Abstract
Antimicrobial peptides (AMPs) are important templates for developing new antimicrobial agents. Previously, we developed a database filtering technology that enabled us to design a potent anti-Staphylococcal peptide DFTamP1. Using this same design approach, we now report the discovery of a new class of bis-indole diimidazolines as AMP small molecule mimics. The best compound killed multiple S. aureus clinical strains in both planktonic and biofilm forms. The compound appeared to target bacterial membranes with antimicrobial activity and membrane permeation ability similar to daptomycin.
Keywords: Antimicrobial peptides, chemical synthesis, database filtering technology, peptide mimics
1. Introduction
Staphylococcus aureus is an antibiotic resistant pathogen that came to our attention as early as in the 1940s owing to the large-scale production of penicillin. The isolation of resistant S. aureus strains implies rapid adaptation of this bacterium to this antibiotic. As a consequence, various derivatives of penicillin such as methicillin were synthesized in an attempt to retain the potency of the medicine.1 Methicillin-resistant S. aureus (MRSA) strains, initially isolated from the clinical setting, have also found their way into the community. Likewise, the community-associated MRSA has set foot into the clinical environments.2 Today, MRSA constitutes a difficult-to-treat superbug, which frequently forms biofilms (communities of pathogens). It is estimated that the total deaths due to S. aureus are now comparable to the total from HIV-1/AIDS.3
Antimicrobial peptides (AMPs) are key components of the innate immune system, which are essential to fend off invading pathogens.4–6 These peptides are usually small with less than 50 amino acids. The majority of these peptides are cationic with a net charge of +1 to +7. They also have a hydrophobic content of ~50%.7,8 Such features satisfy the basic requirement to form an amphipathic structure for recognizing the anionic membranes of pathogenic bacteria. To date, over 2,700 such AMPs have been identified from natural sources, ranging from prokaryotes to eukaryotes (http://aps.unmc.edu/AP). These peptides are interesting templates for developing alternative antimicrobials. Based on the Antimicrobial Peptide Database (APD),9 we previously developed two database approaches for discovery of anti-MRSA peptides.8 First, database screening of representative candidates led to a set of peptides with potent activity against MRSA.10 Second, we also developed a database filtering technology for the de novo design of anti-MRSA peptides. Applying this technology, we identified peptide DFTamP1 which is primarily active against S. aureus.11
There are interests to identify new AMP analogs with improved cell selectivity and protease stability.12,13 We now report the synthesis of small molecule mimics of DFTamP1. Using a modified protocol similar to a previous one,14 several families of diimidazolines were synthesized. Most of the compounds were found to closely mimic the activity of DFTamP1, indicating that the database derived peptide design idea11 can be utilized to design small molecule mimics of AMPs.
2. Results and discussion
2.1. Synthesis of peptide mimics
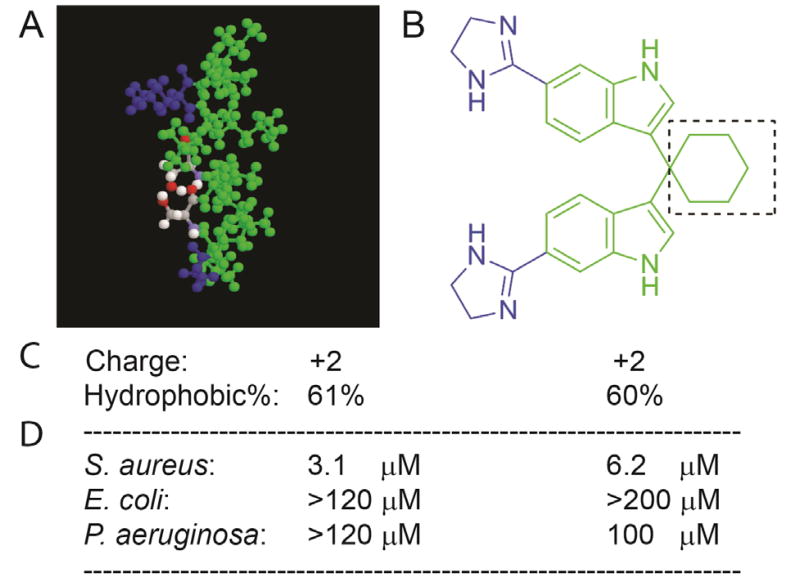
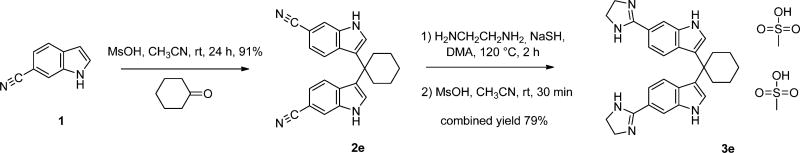
DFTamP1, a peptide designed based on the APD, contains 65% hydrophobic amino acids (green) and two positive charges (blue, Fig. 1A).11 Using this amino acid composition, we initially designed compound 3e (Fig. 1B) as a close mimic of DFTamP1. This design contains five components: one cyclohexyl group (hydrophobic), two indoles (hydrophobic), and two imidazolines (charged). If we regard each component as a mimic of one amino acid, we can calculate hydrophobic percentage for this compound. Thus, compound 3e comprises 60% hydrophobic components and two charged moieties (Fig. 1C), resembling DFTamP1 in terms of the number of positive charges and hydrophobic content (cf., panels A and B). Its synthesis is summarized in Fig. 2. In our three-step synthetic method, methanesulfonic acid was employed as a catalyst in place of iodine to provide advantages of simple work-up and purification, without the need for extraction or chromatography. Remarkably, this compound displayed an activity spectrum (i.e., active against primarily Gram-positive S. aureus but not Gram-negative E. coli and P. aeruginosa), similar to DFTamP1 (Fig. 1D), indicating a successful mimic of the peptide. It should be pointed out that the calculation of hydrophobic content for small molecules are not straight-forward as in the case of peptides. Our fragment-based approach here, however, appears to be useful because the compound indeed resembles DFTamP1 also in terms of antibacterial activity. Based on these encouraging results, we synthesized additional analogs in a similar manner and their detailed characterization by Nuclear Magnetic Resonance (NMR) spectroscopy is provided as supporting information.
Figure 1.
The small molecule compound 3e (B) successfully mimic DFTamP1 (A) in terms of charge, hydrophobic content (C), and activity spectrum (D).
Figure 2.
A scheme for the synthesis of diimidazolines.
2.2. Antimicrobial and hemolytic activity of the mimics
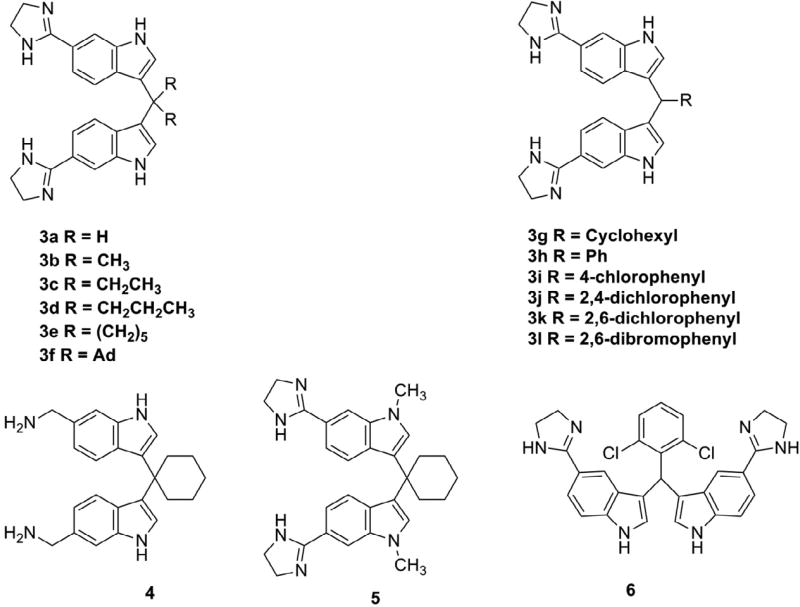
To identify better candidates, several families of bis-indoles were synthesized (Fig. 3). In brief, the first family investigated the size of the central aliphatic group. With an increase in the aliphatic chains from compound 3a to 3d (Fig. 3), compound activity against S. aureus USA300 increased, leading to a reduced minimal inhibitory concentration (MIC) from 50 to 3.1 µM in Table 1. Interestingly, these four compounds had no activity against Gram-negative bacteria, including E. coli ATCC 25922, P. aeruginosa PAO1, and K. pneumoniae ATCC 13883 (MIC >50 µM). The increase in compound activity against MRSA was primarily due to an increase in hydrophobicity of the compounds, consistent with an increase in the HPLC retention time from 9.215 to 12.102 min in Table 1. The increase in hydrophobicity also explained the increased HL50 values (i.e., 50% hemolytic concentration) for this series of compounds. Based on MRSA MIC and HL50 data, we also calculated cell selectivity indices (CSI). In this series, compound 3d had the highest CSI of 48–96 (Table 1).
Figure 3.
Chemical structures of bis-indole dimidazoline and diamine mimics of peptide DFTamP1.
Table 1.
Effects of varying aliphatic hydrophobic groups on antimicrobial activity1
| Compd | MIC (µM) | HL50 (µM) |
CSI | HPLC Rt (min) |
|||
|---|---|---|---|---|---|---|---|
|
S. aureus |
E. coli |
P. aeruginos a |
K. pneumoniae |
||||
| 3a | 50 | >50 | >50 | >50 | 1300 | 26 | 9.215 |
| 3b | 25 | >50 | >50 | >50 | >830 | 33 | 10.005 |
| 3c | 6.2 | >50 | >50 | >50 | 300 | 48 | 11.015 |
| 3d | 1.6–3.1 | >50 | >50 | >50 | 150 | 48–96 | 12.102 |
MIC: minimal inhibitory concentration; HL50, 50% hemolytic concentration of a compound; CSI, cell selectivity index (also called therapeutic index); Rt, retention time.
The second family of the peptide mimics was synthesized to study the effects of cyclic aliphatic moieties (Fig. 3) on antibacterial and hemolytic activities (Table 2). We initially made compound 3e, which had an MRSA MIC of 6.2 µM and an HL50 value of approximately 150–300 µM. To further enhance the activity of the compound, we also synthesized the compound 3g, which contains one more CH2 unit than compound 3e. Compound 3g indeed had an increased activity (MIC 3.1 µM). Unfortunately, this addition made 3g rather hemolytic (HL50 20–40 µM). Likewise, the compound was also highly hemolytic when the cyclohexyl ring was replaced with an adamantylidene group. This substitution did not lead to a further increase in activity. Therefore, these changes are not favorable because the CSI value is reduced. The reduced CSI could be attributed to an increase in hydrophobicity as indicated by a longer HPLC retention time (Table 2).
Table 2.
Effects of cyclic aliphatic groups on antimicrobial activity
| Compd | MIC (µM) | HL50 (µM) |
CSI | HPLC Rt (min) |
|||
|---|---|---|---|---|---|---|---|
| S. aureus | E. coli |
P. aeruginos a |
K. pneumoniae |
||||
| 3e | 6.2 | >50 | >50 | >50 | 150–300 | 24–48 | 11.276 |
| 3g | 3.1 | >50 | >50 | >50 | 20–40 | 7–14 | 11.832 |
| 3f | 3.1 | 50 | >50 | >50 | 20 | 7 | 12.325 |
As a third series, we also compared the differences between a cyclic aliphatic group and the benzene ring. This aromatic ring of compound 3h decreased its activity to 25 µM (Table 3). However, 4-chloro-phenyl substitution is favorable as compound 3i had a CSI comparable to compound 3e. Based on this observation, we decided to compare the effects of halogenation on antibacterial and hemolytic activities of the compounds (Table 3). Surprisingly, the 2,4-dichloro-phenyl group was much more hemolytic while the MIC did not increase. However, the 2,6-dichloro-benzene group was much less hemolytic. As anticipated, incorporation of a 2,6-dibromo-phenyl group, which is slightly more hydrophobic, increased antibacterial potency. It appeared that the pattern and number of halogenation modulated the activity of these aromatic compounds. Halogenations at 2,6 positions were favorable probably due to the effectiveness of this molecular configuration in retarding a deep penetration of the aromatic ring into bacterial membranes. However, moving the imidazoline group from position 6 (compound 3k) to position 5 (compound 6) of the indole ring slightly decreased antibacterial activity (Table 3).
Table 3.
Effects of aromatic hydrophobic groups on antimicrobial activity
| Compd | MIC (µM) | HL50 (µM) |
CSI | HPLC Rt (min) |
|||
|---|---|---|---|---|---|---|---|
| S. aureus |
E. coli |
P. aeruginos a |
K. pneumoniae |
||||
| 3h | 25 | >50 | >50 | >50 | 600 | 24 | 10.643 |
| 3i | 3.1–6.2 | >50 | 25 | >50 | 150 | 24–48 | 11.491 |
| 3j | 3.1–6.2 | 12.5 | 25–50 | >50 | 40 | 12 | 11.924 |
| 3k | 6.2–12.5 | >50 | >50 | >50 | 300 | 24–48 | 10.973 |
| 3l | 3.1–6.2 | >50 | >50 | >50 | 150 | 24–48 | 11.159 |
| 6 | 12.5 | >50 | >50 | >50 | >600 | >48 | 10.537 |
We also compared the effects of nitrogen-containing heterocycles (3e) with aliphatic amines (i.e., charged moiety) on compound activity. The aliphatic amine substitutions (compound 4 in Fig. 3) only slightly affected antibacterial and hemolytic activities (Table 4). Thus, these charged moieties are similar in this case.
Table 4.
Effects of charged heterocycle and linear chain on compound activity
| Compd | MIC (µM) | HL50 | CSI | HPLC Rt (min) |
|||
|---|---|---|---|---|---|---|---|
| S. aureus | E. coli |
P. aeruginosa |
K. pneumoniae |
||||
| 3e | 6.2 | >50 | >50 | >50 | 150–300 | 24–48 | 11.276 |
| 4 | 6.2–12.5 | >50 | >50 | >50 | 75–150 | 12–24 | 10.788 |
2.3. Antimicrobial effects of small molecules on S. aureus clinical strains and biofilms
To further test the efficacy of these compounds, we also tested their potency using a panel of S. aureus clinical strains both in the planktonic and biofilm forms. As shown in Table 5, these compounds are also potent against S. aureus clinical strains mu50, USA200, USA400, and UAMS-1. Except for S. aureus UAMS-1, compound 3d is most active with an MIC in the range of 1.5–3.1, and, in certain cases, was comparable to daptomycin (Table 5).
Table 5.
Antibacterial activity against S. aureus clinical strains
| Compd | MIC (µM) | |||||
|---|---|---|---|---|---|---|
| USA200 | USA300 | USA400 | UAMS-1 | Newman | Mu50 | |
| 3e | 6.2 | 6.2 | 6.2 | 25 | 6.2 | 6.2 |
| 3d | <1.5 | 3.1 | 3.1 | 12.5–25 | 3.1 | 3.1 |
| 5 | 25 | 12.5–25 | 12.5 | >=25 | 12.5 | 25 |
| 3i | 6.2 | 6.2 | 6.2 | 12.5 | 6.2 | 6.2 |
| 3k | 12.5 | 12.5 | 12.5 | >25 | 12.5 | >25 |
| 3l | 3.1 | 12.5 | 3.1 | 6.2 | 3.1–6.2 | 6.2 |
| Daptomycin1 | 0.78 | 0.78 | 0.78 | >12.5 | ND2 | 3.1 |
Taken from ref. 19.
Not determined.
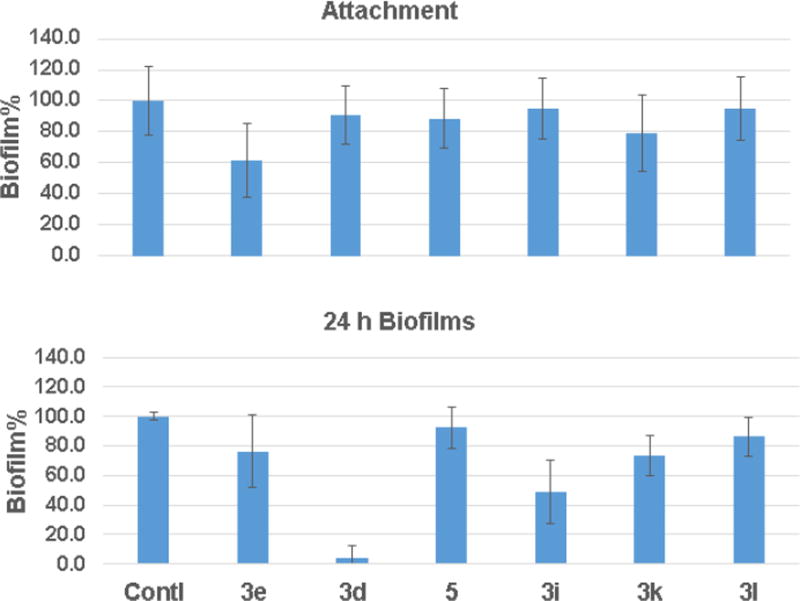
Antibiotic resistance of clinical strains is frequently related to biofilm formation. We then evaluated the inhibitory effects of select compounds on surface attachment of S. aureus USA300. They only had a small effect on the S. aureus attachment, the first step in biofilm formation. The best compound 3e inhibited the process by 40% (Fig. 4A). Although compound 3d only weakly inhibited bacterial attachment, it was more effective in disrupting the 24 h biofilms (Fig. 4B). While compound 3d essentially destroyed all the preformed biofilms, compound 3i, the second best, disrupted biofilms by ~50%.
Figure 4.
Antibiofilm assays of select TP compounds: (A) the attachment of S. aureus USA300 to the 96 well polystyrene plates and (B) disruption of the 24-h formed biofilms of S. aureus. Contl = untreated control.
2.4. Salts and pH effects on activity of select anti-MRSA compounds
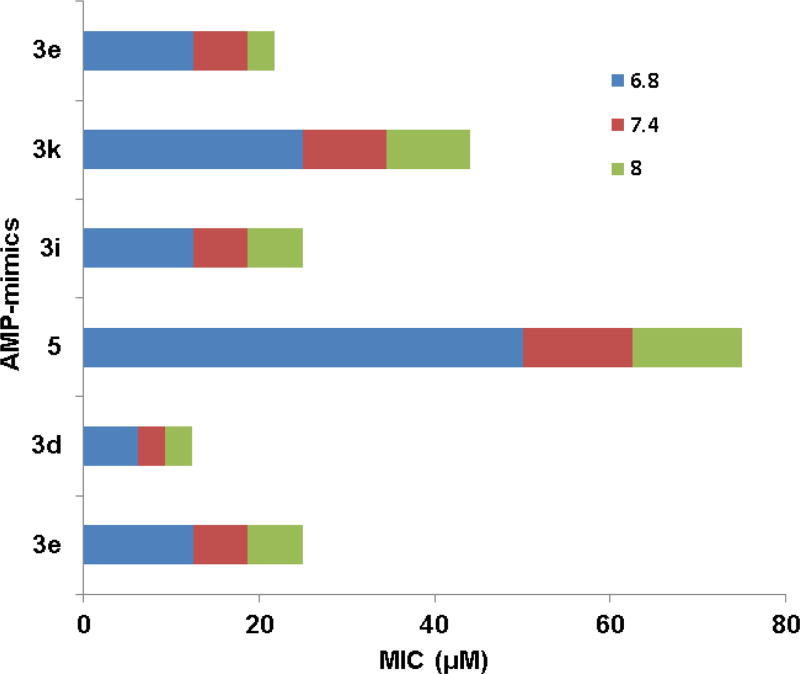
Salts and pH are known to influence the activity of antimicrobial peptides.15 Therefore, it is useful to study the effects of pH and salts on the activity of these synthetic compounds. The MIC changed only up to 2-fold with or without the addition of 100 mM NaCl into the TSB media. Thus, the activity of these compounds was not highly susceptible to the addition of 100 mM NaCl under our assay conditions. We also compared the susceptibility of the compound activity to pH. The MIC values of the six compounds at pH 6.8, 7.4, and 8.0 are plotted in Fig. 5. Interestingly, the MIC of compound 3d was least sensitive to the change in pH, while compound 5 was most sensitive. Taken together, compound 3d appeared to be more robust as a potential antibacterial agent as it could tolerate changes in salt or pH.
Figure 5.
Effects of pH on compound activity against S. aureus USA300.
2.5. Bacterial membrane permeation by TP compounds
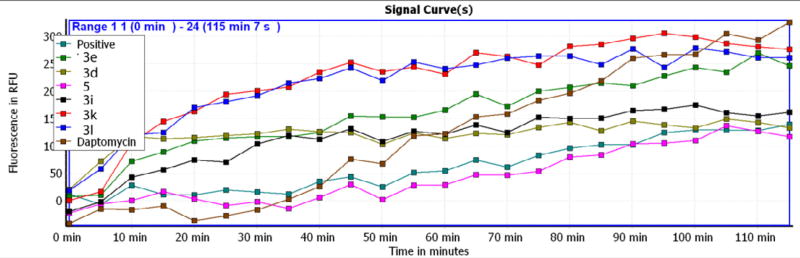
Daptomycin is known to permeate bacterial membranes.16 Using daptomycin as a positive control (Fig. 6), we compared the membrane permeation ability for selected compounds. This experiment was conducted in the presence of a non-membrane permeating dye propidium iodide (PI). No fluorescence increase was detected for S. aureus USA300 (Fig. 6, the dark green curve labeled as positive). However, we did observe fluorescence increase in the presence of selected diimidazolines. The increase in fluorescence could be explained as membrane damage by a TP compound, allowing the PI dye to enter bacterial cells and bind to DNA to emit fluorescence. While compounds 3l and 3K were most efficient, compound 5 appeared to be least effective. compound 3d was slightly better in membrane permeation than daptomycin, a known membrane-permeating peptide, before 1 h and less effective after 1 h. As negative controls, we observed no fluorescence increase when non-membrane-active antibiotics, such as vancomycin (which inhibits cell wall synthesis) or rifamycin (which inhibits RNA polymerase), were tested in the same manner (not shown). This experiment suggests that compound 3d acts on bacterial membranes, similar to daptomycin as well as DFTamP1.11
Figure 6.
Membrane permeation by the TP compounds. S. aureus USA300 was treated with a panel of compounds using a membrane-permeating daptomycin as a positive control. Two negative non-membrane permeating controls (see the text) were not shown here.
3. Conclusion
In order to control the growing problem of Staphylococcal infection, there is a growing need to search for new antibacterials. Based on our database filtering technology, we previously designed a potent anti-MRSA peptide, DFTamP1.11 In this study, we synthesized several families of small molecules AMP mimics of DFTamP1 (Fig. 3). It appears that the linear aliphatic linkers and halogenated aromatic rings are useful substitutes for the aliphatic cyclohexyl ring initially used in our synthesis (Fig. 1). In particular, compound 3d is not only more active in killing MRSA but also more selective than compound 3e (Tables 1 and 2). In addition, its anti-MRSA activity is less influenced by salts and pH. In conclusion, compound 3d constitutes a promising candidate for future studies.
4. Experimental section
4.1. Materials and methods
Strains and Media
The bacterial strains used in this study included Staphylococcus epidermidis, Staphylococcus aureus USA200, USA300, USA400, Mu50, Newman, and UAMS-1, Psudomonas aeruginosa PAO1, Escherichia coli ATCC 25922 and Klebsiella pneumonia. Tryptic soy broth (TSB) growth medium for bacterial growth was obtained from BD Bioscience MD, USA. Daptomycin was obtained from Sigma, USA. In all the assays for daptomycin, the medium was supplemented with 2 mM Ca2+.
4.2. Synthesis
General procedure for the synthesis of dinitriles (TNs)
To a solution of 6-cyanoindole (4 mmol), the corresponding carbonyl compound (2 mmol) in CH3CN (10 ml) was added MsOH (3 mmol). After the mixture was stirred at rt for 24 h, it was quenched with water (50 ml). The resulting precipitate was collected by filtration and purified by crystallization from EtOH/H2O (1:1) to afford the desired dinitrile as a white solid.
General procedure for the synthesis of diimidazolines (TPs)
A suspension of the dinitrile (0.5 mmol), ethylenediamine (1.4 mL), NaHS (68% pure, 80 mg, 1 mmol) in DMA (1.4 mL) was heated at 120°C for 2 h and cooled to rt. After the mixture was diluted with water (50 mL), the resulting precipitate was collected by filtration and purified by crystallization from CH3CN to give the corresponding diimidazoline free base.
A mixture of the free base and methanesulfonic acid (200 mg, 2.1 mmol) in CH3CN (10 mL) was stirred at rt for 30 min. The precipitate was collected by filtration and dried to give the dimesylate salt as a white solid.
4.3. Measurement of minimal inhibitory concentrations (MICs)
The assay was performed as described previously.17 In brief, a fresh culture (second inoculation) was grown until the exponential phase and diluted to 106 CFU/mL. This culture (90 µL) was then mixed with 10 µL of compound solutions in a 96 well microplate (Costar, Corning, NY). After ~20 h incubation at 37°C, bacterial density was measured at 630 nm on a CHROMATE microplate reader. The concentration at which no bacterial growth was detected is defined as the MICs. The experiments were repeated multiple times by different investigators on different dates and always identical or similar results were obtained.
4.4. Measurement of Peptide Hemolytic Concentrations
The experiment was conducted as described.11 In brief, human red blood cells (hRBC) were obtained from the UNMC Blood Bank and washed three times (800 g, 10 min) with normal saline to remove plasma. A 2% hRBC solution was prepared for the hemolytic assay. 90 µL of this 2% solution was mixed with 10 µL of serially diluted compound solutions followed by 1 h incubation at 37°C. After centrifugation at 13,000 rpm for 5 min on an Eppendorf bench-top centrifuge 5415D, 80 µl supernatant was carefully transferred to a fresh 96-well microplate (Costar, Corning, NY) for absorbance measurement at 545 nm. This color is proportional to the amount of hemoglobin released. The extent of hemolysis was calculated by assuming 100% lysis due to the effect of 1% Triton X-100 and 0% lysis without compound treatment.
4.5. Membrane permeation by fluorescence spectroscopy
S. aureus USA300 was grown to the exponential phase from an overnight culture. The cell density was then adjusted to 1 × 105 CFU/mL and treated with small molecules at different concentrations. The effect of the compound on S. aureus membranes was followed by fluorescence spectroscopy in the presence of propidium iodide. This dye does not enter the cell until the membrane is compromised due to the effect of antimicrobial compounds such as daptomycin. The measurement was performed on an Omega Reader for 90 min.15
4.6. Inhibition of bacterial attachment for biofilm formation
The potency of the synthetic compounds to inhibit bacterial attachment was evaluated by following an established protocol with modifications.15,18 In short, S. aureus USA300 cells were inoculated in TSB overnight. After a second inoculation in a fresh media, the bacteria were allowed to grow to the exponential phase (OD600 ~0.5). A bacterial density of 105 CFU/mL was prepared and 180 µL of it was delivered to flat bottom, 96 wells, polystyrene microtiter plates (Corning Costar Cat No. 3595) containing 20 µL of serially diluted 10× compound solution containing 10% DMSO. Media containing bacteria and water were treated as the positive controls while un-inoculated media with water served as the negative controls. The plates were then incubated at 37°C for 4 h. Media were then pipetted out and the wells were washed with normal saline to remove the non-adherent planktonic cells. The amount of biofilms was calorimetrically quantitated by XTT [2,3-bis(2-methyloxy-4-nitro-5-sulfophenyl)-2H-tertazolium-5-carboxanilide] assay. 180 µL of fresh TSB and 20 µL of XTT solution were added to each well and the plates were again incubated for 2 h at 37°C. Absorbance at 450 nm (only media with XTT containing wells served as the blank) was obtained using a ChromateTM microtiter plate reader. Percentage of biofilm growth for the peptide was plotted by assuming 100% biofilm growth in the wells without peptide treatment.
4.7. Disruption of established biofilms
This experiment was conducted similarly to the attachment experiment above except for an extended growth time of 24 h to allow biofilm formation of S. aureus USA300. In addition, the washed biofilms were treated with the compounds for 24 h. The remaining biofilms were finally quantified by the XTT method as described above.
Supplementary Material
Acknowledgments
We thank Dr. Biwajit Mishra for assistance with the use of lab instruments such as HPLC. We also thank Dr. Jonathan L. Vennerstrom for his kind support and helpful suggestions. This study was supported by the NIH R01 grant AI105147 to GW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bayles KW. Trends Microbiol. 2000;8:274. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 2.Carrel M, Perencevich EN, David MZ. Emerg Infect Dis. 2015;21:1973. doi: 10.3201/eid2111.150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. JAMA. 2007;298:1763. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Zasloff M. Nature. 2002;415:389. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 5.Lehrer RI, Lu W. Immuno. Rev. 2012;245:84. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 6.Pletzer D, Coleman SR, Hancock RE. Curr. Opin. Microbiol. 2016;33:35. doi: 10.1016/j.mib.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G. Antimicrobial peptides: Discovery, design and novel therapeutic strategies. CABI; Oxfordshire, UK: 2010. [Google Scholar]
- 8.Wang G. Pharmaceuticals. 2013;6:728. doi: 10.3390/ph6060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Li X, Wang Z. Nucleic Acids Res. 2016;44:D1087. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T, Wang G. Int. J. Antimicrob. Agents. 2012;39:402. doi: 10.1016/j.ijantimicag.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra B, Wang G. J. Am. Chem. Soc. 2012;134:12426. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rotem S, Mor A. Biochim. Biophys. Acta. 2009;1788:1582. doi: 10.1016/j.bbamem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Wang G, Mishra B, Lau K, Lushnikova T, Golla R, Wang X. Pharmaceuticals. 2015;8:123. doi: 10.3390/ph8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandgar BP, Shaikh KA. Tetrahedron Lett. 2003;44:1959. [Google Scholar]
- 15.Mishra B, Golla RM, Lau K, Lushnikova T, Wang G. ACS Med. Chem. Lett. 2016;7:117. doi: 10.1021/acsmedchemlett.5b00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SD, Palmer M. Bioorg Med Chem. 2016 doi: 10.1016/j.bmc.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 17.Wang G. J. Biol. Chem. 2008;283:32637. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 18.Dean SN, Bishop BM, van Hoek ML. BMC Microbiol. 2011;11:114. doi: 10.1186/1471-2180-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra B, Lushnikova T, Wang G. RSC Adv. 2015;5:59758. doi: 10.1039/C5RA07896B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.