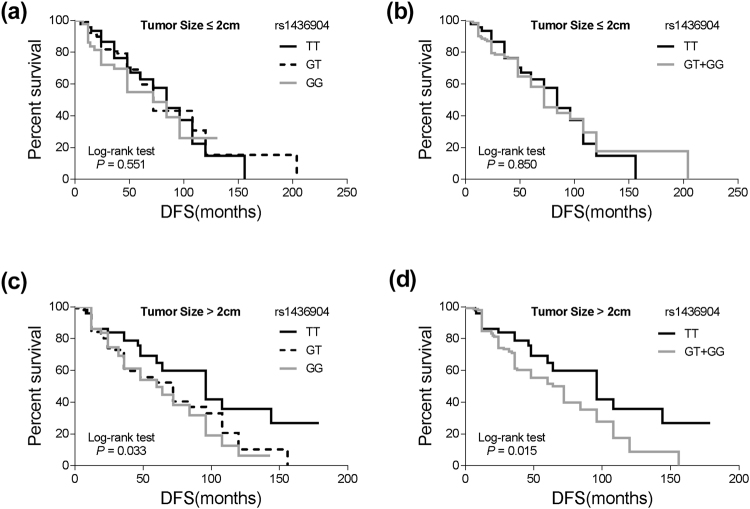
In the subgroup of TNBC patients harboring large tumors (tumor size > 2 cm), the CHST9 rs1436904 G allele (GT and GG genotype) was associated with a significantly increased risk of disease progression (HR = 1.88, 95% CI = 1.06–3.35; P = 0.032) compared to the TT genotype. The mean DFS of the G allele carriers was obviously shorter compared to the cases with the TT genotype (46.4 months vs. 55.9 months; P = 0.015) (Table 2 and Fig. 2). However, such differences were not observed in patients with small tumors (tumor size ≤ 2 cm), indicating that the CHST9 rs1436904 polymorphism was an independent prognostic marker of TNBC cases with large tumors.
Figure 2.
Stratified analyses of DFS for TNBC patients with different genotypes of CHST9 rs1436904 genetic variations according to tumor size. (a,b) DFS for the patients harboring small tumors (≤2 cm; P = 0.551 or 0.850 for different classifications); (c,d) DFS for the patients harboring large tumors (>2 cm; P = 0.033 or 0.015 for different classifications).
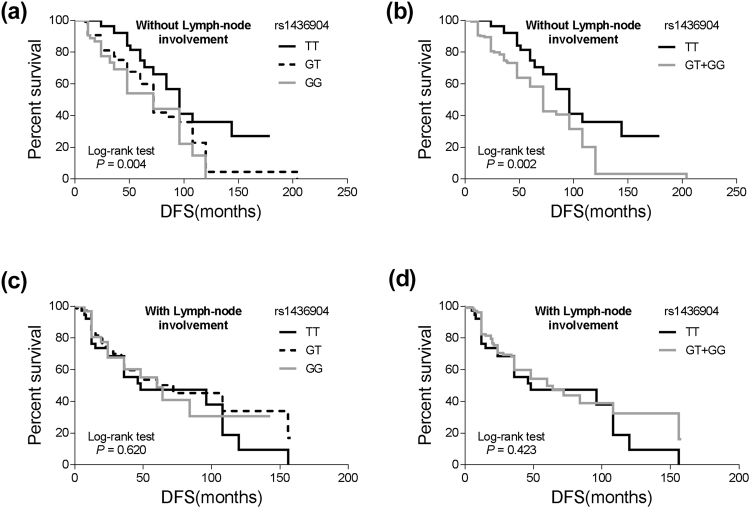
Among the TNBC patients without lymph-node metastasis, the mean DFS of the CHST9 rs1436904 GG genotype was significantly shorter than that of the TT genotype patients (48.1 months vs. 62.6 months; P = 0.033) (Table 2 and Fig. 3). However, there was no such association between the polymorphism and DFS in patients with lymph-node metastasis (mean DFS of the TT, GT and GG genotypes: 48 months, 72 months and 60 months, respectively; P = 0.620). HRs, calculated from the multivariate Cox proportional hazards model, demonstrated that patients without lymph-node metastasis harboring CHST9 rs1436904 GG or GT/GG genotype showed 2.27-fold or 2.01-fold increased risk for disease progression (P = 0.033 or 0.017, respectively) compared to the TT genotype patients (Table 2).
Figure 3.
Stratified analyses of DFS for TNBC patients with different genotypes of CHST9 rs1436904 genetic variations according to lymph-node metastasis. (a,b) DFS for the patients without lymph-node metastasis (P = 0.004 and 0.002 for different grouping mode); (c,d) DFS for the patients with lymph-node metastasis (P = 0.620 and 0.423 for different grouping mode).
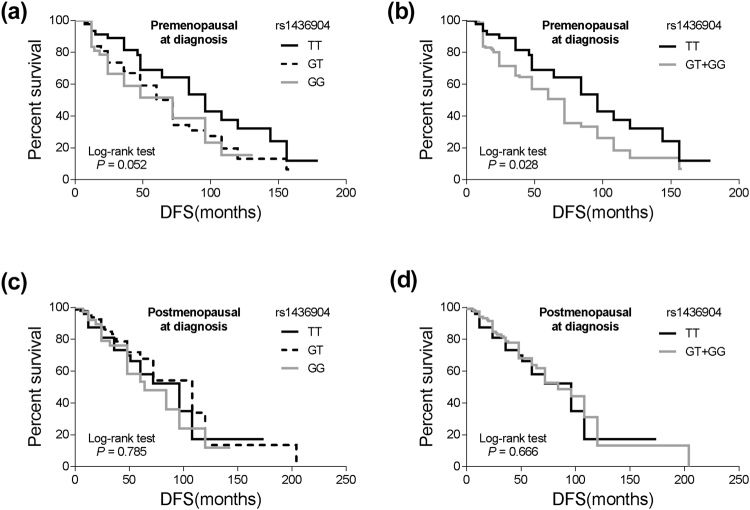
Among the patients who were premenopausal at diagnosis, the median DFS of either the rs1436904 GT or GG genotype patients (48.0 months or 36.0 months) was shorter than that of the rs1436904 TT genotype patients (48.0 months). However, there was no such association between the polymorphism and DFS in patients being postmenopausal at diagnosis (Table 2 and Fig. 4). In the multivariate Cox proportional hazards model, premenopausal patients with the rs1436904 GG genotype showed 2.18-fold increased risk for disease progression (95% CI = 1.12–4.22, P = 0.021) compared to subjects with the TT genotype (Table 2). Similar results were observed among premenopausal patients with the rs1436904 GT genotype (HR = 2.07, 95% CI = 1.18–3.65, P = 0.011) (Table 2).
Figure 4.
Stratified analyses of DFS for TNBC patients with different genotypes of CHST9 rs1436904 genetic variations according to menstrual status. (a,b) DFS for the patients being premenopausalat at diagnosis (P = 0.052 and 0.028 for different grouping mode); (c,d) DFS for the patients being postmenopausalat at diagnosis (P = 0.785 and 0.666 for different grouping mode).
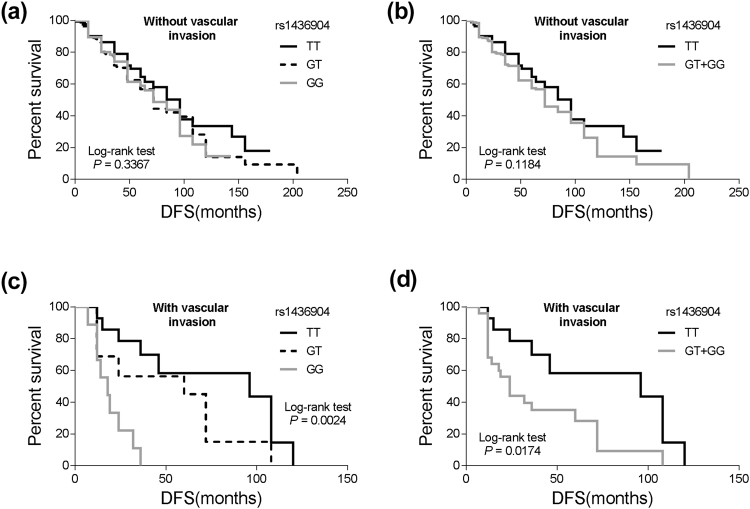
In the subgroup of TNBC patients with vascular invasion, the CHST9 rs1436904 G allele was significantly associated with an increased risk of disease progression compared to the TT genotype (HR = 6.51, 95% CI = 1.89–22.36; P = 0.003). Especially, TNBC patients with GG genotype showed 39.37-fold increase risk for disease progression (P < 0.001). However, such differences were not observed in patients without vascular invasion (Table 2 and Fig. 5).
Figure 5.
Stratified analyses of DFS for TNBC patients with different genotypes of CHST9 rs1436904 genetic variations according to vascular invasion. (a,b) DFS for the patients being premenopausalat at diagnosis (P = 0.337 and 0.1184 for different grouping mode); (c,d) DFS for the patients being postmenopausalat at diagnosis (P = 0.0024 and 0.0174 for different grouping mode).
Discussions
Development of breast cancer are multiple-process consequences of combined genetic and epigenetic changes3,4. About five to ten percent of breast cancer cases are believed to be hereditary and associated with certain gene mutations20–22. Although multiple breast cancer susceptibility genes have been identified, new sets of susceptibility genes should also be identified. TNBC accounts for 12–24% of breast cancers associating with early recurrence and poor outcome. Additional efforts should be made to discover specific loci or genetic variants related to TNBC risk, which will expand our understanding of the etiology of this aggressive breast cancer and improve its prevention and clinical diagnosis. Currently, genome-widely analysis was performed to discover and validate genetic variants that are associated with breast cancer risk using large consortia15–18. Multiple novel breast cancer genetic susceptibility loci were identified and validated base on this approach24. Based on a breast cancer GWAS, which identified AQP4 rs527616 and CHST9 rs1436904 genetic variants, we explored their involvement in early-stage TNBC and concluded that the CHST9 rs1436904 SNP is an independent prognostic genetic variant in Chinese TNBC patients. These results will provide new prevention and diagnosis targets in TNBC therapy.
In Michailidou et al.’s study24, the G (minor allele) in relative to T (major allele) is a relative “protective” risk against breast cancer (OR = 0.96, 95% CI = 0.94–0.98). However, we did observe the significant role of the same SNP on prognosis of TNBC. There might be multiple reasons. First of all, the purpose of our study is to identify prognostic markers, but the GWAS study was aimed to identify susceptibility SNPs. As a result, the observation of different results due to different study purpose might be possible. For example, ERCC1 C118T was associated with lung cancer risk. The OR was 0.90 (95% CI = 0.81–0.99, P = 0.043) in an additive genetic model (C allele vs. T allele) and 0.77 (95% CI: 0.63–0.95, P = 0.013) in a recessive genetic model (CC/CT vs. TT)30. However, ERCC1 C118T was proved to be a risk SNP of overall survival for platinum-based chemotherapy in Asian NSCLC patients (CT + TT versus CC: HR = 1.24, 95% CI = 1.01–1.53)31.
The two SNPs examined in this study locate in AQP4 and CHST9 gene. AQP4 belongs to AQP family and functions in water maintaining and ion homeostasis32. It is located at membrane and cytoplasmic fraction and markedly decreased in tumor tissues compared to paired-adjacent tissue, thus indicating its pathogenic role during cancer development33. CHST9 belongs to the N-acetylgalactosamine 4 sulfotransferase (GalNAc4ST) family, which transfers sulfate to position 4 of nonreducing terminal GalNAc residues34. Sulfate group I carbohydrates play important roles in conferring highly specific functions on glycoproteins, glycolipids, and proteoglycans35. It plays an important role in hematologic malignancies because CHST9 copy number variants (CNV) are associated with acute myelogenous leukemia (AML)36. Additionally, CHST9 CNV and amplification are also found in the brain of schizophrenia patients and gastric cancer patients with metastatic lymph node37,38. Nevertheless, the role of CHST9 as well as its genetic variations in breast cancer, especially TNBC, has not been determined. Our results show that CHST9 rs1436904 SNP significantly contributes to early-stage TNBC progression risk. Notably, our results also show that the CHST9 rs1436904 G allele is a “risk” genetic variant for outcome of TNBC patients, significantly associated with shorten DFS in TNBC patients harboring big tumors (>2 cm), without metastasis, being premenopausal at diagnosis or with vascular invasion.
In all, to the best of our knowledge, our study for the first time identified an inherited variation in CHST9 which was significantly associated with DFS of TNBC patients, especially in TNBC patients harboring big tumors, without lymph-node metastasis, being premenopausal at diagnosis or with vascular invasion. Our findings might have potential clinical implications on precision treatment of TNBC, and eventually affect the therapeutic efficacy.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
Acknowledgements
This study was financially supported by National Natural Science Foundation of China (31671300); the National High-Tech Research and Development Program of China (2015AA020950); Taishan Scholars Program of Shandong Province (tsqn20161060); Natural Science Foundation of Shandong Province (ZR2016HQ48).
Author Contributions
M.Y. and F.M. conceived and designed the experiments; J.Y. and N.Z. performed the experiments; J.Y. and N.Z. analyzed the data; H.Z., J.L. and H.X. contributed materials/analysis tools; M.Y., J.Y. and F.M. wrote the manuscript. All authors reviewed and approved the manuscript prior to submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jupeng Yuan and Nasha Zhang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12306-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fei Ma, Email: mafei2011@139.com.
Ming Yang, Email: aaryoung@yeah.net.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson L. Breast cancer: challenges, controversies, breakthroughs. Nat Rev Clin Oncol. 2010;7:669–670. doi: 10.1038/nrclinonc.2010.192. [DOI] [PubMed] [Google Scholar]
- 4.Ignatiadis M, Sotiriou C. Luminal breast cancer: from biology to treatment. Nat Rev Clin Oncol. 2013;10:494–506. doi: 10.1038/nrclinonc.2013.124. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, J. S., Hickey, T. E., Tarulli, G. A., Williams, M. & Tilley, W. D. Deciphering the divergent roles of progestogens in breast cancer. Nat Rev Cancer (2016). [DOI] [PubMed]
- 6.Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nat Rev Cancer. 2010;10:205–212. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- 7.Narod SA. Modifiers of risk of hereditary breast and ovarian cancer. Nat Rev Cancer. 2002;2:113–123. doi: 10.1038/nrc726. [DOI] [PubMed] [Google Scholar]
- 8.Moran MS. Radiation therapy in the locoregional treatment of triple-negative breast cancer. Lancet Oncol. 2015;16:e113–122. doi: 10.1016/S1470-2045(14)71104-0. [DOI] [PubMed] [Google Scholar]
- 9.Brewster AM, Chavez-MacGregor M, Brown P. Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry. Lancet Oncol. 2014;15:e625–634. doi: 10.1016/S1470-2045(14)70364-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 11.Hudis CA, Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16(Suppl 1):1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- 12.Herold CI, Anders CK. New targets for triple-negative breast cancer. Oncology (Williston Park) 2013;27:846–854. [PubMed] [Google Scholar]
- 13.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Teng W, Qu Y, Wang H, Yuan Q. Sulforaphene inhibits triple negative breast cancer through activating tumor suppressor Egr1. Breast Cancer Res Treat. 2016;158:277–286. doi: 10.1007/s10549-016-3888-7. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, et al. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res Treat. 2013;140:151–157. doi: 10.1007/s10549-013-2615-x. [DOI] [PubMed] [Google Scholar]
- 16.Lv Z, et al. Association of functional FEN1 genetic variants and haplotypes and breast cancer risk. Gene. 2014;538:42–45. doi: 10.1016/j.gene.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 17.Sun T, et al. Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res. 2008;68:7025–7034. doi: 10.1158/0008-5472.CAN-08-0806. [DOI] [PubMed] [Google Scholar]
- 18.Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12:381–394. doi: 10.1038/nrclinonc.2015.73. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen FC, van Overeem Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16:599–612. doi: 10.1038/nrc.2016.72. [DOI] [PubMed] [Google Scholar]
- 20.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 21.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 22.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 23.Fasching PA, et al. Role of genetic polymorphisms and ovarian cancer susceptibility. Mol Oncol. 2009;3:171–181. doi: 10.1016/j.molonc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(353–361):361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CP, et al. Breast cancer risk assessment using genetic variants and risk factors in a Singapore Chinese population. Breast Cancer Res. 2014;16:R64. doi: 10.1186/bcr3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, et al. Integrative Functional Genomics Implicates EPB41 Dysregulation in Hepatocellular Carcinoma Risk. Am J Hum Genet. 2016;99:275–286. doi: 10.1016/j.ajhg.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, et al. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci Rep. 2016;6:31969. doi: 10.1038/srep31969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi M, et al. The Sp1-mediaded allelic regulation of MMP13 expression by an ESCC susceptibility SNP rs2252070. Sci Rep. 2016;6:27013. doi: 10.1038/srep27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, et al. Association of a functional RAD52 genetic variant locating in a miRNA binding site with risk of HBV-related hepatocellular carcinoma. Mol Carcinog. 2015;54:853–858. doi: 10.1002/mc.22156. [DOI] [PubMed] [Google Scholar]
- 30.Xu TP, Shen H, Liu LX, Shu YQ. Association of ERCC1-C118T and -C8092A polymorphisms with lung cancer risk and survival of advanced-stage non-small cell lung cancer patients receiving platinum-based chemotherapy: a pooled analysis based on 39 reports. Gene. 2013;526:265–274. doi: 10.1016/j.gene.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Han Y, et al. A Significant Statistical Advancement on the Predictive Values of ERCC1 Polymorphisms for Clinical Outcomes of Platinum-Based Chemotherapy in Non-Small Cell Lung Cancer: An Updated Meta-Analysis. Dis Markers. 2016;2016:7643981. doi: 10.1155/2016/7643981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulos MC, Saadoun S, Verkman AS. Aquaporins and cell migration. Pflugers Arch. 2008;456:693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J, et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiraoka N, Misra A, Belot F, Hindsgaul O, Fukuda M. Molecular cloning and expression of two distinct human N-acetylgalactosamine 4-O-sulfotransferases that transfer sulfate to GalNAc beta 1>4GlcNAc beta 1–>R in both N- and O-glycans. Glycobiology. 2001;11:495–504. doi: 10.1093/glycob/11.6.495. [DOI] [PubMed] [Google Scholar]
- 35.Hooper LV, Manzella SM, Baenziger JU. From legumes to leukocytes: biological roles for sulfated carbohydrates. FASEB J. 1996;10:1137–1146. doi: 10.1096/fasebj.10.10.8751716. [DOI] [PubMed] [Google Scholar]
- 36.Walter MJ, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai M, et al. Assessment of copy number variations in the brain genome of schizophrenia patients. Mol Cytogenet. 2015;8:46. doi: 10.1186/s13039-015-0144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ema A, et al. Identification of EGFR expression status association with metastatic lymph node density (ND) by expression microarray analysis of advanced gastric cancer. Cancer Med. 2015;4:90–100. doi: 10.1002/cam4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.