Significance
This study systematically identified adenosine to inosine (A-to-I) editing sites in Neurospora crassa and showed the existence of stage-specific editing events at different sexual stages. Unlike in humans, fungal A-to-I editing mainly occurred in coding regions and caused nonsynonymous changes that significantly increased proteome complexity. In general, nonsynonymous editing sites in Neurospora are adaptive and favored by positive selection. RNA editing enables stage-specific functions or expression of proteins important for different sexual developmental processes. Some editing events are well conserved and may affect genes important for other genetic and epigenetic phenomena occurring during sexual reproduction. Overall, our results provide insights into the complex regulation of sexual development and reveal the role of A-to-I editing for adaptive evolution in Neurospora.
Keywords: Neurospora, RNA editing, perithecia, RNA modification, adaptive evolution
Abstract
Although fungi lack adenosine deaminase acting on RNA (ADAR) enzymes, adenosine to inosine (A-to-I) RNA editing was reported recently in Fusarium graminearum during sexual reproduction. In this study, we profiled the A-to-I editing landscape and characterized its functional and adaptive properties in the model filamentous fungus Neurospora crassa. A total of 40,677 A-to-I editing sites were identified, and approximately half of them displayed stage-specific editing or editing levels at different sexual stages. RNA-sequencing analysis with the Δstc-1 and Δsad-1 mutants confirmed A-to-I editing occurred before ascus development but became more prevalent during ascosporogenesis. Besides fungal-specific sequence and secondary structure preference, 63.5% of A-to-I editing sites were in the coding regions and 81.3% of them resulted in nonsynonymous recoding, resulting in a significant increase in the proteome complexity. Many genes involved in RNA silencing, DNA methylation, and histone modifications had extensive recoding, including sad-1, sms-3, qde-1, and dim-2. Fifty pseudogenes harbor premature stop codons that require A-to-I editing to encode full-length proteins. Unlike in humans, nonsynonymous editing events in N. crassa are generally beneficial and favored by positive selection. Almost half of the nonsynonymous editing sites in N. crassa are conserved and edited in Neurospora tetrasperma. Furthermore, hundreds of them are conserved in F. graminearum and had higher editing levels. Two unknown genes with editing sites conserved between Neurospora and Fusarium were experimentally shown to be important for ascosporogenesis. This study comprehensively analyzed A-to-I editing in N. crassa and showed that RNA editing is stage-specific and generally adaptive, and may be functionally related to repeat induced point mutation and meiotic silencing by unpaired DNA.
Adenosine to inosine (A-to-I) RNA editing catalyzed by adenosine deaminase acting on RNA (ADAR) enzymes is the most prevalent type of RNA editing in animals that convert A to I via hydrolytic deamination (1–3). Because I is recognized as guanosine (G) by the cellular machinery, A-to-I editing in protein-coding regions [coding DNA sequences (CDSs)] of mRNAs may cause nonsynonymous codon changes (recoding). However, although A-to-I editing is abundant in mammals, nonsynonymous editing is generally rare because the vast majority of editing sites are in the noncoding regions, including introns and 5′- or 3′-untranslated regions (2, 4). To date, only a small number of animal recoding sites have been experimentally confirmed to affect protein functions (2, 3, 5). Amino acid changes resulting from A-to-I editing are important for the functions of ligand- and voltage-gated ion channels and neurotransmitter receptors in invertebrates and vertebrates (6–9). In the octopus, the isoleucine to valine (I-to-V) change caused by RNA editing in the potassium (K)+ channel is related to temperature adaptation (10). However, other than these few cases, it is not clear or questionable whether vast majority of the observed recoding A-to-I editing events are advantageous. In humans, most of the recoding editing events were nonadaptive and likely resulted from tolerable promiscuous targeting by ADARs (11); however, in cephalopods, recoding RNA editing is often adaptive (12–14).
To date, the ADAR enzymes that typically have one adenosine deaminase domain and one or more dsRNA binding domains are only found in metazoans (15, 16). Recently genome-wide A-to-I RNA editing was identified in the wheat scab fungus Fusarium graminearum, which lacks ADAR orthologs (17). The A-to-I editing in F. graminearum specifically occurred during sexual reproduction, and the majority of the editing sites resulted in protein recoding, including recoding of premature stop codons in the PUK1 and other 69 pseudogenes to express full-length functional proteins (17). PUK1 is a protein kinase gene that is specifically expressed during sexual reproduction, and its orthologs are conserved in filamentous ascomycetes but not in the yeast. The puk1 deletion mutant in F. graminearum had no other defects but ascospore maturation and release (17). Interestingly, the PUK1 ortholog in Neurospora crassa, a model filamentous ascomycete for genetic studies (18, 19), also has one premature stop codon in the ORF that requires A-to-I editing (17). A preliminary analysis with publicly available RNA-sequencing (RNA-seq) data showed that A-to-I editing also specifically occurred during sexual development in N. crassa and Fusarium verticillium. Therefore, it is likely that Sordariomycetes or filamentous fungi in general have novel RNA editing machinery independent of ADARs.
Filamentous fungi are tractable models for understanding the genetic basis of sexual reproduction in multicellular organisms. As a heterothallic fungus, N. crassa has two distinct mating types, A and a (20). Sexual development is initiated when a protoperithecium of one mating type is fertilized via a trichogyne by a male cell of opposite mating type (20). Fertilized protoperithecia develop into perithecia that consist of the perithecium wall, dikaryotic ascogenous hyphae, asci (meiotic cells), and ascospores (meiotic spores). Repeat induced point mutation (RIP), meiotic silencing by unpaired DNA (MSUD), and spore killer are genetic or epigenetic processes that specifically occur during sexual reproduction in N. crassa (21–24). Whereas RIP is a premeiotic hypermutation process that targets duplicated segments of DNA by converting cytidine (C):G to thymidine (T):A, MSUD is an RNAi-based silencing mechanism acting during meiosis and postmeiotic mitosis. Spore killer is a meiotic drive element that distorts genetic ratios.
With the genetic and genomics resources available for N. crassa as a model organism (25), it is an ideal system to study functions and mechanisms of RNA editing and its relationship with other sexual-specific genetic or epigenetic phenomena in fungi. However, currently available RNA-seq data of N. crassa are not suitable for comprehensive analysis of A-to-I editing because they are not strand-specific and sequence variations of the parent strains are not clear. In this study, we profiled the A-to-I editing landscape of N. crassa with strand-specific RNA-seq and showed the existence of stage-specific editing events at different sexual stages. RNA editing occurred before ascus development but became more prevalent during ascosporogenesis. In N. crassa, the majority of editing sites were in coding regions and caused changes in amino acids. In general, nonsynonymous editing events were adaptive in N. crassa, and near half of its nonsynonymous editing sites are conserved and edited in Neurospora tetrasperma. Overall, this study comprehensively analyzed the A-to-I RNA editing in N. crassa, identified evolutionarily conserved editing sites, and revealed the existence of stage-specific editing events at different sexual stages. RNA editing is generally adaptive and may be functionally related to RIP and MSUD.
Results
Genome-Wide A-to-I Editing Occurs Specifically During Sexual Reproduction in N. crassa.
To identify A-to-I editing sites in N. crassa, strand-specific RNA-seq data were generated with RNA isolated from perithecia collected at 6 d postfertilization (dpf) (OR74A × 74-ORS-6a), and vegetative hyphae of OR74A and the genomes of both parental strains were sequenced to ∼100-fold coverage (Table S1). A total of 35,325 A-to-I editing sites were identified in RNA-seq data of perithecia, accounting for 98.7% of total nucleotide variants detected (Table S2). In contrast, no enrichment of A-to-G changes was observed in strand-specific RNA-seq data of vegetative hyphae. Published RNA-seq data of N. crassa hyphae or conidia (26–29) also lack enrichment of A-to-G variants (Fig. S1).
Table S1.
RNA-seq and DNA-seq data generated in this study
| Strain | Stage | Sample | Library | Read, bp | Total reads | Mapped reads | Unduplicated reads |
| N. crassa FGSC2489 | Hyphae 48 h | FGSC2489 | DNA-seq | 101 | 37,849,444 | 37,716,733 | 37,499,966 |
| N. crassa FGSC4200 | Hyphae 48 h | FGSC4200 | DNA-seq | 101 | 54,108,680 | 53,827,878 | 53,440,667 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 3 dpf | P3_1 | Strand-specific RNA-seq | 150 | 27,268,842 | 25,996,890 | 17,200,513 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 3 dpf | P3_2 | Strand-specific RNA-seq | 150 | 32,547,894 | 30,300,681 | 20,030,570 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 4 dpf | P4_1 | Strand-specific RNA-seq | 150 | 35,493,218 | 30,109,571 | 17,610,563 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 4 dpf | P4_2 | Strand-specific RNA-seq | 150 | 37,578,848 | 34,035,599 | 20,644,751 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 5 dpf | P5_1 | Strand-specific RNA-seq | 150 | 35,751,232 | 32,724,442 | 21,075,858 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 5 dpf | P5_2 | Strand-specific RNA-seq | 150 | 28,450,946 | 26,117,987 | 18,585,188 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 6 dpf | P6_1 | Strand-specific RNA-seq | 150 | 36,844,012 | 34,871,471 | 23,259,139 |
| N. crassa FGSC2489 × FGSC4200 | Perithecia 6 dpf | P6_2 | Strand-specific RNA-seq | 150 | 32,861,106 | 30,345,011 | 21,362,699 |
| N. crassa FGSC11152 × FGSC11151 | Perithecia 5 dpf | Δsad-1_1 | Strand-specific RNA-seq | 150 | 29,682,994 | 28,042,537 | 19,011,359 |
| N. crassa FGSC11152 × FGSC11151 | Perithecia 5 dpf | Δsad-1_2 | Strand-specific RNA-seq | 150 | 36,877,166 | 33,066,716 | 21,404,444 |
| N. crassa FGSC17465 × FGSC17464 | Perithecia 5 dpf | Δstc-1_1 | Strand-specific RNA-seq | 150 | 29,836,114 | 28,521,674 | 19,378,626 |
| N. crassa FGSC17465 × FGSC17464 | Perithecia 5 dpf | Δstc-1_2 | Strand-specific RNA-seq | 150 | 26,772,608 | 25,910,706 | 17,395,558 |
| N. crassa FGSC2489 | Hyphae 48 h | Hy2489_1 | Strand-specific RNA-seq | 150 | 32,130,828 | 31,338,489 | 21,821,285 |
| N. crassa FGSC2489 | Hyphae 48 h | Hy2489_2 | Strand-specific RNA-seq | 150 | 38,420,138 | 37,548,932 | 24,998,664 |
| N. tetrasperma FGSC2508 × FGSC2509 | Perithecia 6 dpf | NtP6 | DNA-seq | 150 | 32,926,194 | 29,088,487 | 26,258,514 |
| N. tetrasperma FGSC2508 × FGSC2509 | Perithecia 6 dpf | NtP6 | Strand-specific RNA-seq | 150 | 60,105,658 | 53,153,633 | 50,323,660 |
Table S2.
Nucleotide variant sites identified in RNA-Seq data of N. crassa
| Type | P3 | P4 | P5 | P6 | Δsad-1 | Δstc1 | Hypha |
| A-to-C | 22 | 27 | 26 | 32 | 33 | 25 | 16 |
| A-to-G | 2,938 | 18,599 | 33,579 | 35,325 | 13,086 | 2,450 | 42 |
| A-to-T | 11 | 4 | 9 | 14 | 11 | 8 | 8 |
| C-to-A | 14 | 11 | 15 | 14 | 23 | 20 | 12 |
| C-to-G | 18 | 15 | 16 | 12 | 23 | 15 | 11 |
| C-to-T | 72 | 80 | 120 | 124 | 100 | 94 | 66 |
| G-to-A | 91 | 118 | 110 | 128 | 149 | 150 | 117 |
| G-to-C | 21 | 19 | 21 | 31 | 26 | 29 | 21 |
| G-to-T | 37 | 27 | 30 | 32 | 37 | 31 | 37 |
| T-to-A | 7 | 8 | 6 | 13 | 9 | 14 | 11 |
| T-to-C | 31 | 25 | 29 | 35 | 65 | 57 | 28 |
| T-to-G | 14 | 16 | 26 | 24 | 19 | 14 | 8 |
| Total | 3,276 | 18,949 | 33,987 | 35,784 | 13,581 | 2,907 | 377 |
P, postnatal day.
Fig. S1.
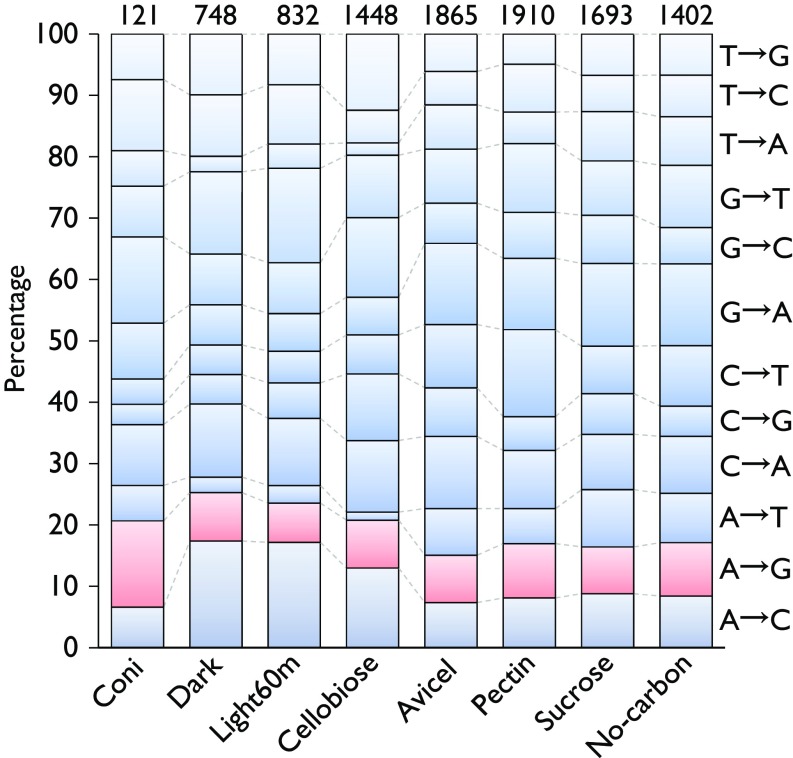
Percentages of each type of RNA variant detected in published RNA-seq data of N. crassa conidia or hyphae under different culture conditions. The total number of RNA variants detected in each sample is shown above each stacked column.
A-to-I Editing Occurs Before Ascus Development but Is More Prevalent During Ascosporogenesis.
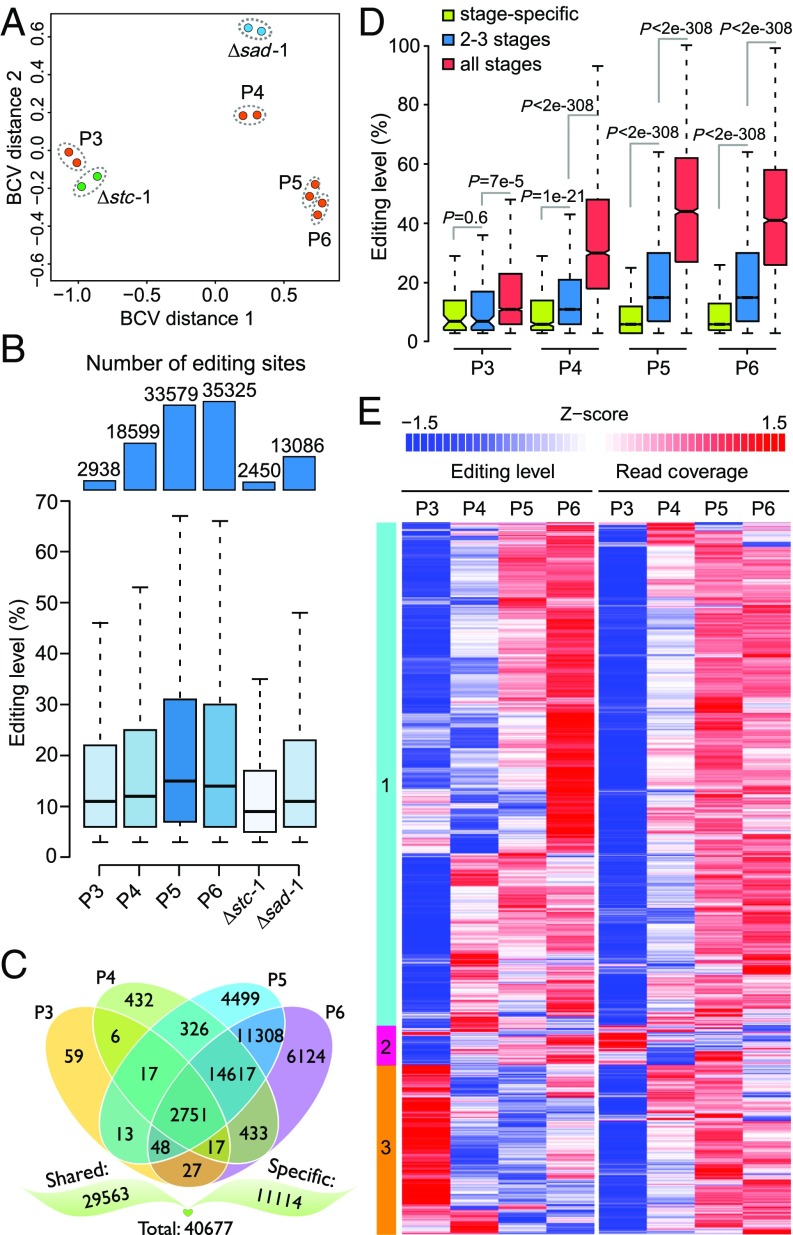
To determine how early RNA editing occurs, strand-specific RNA-seq data were generated with 3-, 4-, and 5-dpf perithecia of the OR74A × 74-ORS-6a cross (Fig. 1A). Although 3-dpf developing perithecia lacked ascus differentiation, 2,938 A-to-I editing sites were identified. A total of 18,599 and 33,579 A-to-I sites were identified in RNA-seq data of 4- and 5-dpf perithecia, respectively (Fig. 1B), in which ascus development and ascospore formation began (30).
Fig. 1.
Profiles of A-to-I RNA editing in different sexual stages of N. crassa. (A) Multidimensional scaling plots of the transcriptome profiles of perithecia of wild type [postnatal day (P) 3 to P6] collected 3, 4, 5, and 6 dpf and 5-dpf perithecia of Δsad-1 or Δstc-1 mutant. Two dots represent two biological replicates of each stage. Distances between samples correspond to the leading biological coefficient of variation (BCV). (B) Numbers of A-to-I editing sites identified in the labeled perithecium samples of the wild-type and mutant strains (Upper) and their corresponding editing levels (Lower). (C) Venn diagram showing the number of A-to-I sites unique to one specific stage or common to at least two different stages in the wild type. In total, 11,114 editing sites were stage-specific (specific) and 29,563 were common to two or more stages (shared). (D) Box plots of the editing level of A-to-I sites that were unique to one specific stage (stage-specific) and common to all or two to three stages. P values are from two-tailed Wilcoxon rank sum tests. (E) Hierarchical clustering of the editing level and read coverage (expression) of the 2,751 editing sites common to all sexual stages. The heat map color of each row depicts various editing levels or read coverages of individual editing sites after being normalized to the Z-score. The color bars on the left show the three clusters of editing sites with similar profiles.
We also conducted RNA-seq analysis with 5-dpf perithecia of the Δstc-1 and Δsad-1 mutants (Fig. 1A). Whereas the Δstc-1 mutant produced normal-sized perithecia that lacked asci, the Δsad-1 mutant formed normal asci, but ascus development was arrested in meiotic prophase after karyogamy (22, 30). A total of 2,450 and 13,086 A-to-I editing sites were identified in Δstc-1 and Δsad-1 perithecia, respectively (Fig. 1B). These results indicate that A-to-I editing occurs before ascus development but increased rapidly during ascus and ascospore development.
Different Sexual Development Stages Have Stage-Specific A-to-I Editing Sites.
For the 40,677 A-to-I sites identified in the wild-type perithecia, only 2,751 were shared across all four stages, but 59, 432, 4,499, and 6,124 were stage-specific for 3, 4, 5, and 6 dpf, respectively (Fig. 1C). Among the 26,812 A-to-I sites that were not detected in at least one stage, approximately a quarter of them had a read coverage (<10) or editing level (<3%) below the cutoff for calling editing events and were filtered out during identification. The majority of them, however, had a read coverage of ≥10 but were found to lack editing events. For example, 1,056 editing sites identified at other stages were not edited in 6-dpf perithecia, although the corresponding transcripts had a read coverage of ≥100. These results indicate that some A-to-I editing sites occur only at specific stages of sexual development in N. crassa.
A-to-I Editing Levels Are Differentially Regulated Across Different Sexual Stages.
The median editing levels of the A-to-I sites were increased during sexual development, with the 5- and 6-dpf perithecia having a relatively higher median editing level than 3- and 4-dpf perithecia (Fig. 1B). The median editing level of the A-to-I sites in the Δstc-1 and Δsad-1 mutants was similar to that of early sexual stages in the wild type, which is consistent with their arrestment in sexual development. While most of the A-to-I sites from each stage had low editing levels, ranging from 5 to 30% (Fig. 1B), the editing sites commonly occurring in different sexual stages tend to have higher editing levels (Fig. 1D), with the highest occurring in editing sites common to all stages.
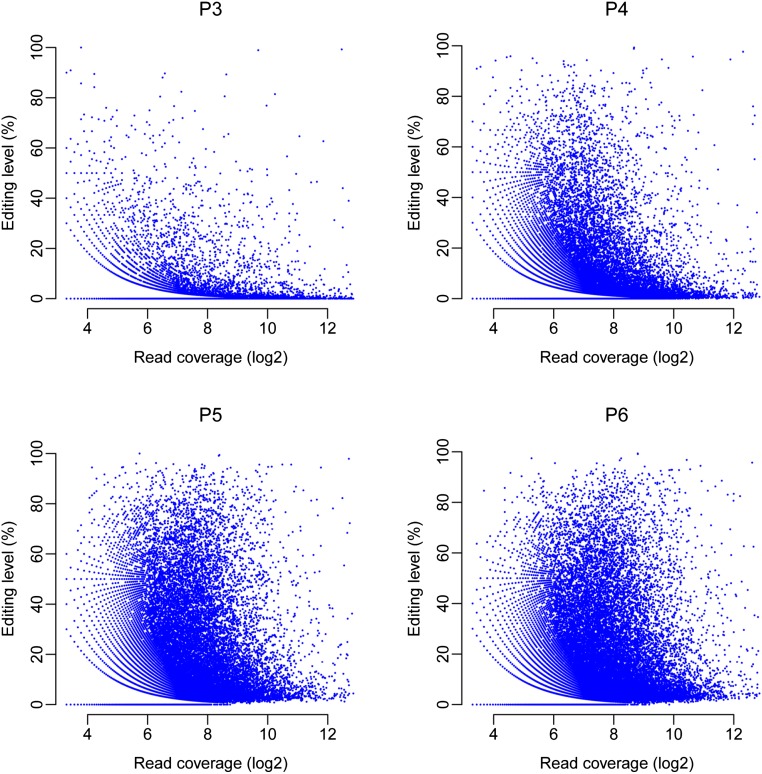
Hierarchical clustering analysis showed that for the majority of the 2,751 A-to-I sites edited at all stages, their editing levels increased during sexual development and generally reached the highest in 5- or 6-dpf perithecia (Fig. 1E). In general, there was no obvious correlation between RNA editing level and read coverage (expression level) (Fig. S2). However, for the editing sites in cluster 1 (Fig. 1E), both gene expression and editing levels increased at later stages of sexual development, suggesting that they are commonly up-regulated, and thus likely important for ascosporogenesis. Gene ontology (GO) enrichment analysis revealed that genes related to the regulation of various biological processes, including mitosis, meiosis, DNA replication, transcription, and translation, were overrepresented in the genes with cluster 1 editing sites (Datasets S1 and S2). Interestingly, cluster 2 editing sites had the highest gene expression but lowest editing levels at 3 dpf (Fig. 1E). In contrast, the editing sites in cluster 3 had the lowest gene expression but highest editing levels in 3-dpf perithecia (Fig. 1E). These data indicate that RNA editing at some of these sites may be related to their stage-specific functions.
Fig. S2.
Scatter plots of the editing level and read coverage (log2) of the A-to-I sites detected in 3- to- 6-dpf perithecia of the wild type (P3 to P6). No obvious positive correlation was observed between read coverages and RNA editing levels, suggesting that the increased expression of a given site does not increase the editing level.
Strong Preference of Uridine at the −1 Position and Recoding Events in CDSs for Editing.
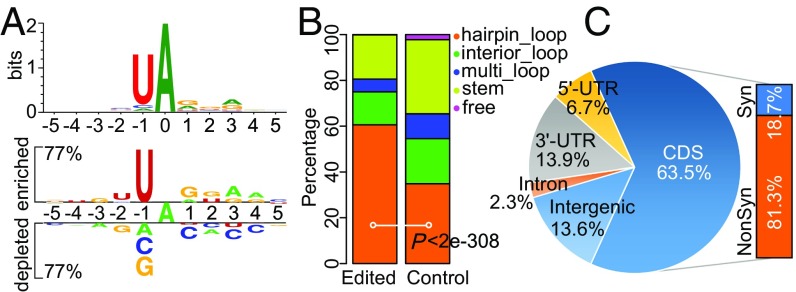
In total, 41,191 bona fide A-to-I editing sites (Dataset S3) were identified in the perithecia of wild type and mutants. Unlike in animals, A-to-I editing in N. crassa had nucleotide preferences from the −2 to +3 positions (Fig. 2A), particularly at the −1 position [96.7% uridine (U)]. Secondary structure analysis showed that editing sites were significantly enriched in the predicted hairpin loops of RNA sequences surrounding the edited A sites (30 nt upstream and 30 nt downstream) (Fig. 2B), which is similar to observations in F. graminearum (17). Furthermore, the largest fraction (63.5%) of editing sites in N. crassa was in the CDSs of 7,039 annotated protein-coding genes (NC12; Broad Institute) (Fig. 2C). Of the 26,526 A-to-I editing sites in CDSs, 81.3% (21,576) are nonsynonymous (recoding) editing sites (Fig. 2C). These observations suggest that RNA editing in N. crassa often causes protein recoding and likely affects protein functions during sexual reproduction.
Fig. 2.
Preference and distribution of the A-to-I editing sites in N. crassa. (A) Sequence conservation and nucleotide preference flanking the A-to-I editing sites. The height of letters in WebLogo (Upper) depicts the information content of the position in bits. Enriched and depleted nucleotides in Two Sample Logo (Lower) were calculated in comparison to a negative control using a t test with P < 0.0001 and Bonferroni correction. For the negative control, an equal number of A’s were randomly selected from cDNA sequences. (B) Stacked columns showing the percentage of edited and control sites located in different types of RNA secondary structure elements. For control sites, an equal number of A’s with similar nucleotide preference at the −2 to +3 positions were randomly selected from cDNA sequences. Prediction of RNA secondary structures is based on 30-nt upstream and 30-nt downstream sequences surrounding the edited or control A sites. The P value is from a χ2 test. (C) Genomic locations of the A-to-I editing sites and percentages of synonymous (Syn) or nonsynonymous (NonSyn) editing sites in CDSs.
Extensive Recoding Sites Are Enriched in Genes Important for Sexual Development.
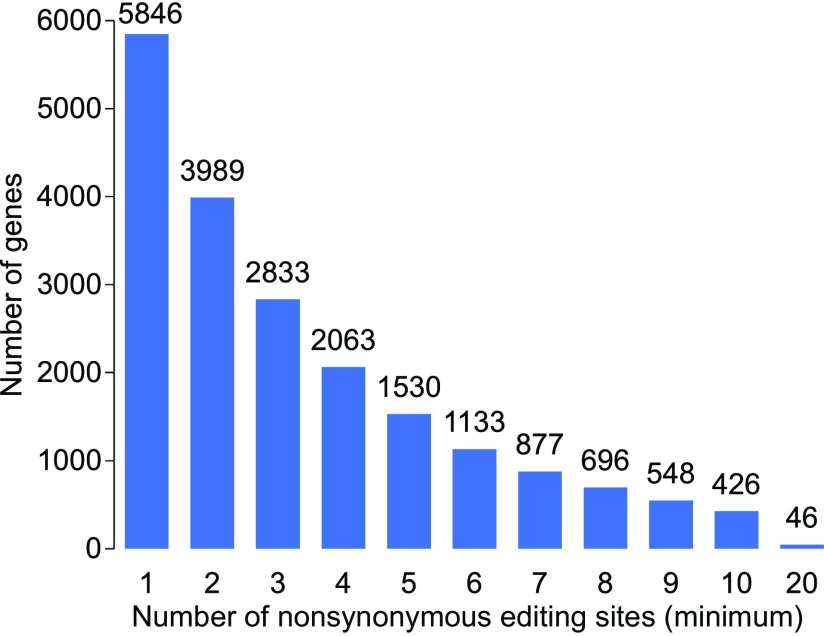
In total, 5,846 (62.8%) of the 9,302 protein-coding genes expressed during sexual development [fragments per kilobase million (FPKM) ≥ 1] in N. crassa had at least one recoding editing site. Among them, 2,833 and 426 had three or more and ≥10 recoding sites, respectively (Fig. S3). To analyze the extent of recoding in these proteins, we calculated the cumulative recoding level for each gene by summing up the editing levels of all of the recoding sites. When recoded genes were ranked by the cumulative recoding level, the top half was significantly enriched for genes involved in sexual reproduction and meiotic cell cycle (Dataset S4). Furthermore, genes functionally related to a variety of biological processes, including chromatin organization and modification, RNA transcription and processing, and protein transport and localization, also were overrepresented in the top half with higher cumulative recoding levels (Dataset S4), further indicating the importance and global effects of RNA editing during sexual development.
Fig. S3.
Numbers of N. crassa genes with marked minimum numbers of nonsynonymous recoding events.
A-to-I Editing May Affect Other Epigenetic Phenomena in N. crassa.
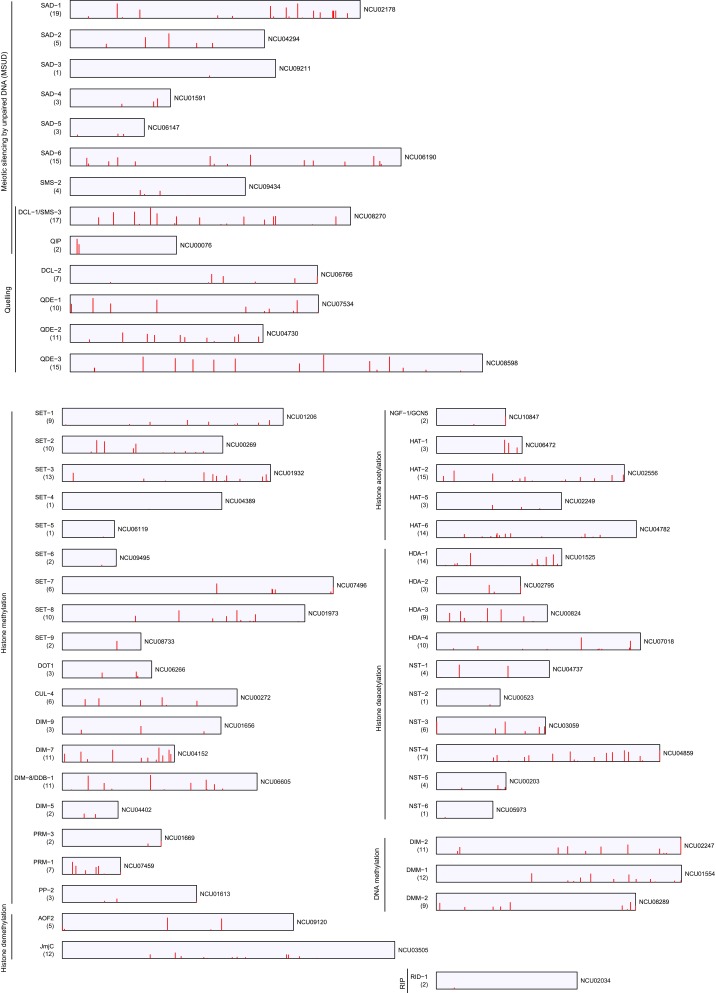
To determine the functional relationship of RNA editing with other epigenetic phenomena, we analyzed the editing events in the genes known to be important for DNA and chromatin modifications in N. crassa (31–33) and found that most of them had extensive nonsynonymous recoding (Fig. S4). The sad-1, sad-6, dcl-1/sms-3, qde-1, qde-2, and qde-3 genes that are required for quelling and/or MSUD all had multiple recoding sites with editing levels over 50%. Similarly, many genes important for DNA methylation and histone modifications also had multiple recoding sites with high editing levels (Fig. S4). For example, the dim-2 DNA methyltransferase gene responsible for cytosine methylation in N. crassa (34) had 11 recoding sites, including stop-loss editing with an editing level of 87%. The dim-7 and dim-8/ddb-1 genes required for histone methylation and DNA methylation (35) both had at least three recoding sites with editing levels >60%. Extensive nonsynonymous recoding in these genes suggests that A-to-I editing may affect RNA silencing, DNA methylation, and histone modification during sexual reproduction in N. crassa.
Fig. S4.
Nonsynonymous recoding events in genes related to MSUD, Quelling, and histone modifications in N. crassa. Each recoding site is marked with a red bar with the height proportional to its editing level. The numbers in parentheses indicate the total number of recoding sites identified in each gene.
A-to-I Editing Enables Pseudogenes with Premature Stop Codons to Encode Full-Length Proteins.
Similar to PUK1 in F. graminearum, its ortholog in N. crassa, stk-21 (NCU03242), also has one premature stop codon UAG in its ORF that was edited to UGG during sexual reproduction (17). These editing events were referred to as premature stop codon correction (PSC) editing in this study. Our RNA-seq data showed that an additional 49 predicted N. crassa pseudogenes had PSC editing events in perithecia (Dataset S5). RNA editing must be essential for these genes to encode full-length proteins specifically during sexual reproduction, particularly for the four of them that had more than one PSC editing event (Dataset S5).
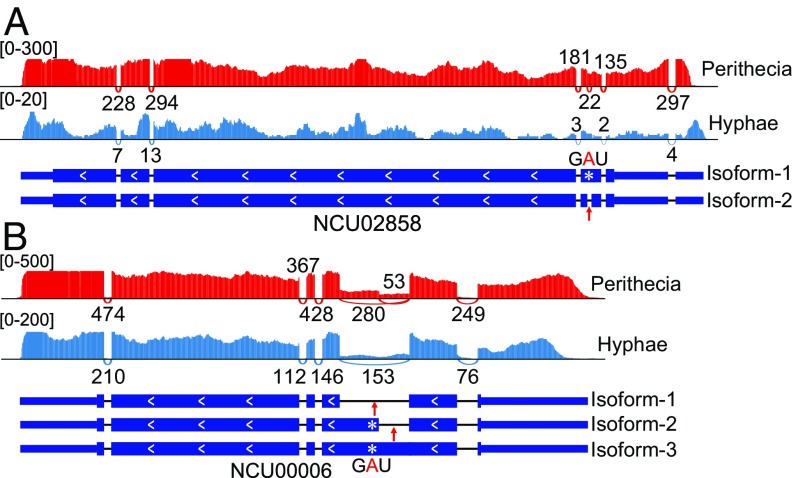
Interestingly, the premature stop codon UAG of NCU02858 and NCU00006 could be removed by either PSC editing or alternative splicing. NCU02858 had two transcript isoforms due to retention events of the third intron that harbors the premature stop codon UA269G (Fig. 3A). During sexual reproduction, isoform-2 transcripts with the third intron properly spliced (encoding a 2,358-aa protein) accounted only for ∼12% of the NCU02858 transcripts. Most of the NCU02858 transcripts (∼88%) were isoform-1 transcripts that had the third intron retained. However, ∼57% of isoform-1 transcripts were subject to PSC editing to code for a 2,377-aa protein. Unlike NCU02858, NCU00006 had three transcript isoforms derived from different alternative splicing events of the second intron that harbors the premature stop codon UA791G (Fig. 3B). Unlike NCU02858, the majority of NCU00006 transcripts had the second intron properly spliced (isoform-1) to encode the full-length protein as predicted. Isoform-2 and isoform-3 with the full-length or part of the second intron retained (Fig. 3B) accounted for ∼15% and ∼20% of the NCU00006 transcripts, respectively, during sexual reproduction. PSC editing of UA791G was observed in both isoform-2 and isoform-3 transcripts. Therefore, the combination of alternative splicing and RNA editing enabled genes like NCU02858 and NCU00006 to code for multiple protein isoforms during sexual development. Although their functional relationship is not clear, both RNA editing and alternative splicing may be involved in the coregulation of proper expression of certain genes during sexual reproduction.
Fig. 3.
Two genes with both PSC editing and alternative splicing events. IGV–Sashimi plots showing the read coverage and transcript isoforms of NCU02858 (A) and NCU00006 (B) in marked RNA-seq data. Numbers in square brackets indicate coverage range. Junction reads are plotted as arcs, and the number of reads aligned to the junction spanning the exons connected by the arc is indicated. A white star marks the edited premature stop codon, and a red arrow marks the intron with alternative splicing events. Isoform 2 of NCU02858 or NCU00006 transcripts derived from alternative splicing is only present in perithecia. A-to-I editing at the marked stop codon was detected in both isoforms 2 and 3 of NCU00006.
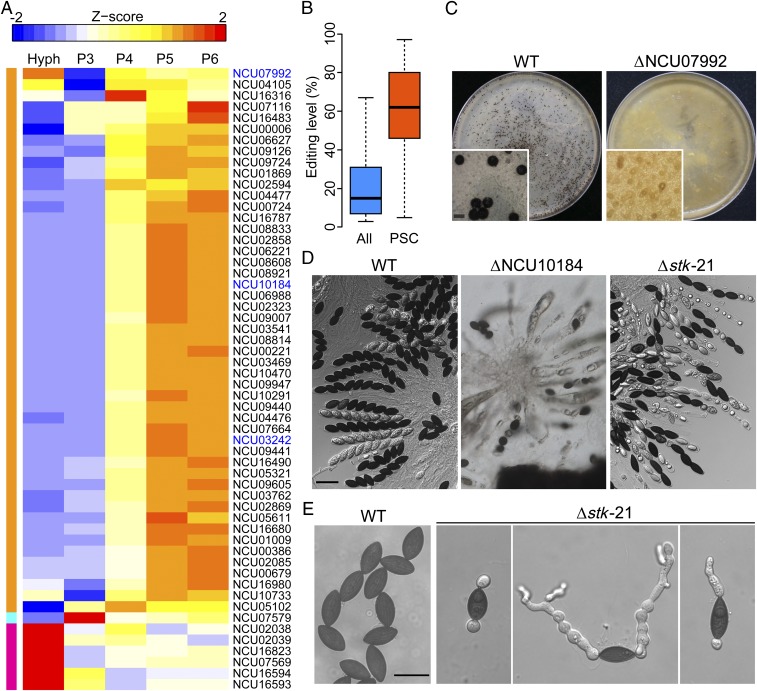
Almost all of the pseudogenes with PSC editing events were up-regulated or specifically expressed during perithecium development (Fig. 4A), and the median editing level of these PSC editing sites was over fourfold higher than that of all of the editing sites in N. crassa (Fig. 4B), suggesting their functional importance during sexual reproduction. Nevertheless, over half of these genes encode hypothetical proteins with no known protein domains or orthologs in the yeast (Dataset S5). Some of them have conserved orthologs in other fungi, including the orthologs of yeast SGF73 (NCU07579) and SPT3 (NCU07992) (36, 37), and putative Rap/ran-GAP (NCU04105) and porphobilinogen deaminase (NCU10291) genes, although their functions during sexual reproduction have not been reported in N. crassa or other filamentous ascomycetes.
Fig. 4.
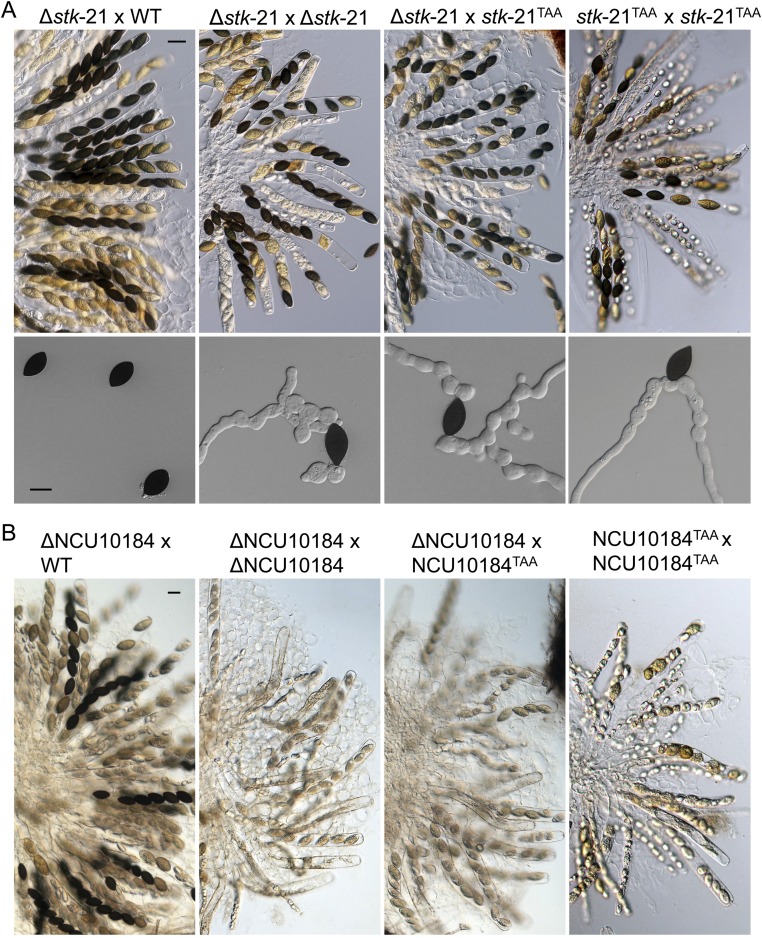
Expression and function of pseudogenes with PSC editing events in N. crassa. (A) Heat map of the expression of pseudogenes with PSC editing events in vegetative hyphae (Hyph) and four sexual stages (P3 to P6). The heat-map color of each row depicts various expressions of pseudogenes after being normalized to the Z-score. The color bars on the left show the clusters of similar profiles. The three genes in blue were experimentally confirmed to be important for sexual development in this study. (B) Box plots comparing the editing levels of the PSC editing sites and all of the A-to-I editing sites. (C) Mating plates and close-up views of the wild type (WT) and ΔNCU07992 mutant (FGSC19620 × FGSC19621) were examined 8 dpf. The ΔNCU07992 mutants formed protoperithecia but not perithecia. (Scale bar: 400 μm.) (D) Asci and ascospores of the WT, ΔNCU10184 mutants (FGSC22605 × FGSC22604), and Δstk-21 mutants in 8-dpf perithecia. (Scale bar: 50 μm.) (E) Ascospores of the WT and Δstk-21 mutants were examined after incubation in YEPD for 12 h without preheating at 65 °C. (Scale bar: 20 μm.)
Three Pseudogenes with PSC Editing Events Are Important for Sexual Development.
For 13 pseudogenes with PSC editing events, knockout mutants of both mating types have been generated in previous studies (38–41). In mating assays with mutants obtained from the Fungal Genetic Stock Center (FGSC) (Table S3), deletion of 11 genes, NCU01009, NCU04476, NCU02869, NCU03469, NCU06221, NCU00006, NCU07579, NCU06627, NCU05321, NCU08921, and NCU03540, had no significant effects on sexual reproduction. However, the NCU07992 deletion mutant failed to produce perithecia, although protoperithecia were formed (Fig. 4C), suggesting its essential role in the early stage of ascogenous tissue development. The NCU10184 deletion mutants were normal in perithecium formation but delayed in ascosporogenesis, and many asci were aborted and contained less than eight ascospores in 8-dpf perithecia (Fig. 4D).
Table S3.
Wild-type and mutant transformants used in this study
| Strain ID* | Description | Gene ID | Source |
| FGSC2489 A | N. crassa OR74A | Wild type | FGSC |
| FGSC4200 a | N. crassa 74-ORS-6a | Wild type | FGSC |
| FGSC2218 A | N. crassa RL3-8A | Wild type | FGSC |
| FGSC2508 A | N. tetrasperma | Wild type | FGSC |
| FGSC2509 a | N. tetrasperma | Wild type | FGSC |
| FGSC11152 A | Sad-1 knockout mutant | NCU02178 | FGSC |
| FGSC11151 a | |||
| FGSC17465 A | Stc-1 knockout mutant | NCU01496 | FGSC |
| FGSC17464 a | |||
| FGSC17940 a | Knockout mutant of gene with PSC editing | NCU03242 | FGSC |
| FGSC12834 A | Knockout mutant of gene with PSC editing | NCU01009 | FGSC |
| FGSC12833 a | |||
| FGSC13863 A | Knockout mutant of gene with PSC editing | NCU01869 | FGSC |
| FGSC17206 a | Knockout mutant of gene with PSC editing | NCU02594 | FGSC |
| FGSC17510 A | Knockout mutant of gene with PSC editing | NCU04476 | FGSC |
| FGSC17509 a | |||
| FGSC20604 a | Knockout mutant of gene with PSC editing | NCU09126 | FGSC |
| FGSC16774 a | Knockout mutant of gene with PSC editing | NCU00724 | FGSC |
| FGSC14160 A | Knockout mutant of gene with PSC editing | NCU02869 | FGSC |
| FGSC14159 a | |||
| FGSC22222 A | Knockout mutant of gene with PSC editing | NCU08608 | FGSC |
| FGSC17301 A | Knockout mutant of gene with PSC editing | NCU03469 | FGSC |
| FGSC17302 a | |||
| FGSC20809 A | Knockout mutant of gene with PSC editing | NCU09605 | FGSC |
| FGSC15170 A | Knockout mutant of gene with PSC editing | NCU06221 | FGSC |
| FGSC15169 a | |||
| FGSC20720 A | Knockout mutant of gene with PSC editing | NCU00006 | FGSC |
| FGSC20719 a | |||
| FGSC19412 A | Knockout mutant of gene with PSC editing | NCU07579 | FGSC |
| FGSC19411 a | |||
| FGSC17985 a | Knockout mutant of gene with PSC editing | NCU07569 | FGSC |
| FGSC12647 A | Knockout mutant of gene with PSC editing | NCU06627 | FGSC |
| FGSC12646 a | |||
| FGSC15357 a | Knockout mutant of gene with PSC editing | NCU06988 | FGSC |
| FGSC21827 A | Knockout mutant of gene with PSC editing | NCU08833 | FGSC |
| FGSC21823 A | Knockout mutant of gene with PSC editing | NCU08814 | FGSC |
| FGSC20686 A | Knockout mutant of gene with PSC editing | NCU05321 | FGSC |
| FGSC20685 a | |||
| FGSC22364 a | Knockout mutant of gene with PSC editing | NCU10291 | FGSC |
| FGSC16758 A | Knockout mutant of gene with PSC editing | NCU04105 | FGSC |
| FGSC22605 A | Knockout mutant of gene with PSC editing | NCU10184 | FGSC |
| FGSC22604 a | |||
| FGSC13928 A | Knockout mutant of gene with PSC editing | NCU07116 | FGSC |
| FGSC12810 a | Knockout mutant of gene with PSC editing | NCU05611 | FGSC |
| FGSC22024 A | Knockout mutant of gene with PSC editing | NCU02323 | FGSC |
| FGSC17485 a | Knockout mutant of gene with PSC editing | NCU02085 | FGSC |
| FGSC13384 A | Knockout mutant of gene with PSC editing | NCU10733 | FGSC |
| FGSC20564 A | Knockout mutant of gene with PSC editing | NCU08921 | FGSC |
| FGSC20563 a | |||
| FGSC20822 a | Knockout mutant of gene with PSC editing | NCU03540 | FGSC |
| FGSC22242 A | NCU03541 | ||
| FGSC21707 A | Knockout mutant of gene with PSC editing | NCU07664 | FGSC |
| FGSC12777 a | Knockout mutant of gene with PSC editing | NCU00386 | FGSC |
| FGSC19620 A | Knockout mutant of gene with PSC editing | NCU07992 | FGSC |
| FGSC19621 a | |||
| FGSC19727 a | Knockout mutant of gene with conserved editing sites | NCU00928 | Michael Freitag |
| FGSC19728 A | |||
| FGSC22440 A | Knockout mutant of gene with conserved editing sites | NCU04187 | Michael Freitag |
| FGSC22441 a | |||
| FGSC14587 a | Knockout mutant of gene with conserved editing sites | NCU05265 | Michael Freitag |
| FGSC14588 A | |||
| FGSC20416 a | Knockout mutant of gene with conserved editing sites | NCU11109 | Michael Freitag |
| FGSC20417 A | |||
| C2-7 A | stk-21 knockout mutant, RL3-8A background | NCU03242 | This study |
| C24 A | stk-21 knockout mutant, RL3-8A background | NCU03242 | This study |
| N9/10-1-2 A | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| N9/10-1-3 A | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| N9/10-1-4 A | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| N9/10-1-8 a | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| N9/10-1-11 a | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| N9/10-1-12 a | Δstk-21/stk-21TAA transformant | NCU03242 | This study |
| 10184s-1-1 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-1-5 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-1-10 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-3-5 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-3-9 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-3-15 A | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-1-14 a | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-1-15 a | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
| 10184s-3-2 a | ΔNCU10184/NCU10184TAA transformant | NCU10184 | This study |
ID, identification.
Mating type A or a is indicated.
For stk-21 and the other 19 pseudogenes with PSC editing events, knockout mutants are only available for one mating type (Table S3). When mating with the wild type as the female, no defects in sexual reproduction were observed in perithecium development and ascosporogenesis. Therefore, their functions during sexual reproduction remain to be determined. Because the editing event in PUK1 was conserved between F. graminearum and N. crassa, we generated the Δstk-21 mutant of a mat A strain in this study and crossed it with strain FGSC17940 (Δstk-21, mat a). The Δstk-21 mutants were defective in late stages of ascospore development and often produced morphologically abnormal ascospores and less than eight ascospores per ascus (Fig. 4D). More interestingly, when ascospores of Δstk-21 mutants were observed under a light microscope, broken ascospores (broken at one end) were often observed, suggesting a weakened ascospore wall or germ pore. When mutant ascospores were incubated in yeast extract–peptone–dextrose (YEPD) medium at 25 °C for 12 h without preheating at 65 °C, ∼6.3% of mutant ascospores germinated and produced bulbous germ tubes, while no germination of wild-type ascospores was observed (Fig. 4E).
To confirm the importance of PSC editing on gene function, we generated the stk-21TAA allele by changing G1575 to A by site-directed mutagenesis and transformed it into the Δstk-21 mutant (mat A). When crossed with strain FGSC17940 (Δstk-21, mat a), the Δstk-21/stk-21TAA transformants, similar to the original Δstk-21 mutant, were defective in ascospore maturation and germination without preheating at 65 °C (Fig. S5A). To rule out possible effects from MSUD, we isolated Δstk-21/stk-21TAA mat a progeny. Similar defects in ascospore maturation and germination were observed in the Δstk-21/stk-21TAA × Δstk-21/stk-21TAA cross (Fig. S5A). These data indicated that expression of stk-21TAA failed to complement the defects of Δstk-21. Unlike editing of TA1574G in the wild-type strain, editing of TA1574A to TGA in stk-21TAA resulted in another stop codon. Therefore, PSC editing of stk-21 must be essential for its function.
Fig. S5.
Functional assays with the stk-21TAA and NCU10184TAA alleles. (A, Upper) Asci and ascospores from the crosses between the mat A Δstk-21 mutant and a compatible (mat a) wild type (WT), Δstk-21 mutant, or Δstk-21/stk-21TAA transformant and the Δstk-21/stk-21TAA homozygous cross. (A, Lower) Ascospores after being incubated in YEPD for 12 h without preheating at 65 °C. (Scale bar: 20 μm.) (B) Asci and ascospores from the crosses between the mat A ΔNCU10184 mutant with a compatible (mat a) WT, ΔNCU10184 mutant, and ΔNCU10184/NCU10184TAA transformant and the ΔNCU10184/NCU10184TAA homozygous cross. (Scale bar: 20 μm.)
We also generated the NCU10184TAA allele by introducing the G660A mutation and transformed it into the ΔNCU10184 mutant (mat A). In crosses between the resulting ΔNCU10184/NCU10184TAA transformants and FGSC22604 (ΔNCU10184; mat a), ascosporogenesis was defective and many asci were aborted (Fig. S5B). When the resulting ΔNCU10184/NCU10184TAA mat a progeny was crossed with ΔNCU10184/NCU10184TAA transformants, similar defects in ascosporogenesis were observed (Fig. S5B). Thus, expression of NCU10184TAA also failed to complement the defects of the ΔNCU10184 mutant, further indicating the importance of PSC editing for these pseudogenes to encode full-length, functional proteins in N. crassa.
Stop-Loss Editing Alters the C Terminus of Conserved Proteins During Ascosporogenesis.
The nonsynonymous editing changed the predicted stop codon UAG of 672 genes to amino acid codon UGG (stop-loss editing) (Dataset S6), resulting in the addition of an extra stretch of peptides to the C terminus of proteins. A number of genes with stop-loss editing are known to be important for sexual development, including the ascospore lethal-1 (asl-1/atf-1), mid-1, pp-2, pre-2, ff-1, and ham-2 genes (31, 42–46). Some are well-conserved genes encoding proteins that are involved in histone modification (e.g., hat-2, hda-2, hat-6, ngf-1) and chromatin assembly or remodeling (e.g., cac-2, cac-3, crf3-1, crf4-3, crf10-1/csw-1). Interestingly, six of the eight subunits of the COP9 signalosome (47), csn-1, csn-3, csn-4, csn-6, csn-7, and NCU08388 genes, are among those genes that are constitutively expressed in N. crassa but targeted for stop-loss editing specifically during sexual reproduction. These observations suggest that stop-loss editing may play an important role in the regulation of expression or stage-specific protein functions.
Nonsynonymous Editing Events Are Generally Beneficial and Under Positive Selection.
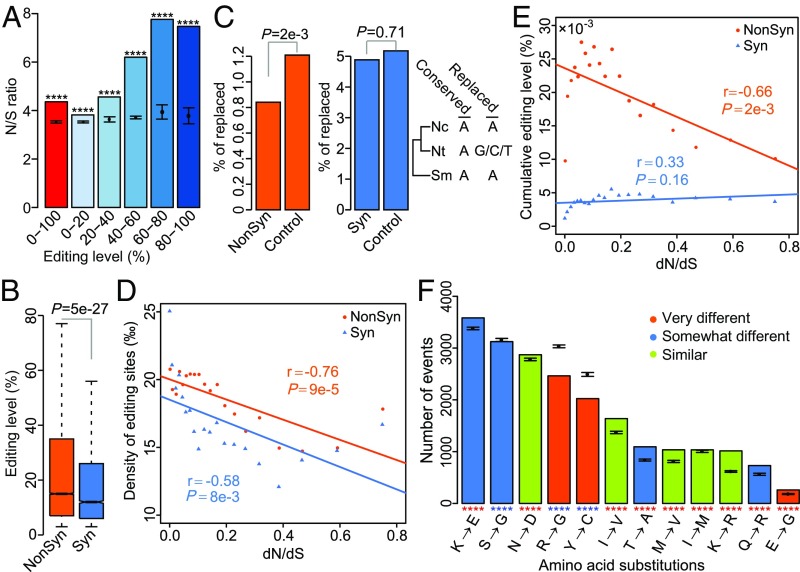
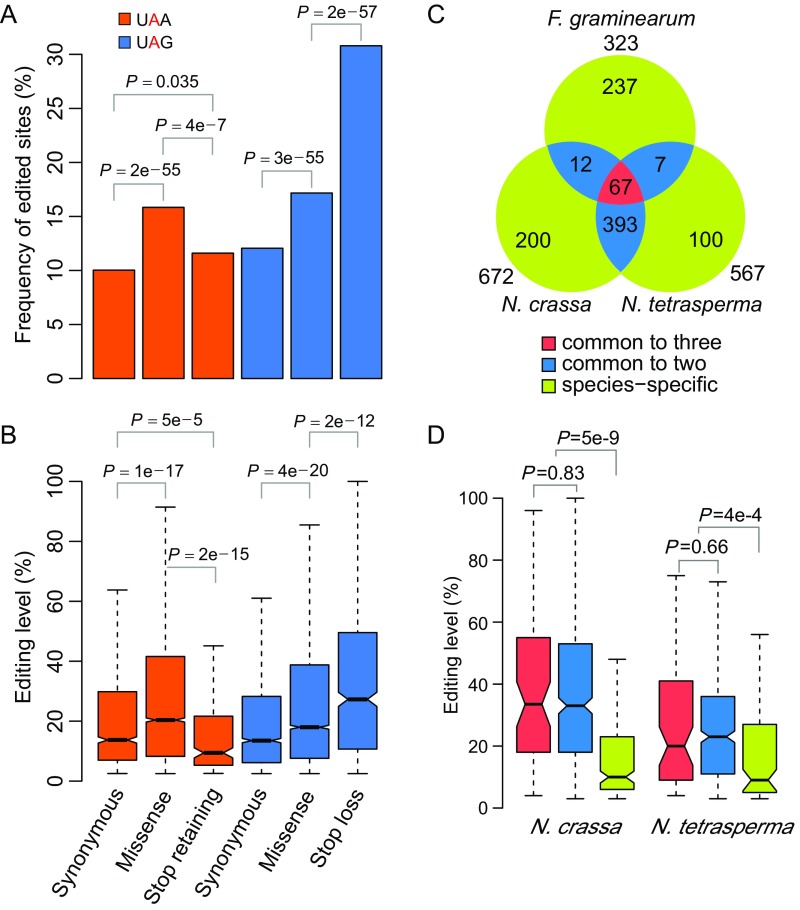
To determine whether the high fraction of nonsynonymous editing is by chance, we estimated the expected nonsynonymous-to-synonymous (N/S) ratio in N. crassa by randomly selecting the same number of A sites from CDSs of edited genes according to the nucleotide preference at the −2 to +3 positions of edited A’s. The observed N/S ratio (4.36) was significantly higher than the expected N/S ratio (3.53) (Fig. 5A), signaling positive selection for the nonsynonymous editing events. The median editing level of nonsynonymous editing sites (15%) also was significantly higher than that of synonymous editing sites (12%) (Fig. 5B). Furthermore, the editing sites with higher editing levels tend to have higher N/S ratios (Fig. 5A), suggesting more advantageous editing events at nonsynonymous editing sites with higher editing levels. Notably, both the frequency and editing level of stop-loss editing were significantly higher than those of missense editing of UAG triplets (Fig. S6 A and B). In contrast, the frequency and editing level of stop-retaining editing sites that changed UAA to UGA were significantly lower than those of missense editing of UAA triplets (Fig. S6 A and B). These results suggest that stop-loss editing is under stronger positive selection. Taken together, our results indicate that nonsynonymous editing events are generally beneficial and favored by positive selection during evolution in N. crassa.
Fig. 5.
Properties of the nonsynonymous editing sites in N. crassa. (A) N/S ratio for the editing sites in each group with marked editing levels. Expected mean and SD were calculated with 100 bootstrap samples of an equal number of A’s with similar nucleotide preference at the −2 to +3 positions of edited A’s randomly selected from coding sequences of edited genes in each group. ****P < 10−8, t test. (B) Box plots comparing the editing levels of nonsynonymous (NonSyn) and synonymous (Syn) editing sites. The P value is from a two-tailed Wilcoxon rank sum test. (C) Percentage of putative ancestral A’s replaced with G/C/T in N. tetrasperma at the NonSyn and Syn editing sites. The P values are from χ2 tests. Nc, N. crassa; Nt, N. tetrasperma; Sm, S. macrospora. The control is an equal number of random A’s with similar nucleotide preference at the −2 to +3 positions of edited A’s from coding sequences of edited genes. (D) Correlation between the density of NonSyn or Syn editing sites and the medians of dN/dS ratios. In each bin, the editing density is calculated by dividing the observed number of NonSyn or Syn editing sites by the total number of NonSyn or Syn editable sites with similar nucleotide sequences at the −2 to +3 positions of edited A’s. (E) Correlation between the cumulative editing level of NonSyn or Syn editing and the medians of dN/dS ratios. In each bin, the cumulative editing level is calculated by summing up the editing levels of all of the NonSyn or Syn sites. In D and E, each data point represents a group of edited genes (5% each) that were categorized based on the dN/dS ratio. The Pearson correlation coefficient and P value are indicated. (F) Distribution of amino acid changes caused by NonSyn editing. Colors indicate amino acid change classification as described elsewhere (48). Expected mean and SD were calculated with 100 bootstrap random samples of an equal number of A’s with similar nucleotide preference at the −2 to +3 positions of edited A’s from coding sequences of edited genes. ****P < 10−5, t test.
Fig. S6.
Statistical features of stop codon editing. The frequency (A) and editing level (B) of stop-loss (UAG to UGG) and stop-retaining (UAA to UGA) editing are compared with those of missense and synonymous editing of the same triplets. P values in A and B are from Fisher’s exact tests and two-tailed Wilcoxon rank sum tests, respectively. The number (C) and editing level (D) of stop-loss editing events that are species-specific (green), common to any two species (blue), or common to all three species (red) are shown. P values in D are from two-tailed Wilcoxon rank sum tests.
Proteome Complexity Increased by Nonsynonymous Editing Confers Adaptive Advantage.
To determine whether the proteome complexity increased by nonsynonymous editing has adaptive advantages, we first identified the orthologous genes that are conserved in N. crassa and its closely related species N. tetrasperma and Sordaria macrospora, and selected the A-to-I editing sites that are also A’s at the corresponding sites in their S. macrospora orthologs. Given the phylogenetic relationship among these three species, their common ancestor likely has A’s at these sites. We then calculated the fraction of nucleotide substitution in N. tetrasperma at these ancestral A sites. If A-to-I editing is adaptive, the nonsynonymous A sites are less likely to be replaced by other nucleotides during evolution because such replacements would reduce the protein complexity and fitness. As a control, we randomly selected the same number of editable A sites from coding sequences of edited genes based on the nucleotide preference at the −2 to +3 positions of edited A sites. Consistent with the adaptive hypothesis, the percentage of nucleotide substitution at the nonsynonymous A sites in N. crassa was significantly lower than that at the control A sites (Fig. 5C). As another control, there was no significant difference between the synonymous A sites and control A sites (Fig. 5C). Therefore, the protein complexity enabled by nonsynonymous editing confers an adaptive advantage that makes the recoding events to be maintained by natural selection during evolution.
Nonsynonymous Editing Tends to Recode Genes Under Stronger Functional Constraints.
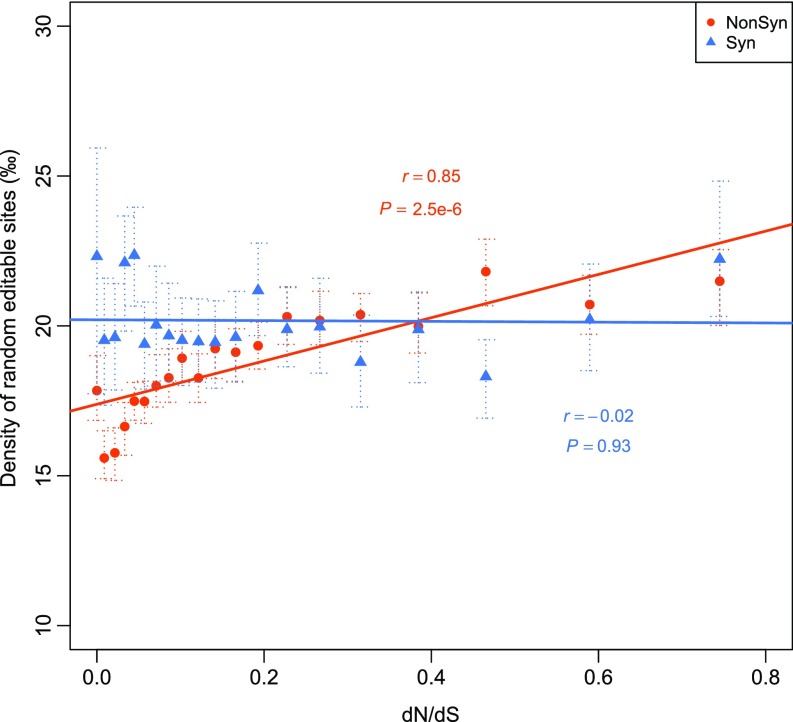
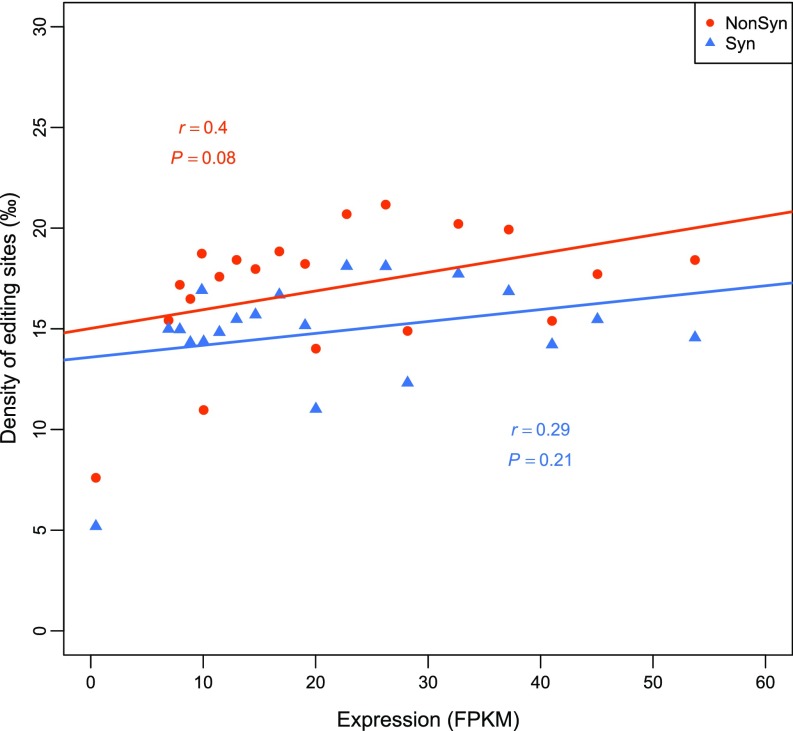
Because the evolutionary rate of proteins is primarily determined by functional constraints, to verify that nonsynonymous editing facilitates adaptive evolution of proteins, we examined the relationship between the selection on nonsynonymous editing and selection on its targeted proteins. The orthologs of N. crassa genes with editing events in N. tetrasperma were identified and divided into 20 bins with an equal number of genes in each bin based on the ranking of the ratio of the nonsynonymous substitution rate (dN) to the synonymous substitution rate (dS). The lower the dN/dS value, the stronger the functional constraint. Remarkably, whereas the density of random nonsynonymous editable A sites had a strong positive correlation with the dN/dS values, the density of nonsynonymous editing sites was strongly negatively correlated with the dN/dS values (Fig. 5D and Fig. S7). A weaker negative correlation was observed for the density of synonymous editing sites, but no significant correlation was observed for the random synonymous editable A sites (Fig. 5D and Fig. S7). Because there was no significant correlation between the editing densities and gene expression values (FPKM) (Fig. S8), it is unlikely that the elevated editing densities in the genes under stronger functional constraints are caused by detection bias due to gene expression levels. Furthermore, a significant negative correlation was identified between the nonsynonymous editing intensity (measured by summing up the editing levels of all nonsynonymous sites in each bin) and the dN/dS values, but not between the synonymous editing intensity and the dN/dS values (Fig. 5E). Altogether, these results suggest that nonsynonymous editing is positively selected to increase the protein diversity of the genes under stronger functional constraints that are less likely accessed through DNA mutations, and therefore provide compelling evidence to support the adaptive advantage of editing in N. crassa.
Fig. S7.
Correlation between the density of nonsynonymous (NonSyn) or synonymous (Syn) random editable sites and the medians of dN/dS ratios. The random editable sites were generated by selecting the same number of A sites from CDSs of edited genes according to the nucleotide preference at the −2 to +3 positions of edited A’s. In each bin, the density is calculated by dividing the number of NonSyn or Syn random editable sites by the total number of NonSyn or Syn editable sites. Error bars show the 95% confidence interval for the mean estimated from 100 random samplings. Each data point represents a group of genes (5% each) that were categorized based on the dN/dS ratio. The Pearson correlation coefficient and P value are indicated.
Fig. S8.
Correlation between the density of nonsynonymous (NonSyn) or synonymous (Syn) editing sites and the median of gene expression values (FPKM). In each bin, the editing density is calculated by dividing the observed number of NonSyn or Syn editing sites by the total number of NonSyn or Syn editable sites with similar nucleotide sequences at the −2 to +3 positions of edited A’s. Each data point represents a group of edited genes (5% each) that were categorized based on the dN/dS ratio. The Pearson correlation coefficient and P value are indicated.
Selection on Nonsynonymous Editing Events Is Codon-Specific.
The majority of the nonsynonymous editing events resulted in changes of amino acid residues with different biochemical properties, including lysine to glutamate (K-to-E), serine to glycine (S-to-G), arginine to glycine (R-to-G), and tyrosine to cysteine (Y-to-C) which are four of the top five most frequent substitution types (Fig. 5F). Although this bias may be mainly caused by editing preferences, for most of these amino acid substitution types, their occurrence was more frequent than expected. The notable exemptions are the R-to-G and Y-to-C changes that occurred far less frequently than expected (Fig. 5F). Notably, these two substitution types cause significant changes in amino acid physicochemical properties (48), which may be deleterious and purged by purifying selection during evolution. These results indicate that nonsynonymous editing is favored by natural selection to fine-tune protein functions but avoid causing dramatic functional changes in N. crassa.
Over Half of the A-to-I Editing Sites Are Conserved Between N. crassa and N. tetrasperma.
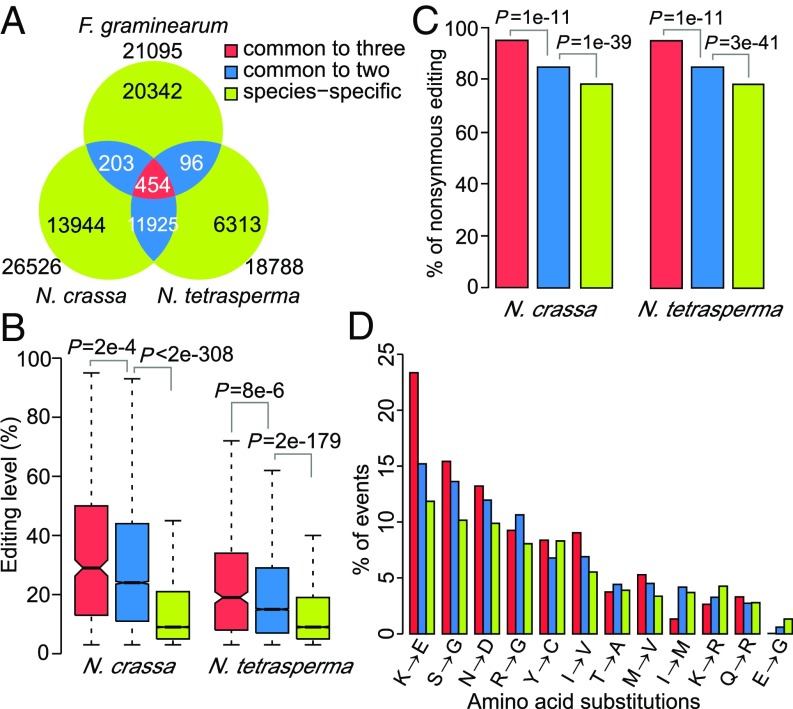
For comparative analysis, we also performed RNA-seq analysis with N. tetrasperma, a pseudohomothallic fungus that produces four heterokaryotic ascospores in each ascus (49). In RNA samples isolated from 6-dpf perithecia, a total of 28,442 bona fide A-to-I editing sites were identified (Dataset S7). Over 66% of them (18,788) were in the CDSs and caused recoding events in 4,752 protein-coding genes. For the 24,001 A-to-I sites in the CDSs of 5,952 N. crassa genes with distinct orthologs in N. tetrasperma, 51.6% of them (12,379) were edited in both Neurospora species (Fig. 6A and Dataset S8). For the 672 N. crassa genes with stop-loss editing, 460 of their orthologs in N. tetrasperma also had stop-loss editing events (Fig. S6C and Dataset S6). For the 50 N. crassa genes requiring PSC editing to encode full-length proteins, all their orthologs in N. tetrasperma also had premature stop codons in their ORFs. PSC editing events at the premature stop codons were detected in 45 of them specifically during sexual reproduction (Dataset S5).
Fig. 6.
Statistical features of evolutionarily conserved A-to-I editing sites. (A) Venn diagram showing the number of A-to-I editing sites that are species-specific (green), common to any two species (blue), or common to all three species (red) among N. crassa, N. tetrasperma, and F. graminearum. (B) Box plots comparing the editing levels of the three color-themed groups of editing sites as shown in A in N. crassa or N. tetrasperma. (C) Percentage of nonsynonymous editing events in these three groups of editing sites in N. crassa or N. tetrasperma. (D) Distribution of amino acid changes caused by missense editing in the three groups of editing sites in N. crassa. P values in B and C are from two-tailed Wilcoxon rank sum tests and χ2 tests, respectively.
Highly Conserved A-to-I Editing Sites Tend to Be Nonsynonymous and Have Higher Editing Levels.
To compare the A-to-I editing sites in N. crassa, N. tetrasperma, and F. graminearum, we first identified the orthologous protein coding genes in these species with OrthoMCL (50). Of 21,935 A-to-I sites in the CDSs of 5,132 N. crassa genes with orthologs in F. graminearum, 657 were also editing sites in the latter (17). In total, 454 A-to-I editing sites were conserved and edited in the orthologs of all three species (Fig. 6A and Dataset S8). Sixty-seven orthologous genes had conserved stop-loss editing events in all three species (Fig. S6C and Dataset S6). In general, the editing sites common to more than one species, particularly the ones conserved in all three fungi, had higher editing levels than species-specific editing sites (Fig. 6B and Fig. S6D). Moreover, the well-conserved editing sites had a higher percentage of nonsynonymous editing events than the less conserved ones (Fig. 6C). These results suggest that the highly conserved nonsynonymous or stop-loss editing sites are under stronger positive selection. Furthermore, the K-to-E, S-to-G, asparagine to aspartate (N-to-D), I-to-V, and methionine to valine (M-to-V) residue changes, particularly the K-to-E change, were obviously overrepresented in substitutions caused by conserved editing events (Fig. 6D). It is likely that the conserved nonsynonymous editing events causing these types of amino acid substitutions are functionally important and have an adaptive advantage in N. crassa.
Two of Four Genes with Conserved Editing Sites Are Important for Sexual Reproduction.
Among the 454 editing sites common to all three fungi, 431 resulted in nonsynonymous editing in 366 protein-coding genes in N. crassa (Dataset S9), including genes important for sexual development, such asl-1/atf-1 and round spore (Rsp/R). GO enrichment analysis showed that the 366 genes with highly conserved editing sites were enriched in genes with a variety of regulatory functions (Dataset S10), such as genes important for epigenetic control of gene expression, cell cycle process, protein phosphorylation, and autophagy. A number of them had more than one conserved editing site, including the set-1 and prm-1 histone methyltransferase genes that each had three conserved nonsynonymous editing sites (Dataset S9).
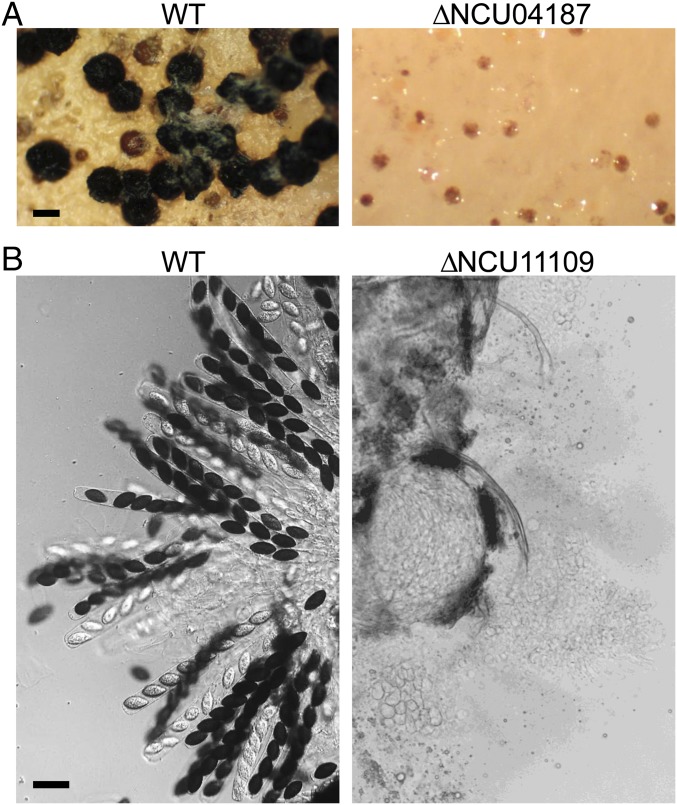
We then randomly selected four uncharacterized genes with conserved editing sites that had deletion mutants of both mating types available (Table S3) for characterizing their functions during sexual reproduction. Deletion of the NCU11109, NCU05265, NCU00928, or NCU04187 gene had no obvious effect on vegetative growth. For the NCU05265 and NCU00928 deletion mutants, no obvious defects in perithecium development and ascospore formation were observed. However, mutants deleted of NCU04187 failed to produce perithecia, although protoperithecia were formed (Fig. 7A). For the NCU11109 mutants, perithecium development appeared to be normal, but over 60% of the mutant perithecia failed to form asci at 8 dpf (Fig. 7B). These results suggest that genes with conserved A-to-I editing sites and their stage-specific editing events likely play important roles in various developmental processes during sexual reproduction.
Fig. 7.
Assays for sexual development in knockout mutants of N. crassa genes with conserved editing sites. (A) Mating cultures of the wild type (WT) and ΔNCU04187 mutant (FGSC22440 × FGSC22441) at 8 dpf were examined under a dissect microscope. The ΔNCU04187 mutants formed protoperithecia but failed to produce perithecia. (B) Asci and ascospores of the WT and ΔNCU11109 mutants (FGSC20416 × FGSC20417). Over 60% of the perithecia formed by ΔNCU11109 mutants had no asci at 8 dpf. (Scale bar: 20 μm.)
Discussion
As a model filamentous fungus, N. crassa is well studied for various genetic and epigenetic phenomena, including sexual-specific RIP, MSUD, and spore killer (20, 32). In this study, a total of 41,191 A-to-I editing sites that affected the transcripts of 7,039 protein coding genes were identified in RNA-seq data of perithecia, suggesting the occurrence of genome-wide A-to-I RNA editing during sexual reproduction in N. crassa. Comparative analysis with 3- to 6-dpf perithecia showed that RNA editing occurs at a low abundance in 3-dpf perithecia but becomes abundant in 5- or 6-dpf perithecia. Because the Δstc-1 mutant did not produce asci and the Δsad-1 mutant was arrested in meiotic prophase, the detection of over 2,000 and 10,000 editing sites in 5-dpf perithecia of the Δstc-1 and Δsad-1 mutants, respectively, suggests that A-to-I editing occurred before ascus development and increased before meiosis. The detection of over 30,000 editing sites in 5- and 6-dpf perithecia indicates the prevalence and importance of A-to-I editing during ascosporogenesis.
Interestingly, whereas some A-to-I editing sites commonly occurred in all four stages (3 to 6 dpf), many others were stage-specific or present only in two or three stages. In general, the commonly occurring A-to-I sites tend to have higher editing levels, but their editing levels often increased during sexual development and reached the highest at 5 or 6 dpf. For the stage-specific editing sites, some of them may be false-positives findings caused by low transcript abundance or editing levels. However, for the editing sites that were present in some but not all stages, the majority of them had sufficient transcripts expressed in the particular stages that lacked RNA editing at these sites. Taken together, both editing levels and the number of editing sites were different at different sexual development stages in N. crassa, suggesting the importance of A-to-I editing in stage-specific expression and functions of edited genes.
Like in F. graminearum, the A-to-I RNA editing in N. crassa has a strong nucleotide preference for U at the −1 position and a secondary structure preference for hairpin loops harboring the editing sites. These observations indicate that the molecular mechanism responsible for RNA editing in these two fungi is conserved. Recently, A-to-I RNA editing was reported to occur during sexual development in S. macrospora and an early-diverging filamentous ascomycete Pyronema confluens, but not in Schizosaccharomyces pombe (51). To date, all of the ascomycetes with A-to-I editing identified during sexual reproduction produce fruiting bodies. Although additional data and further analyses are necessary to conclude which lineages of fungi have A-to-I editing, it is likely that RNA editing is an evolutionarily conserved feature in filamentous ascomycetes and plays a critical role in different sexual developmental processes. Although fungi lack orthologs of animal ADARs that have both a dsRNA binding domain and adenosine deaminase domain, it has been suggested that tRNA-specific adenosine deaminase genes orthologous to yeast TAD2 and TAD3 may be involved in RNA editing in F. graminearum (17, 52). Our RNA-seq data showed that the TAD2 and TAD3 orthologs in N. crassa had increased expression during sexual development, particularly in 5- and 6-dpf perithecia.
Unlike in mammals, over half of the identified editing events in N. crassa (52.4%) caused amino acid changes. The extensive recoding editing in perithecia, including editing of premature stop codons, indicates the importance and functions of A-to-I editing in sexual development. Despite a few cases known to be functionally important (2, 5), the biological significance of nonsynonymous editing events in mammals is not clear or questionable. In humans, A-to-I editing sites identified in the coding regions are largely deleterious rather than beneficial, as both the frequency and level of nonsynonymous editing events are significantly lower than those of synonymous editing events (11). Recently, it has been reported that the nonsynonymous editing sites in brains and evolutionarily constrained editing sites are generally adaptive in Drosophila (53–55). In this study, we showed that the nonsynonymous editing events in N. crassa are generally beneficial and favored by positive selection during evolution. The percentage of nonsynonymous editing sites generally increased as the editing levels increased, suggesting that nonsynonymous editing events with higher editing levels are more likely to be advantageous. The nonsynonymous editing was favored by positive selection to increase the proteome complexity, particularly the protein diversity of genes under stronger functional constraints, therefore supporting the adaptive evolution hypothesis of RNA editing (56). Furthermore, our data showed that selection on nonsynonymous editing is codon-specific. The editing events resulting in changes in residue physicochemical properties, such as K-to-E, are favored by positive selection, whereas those resulting in extremely different amino acid changes, such as R-to-G and Y-to-C, are purged by purifying selection. Therefore, it is possible that RNA editing is adapted to fine-tune protein functions but avoid nonsynonymous editing of the A sites, which may have deleterious effects on protein expression or functions in fungi.
Although it has a tissue or developmental stage preference in animals, A-to-I RNA editing has been identified in virtually all of the tissues examined (4). In contrast, genome-wide A-to-I editing is stage-specific and occurs only during sexual reproduction in N. crassa and N. tetrasperma, which is similar to observations in F. graminearum (17). An intriguing question is why genome-wide A-to-I editing occurs specifically during sexual reproduction in fungi. The advantage of sexual reproduction is to increase genetic variations for adaptation to changing environments by meiotic recombination (57, 58), and possibly by elevated spontaneous mutations during meiosis in fungi (59, 60). RNA editing is hypothesized to facilitate adaptive evolution by expanding proteomic diversity at an epigenetic level (56). In animals, recent studies have showed that RNA editing is used for temperature adaptation by increasing the flexibility of proteins (10, 12, 61). The abundance of recoding events during sexual reproduction may provide great flexibility of proteins for fast acclimation in fungi. Our evolutionary analysis confirmed that A-to-I editing is maintained by natural selection to increase the protein diversity that confers an adaptive advantage in N. crassa. Furthermore, because genetic recombination and DNA mutation are permanent and hardwired, the extent of genetic variations at genes under strong functional constraints is restricted. In N. crassa, nonsynonymous editing tends to recode genes under strong functional constraints (low dN/dS value), confirming that RNA editing may provide amino acid sequence variations in genes or regions that are not accessible for genetic mutations. Taken together, the A-to-I RNA editing during sexual reproduction in fungi serves as a strong driving force for adaptive evolution.
For nonsynonymous editing events with high editing levels, such as the editing events at the premature stop codons of pseudogenes, besides increasing protein diversity, the proteins synthesized from edited transcripts may have higher fitness than those of unedited transcripts because they may be under directional selection to increase the editing level. Nonsynonymous editing events with low editing levels also increase sequence variants that likely confer adaptive advantage. However, they may not improve the fitness of encoded proteins and are maintained at low levels by balancing selection in N. crassa to simply confer a protein heterozygosity advantage.
For genes with distinct orthologs between N. crassa and N. tetrasperma and expressed during sexual reproduction, a total of 12,379 A-to-I editing sites in the CDSs were identified in both Neurospora species, which accounted for 46.7% and 65.9% of the editing sites in CDSs in N. crassa (26,526) and N. tetrasperma (18,788), respectively. Most likely, additional editing sites are conserved in N. tetrasperma but not detected because only 6-dpf perithecia were used for RNA-seq analysis in this study. The conserved nature of these RNA editing events in two Neurospora species suggests their importance during sexual reproduction. Among the editing sites common to N. crassa and N. tetrasperma, 657 of them are conserved in F. graminearum, and they generally had higher editing levels and more frequent nonsynonymous editing. Considering the phylogenetic distance among these three species, it is likely that these highly conserved nonsynonymous editing sites are functionally important for or beneficial to ascosporogenesis in Sordariomycetes. In fact, two of the four uncharacterized genes with conserved editing sites selected for functional characterization were shown to be important for sexual reproduction in N. crassa in this study.
In N. crassa, 50 pseudogenes require PSC editing to encode full-length proteins specifically during sexual reproduction. All of them are conserved, and their orthologs also have the premature stop codon in the ORF in N. tetrasperma, suggesting their specific expression and function during sexual reproduction. Although PSC editing events were only detected in 45 of these pseudogenes in 6-dpf perithecia of N. tetrasperma, the other five genes may have stage-specific editing in early stages of sexual development. Interestingly, among the 18 Neurospora pseudogenes with PSC editing that have distinct orthologs in F. graminearum, three of them also had PSC editing events in the latter (17). One is stk-21, and the other two are the NCU09007 and NCU01009 genes. In this study, we showed that stk-21 is important for ascospore maturation and ascospore integrity. FGRRES_10094, the ortholog of NCU01009, is important for ascospore discharge in F. graminearum (17). It is tempting to speculate that many of these genes with conserved PSC editing sites during sexual reproduction are functionally related to ascosporogenesis and ascospore discharge, and that the expression of their full-length proteins outside perithecia may be detrimental to vegetative growth.
In F. graminearum and many other Fusarium species, the RID ortholog has a premature stop codon in its ORF that requires A-to-I editing to encode a functional protein (17, 52). Therefore, RNA editing is essential for RIP in these fungi. In N. crassa, RID lacks premature stop codons in its ORF but it has two nonsynonymous editing events. The dim-2, dim-8, dmm-1, and dmm-2 genes also had multiple A-to-I editing sites. Similarly, although none of the genes important for MSUD had premature stop codons in their ORFs, the sad-1, sad-6, and dcl-1/sms-3 genes all had multiple nonsynonymous editing sites in N. crassa. Many of them are conserved in N. tetrasperma. These observations suggest that A-to-I editing may be functionally related to RIP and MSUD in N. crassa. It will be important to characterize these conserved recoding editing sites and determine their roles in the functions of RIP- or MSUD-related genes during sexual reproduction.
In summary, this study comprehensively characterized the A-to-I RNA editing in N. crassa during sexual reproduction and revealed the existence of stage-specific editing events at different sexual stages. Our analysis showed that RNA editing is generally adaptive and that it may be functionally related to RIP and MSUD. RNA editing could function together with alternative splicing to increase protein sequence variations and coding capacity of edited genes, and some of these nonsynonymous editing sites are well conserved in Sordariomycetes. As a model organism with unparalleled genetic and genomics resources, N. crassa is uniquely suited for characterizing the molecular mechanism and functions of RNA editing in filamentous fungi.
Materials and Methods
Details of the methods used in this study are provided in SI Materials and Methods, including strain culture conditions, details for strand-specific RNA- or DNA-seq library construction and sequencing, protocols used for read mapping and identification of A-to-I editing sites, evolutionary and statistical methods used for analyzing the adaptation of A-to-I editing, and experimental methods for generation of the NCU10184TAA and stk-21TAA constructs and transformants.
SI Materials and Methods
Strain Information and Culture Conditions.
The wild-type strains of Neurospora crassa OR74A (FGSC2489; mat A) and 74-ORS-6a (FGSC4200; mat a) and Neurospora tetrasperma mat A (FGSC2508) and mat a (FGSC2509) were obtained from the FGSC (www.fgsc.net) (Table S3). Mutant strains of N. crassa were kindly provided by Michael Freitag at Oregon State University, Corvallis, OR, or obtained from the FGSC as described in Table S3. Vogel’s minimal medium and synthetic crossing medium (SCM) were used for vegetative cultures and sexual crosses, respectively (62). For crossing, the mat A strains cultured on SCM plates containing 1% sucrose for 7–8 d were fertilized with conidia of the mat a strains and further incubated as described (62). Vegetative hyphae used for the isolation of genomic DNA and RNA were harvested from 2-d-old YEPD cultures (17).
Library Construction and RNA-Seq or DNA-Seq.
RNA was extracted and purified with the RNAprep Pure Plant Kit (Tiangen Biotech). Poly(A)+ mRNA was then isolated with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) as described by the manufacturer. Strand-specific RNA-seq libraries were prepared with the NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs) following the instructions provided by the manufacturer and sequenced with an Illumina HiSeq 2500 system using the 2 × 150-bp paired-end read mode. For each sample, two independent biological replicates were used for RNA-seq. For genome sequencing, DNA was isolated with the cetyltrimethylammonium bromide (CTAB) method (63) from vegetative hyphae of N. crassa strains OR74A and 74-ORS-6a or 6-dpf perithecia of N. tetrasperma that contain both mat A and mat a nuclei. DNA-seq libraries were prepared with the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) and sequenced with the Illumina HiSeq 2500 system using the 2 × 150-bp paired-end read mode. Both the RNA-seq and DNA-seq were performed at the Novogene Bioinformatics Institute (Beijing, China).
Sequence Mapping and RNA Editing Events Calling.
RNA-seq reads were aligned to the genome of N. crassa OR74A (NC12; Broad Institute) or N. tetrasperma FGSC 2508 (v2; JGI) using HISAT2 (v 2.0.0-beta) (64). DNA-seq reads were aligned using Bowtie 2 (v2.2.3) (65). Duplicated reads in the mapped BAM file of each library were removed using the MarkDuplicates tool from the Picard package (v 1.99) (broadinstitute.github.io/picard/). The unduplicated BAM files of strand-specific RNA-seq were divided into two separate files containing sense-strand and antisense-strand read alignments, respectively, using BamTools (v2.4.0) (66) with its filter utility. A-to-I RNA editing events were called separately from the sense-strand and antisense-strand files using the REDItoolDnaRna.py script included in REDItools package (67) with the following filtering parameters: -m 20,20 -q 25,25 -c 10,10 -v 3 -n 0.03 -a 6-0 -z -e -u. The DNA-seq alignments from both mat A and mat a strains were used during these analyses to support the presence of RNA editing events, excluding potential genomic SNPs. Because the detected A-to-I editing events were highly concordant between two biological replicates of N. crassa, we combined the unduplicated BAM files from the two replicates in our final analysis to maximize the statistical power for identification of A-to-I editing sites. For each A-to-I editing site, the editing level was defined as the percentage of reads with edited events.
The publicly available RNA-seq data of N. crassa OR74A conidia or hyphae grown under different culture conditions (26–29) were downloaded from the National Center for Biotechnology Information Sequence Read Archive database, including conidia (accession no. SRR959443); mycelia under dark (SRR1055985) or light induction for 60 min (SRR1055987); and mycelia exposed to Avicel (cellulose) (SRR627941), pectin (SRR627936), sucrose (SRR627947), cellobiose (SRR447473), or no carbon (SRR627944) conditions. Since these RNA-seq data were derived from single-end and unstranded RNA-seq, detection of A-to-I RNA editing events in these data did not include the process that removes duplicated reads from RNA-seq mappings. A relatively high editing level cutoff value of 10% (-n 0.1) was used for filtration because of their high sequencing and mapping error rates. Additionally, the type of base substitution was inferred based on the strand information of annotated genes rather than the strand direction of reads.
Evolutionary Analysis.
The ortholog families among N. crassa, N. tetrasperma, Sordaria macrospora (Ensembl), and Fusarium graminearum (RR, Ensembl) were identified by OrthoMCL (50) with default parameters. For each ortholog family, their protein sequences were aligned with MUSCLE (68), and codon alignments were generated with PAL2NAL (69). The dN/dS ratios were calculated by yn00 implemented in the PAML package (70) based on the ortholog pairs between N. crassa and N. tetrasperma.
Statistical Analysis.
All statistical tests were performed using R (https://www.r-project.org). Editing site distribution and functional consequence prediction were performed with the Amino Acid Changes tool of the CLC Genomics Workbench 7.5 (CLC Bio). GO enrichment analysis was performed with Blast2GO (https://www.blast2go.com). WebLogo 3 (71) and Two Sample Logo (72) were used to estimate and visualize the sequence preference around the A-to-I editing sites. RNAFold (73) was used to predict the secondary structures of RNA sequences containing the edited A sites (30-bp upstream and 30-bp downstream from the edited A’s). Hierarchical clustering analysis was conducted with Ward’s minimum variance method (Ward.D2) using R with the Pearson correlation coefficient as the distance value. Heat maps were generated by the heatmap.2 function in R. The total editable sites and random sampling of editable sites with similar nucleotide preference at the −2 to +3 positions as edited A’s were generated by Perl scripts developed in this study (https://github.com/wangqinhu/NC.edits) with similar approaches used previously (13).
Generation of the NCU10184TAA and stk-21TAA Transformants.
The mat A Δstk-21 mutants were generated by isolation of ascospore progeny from the crosses between Δstk-21 mutant FGSC17940 (mat a) and wild-type strain RL3-8A as described elsewhere (38). To generate the stk-21TAA allele, the stk-21 gene, including its promoter region, was amplified by overlapping PCR with primers carrying the G1575A mutation (TA1574G to TA1574A). The resulting PCR products were cloned into XhoI-digested plasmid pFL2 (carrying the geneticin resistance marker) (74) by the yeast gap repair approach (75). The stk-21TAA construct was rescued from Trp+ yeast transformants and confirmed by sequencing analysis. The same approach was used to generate the NCU10184TAA allele by introducing the G660A mutation (TA659G to TA659A). The NCU10184TAA and stk-21TAA constructs were transformed into protoplasts of the ΔNCU10184 and Δstk-21 (mat A) mutant strains of mat A, respectively. Geneticin-resistant transformants expressing the NCU10184TAA or stk-21TAA construct were identified by PCR, and homokaryotic transformants were isolated by crossing with ΔNCU10184 or the Δstk-21 mutant of mat a. The NCU10184TAA mat a and stk-21TAA mat a strains were identified by PCR screening of ascospore progeny from crosses of ΔNCU10184/NCU10184TAA and Δstk-21/stk-21TAA transformants, respectively. At least three homokaryotic mat A or mat a transformants with the same phenotype were obtained for the NCU10184TAA or stk-21TAA construct (Table S3).
Supplementary Material
Acknowledgments
We thank Dr. Michael Freitag (Oregon State University) and the FGSC for providing most of the strains used in this study, and an NIH Program Project grant for funding the generation of the Neurospora deletion set. We also thank Dr. Chenfang Wang for fruitful discussions. This work was supported by grants from the National Science Fund for Excellent Young Scholars (Grant 31622045), Purdue University AgSEED, and the Special Fund for Agro-Scientific Research in the Public Interest (Grant 201303016).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data from this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) under accession nos. SRP096774 and SRP097449.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702591114/-/DCSupplemental.
References
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picardi E, et al. Profiling RNA editing in human tissues: Towards the inosinome atlas. Sci Rep. 2015;5:14941. doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA editing. RNA Biol. 2010;7:205–212. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 6.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patton DE, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- 8.Seeburg PH. The role of RNA editing in controlling glutamate receptor channel properties. J Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- 9.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 10.Garrett S, Rosenthal JJ. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science. 2012;335:848–851. doi: 10.1126/science.1212795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu G, Zhang J. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci USA. 2014;111:3769–3774. doi: 10.1073/pnas.1321745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett SC, Rosenthal JJ. A role for A-to-I RNA editing in temperature adaptation. Physiology (Bethesda) 2012;27:362–369. doi: 10.1152/physiol.00029.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alon S, et al. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife. 2015;4:4. doi: 10.7554/eLife.05198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liscovitch-Brauer N, et al. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell. 2017;169:191–202.e111. doi: 10.1016/j.cell.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life. 2009;61:572–578. doi: 10.1002/iub.207. [DOI] [PubMed] [Google Scholar]
- 16.Grice LF, Degnan BM. The origin of the ADAR gene family and animal RNA editing. BMC Evol Biol. 2015;15:4. doi: 10.1186/s12862-015-0279-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, et al. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016;26:499–509. doi: 10.1101/gr.199877.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pöggeler S, Nowrousian M, Kück U. Fruiting-Body Development in Ascomycetes. Growth, Differentiation, and Sexuality. Springer; New York: 2006. pp. 325–355. [Google Scholar]
- 19.Borkovich K, Ebbole DJ. Cellular and Molecular Biology of Filamentous Fungi. American Society for Microbiology Press; Washington, DC: 2010. [Google Scholar]
- 20.Raju NB. Neurospora as a model fungus for studies in cytogenetics and sexual biology at Stanford. J Biosci. 2009;34:139–159. doi: 10.1007/s12038-009-0015-5. [DOI] [PubMed] [Google Scholar]
- 21.Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996;86:103–113. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 22.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 23.Selker EU, Cambareri EB, Jensen BC, Haack KR. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 24.Hammond TM, Rehard DG, Xiao H, Shiu PK. Molecular dissection of Neurospora spore killer meiotic drive elements. Proc Natl Acad Sci USA. 2012;109:12093–12098. doi: 10.1073/pnas.1203267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RH, Perkins DD. Timeline: Neurospora: A model of model microbes. Nat Rev Genet. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- 26.Znameroski EA, et al. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci USA. 2012;109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benz JP, et al. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol. 2014;91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeder AC, Jonkers W, Li J, Glass NL. Early colony establishment in Neurospora crassa requires a MAP kinase regulatory network. Genetics. 2013;195:883–898. doi: 10.1534/genetics.113.156984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, et al. Genome-wide characterization of light-regulated genes in Neurospora crassa. G3 (Bethesda) 2014;4:1731–1745. doi: 10.1534/g3.114.012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryot Cell. 2014;13:154–169. doi: 10.1128/EC.00248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borkovich KA, et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aramayo R, Selker EU. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol. 2013;5:a017921. doi: 10.1101/cshperspect.a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang SS, Zhang Z, Liu Y. RNA interference pathways in fungi: Mechanisms and functions. Annu Rev Microbiol. 2012;66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis ZA, et al. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010;6:e1001196. doi: 10.1371/journal.pgen.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 38.Colot HV, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park G, et al. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot Cell. 2011;10:1553–1564. doi: 10.1128/EC.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park G, et al. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol Biol. 2011;722:179–189. doi: 10.1007/978-1-61779-040-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collopy PD, et al. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol. 2010;638:33–40. doi: 10.1007/978-1-60761-611-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita K, et al. ATF-1 transcription factor regulates the expression of ccg-1 and cat-1 genes in response to fludioxonil under OS-2 MAP kinase in Neurospora crassa. Fungal Genet Biol. 2008;45:1562–1569. doi: 10.1016/j.fgb.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Lew RR, Abbas Z, Anderca MI, Free SJ. Phenotype of a mechanosensitive channel mutant, mid-1, in a filamentous fungus, Neurospora crassa. Eukaryot Cell. 2008;7:647–655. doi: 10.1128/EC.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H, et al. Roles for receptors, pheromones, G proteins, and mating type genes during sexual reproduction in Neurospora crassa. Genetics. 2012;190:1389–1404. doi: 10.1534/genetics.111.136358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCluskey K, et al. Rediscovery by whole genome sequencing: Classical mutations and genome polymorphisms in Neurospora crassa. G3 (Bethesda) 2011;1:303–316. doi: 10.1534/g3.111.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Q, Rasmussen C, Glass NL. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics. 2002;160:169–180. doi: 10.1093/genetics/160.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwechheimer C. The COP9 signalosome (CSN): An evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54. doi: 10.1016/j.bbamcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 48.St Laurent G, et al. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 2013;20:1333–1339. doi: 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
- 49.Ellison CE, et al. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics. 2011;189:55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teichert I, Dahlmann TA, Kück U, Nowrousian M. RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol Evol. 2017;9:855–868. doi: 10.1093/gbe/evx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Xu JR, Liu H. A-to-I RNA editing independent of ADARs in filamentous fungi. RNA Biol. 2016;13:940–945. doi: 10.1080/15476286.2016.1215796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Y, et al. The landscape of A-to-I RNA editome is shaped by both positive and purifying selection. PLoS Genet. 2016;12:e1006191. doi: 10.1371/journal.pgen.1006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang R, Deng P, Jacobson D, Li JB. Evolutionary analysis reveals regulatory and functional landscape of coding and non-coding RNA editing. PLoS Genet. 2017;13:e1006563. doi: 10.1371/journal.pgen.1006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duan Y, Dou S, Luo S, Zhang H, Lu J. Adaptation of A-to-I RNA editing in Drosophila. PLoS Genet. 2017;13:e1006648. doi: 10.1371/journal.pgen.1006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gommans WM, Mullen SP, Maas S. RNA editing: A driving force for adaptive evolution? BioEssays. 2009;31:1137–1145. doi: 10.1002/bies.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goddard MR, Godfray HC, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- 58.Burt A. Perspective: Sex, recombination, and the efficacy of selection–was Weismann right? Evolution. 2000;54:337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 59.Koltin V, Stamberg J, Ronen R. Melosis as a source of spontaneous mutations in Schizophyllum commune. Mutat Res. 1975;27:319–325. doi: 10.1016/0027-5107(75)90288-2. [DOI] [PubMed] [Google Scholar]
- 60.Rattray A, Santoyo G, Shafer B, Strathern JN. Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet. 2015;11:e1004910. doi: 10.1371/journal.pgen.1004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savva YA, et al. Auto-regulatory RNA editing fine-tunes mRNA re-coding and complex behaviour in Drosophila. Nat Commun. 2012;3:790. doi: 10.1038/ncomms1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghosh A, Servin JA, Park G, Borkovich KA. Global analysis of serine/threonine and tyrosine protein phosphatase catalytic subunit genes in Neurospora crassa reveals interplay between phosphatases and the p38 mitogen-activated protein kinase. G3 (Bethesda) 2014;4:349–365. doi: 10.1534/g3.113.008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Picardi E, Pesole G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics. 2013;29:1813–1814. doi: 10.1093/bioinformatics/btt287. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 71.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: A graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 73.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou X, Li G, Xu JR. Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Methods Mol Biol. 2011;722:199–212. doi: 10.1007/978-1-61779-040-9_15. [DOI] [PubMed] [Google Scholar]
- 75.Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.