Abstract
Prevalence of hepatitis C virus (HCV) genotypes in Bosnia and Herzegovina (B&H) is an issue that is not sufficiently researched and there is a need for studies that would explore this in detail.
The aim of this study was to determine the distribution of HCV genotypes in the group of patients with chronic hepatitis C and also in the group of first time blood donors that tested positive for anti HCV antibodies during the blood screening process. Our secondary goal was to compare the proportions of HCV genotypes between these two groups.
We analyzed 75 blood samples of patients with confirmed chronic hepatitis C. We also analyzed 13/16082 blood samples of first time blood donors found to be HCV positive during the blood screening process. We also determined HCV genotype in HCV RNA positive samples.
We have found that genotype 1b was more prevalent in chronic hepatitis C patients (52/75; 69,3%) than in first time blood donors (6/13; 46,1%), however this difference was not statistically significant (χ2=1,721; df=1; p=0,19). Genotype 1a was more prevalent in the group of first time blood donors (3/13; 23,1%) than in the group of chronic hepatitis C patients (3/75; 4%), but this was also with limited statistical significance (χ2=3,71; df=1; p=0,054). We have not found any significant difference in prevalence of genotypes 1a (p=0,2) and genotypes 3 (p=0,70) when compared between chronic patients (3/75 and 16/75; respectively) and first time blood donors (3/13 and 4/13; respectively).
Our study confirmed domination of genotype 1b in the region of northeastern B&H which is in accordance with HCV genotype prevalence in other countries in our part of Europe.
Keywords: hepatitis C virus, genotypes, blood donors, Bosnia and Herzegovina
INTRODUCTION
Hepatitis C virus (HCV) is an RNA virus, a member of Flaviviridae family, that has a size of 55-65 mm and is classified as a member of hepaciviruses. (1) It has been discovered in 1989 as a cause for post transfusion nonA, non-B hepatitis (2). HCV as many other RNA viruses has an impressive mutation capability which allows for a quick adaptation on immune pressure of the host as well as on the antiviral treatment (3,4). Therefore, HCV circulates in serum not as a single species but rather as a population of quasispecies with various differences of up to 1-5% in viral genome (5,6,7). This particular heterogenicity of HCV has an impact on development of chronicity during the natural course of infection since changes in proteins of viral envelope do occur more rapidly under the immunologic pressure. These genetic variations of HCV are also the reason for treatment failure.
HCV classification is based on comparison and grouping of genomic sequences and widely accepted classification is based on phylogenetic analysis of genomic sequence. Simmonds et al have created genetic tree based on nucleotide sequence of 76 isolates from various parts of the World (8). Based on this analysis they defined 4 hierarchical levels in HCV classification: type, subtype, isolate and quasispecies. Sequences are grouped into 6 main genotypes with more than 50 subtypes. It is considered that numerous subtypes of HCV occurred during endemic infection in a single part of World and then spread to other parts. HCV is classified in 6 genotypes and 3 subtypes (a,b,c). Some genotypes (1b, 2a and 2b) are disseminated throughout the World, while some are more frequent in particular areas like genotypes 3 and 6 in India and southeast Asia, genotypes 1,2 and 4 in central and western Africa and genotypes 1,2 and 3a in western Europe and USA (9,10,11). European studies demonstrate change in epidemiologic picture with the increase in frequencies of genotypes 1a and 3a and decrease in frequencies of 2a/2b and 1b, especially within younger age groups which is mainly due to high prevalence of genotypes 1a and 3a among intravenous drug users (12,13,14). Genotypes 1 and 4 are more treatment resistant than genotypes 2 and 3 and some studies show that with genotype 1 b infection a more aggressive and more severe liver disease occurs, when compared with other genotypes (15,16,17). Prevalence of HCV genotypes in Bosnia and Herzegovina is an issue that is not sufficiently researched and there is a need for studies that would explore this problem in detail.
Our goal was to determine the distribution of HCV genotypes in the group of patients with chronic hepatitis C, treated in University Clinical Center Tuzla and also in the group of first time blood donors that tested positive for anti HCV antibodies during the blood screening process. Our secondary goal was to compare the proportions of HCV genotypes between these two groups.
MATERIALS AND METHODS
We analayzed 75 blood samples of patients with chronic hepatitis C treated in our institution within the time period of 2006-2008. A total of 16082 blood donor samples were also routinely analyzed for presence of anti HCV antibodies (anti HCV) as a part of blood screening protocol. We have found 13 blood samples of blood donors that were found to be anti HCV positive during the blood screening process. All patients gave informed consent for participation in study which was also approved by Ethical Committee of University Clinical Center Tuzla.
All patients that were initially found to be positive for HCV antibodies were tested with qualitative PCR HCV RNA test (Cobas Amplicor, Roche diagnostics GmbH, Manheim, Germany). We extracted HCV RNA from 200 μ! of serum by using a lytic agent. Samples with a lytic agent were incubated for 10 minutes on 60o C. RNA was then precipitated with centrifuging on 12000 rounds per minute (rpm) in absolute iso-propranol and in 70-percent alcohol. Isolated RNA was then stored on -20°C until reverse transcription and amplification reaction, but no longer than 7 days.
Frozen RNA samples were left to defrost on room temperature before performing reverse transcription and amplification reaction. Reverse transcription and amplification reaction was performed in one step by using a thermostabile recombinant enzyme Thermus ther-mophilus DNA Polymerase-rTth. In each PCR reaction both positive and negative controls were included. After amplification, PCR product was detected by reverse hybridization with qualitative reading of the end result.
The very same PCR product of the qualitative HCV RNA test was used for determination of the genotype. Genotypes in HCV RNA positive samples were determined by using a method of reverse hybridization with Innolip HCV II test (Innogenetics, Belgium). This particular test allows for distinction of 6 main types and subtypes (1a, 1b, 1a/1b, 2a/2c, 2b i 3a).
Statistical analysis
All statistical analysis procedures were performed using SPSS 12.0 (SPSS, Chicago, IL, USA). Standard test of descriptive statistics have been used for determination of baseline characteristics of groups. Between-group differences in frequencies have been investigated by using chi-squared test with Yates correction where appropriate. Statistical level of 95% (p<0,05) was considered as significant for all performed tests.
RESULTS
After determining HCV genotype in 75 patients with chronic hepatitis C, we detected presence of following 5 genotypes: 1a, 1b, 2, 3 i 4.
Table 1 demonstrates distribution of HCV genotypes in 75 chronic hepatitis C patients. Among 75 chronic patients 52 (69,3%) were males and 23 (30,7%) were females with male to female ratio of 2,26 to 1.
TABLE 1.
Hepatitis C virus genotypes: prevalence of genotypes according to gender and age group in chronic hepatitis C patients
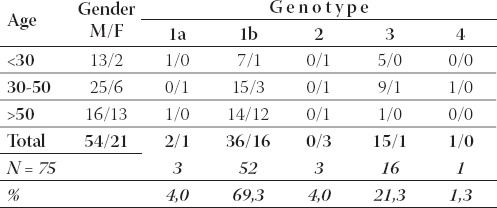
Within the age group of patients under 30 years of age, genotype 1b was the most frequent genotype (8/15; 53,3%), while genotype 3 was found in 5/15 (33,3%) cases. The most frequent genotype within the age group of 30 to 50 years was again 1b (18/31; 58,1%) followed by genotype 3 (10/31; 32,2%). Remaining 3 geno-types (1a, 2, 4) were detected in 3/31 (9,7%) of cases.
Genotype 1b was also found to be the most prevalent in patients over 50 years (26/29; 89,6%). Remaining 3 genotypes (1a, 2, and 3) were found in 9/29 (10,4%) of cases. Prevalence of genotypes in the group of first time blood donors is somewhat different than in chronic hepatitis B patients. Table 2 demonstrates distribution of HCV genotypes in first time blood donors.
TABLE 2.
Hepatitis C virus genotypes: prevalence of genotypes according to gender and age group in first time blood donors
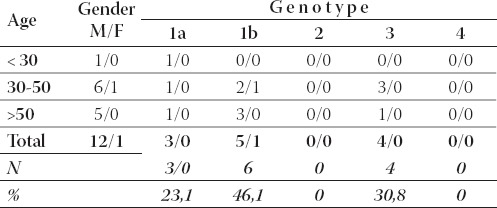
A total of 13 anti HCV positive blood samples were found among 16082 blood donor samples, or 0,81 positives per 1000 samples. Among first time blood donors, males were mostly infected with genotype 1b (5/12; 41,6%), followed by genotype 3 (4/12; 33%) and genotype 1a (3/12; 25,4%). Most of the infected first time blood donors belonged to age group of 30 to 50 years (7/13; 53,8%). Only one female first time blood donor was detected with HCV (genotype 1b).
The most frequent genotype in the complete group of first time blood donors was 1b (6/13; 46,1%), followed by genotype 3 (4/13; 30,7%) and genotype 1a in 3/13 (23,2%) patients. We have found that genotype 1b was more prevalent in chronic hepatitis C patients (52/75; 69,3%) than in first time blood donors (6/13; 46,1%), however this difference was not statistically significant (χ2=1,721; df=1; p=0,19). Genotype 1a was more prevalent in the group of first time blood donors (3/13; 23,1%) than in the group of chronic hepatitis C patients (3/75; 4%), but this was also on the very limit of statistical significance (χ2=3,71; df=1; p=0,054).
We have not found any significant difference in prevalence of genotypes 1a (p=0,2) and genotypes 3 (p=0,70) when compared between chronic patients (3/75 and 16/75; respectively) and first time blood donors (3/13 and 4/13; respectively). Genotypes 2 and 4 were indeed found to be more frequent in chronic hepatitis C patients, but bearing in mind small number of patients in the group of first time blood donors we did not make any statistical comparison.
DISCUSSION
The most frequent HCV genotypes in Bosnia and Herzegovina according to study conducted by Vu- kobrat-Bijedic in 54 treated chronic hepatitis C patients were as follows: genotype 1a in 25 (46%), genotype 1b in 12 (22%), genotype 3 in 12 (22%), genotype 2a in 1(2%) and genotype 4 in 4 (8%) patients (18).
We have found that in our region of Bosnia and Herzegovina genotype 1b was also the most frequent in both chronic hepatitis C patients and first time blood donors. Numerous studies demonstrated dominance of this particular genotype in southern and southeastern Europe (13,16,19).
Genotype 3 was found to be on the second place in prevalence in our sample, mostly in age group from 30 to 50 years of age and in under 30. This is in accordance with other studies which showed that genotype 3 is associated with HCV infection in younger intravenous drug users (7,12,13). Genotype 1a is also associated with younger age in patients with chronic hepatitis C (19,21).
Dominance of genotype 1b in our sample can be explained by fact that HCV genotype 1b is the most frequent in infections transmitted by transfusion of blood and blood products. During the time period of 1992 to 1995 a war in our country resulted with a great number of wounded persons which necessitated the transfusion of blood and blood products. Blood screening for hepatitis C virus did not existed at that time in our country. Prevalence of anti-HCV in patients which received blood transfusion during the war was found to be 4,6%, according to our study that analyzed that particular risk group (20). Those patients had a high prevalence of genotype 1b. Practical implications of genotype 1b are numerous. One of the most important is that patients with both genotype 1a and 1b do not respond to antiviral treatment as good as patients with other HCV genotypes (4,10,13). Additionally, current guidelines recommend that HCV patients with genotype 1 should be treated with pegylated interferon and ribavirin for 48 weeks which significantly increases the cost of treatment.
It was considered that genotype 4 was endemic for regions of Africa and Near East. However, it is becoming more and more frequent in infected patients with hemophilia and intravenous drug users and it has been found in 1.3% patients with chronic hepatitis C (7,13). Croatian researchers revealed similar distribution of genotypes in 203 patients (13).
It appears that there is not a fundamental difference in genotype distribution between HCV positive blood donors and hepatitis C patients. Genotype 1 is predominant and it has been acknowledged in several reports that genotype 1 predominates in the geographic region of the southeast Balkan.(22) As noted in the same paper, (22) the second common prevalence of genotype 3, especially among HCV patients, suggests the increased number of intravenous drug users (IVDU) as the next most frequent mode of HCV transmission. Our results are also showing lower percentage of genotypes 1a, 2 and 4 mainly due to the fact that most of our patients were infected after blood transfusion - a transmission route associated with genotype 1b.
CONCLUSION
Determination of geographic distribution of HCV genotypes provides important information regarding the origin of this particular virus, routes of transmission and it also provides data that is the base for selection of antiviral treatment. Our study confirmed domination of genotype 1b in the region of north-eastern Bosnia and Herzegovina which is in accordance with HCV genotype prevalence in other countries in south-eastern part of the Europe. Genotype distribution in our region suggests that blood transfusion is the main transmission route followed by transmission in IVDU group.
It is also important to acknowledge several limitations of our study. Sample size is the obvious one, particularly among blood donors. We have managed to detect 13 HCV positive samples in the pool of 16082 blood donors. Therefore, to have the sample of more than 70 HCV positive blood donors we would have to screen around 100000 samples, which would significantly lengthen the research. We also did not determined quantitative HCV RNA levels of the patients and therefore were unable to investigate the association between genotype and viral load.
REFERENCES
- 1.Simmonds P, Muttimer D, Follet E. Hepatitis C. Topley and Wilson’s: Microbiology and microbial infections. ¡. London: Virology Arnold; 1998. pp. 717–744. [Google Scholar]
- 2.Choo Q.-L, Kuo G, Weiner A.J, Overby L.R, Bradley D.W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;224:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 3.Martell M, Esteban J.I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes:quasispecies nature of HCV genome distribution. J. Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeuzem S, Lee J.-H, Franke A, Ruster B, Prummer O, Herrmann G, Roth W.K. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology. 1998;27:1149–1156. doi: 10.1002/hep.510270433. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q.L, Richman K.H, Han J.H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P.J, Weiner A.J, Bradly D.W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi N, Higashi H, Kaminaka K, et al. Molecular heterogenecity of the human hepatitis C virus (HCV) genome. J. Hepatol. 1993;17(suppl.3):94–107. doi: 10.1016/s0168-8278(05)80432-5. [DOI] [PubMed] [Google Scholar]
- 7.Zein N.N. Clinical significance of Hepatitis C virus genotypes. Clinical Microbiology Reviews. 2000;13:223–235. doi: 10.1128/cmr.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmonds P, Holmes E.C, Cha T.-A, Chan S.-W, McOmish F, Irvine B, Beall E, Yap P.L, Kolberg J, Urdea MS. Classification of hepatitis C virus into si major genotypes and a series of subtypes by phylogenetic analysis of the NS-5region. J. Gen. Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 9.Anonnymous. Recommendations for Prevention and Control of Hepatitis C virus (HCV) infection and HCV-Related Chronic Disease. CDC MMWR. 1998;47:19. [PubMed] [Google Scholar]
- 10.Maertens G, Stuyver L. Harrison TJ, Zuckerman AJ, editors. Genotiypes and genetic variation of hepatitis C virus. The molecular medicine of viral hepatitis. John Willey&Sons. 1997:183–322. [Google Scholar]
- 11.Vardas E, Sitas F, Seidel K, Casteling A, Sim J. Prevalence of hepatitis C virus antibodies and genotypes in asymptomatic, firsttime blood donors in Namibia. Bulletin of the World Health Organization. 1999;77:965–971. [PMC free article] [PubMed] [Google Scholar]
- 12.Soriano V, Bravo R, Garcia-Samaniego J, Castro A, Caraballo E, Gonzales-Anglada I, Martinez-Odriozola P, Colmenero M, Pedreira J. Circulating hepatitis C virus genotypes in Spain. Vox Sang. 1996;70:181–182. doi: 10.1111/j.1423-0410.1996.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 13.Vince A, Kutela N, Sonicky Z, Jeren T, Radovani M. HCV genotypes in patients with chronic C hepatitis in Croatia. Infection. 1998;26:173–177. doi: 10.1007/BF02771846. [DOI] [PubMed] [Google Scholar]
- 14.Naumov N.V. HCV in Estern Europe. EASL International Consensus Conference on Hepatitis C. 1999:25–28. [Google Scholar]
- 15.Nousbaum J.S.B, Pol B, Nalpas B, et al. Hepatitis C virus type 1b (II) infection in France and Italy. Ann Intern Med. 1995;122:161–168. doi: 10.7326/0003-4819-122-3-199502010-00001. [DOI] [PubMed] [Google Scholar]
- 16.Urbánek P, Mareček P, Brodanova M, Brùha R, Petrtýl J, Dušek L. Large randomised clinical trials and regular practice in a tertiary hepatology centre. Čes a Slov Gastroent a Hepatol. 2008;62(Suppl 2):89–90. [Google Scholar]
- 17.Fried M.W, Shiffman M.L, Reddy R, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl. J. Med. 2002:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 18.Vukobrat-Bijedić Z, Husić-Selimović A, Radović S, Gornjaković S, Gogov B, Zubčević N, Bilalović N, Koluder N. Evaluacija genotipa hepatitis C virusa i njegov uticaj na terapijski odgovor pegiliranim interferonom alfa 2a(40) kD u kombinaciji sa Riba-virinom. Med Arh. 2007;61(4):221–224. [PubMed] [Google Scholar]
- 19.Fabri M, Klašnja B, Ružić M, Pobor M, Tomislav P. Epidemiološke karakteristike hepatitis C virusne infekcije anti-HCV pozitivnih osoba lečenih u Klinici za infektivne bolesti u Novom Sadu. Acta Infectologica Yugoslavica. 2003;8:19–24. [Google Scholar]
- 20.Ahmetagić S, Muminhodžić K, Čičkušić E, Stojić V, Petrović J, Tihić N. Hepatitis C infection in risk groups. Bosnian Journal of Basic Medical Sciences. 2006;6(4):13–17. doi: 10.17305/bjbms.2006.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svirtlih N, Delic D, Simonovic J, Jevtovic D, Dokic L, Gvozde-novic E, Boricic I, Terzic D, Pavic S, Neskovic G, Zerjav S, Urban V. Hepatitis C virus genotypes in Serbia and Montenegro: The prevalence and clinical significance. World J Gastroenterol. 2007;21-13(3):355–360. doi: 10.3748/wjg.v13.i3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]


