Abstract
Sudden unexplained nocturnal death syndrome (SUNDS) is a perplexing disorder to both forensic pathologists and clinic physicians. Clinical features of SUNDS survivors suggested that SUNDS is similar to Brugada syndrome (BrS). Leucine-rich repeat containing 10 (LRRC10) gene was a newly identified gene linked to dilated cardiomyopathy, a disease associated with sudden cardiac death. To investigate the prevalence and spectrum of genetic variants of LRRC10 gene in SUNDS and BrS, the coding regions of LRRC10 were genetically screened in 113 sporadic SUNDS victims (from January 2005 to December 2015, 30.7 ± 7.5 years) and ten BrS patients (during January 2010 to December 2014, 38.7 ± 10.3 years) using direct Sanger sequencing. Afterwards, LRRC10 missense variant carriers were screened for a panel of 80 genes known to be associated with inherited cardiac arrhythmia/cardiomyopathy using target-captured next-generation sequencing. In this study, an in silico-predicted malignant LRRC10 mutation p.E129K was detected in one SUNDS victim without pathogenic rare variant in a panel of 80 arrhythmia/cardiomyopathy-related genes. We also provided evidence to show that rare variant p.P69L might contribute to the genetic cause for one SUNDS victim and two BrS family members. This is the first report of genetic screening of LRRC10 in Chinese SUNDS victims and BrS patients. LRRC10 may be a new susceptible gene for SUNDS, and LRRC10 variant was initially and genetically linked to BrS-associated arrhythmia.
Keywords: Forensic pathology, Sudden cardiac death, Genetics, Leucine-rich repeat containing 10 (LRRC10) gene
Introduction
Sudden unexplained nocturnal death syndrome (SUNDS), also called Lai Tai (died during sleep) in Thailand [1], Pokkuri death syndrome (sudden unexplained death at night) in Japan [2], and Bangungut (moaning and dying during sleep) in the Philippines [3], is most prevalent in Southeast Asia and is characterized by sudden unexplained death during sleep or at rest in apparently healthy young people, most of whom are males [4]. Forensic autopsy, histopathology examination, toxicological analysis, and death-scene investigation have no identifiable abnormalities to explain the cause of death. In southern China, the annual incidence of SUNDS is as high as 2 per 100,000 person-years [4].
Clinical phenotype and functional characterization of SUNDS survivors suggest that some, but not all, SUNDS are most likely the same allelic disorder as Brugada syndrome (BrS), affecting younger males with structurally normal hearts [1, 5]. Similarly, our molecular autopsy of the BrS-associated genes (SCN5A, SCN1B-4B, SCN10A, MOG1, and GPD1-L) could only account for a small part of the genetic cause of SUNDS victims [6, 7], indicating that a majority of SUNDS may be due to other genetic bases.
Leucine-rich repeat containing 10 (LRRC10) is a cardiac-specific and highly conserved protein exclusively expressed in cardiomyocytes [8, 9].Deletion of LRRC10 in mice results in dilated cardiomyopathy (DCM) [9], and mutations in LRRC10 gene have been recently linked to human DCM [10]. Because mutations causing cardiomyopathy (such as arrhythmogenic right ventricular cardiomyopathy (ARVC) and DCM) have been shown to cause arrhythmogenic heart disease without overt structural defects (such as BrS), and vice versa. For example, PKP2 (gene related to ARVC) missense mutations that cause sodium current deficit could yield a BrS phenotype, even in the absence of overt structural feature characteristic of ARVC [11], and SCN5A (gene linked to BrS) missense mutations have been reported in DCM patients [12, 13]. Moreover, LRRC10 has been recognized as a cardiac-specific transcriptional target gene of Nkx2.5 [14], which transcriptionally regulates cardiac ion channels, including Nav1.5, Cav1.2, and ERG [15]. Dysfunction of these cardiac ion channels are all associated with BrS and even sudden cardiac death [16–21]. Here, we tested the hypothesis that genetic variants of LRRC10 may underlie some cases of SUNDS and BrS.
Materials and methods
Study population
From January 2005 to December 2015, 113 sporadic SUNDS cases were enrolled at the Department of Forensic Pathology, Zhongshan School of Medicine, Sun Yat-sen University, in this study. The inclusion criteria for SUNDS are as follows [6]: (1) a southern Han Chinese (most of whom are male) older than or equal to 15 years of age who was (2) healthy without any significant disease (3) prior to experiencing a sudden unexplained death during sleep or at rest and (4) had a negative forensic autopsy, toxicological analysis, and death-scene investigation, resulting in an unexplained death.
Ten BrS patients collected during January 2010 to December 2014 from the Department of Cardiology at the First Affiliated Hospital of Sun Yat-sen University were included. The inclusion criteria for BrS in this study were patients with (1) a basal ECG showing a BrS type I pattern, (2) at least one clinical criterion (documented family history of sudden cardiac death or BrS, and/or symptoms secondary to arrhythmia), and (3) no structural heart disease.
A control population of 220 age- and ethnic-matched unrelated healthy southern Chinese (440 alleles) were provided by the First Affiliated Hospital of Sun Yat-sen University. None of the control subjects had a history of syncope or cardiovascular disease. Genomic data of East Asian (EAS) from the 1000 Genomes Project Phase 3 (1000G, http://browser.1000genomes.org/) and Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/) were also used as a population control.
Genomic DNA extraction
Genomic DNA was extracted from blood samples using DNA IQ™ Casework Pro Kit for Maxwell® 16 (Promega Corporation, Madison, WI, USA), according to standard procedures.
Genetic screening
Genetic screening of the LRRC10 gene (Ensembl Transcript ID: ENST00000361484) was performed on all SUNDS cases and BrS patients using direct Sanger sequencing. PCR products were then direct sequenced on a 3730XL DNA Analyzer with the use of BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Sequence chromatograms were analyzed by Sequencing Analysis 5.2 software (Applied Biosystems, Foster City, CA, USA). To investigate the importance of missense variants in LRRC10 gene, a total of 80 genes associated with inherited cardiac arrhythmia/cardiomyopathy were genetically screened in LRRC10 missense variant carriers using target-captured next-generation sequencing, and then the identified variants were filtered for candidate pathogenic rare variants and confirmed by direct Sanger sequencing as we previously described [22].
Genetic variant analysis
The sequences were compared with the corresponding reference cDNA sequence of the LRRC10 gene using SeqMan™I I exper t sequence analysis software (DNASTAR, Inc., Madison, WI, USA). All suspicious variants were sequenced in both sense and antisense directions. Any variant observed in only SUNDS cases or BrS patients and absent in EAS population from 1000G and ExAC was termed as a mutation. Variant identified in EAS population was annotated as a polymorphism. Polymorphism identified with a minor allele frequency (MAF) less than 0.01 was termed as a rare polymorphism. If the MAF were higher than 0.01, the polymorphism was regarded as a common polymorphism.
Bioinformatic analyses
PROVEAN (http://provean.jcvi.org/index.php) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) were used to evaluate the impact of amino acid substitutions on the structure and function of LRRC10. Evolutionary conservation of P69 and E129 residues of LRRC10 was generated by MUSCLE [23] among different species.
Statistical analysis
MAF were calculated for each SNP site by the allele counting method using the data from direct Sanger sequencing of SUNDS, control, 1000G, and ExAC.
Results
Demographics of study population
The average death age of 113 unrelated SUNDS victims (three cases were females) was 30.7 ± 7.5 years (range 15–50 years). There was no clinic record for all these apparently healthy SUNDS cases. After comprehensive forensic pathological examination, there were no significant morphological changes found to explain the sudden death of SUNDS victims.
All but two of the ten BrS cases were unrelated males; the related cases were from the same family (father and son). The average age at the time of diagnosis was 38.7 ± 10.3 years (range 25–58 years). Seven of ten BrS had suffered previous syncope and seizures. Six of ten BrS cases had suffered an episode of spontaneous ventricular fibrillation and had an implantable cardioverter defibrillator implanted. Eight of ten BrS had a previous family history of sudden unexpected death or BrS. The blood sample and detailed clinic information of family member of all 113 SUNDS and eight BrS cases were not available.
Genetic variants of LRRC10 in SUNDS and BrS
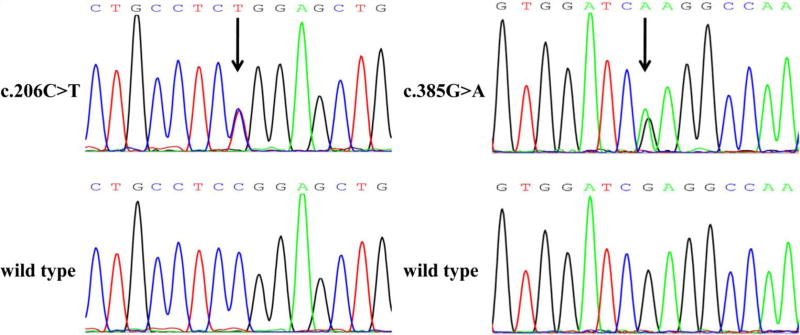
Four LRRC10 genetic variants were found in SUNDS and BrS, including two missense variants, one noncoding region variant, and one synonymous variant (Table 1). The details of these variants are as follows (Fig. S1 in Electronic Supplementary Material online): c.−2G>T was a G to T transition at 5′ UTR of LRRC10 gene and was identified in 1 of 88 SUNDS victims; c.206C>T was a C to T transition at the leucine-rich repeat 1 (LRR1) of LRRC10 leading to the missense exchange of a proline to a leucine residue (p.P69L) and was detected in 1 of 88 SUNDS cases and 2 of 10 BrS patients (Fig. 1); c.385G>A was a G to A transition at the LRR4 of LRRC10 leading to the missense exchange of a glutamic acid to a lysine residue (p.E129K) and was found in 1 of 113 SUNDS victims (Fig. 1); and c.453C>T was a synonymous variant (p.A151A) within the LRR5 of LRRC10 and was identified in 2 of 113 SUNDS victims.
Table 1.
Genetic variants of LRRC10 gene in southern Han Chinese sporadic SUNDS cases and BrS patients
| Region | Nucleotide changea | Amino acid change | Disease | Cases harboring variants/allb |
dbSNP | Tape of variant | Position | PROVEAN | PolyPhen-2 |
|---|---|---|---|---|---|---|---|---|---|
| 5′ UTR | c.−2G>T | Noncoding | SUNDS | 1/88 | rs762951776 | Noncoding | – | Unavailable | Unavailable |
| Exon | c.206C>T | p.Pro69Leu (p.P69L) | SUNDS | 1/88 | rs140389574 | Missense | LRR1 | Deleterious | Benign |
| Exon | c.206C>T | p.Pro69Leu (p.P69L) | BrS | 2/10 | rs140389574 | Missense | LRR1 | Deleterious | Benign |
| Exon | c.385G>A | p.Glu129Lys (p.E129K) | SUNDS | 1/113 | rs762144273 | Missense | LRR4 | Deleterious | Probably damaging |
| Exon | c.453C>T | p.Ala151Ala (p.A151A) | SUNDS | 2/113 | rs12229290 | Synonymous | LRR5 | Neutral | Unavailable |
LRR leucine-rich repeat
Nucleotide number relative to the translation start site
Due to the quality and quantity limit of the SUNDS samples, the sample size of each site that can be successfully detected is different
Fig. 1.
Sequence electropherogram of missense variants in LRRC10 gene. Numbering of the nucleotide starts at the ATG codon. Genetic variants are indicated by an arrow
Of these two missense variants, functional prediction by PROVEAN indicated that p.P69L may affect the function of LRRC10, which was not consistent with PolyPhen-2 (Table 1). p.E129K was predicted to be pathogenic by both PROVEAN and PolyPhen-2 (Table 1). Both P69 and E129 are evolutionarily conserved among different species (Fig. S2 in Electronic Supplementary Material online).
Target-captured next-generation sequencing of LRRC10 missense variant carriers
p.P69L was detected in one SUNDS and two BrS (Table 1). Of note, these two BrS patients (father and son) are from the same family. p.E129K was found only in one SUNDS victims (Table 1). To test the importance of p.P69L and p.E129K, the above carriers of LRRC10 missense variants were genetically screened using target-captured next-generation sequencing for pathogenic rare variants that might be involved in SUNDS and BrS.
In two BrS family members with p.P69L, only two genetic variants were identified among these candidate genes (Table 2). The first variant c.667C>G was a C to G transition in SCN1B gene leading to the missense exchange of a histidine to an aspartic acid residue (p.H223D) and it was detected in both father and son. The second variant c.536A>G was an A to G transition in KCNJ5 gene leading to the missense exchange of a asparagine to a serine residue (p.N179S) and it was only detected in the son. Functional prediction in silico suggested that p.H223D may not alter the function of SCN1B, while p.N179S was predicted to alter the function of KCNJ5 (Table 2). The results suggested that these variants were unlikely underlying BrS. Moreover, there was no pathogenic rare variant found in SUNDS cases with p.P69L or p.E129K in all candidate genes.
Table 2.
Genetic variants in two p.P69L BrS family members
| Patient | Age | Gene | Gene-associated disease |
Nucleotide changea | Amino acid change | dbSNP | Tape of variant | PROVEAN | PolyPhen-2 |
|---|---|---|---|---|---|---|---|---|---|
| Father | 58 years | SCN1B | BrS | c.667C>G | p.His223Asp (p.H223D) | rs754870200 | Missense | Neutral | Benign |
| Son | 40 years | SCN1B | BrS | c.667C>G | p.His223Asp (p.H223D) | rs754870200 | Missense | Neutral | Benign |
| KCNJ5 | LQTS | c.536A>G | p.Asn179Ser (p.N179S) | rs147070381 | Missense | Deleterious | Probably damaging |
Nucleotide number relative to the translation start site
Minor allele frequency of LRRC10 genetic variants
Here, the MAF of four genetic variants in SUNDS were compared with our control and the population data from 1000G and ExAC (Table 3). c.−2G>Twas detected in 1 of 88 SUNDS victims (MAF = 0.00568) while it was either absent in our control, EAS, South Asian (SAS), African, and Latino population or rare in European (EUR) population (1 of 17,998, MAF = 0.00003). c.385G>A was identified in 1 of 113 SUNDS victims (MAF = 0.00442) while it was either absent in our control, EAS, EUR, African, and Latino population or detected in 1 (with unknown clinic phenotype) of 8241 SAS population (MAF = 0.00006). c.206C>T was detected in 1 of 88 SUNDS victims (MAF = 0.00568) and 2 of 10 BrS patients. Its MAF was less than 0.01 in our control, EAS, EUR, and Latino but higher than 0.01 in SAS population. MAF of c.453C>T was 0.00885 (1 of 113) in SUNDS victims and higher than 0.01 in our control, EAS population. In contrast, the MAF of this variant was less than 0.01 in EUR and Latino population. Taken together, these four genetic variants were termed as mutations (c.−2G>T and c.385G>A), rare variant (c.206C>T), and common polymorphism (c.453C>T) in EAS population.
Table 3.
Minor allele frequency of genetic variants in LRRC10 gene
| Nucleotide change | MAF (M/m) SUNDS | Control | EAS-CHSa | EASa | EASb | SASa | SASb |
| Mutations in EAS | |||||||
| c.−2G>T (rs762951776) | 0.00568 (175/1) | 0.00000 (440/0) | Undetected | Undetected | 0.00000 (4028/0) | Undetected | 0.00000 (6256/0) |
| c.385G>A (rs762144273) | 0.00442 (225/1) | 0.00000 (440/0) | Undetected | Undetected | 0.00000 (8640/0) | Undetected | 0.00006 (16,481/1) |
| Rare polymorphism in EAS | |||||||
| c.206C>T (rs140389574) | 0.00568 (175/1) | 0.00000 (440/0) | 0.00000 (210/0) | 0.00000 (1008/0) | 0.00208 (8630/18) | 0.02249 (956/22) | 0.01836 (16,197/303) |
| Common polymorphism in EAS | |||||||
| c.453C>T (rs12229290) | 0.00885 (224/2) | 0.02045 (431/9) | 0.01905 (206/4) | 0.02778 (980/28) | 0.02953 (8379/255) | 0.00000 (978/0) | 0.00000 (16,470/0) |
| Nucleotide change | MAF (M/m) EURa | EURb | AFRa | AFRb | AMRa | Latinob | |
| Mutations in EAS | |||||||
| c.−2G>T (rs762951776) | Undetected | 0.00003 (35,995/1) | Undetected | 0.00000 (5452/0) | Undetected | 0.00000 (4998/0) | |
| c.385G>A (rs762144273) | Undetected | 0.00000 (73,188/0) | Undetected | 0.00000 (10,324/0) | Undetected | 0.00000 (11,560/0) | |
| Rare polymorphism in EAS | |||||||
| c.206C>T (rs140389574) | 0.00000 (1006/0) | 0.00014 (73,176/10) | 0.00000 (1322/0) | 0.00000 (10,376/0) | 0.00000 (694/0) | 0.00009 (11,551/1) | |
| Common polymorphism in EAS | |||||||
| c.453C>T (rs12229290) | 0.00000 (1006/0) | 0.00001 (73,131/1) | 0.00000 (1322/0) | 0.00000 (10,284/0) | 0.00000 (694/0) | 0.00017 (11,548/2) |
MAF minor allele frequency, M major allele, m minor allele
1000 Genomes Project Phase 3
ExAC
Discussion
LRRC10 is a cardiac-specific and highly conserved protein in embryonic and adult cardiomyocytes [8, 9]. Knockdown of LRRC10 caused severe cardiac morphological and functional defects [24]. Human LRRC10 contains no known functional domains other than its seven leucine-rich repeat (LRR) motifs (LRR1–7) [8], which function as protein interaction domains [25, 26]. LRRC10 can physically and endogenously interact with α-actinin and α-actin in the heart [9], forming a cytoskeletal complex possibly through its LRR motifs. Mutations in α-actinin and α-actin have been linked to human idiopathic DCM [27, 28]. In mice, deletion of LRRC10 results in DCM [9]. Most recently, the identification of rare variants of LRRC10 in unrelated idiopathic DCM patients confirmed LRRC10 as a novel DCM-susceptibility gene [10]. LRRC10 is also needed to maintain cardiac function in response to pressure overload [29]. Taken together, LRRC10 is necessary for normal cardiac development and function.
Does LRRC10 also play role in maintaining normal cardiac rhythm? LRRC10 has been recognized as a cardiac-specific transcriptional target gene of Nkx2.5 [14], which regulates cardiac ion channels, including Nav1.5 (encoded by SCN5A gene), Cav1.2 (encoded by CACNA1C gene), and ERG (encoded by KCNH2 gene) [15]. Dysfunction of these cardiac ion channels are all linked to BrS and even sudden cardiac death [16–21]. Based on these facts, we propose that LRRC10 may be associated with cardiac ion channel disease. Here, we identified two rare LRRC10 missense variants in SUNDS and BrS, which linked LRRC10 to primary arrhythmia-associated diseases for the first time.
c.385G>A (p.E129K) was detected in one SUNDS and absent in EAS population and, herein, was annotated as a mutation in EAS population. p.E129K was found in a 26-year-old male who died suddenly during sleep with moaning, which was confirmed by his family member. The main features of the heart of this case were heart weight 320 g, left ventricular wall 1.1 cm thick, right ventricular wall 0.2 cm thick, and no identifiable abnormalities (including luminal narrowing and coronary atherosclerosis) in coronary arteries and cardiac electrical conduction system. Forensic autopsy and histopathology examination of this case revealed a structurally normal heart. Additional toxicological analysis and death-scene investigation provided no apparent abnormalities to explain the cause of death, supporting the forensic pathologic diagnosis of SUNDS. Functional prediction in silico by PROVEAN and PolyPhen-2 suggest that this mutation is deleterious to the function of LRRC10, which was also confirmed by Mutation Taster (http://www.mutationtaster.org/) and SIFT (http://sift.jcvi.org/). Additionally, the altered amino acid of p.E129K is conserved evolutionarily among different species in LRR4 motif, which function as protein interaction domain. What is more, we found no reported or in silico-predicted pathogenic rare variants in candidate genes in this p.E129K carrier. Although we cannot exclude the possibility that pathogenic mutation(s) in genes not included in this screen may be the genetic cause of this SUNDS case, our analysis strongly suggests the pathogenic potential of p. E129K, which mechanism warrants further investigation.
c.206C>T (p.P69L) was detected not only in one SUNDS (MAF < 0.01) and two BrS but also in EAS population; therefore, it was termed as a rare variant in EAS population. p.P69L was found in a 37-year-old male who died suddenly during sleep, which was confirmed by his wife. The main features of the heart of this case were heart weight 360 g, left ventricular wall 1.3 cm thick, right ventricular wall 0.3 cm thick, and no identifiable abnormalities (including luminal narrowing and coronary atherosclerosis) in coronary arteries and cardiac electrical conduction system. Forensic autopsy, microscopic examination, toxicological analysis, and death-scene investigation all support the forensic pathologic diagnosis of SUNDS. Functional prediction by PROVEAN indicated that p.P69L may affect the function of LRRC10, which was not consistent with PolyPhen-2. Although in silico prediction produced inconsistent results, we cannot exclude the potentially deleterious effect of this p.P69L variant for (1) this variant is absent in our control (440 alleles) and rare in EAS population (MAF < 0.01), (2) there is no pathogenic rare variant found in candidate genes in this p.P69L carrier, (3) the altered amino acid of p.P69L is completely conserved evolutionarily among different species in LRR1 motif, which function as protein interaction domain, and (4) there is no cellular electrophysiological evidence showing p.P69L exerts a benign effect.
Furthermore, p.P69L was also found in two BrS family members (father and son) with a history of syncope and implantable cardioverter defibrillator implanted. Target-captured next-generation sequencing in these two BrS family members identified one genetic variant each in SCN1B and KCNJ5, which are genes associated with BrS and long QT syndrome (LQTS), respectively. However, our analysis suggested that neither variant could be the cause of BrS in these patients: p.H223D in SCN1B was predicted to be benign by in silico analysis. On the other hand, although p.N179S may alter the function of KCNJ5, it was found only in the son. Moreover, KCNJ5 is a LQTS but not BrS susceptible gene. Combining the finding in SUNDS, p.P69L was evaluated to be likely contributed to one SUNDS and two BrS family members. The likely pathogenic potential and mechanism of p.P69L warrants further investigation.
In present study, we identified mutation p.E129K in 1 of 113 SUNDS, revealing the prevalence of mutation was approximately 0.89%. Rare polymorphism p.P69L was detected in 1 of 88 SUNDS with prevalence of 1.14% and two BrS family members. Besides p.P69L in two BrS family members, missense variants (p.E129K and p.P69L) with prevalence around 1% in SUNDS cases, revealing LRRC10 gene variant may contribute to the genetic cause of some SUNDS victims and BrS patients. The electrophysiological study of these plausible pathogenic variants and the genetic studies with larger SUNDS cases and healthy controls are in the process by our group.
For forensic pathological investigation, forensic autopsy, histopathology examination, toxicological analysis, and death-scene investigation are all extremely important to establish the cohort of SUNDS. Unfortunately, absence of clinical data and family disease history often made it difficult to explain the cause of death. With the development of genomics, molecular autopsy using Sanger direct sequencing or with next-generation sequencing can be a very powerful technique in forensic pathological investigation of sudden unexplained death with negative autopsy, including SUNDS. Reasonably, a combination of current forensic pathological investigation, molecular autopsy, and cautious forensic pathologic evaluation could provide a reliable forensic diagnosis for SUNDS. Thus, molecular autopsy should be involved in all SUNDS cases as a routine postmortem genetic testing, and the victim’s or survivor’s genetic information could help the family members or survivors to prevent sudden cardiac death.
Conclusion
In summary, we identified p.E129K might be a potential pathogenic mutation underlying the death of one SUNDS. We also provide evidence to show that rare variant in East Asia, p.P69L, might contribute to the genetic cause for one SUNDS and BrS for two family members. This is the first report of genetic screening of LRRC10 in Chinese SUNDS and BrS. Our investigation implies that LRRC10 may be a new susceptible gene for SUNDS and genetically links LRRC10 variants to primary arrhythmia-associated BrS and SUNDS. We are currently carrying out electrophysiological study to test the functional impact of these putative pathogenic mutations of LRRC10 and confirm and support their role as pathogenic variants in Chinese SUNDS and BrS.
Supplementary Material
Acknowledgments
This work was supported by the State Key Program from the National Natural Science Foundation of China (81430046) and the grants from the National Institutes of Health (R56 HL71092, R01HL128076-01). We thank Dr. Robert F. Corliss (Department of Pathology and Laboratory Medicine and Waisman Center, University of Wisconsin, Madison, WI) for his assistance in pathological analysis.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00414-016-1516-z) contains supplementary material, which is available to authorized users.
Compliance with ethical standards The patients or legal representatives of the victims gave written informed consent, and the principles outlined in the Declaration of Helsinki were followed. The project was approved for human study by the Ethics Committee of Sun Yat-sen University.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Nademanee K, Veerakul G, Nimmannit S, et al. Arrhythmogenic marker for the sudden unexplained death syndrome in Thai men. Circulation. 1997;96:2595–2600. doi: 10.1161/01.cir.96.8.2595. [DOI] [PubMed] [Google Scholar]
- 2.Nakajima K, Takeichi S, Nakajima Y, et al. Pokkuri death syndrome; sudden cardiac death cases without coronary atherosclerosis in South Asian young males. Forensic Sci Int. 2011;207:6–13. doi: 10.1016/j.forsciint.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Gaw AC, Lee B, Gervacio-Domingo G, et al. Unraveling the enigma of Bangungut: is sudden unexplained nocturnal death syndrome (SUNDS) in the Philippines a disease allelic to the Brugada syndrome? Philipp J Intern Med. 2011;49:165–176. [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J, Huang E, Tang S, et al. A case-control study of sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Am J Forensic Med Pathol. 2015;36:39–43. doi: 10.1097/PAF.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 5.Sangwatanaroj S, Prechawat S, Sunsaneewitayakul B, et al. New electrocardiographic leads and the procainamide test for the detection of the Brugada sign in sudden unexplained death syndrome survivors and their relatives. Eur Heart J. 2001;22:2290–2296. doi: 10.1053/euhj.2001.2691. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Tester DJ, Hou Y, et al. Is sudden unexplained nocturnal death syndrome in southern China a cardiac sodium channel dysfunction disorder? Forensic Sci Int. 2014;236:38–45. doi: 10.1016/j.forsciint.2013.12.033. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Zhou F, Huang L, et al. Association of common and rare variants of SCN10A gene with sudden unexplained nocturnal death syndrome in Chinese Han population. Int J Legal Med. 2016 doi: 10.1007/s00414-016-1397-1. [DOI] [PubMed] [Google Scholar]
- 8.Kim KH, Kim TG, Micales BK, et al. Dynamic expression patterns of leucine-rich repeat containing protein 10 in the heart. Dev Dyn. 2007;236:2225–2234. doi: 10.1002/dvdy.21225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brody MJ, Hacker TA, Patel JR, et al. Ablation of the cardiac-specific gene leucine-rich repeat containing 10 (Lrrc10) results in dilated cardiomyopathy. PLoS One. 2012;7:e51621. doi: 10.1371/journal.pone.0051621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu XK, Yuan F, Li RG, et al. Prevalence and spectrum of LRRC10 mutations associated with idiopathic dilated cardiomyopathy. Mol Med Rep. 2015;12:3718–3724. doi: 10.3892/mmr.2015.3843. [DOI] [PubMed] [Google Scholar]
- 11.Cerrone M, Lin X, Zhang M, et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckermann TM, McLeod K, Murday V, et al. Novel SCN5A mutation in amiodarone-responsive multifocal ventricular ectopy-associated cardiomyopathy. Heart Rhythm. 2014;11:1446–1453. doi: 10.1016/j.hrthm.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amin AS. SCN5A-related dilated cardiomyopathy: what do we know? Heart Rhythm. 2014;11:1454–1455. doi: 10.1016/j.hrthm.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 14.Brody MJ, Cho E, Mysliwiec MR, et al. Lrrc10 is a novel cardiac-specific target gene of Nkx2-5 and GATA4. J Mol Cell Cardiol. 2013;62:237–246. doi: 10.1016/j.yjmcc.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furtado MB, Wilmanns JC, Chandran A, et al. A novel conditional mouse model for Nkx2-5 reveals transcriptional regulation of cardiac ion channels. Differentiation. 2016;91:29–41. doi: 10.1016/j.diff.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Calloe K, Refaat MM, Grubb S, et al. Characterization and mechanisms of action of novel NaV1.5 channel mutations associated with Brugada syndrome. Circ Arrhythm Electrophysiol. 2013;6:177–184. doi: 10.1161/CIRCEP.112.974220. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita K, Takahashi H, Hata Y, et al. SCN5A(K817E), a novel Brugada syndrome-associated mutation that alters the activation gating of NaV1.5 channel. Heart Rhythm. 2016;13:1113–1120. doi: 10.1016/j.hrthm.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Fukuyama M, Ohno S, Wang Q, et al. Nonsense-mediated mRNA decay due to a CACNA1C splicing mutation in a patient with Brugada syndrome. Heart Rhythm. 2014;11:629–634. doi: 10.1016/j.hrthm.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Béziau DM, Barc J, O’Hara T, et al. Complex Brugada syndrome inheritance in a family harbouring compound SCN5A and CACNA1C mutations. Basic Res Cardiol. 2014;109:446. doi: 10.1007/s00395-014-0446-5. [DOI] [PubMed] [Google Scholar]
- 20.Wilders R, Verkerk AO. Role of the R1135H KCNH2 mutation in Brugada syndrome. Int J Cardiol. 2010;144:149–151. doi: 10.1016/j.ijcard.2008.12.177. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Ohno S, Ding WG, et al. Gain-of-function KCNH2 mutations in patients with Brugada syndrome. J Cardiovasc Electrophysiol. 2014;25:522–530. doi: 10.1111/jce.12361. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Q, Chen Y, Peng L, et al. Identification of rare variants of DSP gene in sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Int J Legal Med. 2016;130:317–322. doi: 10.1007/s00414-015-1275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim KH, Antkiewicz DS, Yan L, et al. Lrrc10 is required for early heart development and function in zebrafish. Dev Biol. 2007;308:494–506. doi: 10.1016/j.ydbio.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 26.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 27.Mohapatra B, Jimenez S, Lin JH, et al. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- 28.Olson TM, Michels VV, Thibodeau SN, et al. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 29.Brody MJ, Feng L, Grimes AC, et al. LRRC10 is required to maintain cardiac function in response to pressure overload. Am J Physiol Heart Circ Physiol. 2016;310:269–278. doi: 10.1152/ajpheart.00717.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.