Significance
Denitrification, a form of microbial anaerobic respiration where nitrate is sequentially reduced (NO3− → NO2− → NO → N2O → N2) is environmentally, biologically, and chemically interesting, as well as being medically significant. Some pathogenic bacteria, including the major opportunistic pathogen Pseudomonas aeruginosa, can survive in oxygen-limited environments such as biofilms and the lungs of cystic fibrosis patients, owing to denitrification. The current proposal of a complex formation of NO-generating nitrite reductase and NO-decomposing nitric oxide reductase for rapid elimination of NO, a cytotoxic intermediate, in denitrification contributes to further understanding of denitrification and to the design of antimicrobial drugs. This paper also provides an idea of how biological systems control the dynamics of cytotoxic diffusible compounds such as NO in cells.
Keywords: nitric oxide, denitrification, protein–protein complex, NOR, NiR
Abstract
Nitric oxide (NO) plays diverse and significant roles in biological processes despite its cytotoxicity, raising the question of how biological systems control the action of NO to minimize its cytotoxicity in cells. As a great example of such a system, we found a possibility that NO-generating nitrite reductase (NiR) forms a complex with NO-decomposing membrane-integrated NO reductase (NOR) to efficiently capture NO immediately after its production by NiR in anaerobic nitrate respiration called denitrification. The 3.2-Å resolution structure of the complex of one NiR functional homodimer and two NOR molecules provides an idea of how these enzymes interact in cells, while the structure may not reflect the one in cells due to the membrane topology. Subsequent all-atom molecular dynamics (MD) simulations of the enzyme complex model in a membrane and structure-guided mutagenesis suggested that a few interenzyme salt bridges and coulombic interactions of NiR with the membrane could stabilize the complex of one NiR homodimer and one NOR molecule and contribute to rapid NO decomposition in cells. The MD trajectories of the NO diffusion in the NiR:NOR complex with the membrane showed that, as a plausible NO transfer mechanism, NO released from NiR rapidly migrates into the membrane, then binds to NOR. These results help us understand the mechanism of the cellular control of the action of cytotoxic NO.
Nitric oxide (NO) is a diffusible radical gas molecule that has been recognized as an integral signaling molecule in eukaryotes and bacteria (1, 2). NO plays pivotal roles in vasodilation, smooth muscle relaxation, neurotransmission, and the immune system in mammals. In bacteria, NO is also involved in several biological processes such as biofilm formation, quorum sensing, and symbiosis. NO is synthesized from l-arginine by an enzyme called nitric oxide synthase (NOS). In mammals, NO activates an NO receptor, soluble guanylate cyclase (sGC), upon the binding to heme in sGC, which induces the formation of signaling molecule, cGMP, from GTP. Currently, an NO binding protein homologous to the heme domain of sGC is thought to participate in bacterial NO signaling pathways (2). However, NO is a highly cytotoxic gas and easily reacts with biomolecules, despite its essential function. Thus, regulation of the cellular action of NO is crucial.
Some bacteria would be exposed to large amounts of NO during denitrification, which implies that the control of NO dynamics is indispensable to minimize the cytotoxic effects of NO in such bacteria. Denitrification is a form of microbial anaerobic respiration by bacteria living in oxygen-limited environments in which the sequential reduction of nitrate to dinitrogen (NO3− → NO2− → NO → N2O → N2) is coupled to bioenergy production. Denitrification is also crucial for the survival of some pathogenic bacteria inside host cells (3, 4). For example, Pseudomonas aeruginosa is a major opportunistic pathogen that survives through denitrification in hypoxic and anoxic environments such as the lungs of cystic fibrosis patients. As described above, cytotoxic NO is produced as an intermediate product of denitrification, yet denitrifying bacteria can grow unaffected by NO. This suggests that denitrifying bacteria such as P. aeruginosa can effectively decompose NO during denitrification. Such an effective decomposition system in denitrification is supported by the fact that it was unclear whether cytotoxic NO is formed during denitrification, before the identification of NO reductase, since little NO is detected (5). Elucidation of the efficient NO decomposition mechanism in denitrification will provide insights into how nature controls NO dynamics in biological processes, including the NO signaling process. We therefore focus on the enzymes involved in NO dynamics to elucidate the mechanism for effective metabolism of cytotoxic intermediates.
In denitrification, highly cytotoxic diffusible NO is produced from NO2− by periplasmic nitrite reductase (NiR) (Eq. 1) and is decomposed into less-cytotoxic nitrous oxide (N2O) by membrane-integrated nitric oxide reductase (NOR) (Eq. 2):
| [1] |
| [2] |
NO produced from nitrite by NiR is the substrate for NOR, and thus it is of great interest to examine whether NiR can interact with NOR to channel NO and prevent NO-induced cellular damage. In this study, we focused on cd1NiR and cNOR from P. aeruginosa, a unique pair of enzymes whose atomic-level crystal structures are available (6–8), to understand the molecular mechanism by which NO is rapidly decomposed. The structural genes for cd1NiR and cNOR are clustered on the P. aeruginosa chromosome (9), indicating that the expression of cd1NiR and cNOR is coordinated. In addition, the number of cd1NiR could be similar to that of cNOR in P. aeruginosa, since the cd1NiR and cNOR activities are comparable both in vivo (10) and in vitro (purified enzymes showed 182 ± 12 μM NO generation per min per micromolar NiR and 206 ± 12 μM NO consumption per min per micromolar cNOR). Taken together, the evidence to date raises the possibility of the formation of a cd1NiR:cNOR complex.
Here, we successfully obtained crystals of the cd1NiR:cNOR complex and solved its structure at a resolution of 3.2 Å. All-atom molecular dynamics (MD) simulations using the current structure of the cd1NiR:cNOR complex embedded into a model biological membrane provide insights into the essential factors for cd1NiR:cNOR complex formation. Structure-guided mutagenesis suggests the importance of cd1NiR:cNOR complex formation in allowing rapid NO decomposition in vivo. The data obtained in this work allow us to discuss how the cellular dynamics of NO is controlled by denitrification enzymes, without causing cellular damage.
Results and Discussion
Structural Properties of the cd1NiR:cNOR Complex.
To test our hypothesis that cd1NiR interacts with cNOR, cNOR-binding proteins were explored by pull-down analysis. The lysate from anaerobically cultured P. aeruginosa was loaded onto a column that immobilizes cNOR, and possible cNOR-binding proteins were eluted by washing with NaCl. The peptide mass fingerprint analysis on the eluted proteins suggested that cd1NiR is a possible binding partner of cNOR (SI Appendix, Fig. S1).
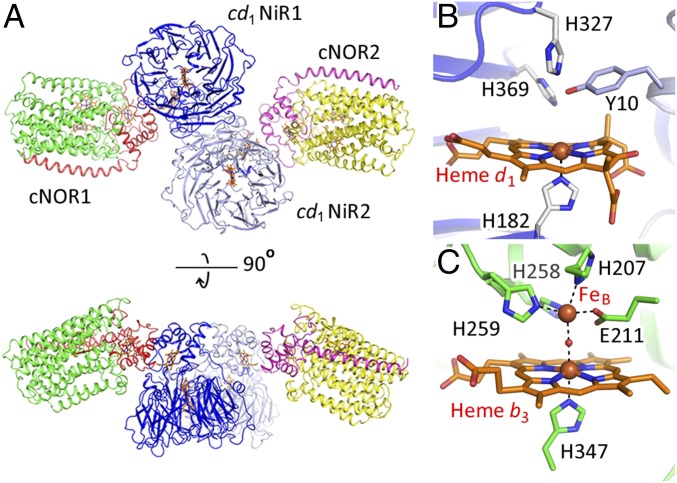
Crystallization screening with a solution mixture of cd1NiR and cNOR yielded brown crystals (SI Appendix, Fig. S2A). A visible absorption spectrum of a single brown crystal is superimposable with that of the solution mixture of cd1NiR and cNOR (SI Appendix, Fig. S2B). A structure determined at a resolution of 3.2 Å shows a one cd1NiR (a functional homodimer):two cNOR (two NorB and NorC heterodimers) complex within the asymmetric unit (Fig. 1A and SI Appendix, Table S1). The structures of cd1NiR and cNOR in the complex are almost identical to those of the individual, noncomplexed enzymes. In addition to the overall structures, the structures of the heme d1 active site of cd1NiR and the heme b3/nonheme iron (FeB) active center of cNOR in the complex (Fig. 1 B and C) are essentially indistinguishable from those of the individual enzymes in the resting state.
Fig. 1.
(A) The overall crystal structure of the cd1NiR:cNOR complex in the asymmetric unit is shown by ribbons and heme cofactors are shown by orange sticks. Blue and light blue ribbons represent each subunit of the cd1NiR homodimer (cd1NiR1 and cd1NiR2). Red and green ribbons represent the NorC and NorB subunits, respectively, of one cNOR molecule (cNOR1). Magenta and yellow ribbons represent the other cNOR molecule (cNOR2). Two pairs of a cNOR molecule and a cd1NiR monomer subunit (cNOR1-cd1NiR1 and cNOR2-cd1NiR2) are related by a noncrystallographic twofold symmetry axis. (B) Heme d1 active site of cd1NiR and (C) Heme b3/nonheme FeB active center of cNOR in the crystal structure of the cd1NiR:cNOR complex.
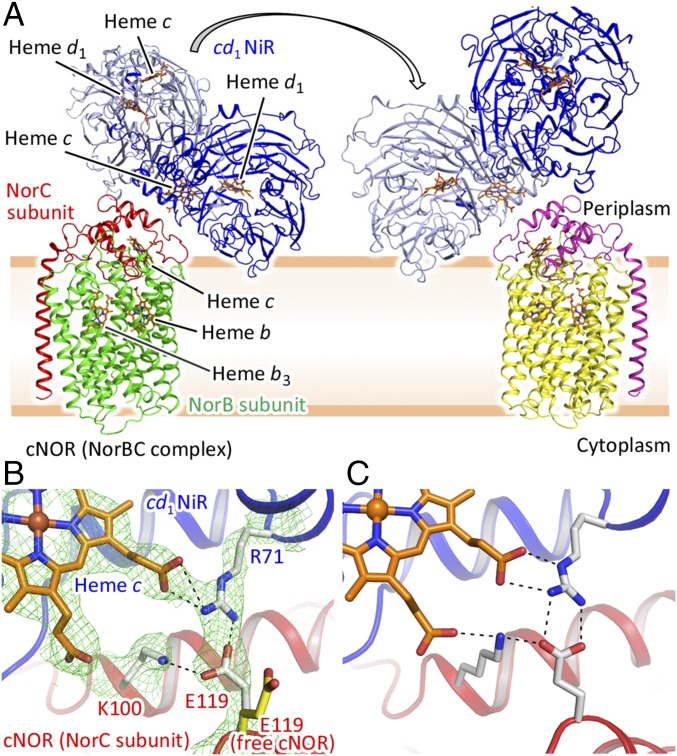
The interfaces between the heme c domain of cd1NiR and NorC of cNOR of each pair are essentially the same, since these are related by a noncrystallographic twofold symmetry axis (Fig. 1A). Given that cNOR is a membrane-integrated protein, it is therefore plausible that formation of the complex observed in the crystalline state, a one cd1NiR (a functional homodimer):two cNOR (two NorB and NorC heterodimers) complex, is unlikely in cells due to the topology of the biological membrane. Therefore, we propose that cd1NiR and cNOR form a one (a cd1NiR homodimer):one (a NorBC heterodimer) complex in vivo through the interaction observed in the crystal structure (Fig. 2A). This view was supported by the fact that the interface area of each interaction site in the cd1NiR:cNOR complex (about 1,700 Å2) is in the range of known protein:protein complexes (1,940 ± 760 Å2) (11). Furthermore, the MD simulation and the mutagenesis study also supported the complex formation of one cd1NiR homodimer and one molecule of cNOR in vivo (more details are given below). Of note, it might be possible that a dissociation of cd1NiR from cNOR could allow the rotational rearrangement of cd1NiR dimer so that the other side of cd1NiR interacts with the other cNOR in vivo (Fig. 2A). It is noteworthy that a portion of cd1NiR dips into the membrane in the proposed structure of the cd1NiR:cNOR complex in cells. Possible interactions between cd1NiR and the membrane might contribute to formation of the cd1NiR:cNOR complex (more discussion is given below).
Fig. 2.
(A) Presumed structure of the cd1NiR:cNOR complex in vivo. Due to the membrane topology, it is likely that one homodimer of cd1NiR and one molecule of cNOR could form the complex in vivo. The structural model shown here was created by omitting one cNOR molecule from the crystal structure and adding the biological membrane on the basis of the cNOR structure. A portion of cd1NiR which contains several charged residues possibly interacts with the membrane (Fig. 3, Movie S1, and SI Appendix, Fig. S6). The overall structure is shown by ribbons and the heme cofactors are represented by orange sticks. Blue and light blue ribbons represent the two monomers of the cd1NiR homodimer. Red and green ribbons represent the NorC and NorB subunits, respectively, of cNOR. The arrow represents possible rearrangement of cd1NiR. As a result, the monomer which does not interact with cNOR (red and green ribbons) could interact with the other cNOR molecule shown with magenta (NorC) and yellow (NorB) ribbons. (B) Salt-bridge formation observed at the interface of the complex. cd1NiR and cNOR are shown by blue and red ribbons, respectively. Interaction residues and heme c of cd1NiR are represented by white and orange sticks, respectively. Green mesh represents the 2Fo − Fc electron density map contoured at the 1.5σ level. Dashed lines indicate hydrogen bonds and the salt bridge. Salt-bridge formation between E119 of cNOR and R71 of cd1NiR is clearly seen in the structure. For comparison, free cNOR (cNOR of its complex with Fab, PDB ID code 3O0R) is superimposed on the cd1NiR:cNOR complex and E119 of cNOR is shown as a yellow stick. (C) Typical snapshot of the cd1NiR:cNOR interface from the MD simulation. K100 of cNOR is located >4.0 Å from the propionate group of heme c in cd1NiR in the crystal structure, yet interactions between these groups were frequently observed in the MD trajectory (Movie S2 and SI Appendix, Table S2 and Fig. S5).
One salt bridge between E119 (NorC of cNOR) and R71 (cd1NiR) and several van der Waals contacts apparently contribute to the formation of the cd1NiR:cNOR complex (Fig. 2B and SI Appendix, Table S2). The cd1NiR binding site on cNOR is different from the binding site of Fab which is required for the crystallization of cNOR itself, while overlapping these binding sites (SI Appendix, Fig. S3). Therefore, the structural feature of the binding interface of the cd1NiR:cNOR complex differs from those in tight-binding complexes such as the cNOR:Fab complex in which several hydrogen-bonding interactions are observed and is rather typical of loose-binding complexes, such as electron transfer complexes (12, 13). Indeed, the affinity between cd1NiR and cNOR seemed to be low in solution based on the fact that the cd1NiR:cNOR complex could not be trapped by gel filtration (however, the presence of the biological membrane would stabilize the cd1NiR:cNOR complex through the interaction with cd1NiR in vivo, as discussed below).
Compared with the structure of free cNOR (cNOR:Fab complex), the side chain of E119 in the complex is flipped toward R71 of cd1NiR for salt-bridge formation (Fig. 2B), signifying the vital role of the R71–E119 salt bridge in complex formation. Furthermore, no crystals were obtained from a mixture of purified E119R variant of cNOR and cd1NiR under various crystallization conditions, which is indicative of the importance of the R71–E119 interaction in formation of the complex (further characterization of the E119R variant is given below). The negatively charged character of the residue at position 119 in NorC of cNOR is highly conserved: 40% of the residues at position 119 are E and 54% of them are D (SI Appendix, Fig. S4). Similarly, the positively charged character at position 71 of cd1NiR is also dominant: 32% of the residues are R and 32% are K (SI Appendix, Fig. S4). These alignment data suggest that the corresponding salt-bridge interaction is essential for the formation of a cd1NiR:cNOR complex in other denitrifying bacteria.
MD Simulations Provide Insights into Formation of the cd1NiR:cNOR Complex in Cells.
Further insights into the elements responsible for the formation of the cd1NiR:cNOR complex (one cd1NiR homodimer:one cNOR molecule complex) in vivo were obtained through all-atom MD simulations (210 ns) using present crystallographic structure without one cNOR molecule (e.g., cNOR2 in Fig. 1A) embedded into a model biological membrane (Movie S1). The membrane comprised a mixture of phosphatidylethanolamine (POPE), phosphatidylglycerol (POPG), and cardiolipin (PVCL2), which mimics the plasma membrane of P. aeruginosa. The multiple MD trajectories, trajectories 1 and 2, showed several transient hydrogen bonds and salt bridges, which were not observed in the crystal structure, as well as the continuous R71–E119 salt bridge at the interface of cd1NiR and cNOR (Fig. 2C, Movie S2, and SI Appendix, Fig. S5 and Table S2). In particular, K100 of cNOR, which forms a hydrogen bond to E119 of cNOR, but does not interact with any cd1NiR residue in the crystal structure of the complex, frequently formed a hydrogen bond to the propionate group of heme c and/or Y75 of cd1NiR during the MD simulations. Thus, several transient interactions in addition to the stable R71–E119 salt bridge could assist in cd1NiR:cNOR complex formation.
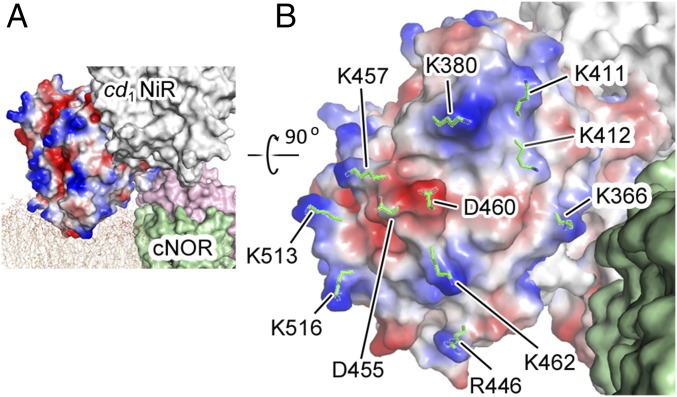
As seen in the proposed structure of the cd1NiR:cNOR complex with the membrane (Fig. 2A), the MD simulations also provide information on the interaction between the d1 domain of cd1NiR and the biological membrane for complex formation (Movie S1 and SI Appendix, Fig. S6). The initial structure of the cd1NiR:cNOR system for the MD simulations could be modeled with the crystal structure of the complex and the biological membrane. In this structure, the presumed membrane interaction site of cd1NiR pushes the membrane, and the head groups of the lipids in this area are slightly deviated from the averaged position in the entire membrane system. Then, the MD simulations show that the lipids in this area push the d1 domain of cd1NiR to locate close to the averaged position while retaining the cd1NiR-cNOR and cd1NiR-membrane interactions (Movie S1). It is also noteworthy that the putative membrane interaction site of the d1 domain of cd1NiR in the complex includes several positively charged residues (Fig. 3), and these positively charged residues can electrostatically interact with the negatively charged phosphate groups of lipid bilayer (14, 15). In addition, the MD snapshots suggest that negatively charged residues such as D455 and D460 could form salt bridges with the positively charged amine groups of the POPE molecules (SI Appendix, Fig. S6). Thus, the MD results imply that the interactions of cd1NiR with the biological membrane, which was predicted from current crystal structure of the cd1NiR:cNOR complex, would be possible in vivo. Furthermore, continuous interactions of cd1NiR with the biological membrane is consistent with a previous view that cd1NiR can be associated with the plasma membrane in P. aeruginosa (16), while cd1NiR is a soluble protein. Therefore, the interactions between the d1 domain in cd1NiR and the biological membrane would help the formation of the cd1NiR:cNOR complex in vivo, despite only one salt bridge at the interface of cd1NiR and cNOR.
Fig. 3.
Presumed interaction between the d1 domain of cd1NiR and the biological membrane upon formation of the cd1NiR:cNOR complex. (A) Location of the cd1NiR:cNOR complex in the biological membrane. A biological membrane mimicking the plasma membrane of P. aeruginosa and consisting of a mixture of POPE, POPG, and PVCL2 was computed together with the crystal structure of the cd1NiR:cNOR complex. The computed membrane is shown by narrow beige lines. The cd1NiR subunit that may interact with the biological membrane is shown as a surface model with electrostatic potential (blue indicates positive and red indicates negative) calculated using the program APBS. (B) Presumed interaction site of the d1 domain of cd1NiR with the lipid membrane. Charged residues that could interact with the membrane are represented by green sticks.
Formation of the cd1NiR:cNOR Complex Contributes to Rapid NO Decomposition in Cells.
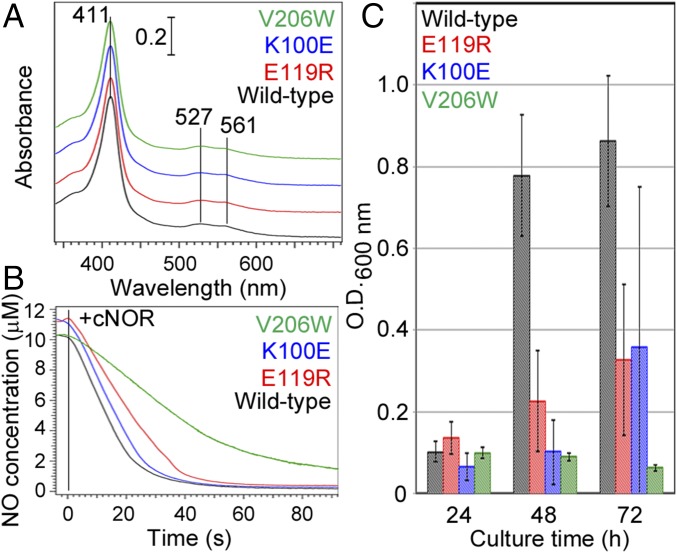
Based on the crystal structure and the MD results we eliminated the specific interaction between cd1NiR and cNOR by mutating residues E119 and K100 at the cd1NiR binding site in cNOR to examine the functional significance of the formation of the complex. All purified variants (K100E and E119R) obtained using the cNOR-deficient P. aeruginosa strain (ΔcNOR) (17) exhibited essentially the same visible absorption spectra as for wild-type cNOR (Fig. 4A). In addition, the NO reduction rates of the cNOR variants with a physiological electron donor, cytochrome c551, were comparable to that of wild-type cNOR (Fig. 4B). Therefore, the mutations did not seriously affect the overall and active site structures, and the variants of cNOR were expressed as active forms in ΔcNOR P. aeruginosa.
Fig. 4.
Effects of the mutations of K100 and E119 located at cd1NiR binding site and the mutation of V206 in the NO transfer channel in cNOR. (A) Visible absorption spectra of the K100E, E119R, and V206W variants and wild-type cNOR in the ferric state. (B) NO consumption activity of wild type and the variants of cNOR. Enzymatic NO consumption was evaluated using a Clark-type electrode equipped with an ISO-NO mark II system (WPI). The details of the experimental condition are given in Methods. (C) Bacterial growth under anaerobic denitrification conditions. Bacterial growth was monitored at OD600 nm during anaerobic static culture in the presence of 100 mM NaNO3 at 30 °C. Error bars indicate the SD calculated from four independent experiments.
To observe the mutational effects on denitrification we monitored the growth rate of the ΔcNOR strain of P. aeruginosa expressing each cNOR variant. While the ΔcNOR strain cannot grow at all under denitrification conditions due to the accumulation of cytotoxic NO (17), the expression of wild-type cNOR rescued the bacterial growth (Fig. 4C). However, ΔcNOR strains expressing any of the cNOR variants grew significantly slower compared with that expressing wild-type cNOR under denitrification conditions; for example, the growth of bacteria expressing the cNOR variants was 10–30% of that with wild type at 48 h after the initiation of anaerobic culture (Fig. 4C). Taken together with the observation that the mutations did not affect the enzymatic activity, the bacterial growth results suggested that NO diffused into the cellular environment and inhibited bacterial growth when the cd1NiR binding site variants were expressed in the ΔcNOR strain. Therefore, the salt bridge could be crucial for the formation of the cd1NiR:cNOR complex and for the elimination of NO diffusion in vivo.
Rapid NO Decomposition Mechanism in cd1NiR:cNOR Complex.
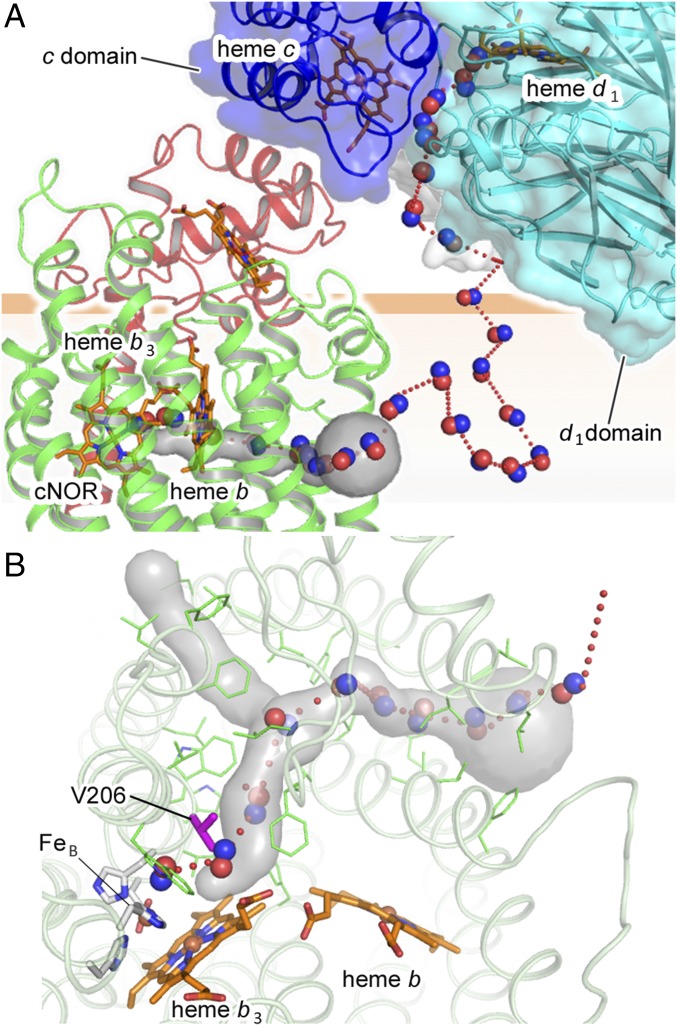
Despite the contribution of cd1NiR:cNOR complex formation toward rapid NO elimination, no direct NO channeling pathway is apparent in the current complex structure. However, 100-ns MD simulations for NO diffusion in the cd1NiR:cNOR complex with the membrane suggested a possible NO transfer mechanism in the complex (Fig. 5A, Movie S3, and SI Appendix, Fig. S7 A–C). Of 30 NO trails obtained from 20 independent MD simulations, 18 NO trails showed that the NO escaped near the surface of the biological membrane through a cavity located at the interface of the d1 and c domains of cd1NiR (Fig. 5A and Movie S3) and then rapidly migrated into the biological membrane (Fig. 5A and Movie S3). Rapid migration of the NO into the biological membrane after its release from cd1NiR is highly plausible, because hydrophobic NO is four- to fivefold more soluble in a biological membrane than in aqueous solution (18).
Fig. 5.
Possible mechanism for rapid NO decomposition by the cd1NiR:cNOR complex. (A) Proposed NO transfer pathway based on the MD simulations (Movie S3 and SI Appendix, Fig. S7). The NO trail is represented by a red dotted curve, and some NO molecules on the trail are represented by red (oxygen atom) and blue (nitrogen atom) balls. The NO transfer channel in cNOR deduced from CAVER analysis is shown by a gray surface. The NO molecule produced at the d1 active center of cd1NiR could escape through a cavity located at the interface of the d1 (blue surface) and c (cyan surface) domains of cd1NiR then migrate into the biological membrane and reach the active center of cNOR through a hydrophobic NO transfer channel. (B) Hydrophobic NO channel in cNOR viewed from the periplasmic side. The computed NO trail (red dotted curve) overlaps with the NO transfer channel (gray surface). Some selected NO molecules on the trail are represented by red (oxygen atom) and blue (nitrogen atom) balls. The residues lining the hydrophobic NO transfer channel are shown by stick models in green. V206 in the NO transfer channel, which was substituted with W in this study, are shown by magenta stick. Almost all of the residues in the NO transfer channel are hydrophobic residues.
In four NO trails obtained by the MD simulations NO migrated into the biological membrane eventually reached the active center of cNOR through the entrance of a hydrophobic channel located in the middle of the biological membrane within the entire simulation time (Fig. 5 and SI Appendix, Fig. S7 A–C). Since the NO transfer channel did not form any bottleneck during the MD simulations, a well-formed static hydrophobic NO channel could provide a ready void for NO entering into the NO transfer channel in cNOR. Indeed, the fractions of available cavity volume for NO in the cNOR (PAcNOR = 0.66%), which is defined in SI Appendix, Fig. S8, are much higher than that of the biological membrane (PAmembrane = 0.023%). Such differences in the fractions of available cavity volume between the membrane and cNOR could be an entropic driving force for NO entering into the hydrophobic channel in cNOR from the biological membrane.
The experimental results on continuous xenon binding to the hydrophobic channel in cNOR (SI Appendix, Fig. S7D) suggest that the proposed NO transfer channel is suitable for NO binding, since in cytochrome c oxidase, which is an evolutionary close relative to cNOR, the hydrophobic channel corresponding to the NO transfer channel of cNOR functions as a substrate O2 transfer channel (19, 20) (SI Appendix, Fig. S7E). Furthermore, the substitution of highly conserved V206 in the possible NO transfer channel (Fig. 5B) with bulky Trp residue lowered NO reduction activity and inhibited the bacterial growth (Fig. 4 B and C), while the substitution did not affect the visible absorption spectrum (Fig. 4A). Although further mutations at different position in the proposed NO transfer channel are required to reach a solid conclusion, these mutational results shown here are consistent with the results obtained from the MD simulations for NO diffusion in the cd1NiR:cNOR complex. Taken together with the fact that NO binding to cNOR was reported to be barrierless with a fast rate constant (i.e., a bimolecular rate constant of ∼109 M−1⋅s−1) (21), cNOR could readily capture NO migrated into the membrane. In other words, cd1NiR:cNOR complex formation permits cd1NiR to produce NO in the vicinity of cNOR, thereby facilitating NO decomposition by cNOR.
Summary
We found from the X-ray structural analysis that one cd1NiR homodimer forms a complex with two membrane-integrated cNOR molecules, which suggest that one cd1NiR and one cNOR could form a complex in cells on the basis of the topology of the membrane. Structure-based MD simulations and mutagenesis support the view that the one cd1NiR homodimer:one cNOR molecule complex could be formed in vivo and suggest that cNOR could capture NO immediately after its production by cd1NiR owing to formation of the cd1NiR:cNOR complex. The finding on the cellular control of NO dynamics by the related protein complex provides an idea on how the denitrification proteins effectively metabolize cytotoxic intermediate products. Another cytotoxic intermediate, nitrite NO2−, produced from nitrate by nitrate reductase in the cytoplasm is transferred to the periplasmic space via the membrane-integrated nitrate/nitrite exchanger, followed by its decomposition by NiR in denitrification. Hence, it is highly plausible that nitrate reductase, nitrate/nitrite exchanger, and NiR could form a supracomplex to suppress the diffusion of nitrite. This view is supported by the very recent finding that the denitrification proteins of P. aeruginosa can form a megacomplex (22).
The structural evidence of the formation of protein complexes for efficient reaction processes has been reported in several biological processes. For example, the presence of the supercomplex of aerobic respiratory complexes, complexes I, III, and IV, was demonstrated and the structure was characterized by cryoelectron microscopy (23, 24). It is likely that the supercomplex formation of the respiratory complexes contributes to effective electron transfer for aerobic respiration. During the NO signaling process in the brain, it was suggested that an adaptor protein mediates the interaction between neuronal NOS (NO-donor) and a small G protein called Dexras1, a potential NO-receptor, to enhance the ability of neuronal NOS to activate the receptor protein (25). Our current finding will lead to further understanding of how biological molecules effectively conduct consecutive reactions in biological processes, as well as of how cells control cytotoxic NO in NO signaling processes.
Methods
Sample Preparation.
cNOR was obtained from P. aeruginosa cultured under anaerobic conditions. The membrane fractions were isolated and active cNOR was purified as described in previous studies (6, 7). Purified cNOR with an A410/A280 ratio greater than 1.4 was used for the experiments. The concentration of cNOR was determined using a molar extinction coefficient of 311 mM−1⋅cm−1 at the Soret peak. Purified cNOR was dissolved in 20 mM Hepes buffer, pH 7.0, 150 mM NaCl, and 0.02% (wt/vol) n-dodecyl-β-d-maltoside (DDM; Dojindo) and was stored at −80 °C until use. cd1NiR was purified from a soluble fraction of anaerobically cultured P. aeruginosa as described in a previous report (26) and purified cd1NiR with both A410/A280 and A640/A520 ratios greater than 1.2 was used for the experiments. The concentration of cd1NiR was determined using a molar extinction coefficient of 282 mM−1⋅cm−1 (for the functional dimer) at the Soret peak. Purified cd1NiR was dissolved in 40 mM potassium phosphate buffer, pH 6.9, and was stored at −80 °C until use. The physiological electron donor cytochrome c551 (cyt-c551) was purified from a soluble fraction of anaerobically cultured P. aeruginosa using a published method (26) and was stored at −80 °C until use.
Protein Crystallization.
Before crystallization, purified cNOR was applied to a Superdex 200 column (GE Healthcare) preequilibrated with 20 mM Hepes buffer, pH 7.0, 150 mM NaCl, and 0.1% (wt/vol) n-decyl-β-d-thiomaltoside (DTM; Anatrace). cNOR was mixed with cd1NiR at a stoichiometric molar ratio of 1:1 (a NorBC complex:a cd1NiR homodimer). Crystals were grown using the sitting drop vapor diffusion method in drops containing 0.1 μL of 50 μM cd1NiR and 50 μM cNOR mixed with an equal volume of the precipitant solution, 0.1 M MES buffer, pH 6.5, 0.2 M CsCl, and 12% (wt/vol) PEG 3,350. Plate-like brown crystals 150 × 40 × 20 μm3 in size were formed within 1 wk at 20 °C. The crystals were dehydrated and cryoprotected by gradually increasing the concentration of PEG 3,350 in the reservoir to 35% in 2% increments by incubating the crystals for about 1 h at 20 °C for each condition. The dehydrated crystals were flash-cooled in liquid nitrogen.
Data Collection and Refinement.
X-ray diffraction data were collected at 100 K at a wavelength of 1.0 Å on beamline BL41XU at SPring-8 using an MX225HE CCD detector (Rayonix). The data were processed using HKL2000 (27). Data collection statistics are summarized in SI Appendix, Table S1. An initial model was obtained by molecular replacement using Phaser (28) with the oxidized forms of cNOR [Protein Data Bank (PDB) ID code 3O0R] and cd1NiR (PDB ID code 1NIR) as search models. The structure was refined using Refmac5 (29). Model building was carried out using Coot (30). The detailed refinement parameters are summarized in SI Appendix, Table S1. All structural figures were prepared using PyMOL (www.pymol.org). UV/visible absorption spectra of the crystalline sample before and after the diffraction measurements were recorded on a linear CCD-array spectrometer (USB2000; Ocean Optics) at 100 K. The optical system consisted of a deuterium tungsten halogen lamp (DT-MINI; Ocean Optics), Cassegrain mirrors (Bunkoh-Keiki), and an optical fiber (31).
NO Reductase Activity Measurements.
Enzymatic NO consumption measurements were evaluated using a Clark-type electrode equipped with an ISO-NO mark II system (WPI). All reagents used in the activity measurement were prepared under an N2 atmosphere. The assay mixture contained 10 mM ascorbate, 10 μM cyt-c551, and 10 μM substrate NO in 50 mM Hepes buffer, pH 7.0, and 0.1% (wt/vol) DTM. An oxygen scavenging system consisting of 10 mM d-glucose, 10 μg/mL glucose oxidase, and 10 μg/mL catalase was added to ensure anaerobic conditions during the measurements. The reaction was initiated by the addition of the enzymes at a final concentration of 0.1 μM. The measurements were performed at 20 °C.
Pull-Down Analysis.
Possible interaction proteins for cNOR were explored using a Pierce Pull-Down Biotinylated Protein:Protein Interaction Kit. Cys86 of the NorB subunit of cNOR, the only free cysteine residue in cNOR, was biotinylated using a Biotin Labeling Kit-SH (Dojindo). Biotinylated cNOR was loaded onto a streptavidin column to immobilize cNOR through biotin–streptavidin interactions, then the soluble fraction from anaerobically cultured P. aseruginosa was loaded onto the cNOR-immobilized column. After the column was washed with 20 mM Hepes buffer, pH 7.0, containing 0.1% DTM, bound proteins were eluted by stepwise increases in NaCl concentration (10, 50, 100, and 500 mM). All fractions were analyzed by SDS/PAGE. The protein bands detected in the SDS/PAGE gel were analyzed using the peptide mass fingerprint method.
Bacterial Growth Experiments.
The DNA fragment encoding NorCBD of P. aeruginosa was cloned into the EcoRI and HindIII sites of pETDuet-1 (Novagen). NorCBD with an N-terminal His-tag in pETDuet-1 was digested by XbaI and HindIII, and the fragment was inserted into the multicloning site of pMMB67EH for expression in P. aeruginosa (10). For mutations of cNOR the DNA fragment encoding NorCB and a part of following NorD in pETDuet-1 was digested by EcoRI and XhoI, and the fragment was inserted into the multicloning site of pRSET C (Invitrogen). Mutations were introduced into cNOR using a QuikChange mutagenesis kit (Agilent Technologies) and the pRSET C vector as a template. The fragment containing mutated NorCB and a part of NorD was obtained using EcoRI and XhoI restriction enzymes and was inserted into the pMMB67EH cNOR expression vector instead of wild-type cNOR. Transformation of cNOR expression vector into the ΔcNOR strain of P. aeruginosa was performed by electroporation using a MicroPulser (Bio-Rad). Preculture of the ΔcNOR strain bearing the cNOR expression vector was carried out overnight in LB medium (2 mL) under aerobic conditions. Then, anaerobic culture was initiated by the addition of 100 μL of the aerobic preculture to anaerobically prepared LB medium (5 mL) containing 50 mM KNO3. Bacterial growth was monitored by measuring OD600 nm every 24 h during static anaerobic culture at 30 °C.
Wild-type and the cd1NiR binding-site variants of cNOR expressed in the ΔcNOR strain of P. aeruginosa were obtained from anaerobic culture of the bacteria in 5 L of medium containing 50 mM KNO3. The membrane fraction was isolated and solubilized by the same method for cNOR expressed in P. aeruginosa. The solubilized fraction was loaded onto a Ni-NTA column preequilibrated with 20 mM Tris⋅HCl buffer, pH 8.0, 150 mM NaCl, and 30 mM imidazole containing 0.02% (wt/vol) DDM and the cNOR fraction was eluted by the buffer containing 150 mM imidazole. The cNOR fraction was then loaded onto a Superdex 200 size-exclusion column (GE Healthcare) preequilibrated with 20 mM Hepes buffer, pH 7.0, 150 mM NaCl, and 0.02% (wt/vol) DDM. Since the ΔcNOR strain expressing V206W could not grow under denitrification conditions, V206W was expressed in the wild-type strain of P. aeruginosa. Then, the expressed V206W variant was separated from endogenous native cNOR using an Ni-NTA affinity column. Purified samples with an A410/A280 ratio greater than 1.4 were used for the experiments.
Identification of the Xenon Binding Sites.
The crystals of the cNOR:Fab complex were prepared according to a previous report (6, 7). The xenon derivatives were produced by pressurizing crystals of the cNOR:Fab complex using Cryo-Xe-Siter (Rigaku) at room temperature. Cryoprotected crystals were mounted in a loop and placed in the pressure vessel. After incubation of 5 min at xenon pressure of 150 psi the crystal was flash-cooled by being plunged into liquid tetrafluoromethane. X-ray diffraction data were collected and processed as for the cd1NiR:cNOR complex. Data collection statistics are summarized in SI Appendix, Table S1. An initial model was obtained by molecular replacement with Molrep (32) using the structure of the cNOR:Fab complex (PDB ID code 3O0R) as a search model. The structure was refined using Refmac5 (29). Xenon molecules were added to the structural model at the anomalous peaks greater than 2.5σ and at position-appropriate hydrophobic environments. Model building was carried out using Coot (30). The refinement parameters are summarized in SI Appendix, Table S1.
Modeling of the cd1NiR:cNOR Complex for MD Simulation.
The cd1NiR:cNOR complex with the model biological membrane was constructed for the MD simulations. The plasma membrane of P. aeruginosa was mimicked by preparing a mixture of POPE (3-palmitoyl-2-oleoyl-d-glycero-1-phosphatidylethanolamine), POPG (3-palmitoyl-2-oleoyl-d-glycero-1-phosphatidylglycerol), and PVCL2 (1,1′-palmitoyl-2,2′-vacenoyl cardiolipin) at a ratio POPE:POPG:PVCL2 = 70:15:15 using CHARMM-GUI Membrane Builder (33). More details on the modeling of the cd1NiR:cNOR complex can be found in SI Appendix, SI Materials and Methods.
MD Simulation of the cd1NiR:cNOR Complex.
After the 24.8-ns equilibration we conducted two 210-ns production runs, each of which started with different velocities from the Maxwell–Boltzmann distribution in the NPT ensemble at 300 K and 1 atm without any restraints. For the analysis of hydrogen-bonding interactions coordinates were stored at every 10 ps. The GENESIS package (34) was used for the MD simulations. The detailed methods can be found in SI Appendix, SI Materials and Methods.
NO Diffusion Simulation Between cd1NiR and cNOR.
To examine NO diffusion in the cd1NiR:cNOR complex we carried out another MD simulation including explicit NO molecules in the system. In total, 20 replicas of the cd1NiR-NO-cNOR system were modeled: 10 replicas with two NO molecules, each at the active site pocket of cd1NiR, and 10 replicas with four NO molecules, with two more NO molecules placed at protein cavities close to the active sites. Each system was simulated for 100 ns in the NPT ensemble at 300 K and 1 atm using GROMACS 5.1.2 (35). Detailed methods and analysis of the MD simulations on the NO diffusion are given in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank K. Takatsu (RIKEN) and M. Suematsu (RIKEN) for support in sample preparation, Dr. Naoshi Dohmae for mass analysis, and the staff of the SPring-8 beamline for their help with diffraction measurements and microphotometry. The diffraction measurements were performed at SPring-8 BL41XU (Proposals 2012B1526, 2013A1221, 2013B1427, 2014A1424, and 2014B1528). The MD simulations were carried out on TSUBAME2.5 at the Tokyo Institute of Technology through the HPCI System Research Project (Project ID hp160184) and on HOKUSAI GreatWave at RIKEN. This work was supported by Grants JP15H00965, JP17H03092, JP26220807, and JP26620140 from the Ministry of Education, Culture, Sports Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.S.M. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 5GUW for the cd1NiR:cNOR complex and 5GUX for the xenon derivative of cNOR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621301114/-/DCSupplemental.
References
- 1.Bredt DS, Snyder SH. Nitric oxide, a novel neuronal messenger. Neuron. 1992;8:3–11. doi: 10.1016/0896-6273(92)90104-l. [DOI] [PubMed] [Google Scholar]
- 2.Nisbett LM, Boon EM. Nitric oxide regulation of H-NOX signaling pathways in bacteria. Biochemistry. 2016;55:4873–4884. doi: 10.1021/acs.biochem.6b00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevanin TM, Moir JW, Read RC. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun. 2005;73:3322–3329. doi: 10.1128/IAI.73.6.3322-3329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakishima K, Shiratsuchi A, Taoka A, Nakanishi Y, Fukumori Y. Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem Biophys Res Commun. 2007;355:587–591. doi: 10.1016/j.bbrc.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Braun C, Zumft WG. Marker exchange of the structural genes for nitric oxide reductase blocks the denitrification pathway of Pseudomonas stutzeri at nitric oxide. J Biol Chem. 1991;266:22785–22788. [PubMed] [Google Scholar]
- 6.Hino T, et al. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 7.Sato N, et al. Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes. Proteins. 2014;82:1258–1271. doi: 10.1002/prot.24492. [DOI] [PubMed] [Google Scholar]
- 8.Nurizzo D, et al. N-terminal arm exchange is observed in the 2.15 A crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa. Structure. 1997;5:1157–1171. doi: 10.1016/s0969-2126(97)00267-0. [DOI] [PubMed] [Google Scholar]
- 9.Arai H, Igarashi Y, Kodama T. The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1995;1261:279–284. doi: 10.1016/0167-4781(95)00018-c. [DOI] [PubMed] [Google Scholar]
- 10.Arai H, Kodama T, Igarashi Y. The role of the nirQOP genes in energy conservation during anaerobic growth of Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1998;62:1995–1999. doi: 10.1271/bbb.62.1995. [DOI] [PubMed] [Google Scholar]
- 11.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 12.Nojiri M, et al. Structural basis of inter-protein electron transfer for nitrite reduction in denitrification. Nature. 2009;462:117–120. doi: 10.1038/nature08507. [DOI] [PubMed] [Google Scholar]
- 13.Antonyuk SV, Han C, Eady RR, Hasnain SS. Structures of protein-protein complexes involved in electron transfer. Nature. 2013;496:123–126. doi: 10.1038/nature11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macedo-Ribeiro S, et al. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 1999;402:434–439. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 16.Coyne MS, Arunakumari A, Pankratz HS, Tiedje JM. Localization of the cytochrome cd1 and copper nitrite reductases in denitrifying bacteria. J Bacteriol. 1990;172:2558–2562. doi: 10.1128/jb.172.5.2558-2562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arai H, Kodama T, Igarashi Y. Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1999;170:19–24. doi: 10.1111/j.1574-6968.1999.tb13350.x. [DOI] [PubMed] [Google Scholar]
- 18.Möller M, et al. Direct measurement of nitric oxide and oxygen partitioning into liposomes and low density lipoprotein. J Biol Chem. 2005;280:8850–8854. doi: 10.1074/jbc.M413699200. [DOI] [PubMed] [Google Scholar]
- 19.Salomonsson L, Lee A, Gennis RB, Brzezinski P. A single-amino-acid lid renders a gas-tight compartment within a membrane-bound transporter. Proc Natl Acad Sci USA. 2004;101:11617–11621. doi: 10.1073/pnas.0402242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the heme-Cu oxidases. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 21.Kapetanaki SM, et al. Ultrafast ligand binding dynamics in the active site of native bacterial nitric oxide reductase. Biochim Biophys Acta. 2008;1777:919–924. doi: 10.1016/j.bbabio.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Borrero-de Acuña JM, et al. Protein network of the Pseudomonas aeruginosa denitrification apparatus. J Bacteriol. 2016;198:1401–1413. doi: 10.1128/JB.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 24.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 25.Fang M, et al. Dexras1: A G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 26.Parr SR, Barber D, Greenwood C. A purification procedure for the soluble cytochrome oxidase and some other respiratory proteins from Pseudomonas aeruginosa. Biochem J. 1976;157:423–430. doi: 10.1042/bj1570423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin AA, et al. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Chiu YC, et al. Kinetic and structural studies on the catalytic role of the aspartic acid residue conserved in copper amine oxidase. Biochemistry. 2006;45:4105–4120. doi: 10.1021/bi052464l. [DOI] [PubMed] [Google Scholar]
- 32.Vagin AA, Teplyakov A. MOLREP: An atuomated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 33.Wu EL, et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung J, et al. GENESIS: A hybrid-parallel and multi-scale molecular dynamics simulator with enhanced sampling algorithms for biomolecular and cellular simulations. Wiley Interdiscip Rev Comput Mol Sci. 2015;5:310–323. doi: 10.1002/wcms.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.