Summary
The RNA-degrading exosome mediates the processing and decay of many cellular transcripts. In the yeast nucleus, the ubiquitous 10-subunit exosome core complex (Exo-9–Rrp44) functions with four conserved cofactors (Rrp6, Rrp47, Mtr4, and Mpp6). Biochemical and structural studies to date have shed insights into the mechanisms of the exosome core and its nuclear cofactors, with the exception of Mpp6. We report the 3.2-Å resolution crystal structure of a S. cerevisiae Exo-9–Mpp6 complex, revealing how linear motifs in the Mpp6 middle domain bind Rrp40 via evolutionary conserved residues. In particular, Mpp6 binds near a tryptophan residue of Rrp40 that is mutated in human patients suffering from pontocerebellar hypoplasia. Using biochemical assays, we show that Mpp6 is required for the ability of Mtr4 to extend the trajectory of an RNA entering the exosome core, suggesting that it promotes the channeling of substrates from the nuclear helicase to the processive RNase.
Keywords: X-ray crystallography, exosome, ribosome biogenesis, KH domain, helicase, pontocerebellar hypoplasia
Graphical Abstract
Highlights
-
•
Yeast Mpp6 is stably bound to the nuclear exosome core both in vivo and in vitro
-
•
The Mpp6 middle domain binds the Rrp40 exosome subunit with conserved interactions
-
•
Mpp6 enhances the ability of the Mtr4 helicase to channel RNA into the exosome core
-
•
The pontocerebellar W238R mutation in human EXOSC3 affects the hMPP6-binding site
Falk et al. provide insights into the structure and function of the nuclear RNA exosome. The authors elucidate how the nuclear cofactor Mpp6 is recruited to the exosome core complex and show that it facilitates the threading of RNA substrates from the Mtr4 helicase to the Rrp44 RNase.
Introduction
The eukaryotic RNA exosome is a RNase complex that completely degrades or partially trims RNA substrates by successively cleaving their 3′ end nucleotides. The exosome is conserved from yeast to humans and acts on a wide variety of RNAs in turnover, surveillance, and processing pathways (Chlebowski et al., 2013, Kilchert et al., 2016). In the nucleus, the yeast exosome eliminates, for example, transcripts that are in excess or are defective, such as pre-mRNAs that failed to complete splicing and tRNAs that lack specific base modifications, and also destroys non-coding RNAs that are produced from leaky transcription initiation (reviewed in Chlebowski et al., 2013, Kilchert et al., 2016). Besides these destructive functions, the nuclear exosome is also involved in the maturation of rRNAs and small nuclear or small nucleolar RNAs (sn[o]RNAs) (Allmang et al., 1999). Defects in the nuclear functions of the exosome have been recently linked to a mutation in one of the exosome core subunits (yeast Rrp40/human EXOSC3) that is associated with a motor neuron degeneration disease (Fasken et al., 2017, Gillespie et al., 2017).
The processive core of the RNA exosome is a 10-protein complex (Exo-10) containing a single RNase subunit (Rrp44) and a catalytically inactive barrel (Exo-9), formed by six RNase PH-like proteins and three S1-KH “cap” proteins (reviewed in Zinder and Lima, 2017). Using biochemical and structural studies on the S. cerevisiae complex, we have previously shown that an RNA substrate is channeled from the entry pore at the top of Exo-9 to the Rrp44 exoribonuclease site via an internal conduit that is spanned by about 30 nucleotides (Bonneau et al., 2009, Makino et al., 2013a). The 30-nucleotide footprint of Exo-10 in vitro is reminiscent of a pre-rRNA processing defect common to yeast strains lacking functional nuclear exosome cofactors, namely the accumulation of a 5.8S rRNA precursor with a 3′ extension of 30 nucleotides (reviewed in Butler and Mitchell, 2011).
Studies over the last two decades have converged on four conserved nuclear exosome cofactors: Rrp6; Rrp47; Mtr4; and Mpp6 (reviewed in Butler and Mitchell, 2011, Kilchert et al., 2016). Rrp6 and its interacting partner Rrp47 are constitutively bound nuclear exosome subunits. Rrp6 contains a distributive 3′-5′ RNase domain that is positioned at the top of Exo-9 barrel (Makino et al., 2013a, Wasmuth et al., 2014, Zinder et al., 2016). Rrp47 does not have enzymatic activity but, together with Rrp6, forms a binding platform for recruiting Mtr4 (Schuch et al., 2014). Mtr4 is an essential helicase believed to assist the exosome by presenting it with suitably remodeled substrates that can be threaded with their unwound 3′ end into the degradation core (Johnson and Jackson, 2013, Makino et al., 2013b). In contrast to Rrp6, Rrp47, and Mtr4, there is currently no mechanistic structural information on Mpp6, a small basic protein lacking recognizable domains. Yeast Mpp6 is physically associated with the nuclear exosome in vivo (Milligan et al., 2008) and in vitro (Schuch et al., 2014) and is expected to bind near the cap protein Rrp40 (Shi et al., 2015). The precise role of Mpp6 in the nuclear functions of the exosome has remained elusive. In this work, we set out to shed light on the mechanisms with which yeast Mpp6 binds to and cooperates with the exosome.
Results and Discussion
Yeast Mpp6 Binds the Exosome Core with High Affinity via the Middle Domain
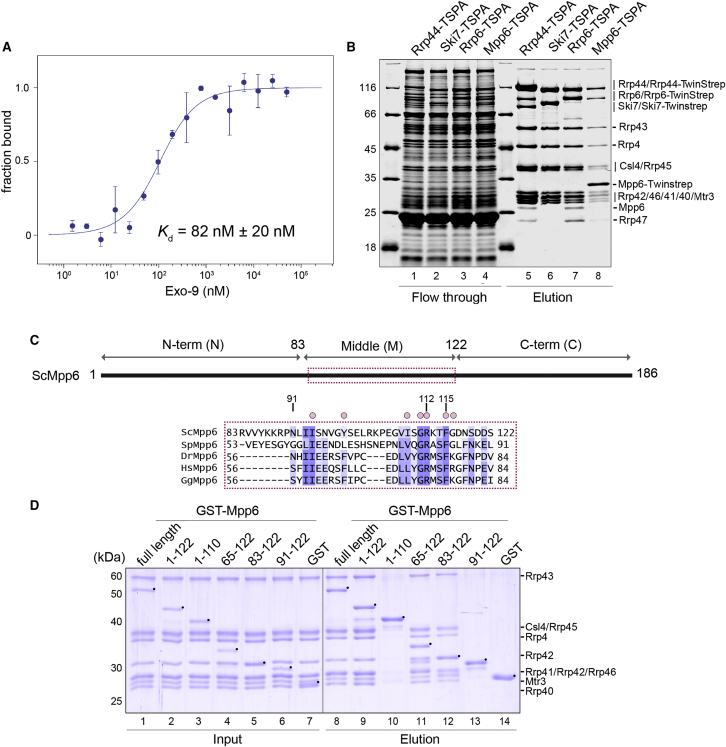
We had previously shown that S. cerevisiae Mpp6 binds Exo-9 independently of Rrp6-Rrp47 and Rrp44 (Schuch et al., 2014). To obtain a quantitative measure of the binding affinity, we introduced a single cysteine residue near the C terminus of full-length Mpp6 (Mpp6FL S184C substitution), labeled the purified protein with a cysteine-reactive fluorophore (red-maleimide) and performed microscale thermophoresis (MST) to analyze in the interaction with Exo-9. The MST measurements revealed that the Mpp6–Exo-9 dissociation constant is in the high nanomolar range (Kd ∼82 nM; Figure 1A), indicating a relatively tight binding, albeit one order of magnitude weaker than that of Rrp6 (Kowalinski et al., 2016). Next, we compared endogenous nuclear exosomes purified from yeast strains genetically engineered to contain a Twin Strep-ProtA tag (TSPA) at the C terminus of either Mpp6 or Rrp6. In addition to tagging the nuclear cofactors, we also engineered strains with TSPA-tagged Ski7 (the cytoplasmic cofactor whose binding to Exo-9 is mutually exclusive with that of Rrp6; Kowalinski et al., 2016) and TSPA-tagged Rrp44 (the ubiquitous RNase that is present both in the nuclear and cytoplasmic exosome complexes). As expected, tandem-affinity purifications from the strain expressing Rrp44-TSPA yielded the exosome subunits, the cytoplasmic cofactor Ski7, and the nuclear cofactors Rrp6, Rrp47, and Mpp6 (Figure 1B, lane 5), whereas purifications from a strain expressing Ski7-TSAP precipitated the exosome core, but not the nuclear cofactors (Figure 1B, lane 6). In purifications from the Rrp6-TSPA-tagged strain, we detected the exosome subunits, Rrp47 and Mpp6 (Figure 1B, lane 7). Vice versa, purification using Mpp6-TSPA yielded the exosome, Rrp6 and Rrp47 (Figure 1B, lane 8). Thus, Mpp6 and Rrp6-Rrp47 can be part of the same endogenous exosome complexes. As a note, Mpp6 appears to be sub-stoichiometric with respect to other exosome components, either due to technical issues or its estimated lower abundance (Kulak et al., 2014). None of the purifications resulted in a significant co-precipitation of Mtr4 in the conditions we used (Figure 1B, lanes 5–8), suggesting that this is a transiently associated rather than a constitutively bound cofactor of the nuclear exosome.
Figure 1.
Mpp6 Binds with High Affinity to the Exo-9 Core via the Middle Domain
(A) The binding of Mpp6 to the Exo-9 core was measured with MST. The titration of Exo-9 ranged from 1.5 nM to 50 μM with a constant concentration of labeled Mpp6-S184C at 50 nM. Data were analyzed by thermophoresis and temperature jump mode. The error bars represent the SD of each data point calculated from three independent measurements.
(B) Tandem-affinity purification of exosome complexes using different bait proteins C-terminally tagged with a Twin Strep-ProtA tag (TSPA). Eluates were TCA precipitated and analyzed on 12% SDS-PAGE and stained with Instant Blue (Expedeon). Lanes 1–4 correspond to the flowthrough, and lanes 5–8 correspond to the elution.
(C) Schematic domain organization of yeast Mpp6, with the three domains (N-terminal, middle, and C-terminal) indicated. The conserved sequence motif of the middle domain is shown below the schematic representation (red box). The sequence alignment includes orthologs from the representative species. The level of conservation is indicated by color, from dark blue (high conservation) to white (no conservation). Above the sequence, residues of Mpp6M that interact with the Rrp40 are labeled with a salmon circle.
(D) Protein co-precipitations by GST pull-down assays. GST-tagged yeast Mpp6FL and truncations were purified and mixed with purified Exo-9. Pull-down assays were carried out using GSH-Sepharose beads in a buffer containing 100 mM NaCl. The Coomassie-stained 12% SDS-PAGE gels show the input (lanes 1–7) and the pulled-down protein precipitates (lanes 8–14). Black dots denote the bait protein.
Next, we mapped the domain of Mpp6 responsible for binding Exo-9. S. cerevisiae Mpp6 (186 residues) is an intrinsically disordered protein (Figures S1A and S1B). The N-terminal two-thirds of Mpp6 are evolutionary conserved, particularly at two hotspots between residues 11–21 and 111–115 (Milligan et al., 2008; Figures 1C and S1C). The C-terminal third is generally less conserved and contains a high percentage of positively charged residues. We purified several versions of glutathione S-transferase (GST)-tagged Mpp6 proteins and tested their interaction with untagged Exo-9 in pull-down assays with glutathione-agarose beads (Figure 1D). Removal of the C-terminal region of Mpp6 still allowed efficient binding to Exo-9 (Mpp61–122; Figure 1D, lane 9). However, a further C-terminal truncation (Mpp6 residues 1–110) impaired binding in the pull-down assay (Figure 1D, lane 10), suggesting the conserved 111- to 115-residue hotspot (Milligan et al., 2008) is involved in Exo-9 binding. Next, we truncated Mpp6 from the N terminus to progressively remove patches of conserved residues (Figures 1D, lanes 11–13, and S1C). We found that Mpp6 could be truncated to residues 83–122 and still interact with Exo-9 (Figure 1D, lane 12). However, a further truncation to residues 91–122 impaired binding (Figure 1D, compare lanes 12 and 13), suggesting that the 111- to 115-residue hotspot is not sufficient for Exo-9 binding. We concluded that the middle domain of Mpp6 (residues 83–122 or Mpp6M) contains the major Exo-9-binding determinants (Figures 1C and S1C). We proceeded to obtain the three-dimensional structure of a minimal Exo-9–Mpp6 complex.
Overall Structure of Exo-9 Bound to Mpp6
Although we could obtain several crystal forms from Mpp6-containing exosome samples, upon solving these structures, we could only observe density for Exo-9. We reasoned that crystal-packing interactions might interfere with Mpp6 binding. From previous crosslinking and mass-spectrometry experiments, Mpp6 was expected to reside in close proximity to Rrp40 (Shi et al., 2015). Inspection of all crystal forms we had obtained indeed revealed that a surface of Rrp40 was involved in lattice interactions with an Rrp4 subunit from an adjacent complex in the crystal lattice (Figure S2A). To disrupt this contact without affecting Rrp40, we introduced mutations on the surface of Rrp4 involved in crystal packing (I66E, M68E, or Rrp4mut). The Exo-9Rrp4mut–Mpp6 complex yielded diffracting crystals with four copies of the complex in the asymmetric unit (Figure S2B). Inspection of the map showed density for Mpp6M in two of the four copies. The final model was refined to 3.2 Å resolution with Rfree of 26.2% and good stereochemistry (Table 1).
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Dataset | Exo-9Rrp4mut–Mpp6M |
|---|---|
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 166.8, 237.4, 201.9 |
| α, β, γ (°) | 90.0, 110.4, 90.0 |
| Data Collection | |
| Wavelength (Å) | 1.25 |
| Resolution (Å) | 151.20–3.20 |
| No. of reflections | 996,533 |
| No. of unique reflection | 229,497 |
| Rmerge (%) | 24.3 (229.9) |
| Rpim (%) | 11.4 (129.8) |
| I/σI | 5.7 (0.6) |
| Completeness (%) | 97.7 (79.9) |
| Multiplicity | 4.3 |
| CC1/2 | 98.5 (24.3) |
| Refinement | |
| Resolution (Å) | 80.76–3.20 (3.31–3.20) |
| No. of unique reflections | 229,411 |
| Rwork/Rfree (%) | 22.5/26.2 |
| Wilson B factor (Å2) | 72.27 |
| Average B factors (Å2) | 95.3 |
| No. of Atoms | |
| Proteins | 66,857 |
| Ligands | 53 |
| Stereochemistry | |
| Root-mean-square deviation (RMSD) bond lengths (Å) | 0.003 |
| RMSD bond angles (°) | 0.52 |
| Ramachandran favored (%) | 96.0 |
| Ramachandran outliers (%) | 0.4 |
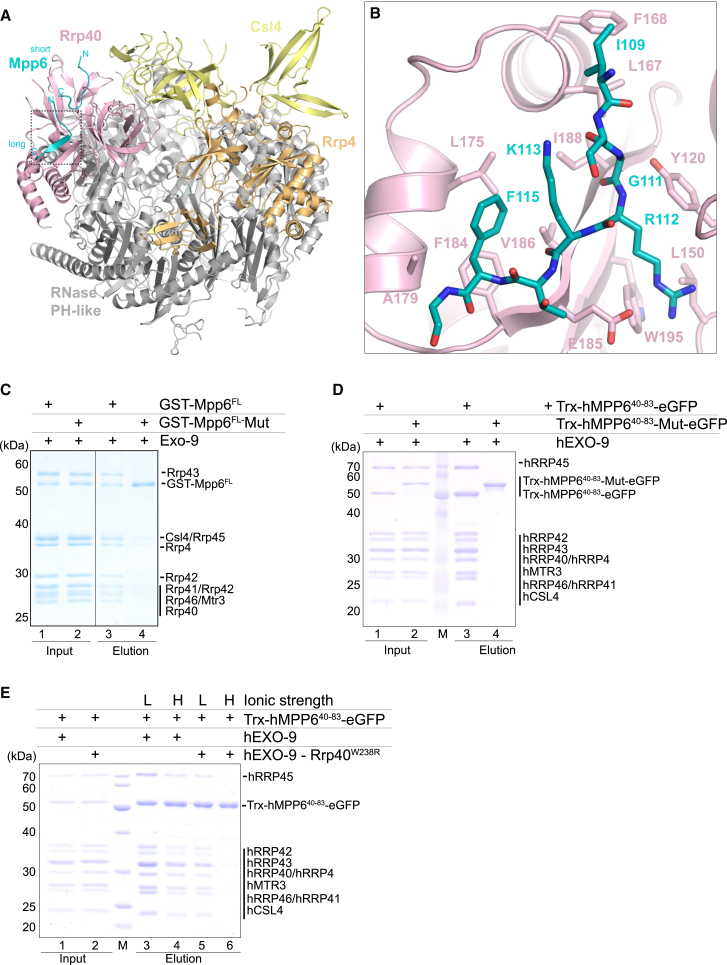
In the Mpp6-bound structure, Exo-9 has the typical barrel-like architecture that has been previously described for other exosome complexes, with a ring of six RNase PH-like proteins (Mtr3-Rrp42-Rrp41-Rrp45-Rrp46-Rrp43) capped by a ring of three S1/KH proteins (Rrp4, Rrp40, and Csl4; Kowalinski et al., 2016, Liu et al., 2006, Makino et al., 2013a, Makino et al., 2015, Wasmuth et al., 2014, Zinder et al., 2016). The most significant difference with previous X-ray structures is that the density for Csl4 is generally not well defined, in line with the increased flexibility of this subunit in the absence of Rrp6 (Makino et al., 2013a, Wang et al., 2007). Mpp6M binds on the surface of the Rrp40 subunit with two extended segments (Figure 2A).
Figure 2.
Crystal Structure of Mpp6 Middle Domain Bound to Exo-9 (to Rrp40)
(A) Crystal structure of the S. cerevisiae Exo-9–Mpp6 complex (shown in a “front” view). The RNase PH-like core is in gray and the cap proteins Csl4, Rrp4, and Rrp40 in yellow, orange, and salmon, respectively. Mpp6 is shown in cyan, and the N- and C-termini of Mpp6 are indicated.
(B) Zoom-in views of the Mpp6-Rrp40 interaction interface at the long region in the same colors as in (A). Conserved residues involved in the interaction are shown in stick representation.
(C) Protein co-precipitations with GST-tagged yeast Mpp6FL and Mpp6 R112E/F115A (Mpp6-Mut). The pull-down assays were carried out as in Figure 1D. The Coomassie-stained 12% SDS-PAGE gels show the input (lanes 1 and 2) and the pulled-down protein precipitates (lanes 3 and 4).
(D) Protein co-precipitations with Trx-eGFP-tagged human MPP6 residues 40–83 (Trx-hMPP640–83-eGFP) and MPP6 R74E/F77A (Trx-hMPP640–83-Mut-eGFP). The pull-down assays were carried out using GFP-binder Sepharose beads in a buffer containing 100 mM NaCl. The Coomassie-stained 12% SDS-PAGE gels show the input (lanes 1 and 2) and the pulled-down protein precipitates (lanes 3 and 4).
(E) Protein co-precipitations with Trx-eGFP-tagged human MPP6 residues 40–83 (Trx-hMPP640–83-eGFP) and purified hEXO-9 (either wild-type or hExo-9-Rrp40W238R). Pull-down assays were carried out using GFP-binder Sepharose beads in either 100 mM or 500 mM NaCl. The Coomassie-stained 12% SDS-PAGE gels show the input (lanes 1 and 2) and the pulled-down protein precipitates (lanes 3–6).
The Mpp6 Middle Domain Wraps around Rrp40 with Evolutionary Conserved Interactions
Rrp40 contains three domains. The N-terminal domain is an all β-fold that binds on top of the Rrp46 subunit (Liu et al., 2006). The central S1 domain is a β-barrel characteristic of oligonucleotide/oligosaccharide-binding (OB)-fold proteins. The C-terminal domain is a type 1 KH domain, with a 3-stranded β-sheet packed against an α-helical bundle (Oddone et al., 2007). The KH β-sheet packs against the S1 domain, forming a single structural module that binds across the top of Rrp45, with the KH domain positioned on the outside of the ring and the S1 domain on the inside.
The longer segment of Mpp6 binds in a pocket formed between the S1 and KH domains of Rrp40 and could be unambiguously assigned to residues 108–117 (Figures 2B and S2C). This segment starts with van der Waal contacts of Ile109Mpp6 with the first helix of the KH domain (at Leu167Rrp40 and Phe168Rrp40). It continues with a sharp 90° turn at Gly111Mpp6 and ends with a β-strand that packs against and extends the β-sheet of the KH domain. The side chains of Lys113Mpp6 and Phe115Mpp6 protrude on one side of the β-strand toward the KH helical bundle and bind in a hydrophobic pocket lined by Phe184Rrp40, Val186Rrp40, Ile188Rrp40 and Leu175Rrp40, Ala179Rrp40. The side chain of Arg112Mpp6 protrudes on the other side of the β- strand toward the S1 domain, engaging the aliphatic portion in apolar contacts with Tyr120Rrp40 and Leu150Rrp40 and engaging the guanidinium group in an electrostatic interaction with Glu185Rrp40. Consistent with the structural analysis, mutation of Arg112Mpp6 and Phe115Mpp6 (R112E, F115A mutant) impaired the interaction of Mpp6FL with Exo-9 in pull-down assays with recombinant proteins (Figure 2C, compare lanes 3 and 4).
In the case of the shorter segment of Mpp6, the interpretation of the density was rather ambiguous. This segment extends on a conserved surface of the Rrp40 N-terminal domain (above the Leu21-Gly22-Pro23-Gly24 loop) and contacts the edge of the KH helical bundle (at Arg164, Phe168). Guided by the density features of Mpp6, sequence conservation, results of the mapping experiments (Figure 1D), and by chemical considerations, we tentatively assigned the sequence of the Mpp6 shorter segment to residues 90–99 (Figure S2D). The structural model is consistent with geometric restraints deduced from previous mass spectrometry-crosslinking data (e.g., a distance of less than 30 Å between crosslinked Mpp6 Lys104–Rrp40 Lys49 and between crosslinked Mpp6 Lys113–Rrp40 Lys176; Figure S2E; Shi et al., 2015).
The Human EXOSC3 W238R Disease Mutation Affects Binding of Mpp6
The evolutionary conservation of the S. cerevisiae Mpp6-Rrp40 interactions suggested that the human orthologs hMPP6-EXOSC3 engage in a similar binding mode. Mutations of human Rrp40 (EXOSC3) are associated with pontocerebellar hypoplasia, a severe and often deadly motor neuron disease in children (Wan et al., 2012). The most severe phenotypes are linked to the mutation of Trp238EXOSC3 to arginine (Wan et al., 2012). Previous studies have shown that the corresponding mutation of yeast Trp195Rrp40 to arginine (W195R) affects cell growth and impacts specifically on the nuclear functions rather than the cytoplasmic functions of the exosome (Fasken et al., 2017), particularly rRNA processing (Gillespie et al., 2017). From the Exo-9–Mpp6 structure, the yeast Rrp40 W195R mutation is predicted to cause an unfavorable and likely incompatible electrostatic environment for accommodating Arg112Mpp6 (corresponding to human hMPP6 Arg74). We tested the effect of mutating hMPP6 and EXOSC3 in in vitro binding experiments with recombinant proteins. Whereas the middle domain of hMPP6 interacted with human EXO-9 in the pull-down assays, an hMPP6 mutant containing the R74E, F77A substitution failed to do so (Figure 2D), similarly to the results we had obtained with the corresponding yeast Mpp6 R112E, F115A mutant (Figure 2C). Next, we reconstituted a human hEXO-9 complex containing the W238R disease mutation. Under low ionic strength conditions (100 mM NaCl), the interaction between hMPP6 and hEXO-9 mutant was weakened (Figure 2E, compare lanes 3 and 5). Applying more stringent washing conditions (e.g., increasing the ionic strength to 500 mM NaCl) strongly reduced the binding of hMPP6 to the hEXO-9 mutant (Figure 2E, compare lanes 4 and 6). This suggests that the EXOSC3 W238R mutant destabilizes the hMPP6-binding surface of EXOSC3, providing a mechanistic explanation for the impact of this mutation on the functions of the nuclear exosome. Whether hMPP6 recruitment is the ultimate deficit in pontocerebellar hypoplasia patients remains unclear, because in human cells, the hMPP6-binding surface could in principle be used to recruit another nuclear cofactor and destabilization of this surface might also lead to instability of the protein in vivo (Fasken et al., 2017).
The Presence of Mpp6 in the Nuclear Exosome Supports Efficient RNA Channeling through Mtr4
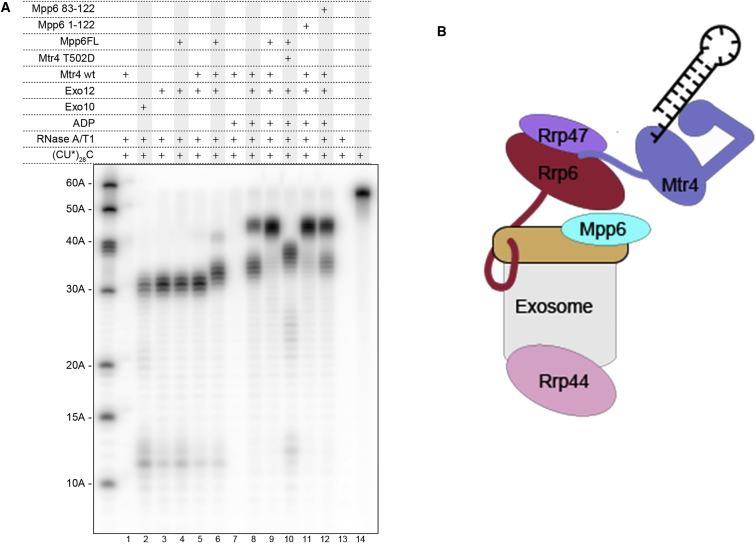
How does Mpp6 binding on top of the exosome core impact on the biochemical properties and functions of the nuclear exosome? Because Mpp6 has been shown to have RNA-binding properties (Milligan et al., 2008, Schilders et al., 2005), we analyzed its contribution to the nuclear exosome complex using RNase protection assays (Figures 3A and S3). Using these assays, we have previously shown that yeast Exo-10 has an RNA-binding footprint of about 30 nucleotides (Bonneau et al., 2009), reflecting the RNA-binding path in the internal channel of Exo-10 (Makino et al., 2013a). The presence of Rrp6-Rrp47 on Exo-10 intensified but did not shift the size of the protected fragments (Figure 3A, lanes 2 and 3). This is consistent with the stabilizing effects of Rrp6 (Makino et al., 2013a) and with the features of the Exo-10–Rrp6–Rrp47 (Exo-12) structure (Makino et al., 2015). Addition of the Mtr4 helicase led to a partial shift to 45-nucleotide-long RNA fragments but only in the presence of ADP (compare lane 8 and lane 5). Further addition of Mpp6 completely shifted the protection pattern to the 45-nucleotide fragments in the presence of ADP (lane 9). Because Mtr4 is an ATP-dependent RNA helicase, we speculate that productive RNA binding and channeling depends on the presence of nucleotides.
Figure 3.
Mpp6 Supports Efficient RNA Channeling through the Mtr4–Exosome Complex
(A) RNase protection patterns of the nuclear exosome. A single-stranded (C∗U)28C RNA, internally 32P-labeled at the uridine phosphate, was incubated with different combinations of components of the nuclear exosome as indicated and treated with RNase A/T1 (as in Bonneau et al., 2009). The reaction products were analyzed by denaturing PAGE and phosporimaging. RNA fragments of ∼30-nt length accumulated in the presence of Exo-10 and Exo-12. Longer RNA fragments of ∼45 nt accumulated in the presence of Exo-12 and Mtr4, ADP, and Mpp6.
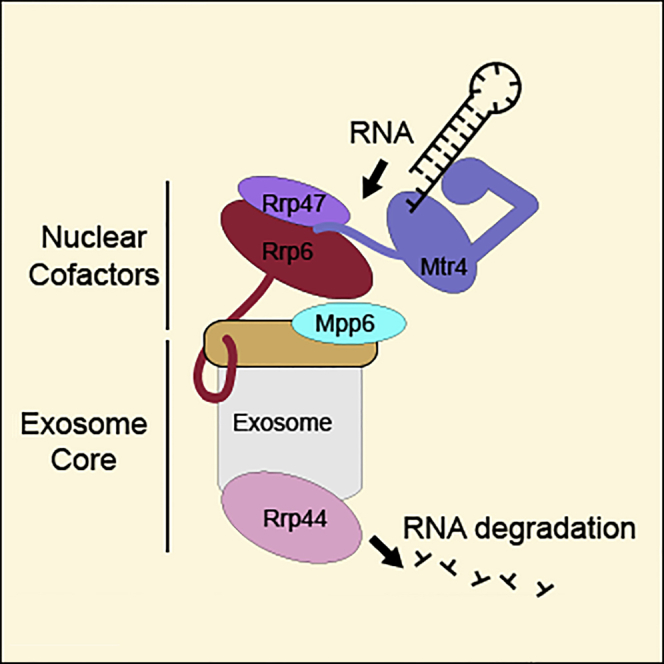
(B) Model of Mtr4 recruitment and RNA targeting to the nuclear exosome complex. The model recapitulates structural features and interactions of nuclear exosome components from this work and from previous studies (Weir et al., 2010, Jackson et al., 2010, Makino et al., 2013a, Makino et al., 2015, Schuch et al., 2014, Wasmuth et al., 2014, Zinder et al., 2016).
These results suggested that, in the context of 14-subunit complex, Mpp6 helps recruiting or holding Mtr4 in a conformation whereby the helicase channel can form a continuous conduit with the exosome core. To confirm the presence of an Mtr4-channel-dependent path to the exosome, we introduced an unfavorable negative charge by substituting a conserved RNA-binding residue in the helicase channel (Thr502Asp; Weir et al., 2010). The presence of the Mtr4 T502D mutant indeed impaired the 45-nucleotide pattern (lane 10), leading to an intermediate shift (whose significance is currently unclear). Interestingly, the C-terminal domain of Mpp6 could be removed without affecting the accumulation of the 45-nucleotide fragments (Mpp61–122; lane 11). However, the Exo-9-binding domain of Mpp6 (Mpp6M; residues 83–122) was not as efficient as the fragment encompassing residues 1–122 in promoting the accumulation of the 45-nucleotide fragments, suggesting that the N-terminal domain contributes to hold Mtr4 in the RNA-channeling conformation on top of the exosome core, although the detailed mechanisms are currently unclear.
In summary, the emerging picture from our study is that Mpp6 stably binds Exo-12 to form a nuclear exosome population (Exo-13) that can effectively recruit the more transient fourteenth subunit, Mtr4 (Figure 3B). We speculate that, in an endogenous context, protein-protein interactions might not be sufficient to recruit Mtr4 and that the helicase might have to be productively engaged with an RNA substrate to be efficiently targeted to the nuclear exosome. Such targeting would in turn result in the channeling of the RNA substrate from the unwinding helicase to the processive RNase of the nuclear exosome complex.
Experimental Procedures
See the Supplemental Information for detailed methods.
Recombinant Protein Expression and Purification
Exosome proteins and cofactors were expressed and purified as previously described (Falk et al., 2014, Greimann and Lima, 2008, Makino et al., 2013a). The yeast Exo-9–Mpp6 complex was reconstituted by mixing Exo-9 with 1.2-fold molar excess of Mpp6 full-length followed by gel filtration. Human MPP6 residues 40–83 were tagged with an N-terminal His6-thioredoxin-tag (His-Trx) and a C-terminal eGFP-StrepII-tag.
Endogenous Protein Purification
All yeast strains generated here are derivatives of the base strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). General yeast manipulations were conducted by standard methods.
Biochemical Assays
Pull-down assays with GST-tagged S. cerevisiae Mpp6 were carried out with GSH Sepharose (GE Healthcare). Pull-down assays with GFP-tagged human MPP6 were carried out with GFP binder resin. The tagged bait proteins were incubated with 1.2 molar excess of untagged prey (Exo-9) and washed three times with a buffer containing either 100 mM or 500 mM NaCl. RNase protection was carried out as previously described (Bonneau et al., 2009).
Biophysical Assays
The microscale thermophoresis experiments were carried out with Mpp6 S184C labeled with red maleimide incubated with increasing concentrations of unlabeled Exo-9 in a buffer containing 150 mM NaCl. Thermophoresis was measured with an light-emitting diode (LED) power of 40% and standard parameters on a NanoTemper Monolith NT.115 machine. Titrations were performed in triplicates, and the data were analyzed using the Thermophoresis and T-Jump strategy option with the MO software (NanoTemper Technologies).
Crystallization and Structure Determination
The best diffracting crystals of the Exo-9–Mpp6 complex were obtained at 12 mg/mL in 0.1 M Tris/Mops (pH 7.5), 30 mM MgCl2, 30 mM CaCl2, and 30% polyethylengylcol (PEG) 8000/ethylene glycol. X-ray data were collected at 100 K at the beamline PXII (X10SA) of the Swiss Light Source (SLS) (Villigen, Switzerland). The structure was solved by molecular replacement of the coordinates of Exo-9 from PDB 5JEA (Kowalinski et al., 2016). Data processing, phasing, model building, and refinement were carried out with standard programs (as detailed in Supplemental Information).
Author Contributions
S.F. and E.C. initiated the project; J.E. obtained crystals; A.K. performed the work in Figures 2E and 2F; F.B. performed the work in Figures 2B and 3; S.F. carried out all other experiments in the paper; and S.F. and E.C. wrote the manuscript.
Acknowledgments
We would like to thank Jérôme Basquin, Karina Valer-Saldaña, and Sabine Pleyer at the MPI-Martinsried crystallization facility and the staff of the PX beamlines of the SLS synchrotron for assistance in data collection. This study was supported by the Max Planck Gesellschaft, the European Research Council (ERC Advanced Investigator Grant 294371), and the Deutsche Forschungsgemeinschaft (SFB1035, GRK1721, and Cluster of Excellence CIPSM).
Published: September 5, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.08.033.
Accession Numbers
The accession number for the coordinates and structure factors for the Exo-9–Mpp6 complex reported in this paper is PDB: 5OKZ.
Supplemental Information
References
- Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F., Basquin J., Ebert J., Lorentzen E., Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Butler J.S., Mitchell P. Rrp6, rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 2011;702:91–104. doi: 10.1007/978-1-4419-7841-7_8. [DOI] [PubMed] [Google Scholar]
- Chlebowski A., Lubas M., Jensen T.H., Dziembowski A. RNA decay machines: the exosome. Biochim. Biophys. Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Falk S., Weir J.R., Hentschel J., Reichelt P., Bonneau F., Conti E. The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol. Cell. 2014;55:856–867. doi: 10.1016/j.molcel.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Fasken M.B., Losh J.S., Leung S.W., Brutus S., Avin B., Vaught J.C., Potter-Birriel J., Craig T., Conn G.L., Mills-Lujan K. Insight into the RNA exosome complex through modeling pontocerebellar hypoplasia type 1b disease mutations in yeast. Genetics. 2017;205:221–237. doi: 10.1534/genetics.116.195917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie A., Gabunilas J., Jen J.C., Chanfreau G.F. Mutations of EXOSC3/Rrp40p associated with neurological diseases impact ribosomal RNA processing functions of the exosome in S. cerevisiae. RNA. 2017;23:466–472. doi: 10.1261/rna.060004.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greimann J.C., Lima C.D. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.N., Klauer A.A., Hintze B.J., Robinson H., van Hoof A., Johnson S.J. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.J., Jackson R.N. Ski2-like RNA helicase structures: common themes and complex assemblies. RNA Biol. 2013;10:33–43. doi: 10.4161/rna.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C., Wittmann S., Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- Kowalinski E., Kögel A., Ebert J., Reichelt P., Stegmann E., Habermann B., Conti E. Structure of a cytoplasmic 11-subunit RNA exosome complex. Mol. Cell. 2016;63:125–134. doi: 10.1016/j.molcel.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Makino D.L., Baumgärtner M., Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- Makino D.L., Halbach F., Conti E. The RNA exosome and proteasome: common principles of degradation control. Nat. Rev. Mol. Cell Biol. 2013;14:654–660. doi: 10.1038/nrm3657. [DOI] [PubMed] [Google Scholar]
- Makino D.L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., Conti E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015;524:54–58. doi: 10.1038/nature14865. [DOI] [PubMed] [Google Scholar]
- Milligan L., Decourty L., Saveanu C., Rappsilber J., Ceulemans H., Jacquier A., Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol. Cell. Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddone A., Lorentzen E., Basquin J., Gasch A., Rybin V., Conti E., Sattler M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007;8:63–69. doi: 10.1038/sj.embor.7400856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G., Raijmakers R., Raats J.M., Pruijn G.J. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch B., Feigenbutz M., Makino D.L., Falk S., Basquin C., Mitchell P., Conti E. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 2014;33:2829–2846. doi: 10.15252/embj.201488757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Pellarin R., Fridy P.C., Fernandez-Martinez J., Thompson M.K., Li Y., Wang Q.J., Sali A., Rout M.P., Chait B.T. A strategy for dissecting the architectures of native macromolecular assemblies. Nat. Methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Yourshaw M., Mamsa H., Rudnik-Schöneborn S., Menezes M.P., Hong J.E., Leong D.W., Senderek J., Salman M.S., Chitayat D. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 2012;44:704–708. doi: 10.1038/ng.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-W., Wang J., Ding F., Callahan K., Bratkowski M.A., Butler J.S., Nogales E., Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc. Natl. Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth E.V., Januszyk K., Lima C.D. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511:435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J.R., Bonneau F., Hentschel J., Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc. Natl. Acad. Sci. USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder J.C., Lima C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31:88–100. doi: 10.1101/gad.294769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder J.C., Wasmuth E.V., Lima C.D. Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol. Cell. 2016;64:734–745. doi: 10.1016/j.molcel.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.