Significance
Our understanding of how primary producers at the base of aquatic ecosystems respond to complex environmental change currently depends on studies using few environmental drivers, or scenarios where drivers covary. However, we lack a general understanding of evolution in multidriver environments. We evolve a microbial primary producer in 96 different multidriver environments and find that evolutionary responses in growth are largely driven by a few drivers but that the intensity of selection is, on average, higher in multidriver environments. Functional traits (cell size, chlorophyll content) often revert to ancestral values during adaptation in multidriver environments. This expands the framework for understanding how microbial primary producers evolve under global change and the potential ramifications for their function in aquatic ecosystems.
Keywords: multiple environmental drivers, adaptation, microbial evolution, Chlamydomonas, ocean global change biology
Abstract
Climate change is altering aquatic environments in a complex way, and simultaneous shifts in many properties will drive evolutionary responses in primary producers at the base of both freshwater and marine ecosystems. So far, evolutionary studies have shown how changes in environmental drivers, either alone or in pairs, affect the evolution of growth and other traits in primary producers. Here, we evolve a primary producer in 96 unique environments with different combinations of between one and eight environmental drivers to understand how evolutionary responses to environmental change depend on the identity and number of drivers. Even in multidriver environments, only a few dominant drivers explain most of the evolutionary changes in population growth rates. Most populations converge on the same growth rate by the end of the evolution experiment. However, populations adapt more when these dominant drivers occur in the presence of other drivers. This is due to an increase in the intensity of selection in environments with more drivers, which are more likely to include dominant drivers. Concurrently, many of the trait changes that occur during the initial short-term response to both single and multidriver environmental change revert after about 450 generations of evolution. In future aquatic environments, populations will encounter differing combinations of drivers and intensities of selection, which will alter the adaptive potential of primary producers. Accurately gauging the intensity of selection on key primary producers will help in predicting population size and trait evolution at the base of aquatic food webs.
A growing body of evidence from experiments shows that functional traits in aquatic primary producers can be altered by evolution in the face of global change, which has to date been explored in terms of drivers such as temperature (1, 2) or CO2 levels (3). These studies investigate the responses of primary producers to single aspects of global change (4), and the results are often used to understand how changes to the biological component of nutrient cycling, including air–water carbon exchange, will be impacted (5, 6). Recent short-term studies show that in multidriver environments, the majority of the organismal response is often explained by one or two drivers (7, 8). We call these dominant drivers (7). This study investigates how the evolutionary responses to dominant environmental drivers depend on the other drivers present. This helps link patterns of complex (multidriver) environmental change to the evolutionary potential of primary producers and informs the design of future experiments. We do this using an evolution experiment that disentangles the effects of driver number and identity on trait evolution in multidriver environments in a single-celled alga.
A small number of experiments have investigated the plastic and evolutionary responses to multiple drivers thus far (7, 9–12). These studies consistently show that both plastic and evolutionary responses to pairs of drivers differ from responses to either single driver. In the short term, the effect of multidriver environments on plastic responses (changes in phenotype in response to an environmental cue that does not require change in the genetic composition of the population) can be understood through the mode of interaction of the drivers (7, 13). However, we previously showed that when more than three drivers co-occur, average plastic responses in growth are explained by dominant drivers (7), because of either small interactions between nondominant or zero-sum interactions between drivers.
Aquatic primary producers will evolve under global change, due to their rapid cell division rates, and high standing genetic variation and ability to generate genetic variation (4, 14, 15). We do not currently have an empirically supported, general understanding of how natural selection differs between single- and multidriver environments. However, complex environmental change will be common in aquatic environments, with combinations and intensities of drivers having significant regional variation (8), so it is vital that we understand the joint contributions of the identity, number, and intensity of drivers to trait evolution in primary producers. Two nonexclusive mechanisms could cause natural selection to act differently in cases of complex (multidriver) versus simple (single driver) environmental change. First, if the number of independent traits under selection increases as the number of drivers in the environment increases, pleiotropic interactions could limit adaptation in complex environments more than in simple ones (16). Second, if selection intensity increases as the number of drivers increases, then the probability of population extinction increases with the number of drivers, but surviving populations will adapt more and more rapidly in environments with more drivers. These two mechanisms have different ecological consequences. If differences in evolutionary responses are mainly due to differences in pleiotropy under multidriver change, we expect that shifts in communities result mainly from changes in interactions between groups, with less trait evolution within groups than predicted based on single-driver experiments. In contrast, if the intensity of selection increases with the number of drivers, we expect that in addition to shifts in the taxonomic composition of communities, there will be changes in functional trait values within groups (17). Trait evolution in primary producers can in turn affect how food webs and aquatic nutrient cycles are impacted (18). Finally, if there are increased pleiotropic limitations as well as stronger selection in multidriver environments, then we expect more local extinctions and less trait evolution in surviving populations in multidriver relative to single-driver environments.
We use experimental evolution to measure how the number and identity of environmental drivers affect evolution in an initially isogenic population. Using many driver combinations allows us to disentangle the effects of driver number and identity on trait evolution, though some driver combinations are unrealistic (19, 20). The strength of this approach is that it builds a general understanding of how natural selection acts in multidriver environments. However, the model system and environments suitable for this experimental design mean that our findings cannot be applied directly to the immediate debate on how marine life will respond to global change; the fundamental insights gained here must be translated to the appropriate systems and environments, which can be done in smaller targeted studies. Replicate clonal populations of Chlamydomonas reinhardtii were grown in 96 unique environments (each unique environment is referred to as a regime) with up to eight simultaneous drivers including elevated temperature, elevated CO2, periodic UVB exposure, reduced light intensity, reduced phosphate, acidification, reduced nutrients, and the addition of herbicide, for ∼450 asexual generations. See Fig. 1 for a schematic and SI Appendix, Table S1 for drivers in each regime. See SI Appendix for a discussion of drivers and driver intensities. Discussions of plastic responses to single drivers were previously published in ref. 7. Because the populations were initially isogenic, genetic variation in the evolved populations is from novel mutations or other heritable changes (epigenetic mutations or transgenerational plasticity). Since the role of primary producers in aquatic systems is determined not only by their population growth rates but also by their trait values (21, 22), we measured evolutionary change in cell size and a commonly used proxy for primary production (chlorophyll) (23). Cell size is a “master trait” that constrains several organismal characteristics and biotic interactions for single-celled organisms, such as growth and metabolic rates (17), nutrient affinity (17), light absorption affinity (24), and predation (25).
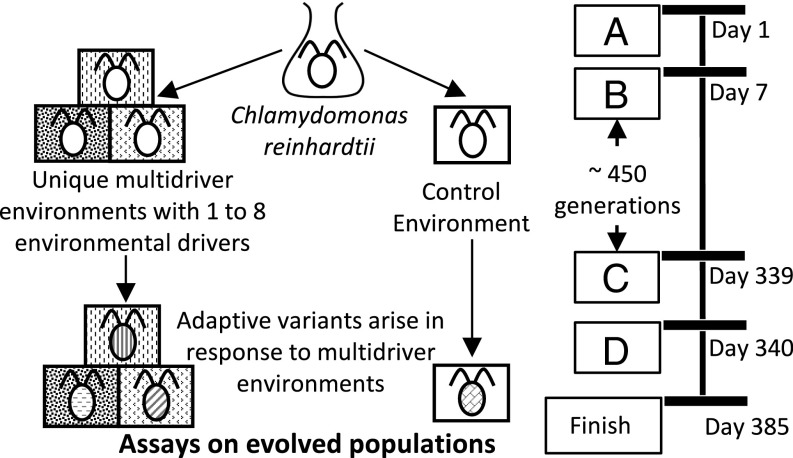
Fig. 1.
Schematic diagram illustrates the experimental design of the study. (A) The founding population was established from one colony of C. reinhardtii, grown from a single cell. (B) The founding population was grown for 1 wk under control conditions, then split into 96 different regimes (square boxes) with one to eight environmental drivers (regimes are shown as different pattern backgrounds), and a control environment (white background). (C) Populations evolved in each regime for 95 transfers. This provides enough time for adaptive variants to arise and increase in frequency. (D) After 95 transfers, the multidriver-evolved populations were assayed in their regime and the control environment. The control populations were assayed in all test regimes.
Results
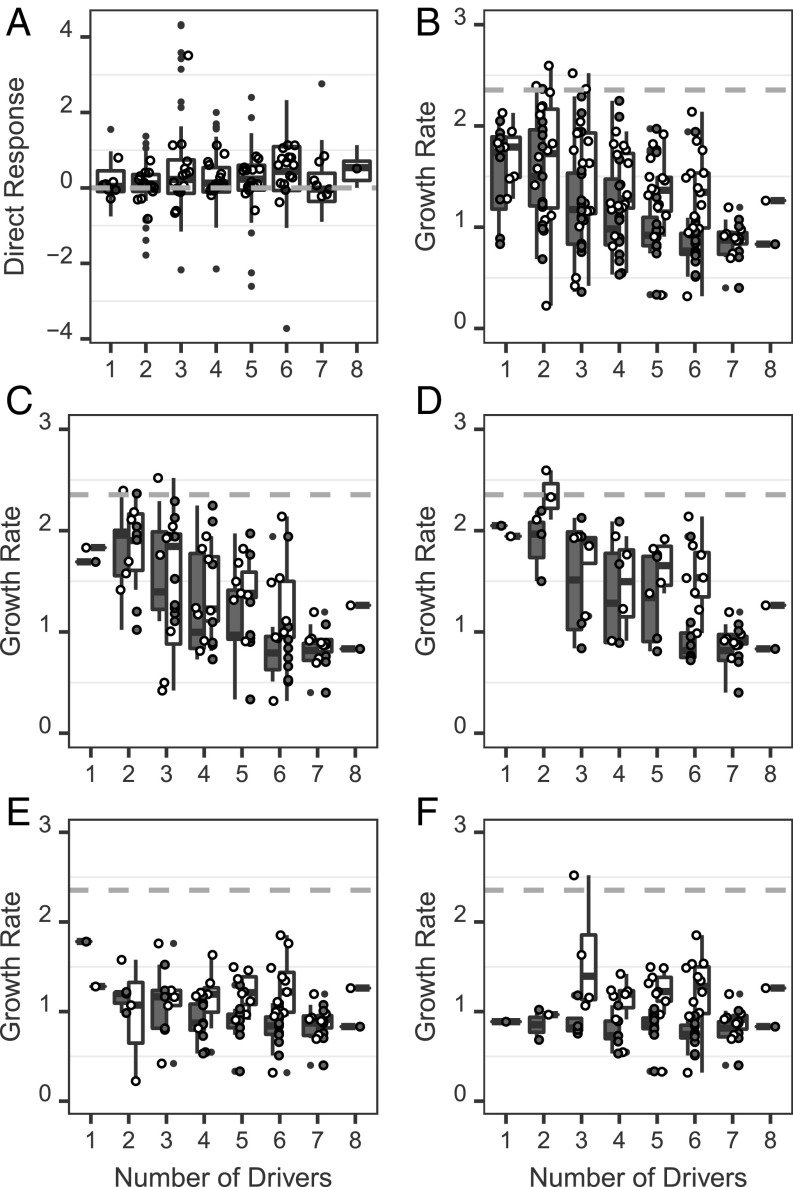
Following ∼450 generations of evolution in 96 regimes, we compared the endpoints of evolution by measuring the growth rates of the evolved populations. Populations evolved in multidriver environments all reached similar growth rates by the end of the experiment. However, the small amount of variation in evolved growth rate was explained by a few individual drivers, regardless of which other drivers were present. These dominant drivers were elevated CO2 (Fig. 2C; F1, 77 = 5.454, P = 0.022), elevated temperature (Fig. 2D; F1, 78 = 10.042, P = 0.002), reduced phosphate (Fig. 2E; F1, 77 = 20.686, P < 0.0001), and herbicide (Fig. 2F; F1, 77 = 22.036, P < 0.0001). Here, CO2 increased growth rates, while the other dominant drivers decreased growth (SI Appendix, Fig. S4). The selection regimes themselves explained only 5% of the variation in growth rates of populations in their selection regimes. Thus, the dominant drivers drive growth rate evolution. The overall effect of the number of environmental drivers in a selection regime on evolved population growth rate is not significant (Fig. 2B, white boxplots; F1, 73 = 0.043, P = 0.837), and the small amount present was explained by variation among evolved replicate populations within regimes (43%). Thus, increasing the number of drivers in multidriver environments does not constrain the endpoint of evolution on average, at least in terms of population growth rates.
Fig. 2.
The response of evolved populations under increasing numbers of drivers. Boxes show the (A) direct response to selection measured as the average number of cell divisions (d−1) relative to control populations assayed in the same regime. Open circles show the average of evolved populations within each regime. The dashed line indicates that there is no difference between the growth rate of the evolved control and the multidriver-evolved populations, in the same selection regime. Average cell divisions (d−1) of evolved populations assayed in (B) all regimes, (C) regimes with elevated CO2, (D) regimes with elevated temperature, (E) regimes with reduced phosphate, and (F) regimes with herbicide. White symbols show multidriver populations assayed in their selection regimes, and gray symbols show control populations assayed in the same regimes. The dashed line (B–F) shows the average growth rate of control populations in the control environment.
Evolved populations have undergone ∼450 generations of evolution in their selection regimes (Fig. 1, time C). The direct response to selection compares the population growth rate of a population evolved in a given regime with the plastic response of a control population to that same regime. This measures the difference in plastic and evolutionary responses to an environment and estimates net adaptive change over the experiment. A positive response to selection indicates that evolution increases growth rates beyond the plastic response, and a negative direct response to selection indicates that evolution slows growth relative to the plastic response.
While populations converge on similar growth rates, the direct response to selection is larger when there are more drivers in selection regimes, so that populations in environments with more drivers evolve more to arrive at the same endpoint. This is because in environments with more drivers, populations tend to have lower initial growth rates (Fig. 2 B–F), which indicates stronger selection. This is consistent with extinctions occurring in seven and eight driver environments (Fig. 2B and SI Appendix, Fig. S1). Across all regimes, variation in the initial drop in growth rate explains variation in the direct response to selection, regardless of the number of drivers (Fig. 2A; effect of plastic response on direct response, F1, 341 = 69.356, P < 0.0001). The larger direct response to selection in regimes with more drivers is thus attributable to stronger selection in these regimes. Intermediate timepoints were not characterized, so we cannot draw conclusions about the timing of adaptation. In addition to the average size of the direct response increasing with the number of drivers, a higher proportion of regimes contained populations with a positive direct response to selection when more drivers were present (SI Appendix, Fig. S2). This is unsurprising, because regimes with more drivers are more likely to contain a dominant driver, such that selection is strong enough to drive adaptation (7) (Fig. 2 B–F, gray boxplots and SI Appendix, Fig. S3). Rather than constraining evolution, as predicted by the pleiotropy hypothesis, increasing the number of drivers in an environment leads to more adaptation due to stronger selection. Pleiotropic constraints may be present but do not override the effects of stronger selection here.
A few dominant drivers affected the direct response to selection. These were reduced phosphate (F1, 329 = 26.197, P < 0.0001), herbicide (F1, 334 = 7.862, P = 0.005), and elevated CO2 (F1, 346 = 7.83, P = 0.005) (see SI Appendix, Table S1 for regimes). The number of drivers in regimes explained less than 1% of the variation in the direct response to selection once the initial drop in growth rate and the identity of the drivers were taken into account (SI Appendix, Table S5; F1, 329 = 0.89, P = 0.346). The initial drop in growth rate and the identity of the selection regimes explained 27% of the variation in the direct response, but there was divergence among replicate populations within regimes, which explained most of the variation in the direct response of the evolved populations (40%).
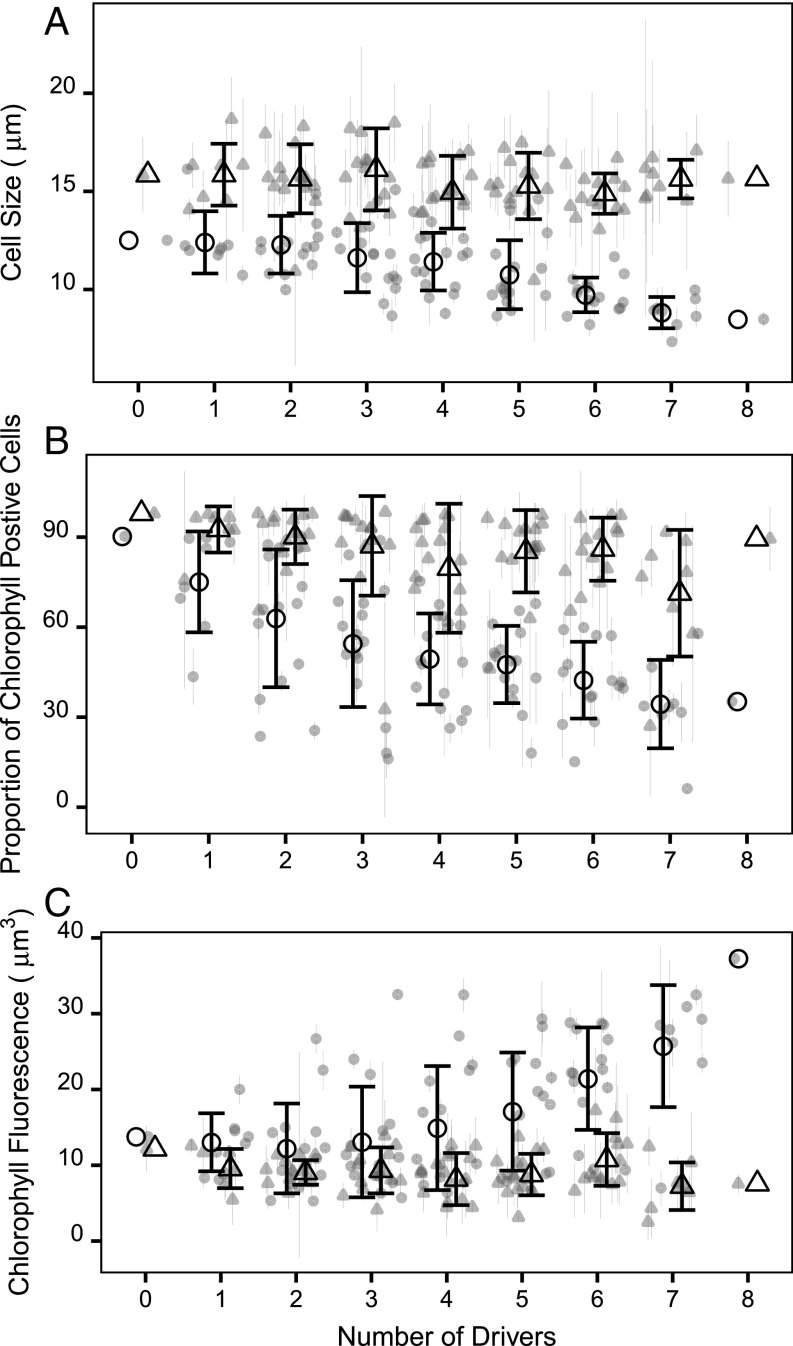
Before evolution, the trait values for cell size and chlorophyll content showed a large plastic response to the multidriver environments, but the plastic response was fully or nearly reversed by the end of the evolution experiment. Because traits converged on similar values across regimes after ∼450 generations, the majority of variation must be explained by variation between replicate populations. This variation, though statistically significant, is extremely low.
Following evolution, there was little variation in cell size when populations were grown in their own regimes (Fig. 3A, triangles; 15.5 µm ± 0.41 µm; mean ± SD over all environments). The number of drivers in regimes explained less than 2% of the variation in cell size of the evolved populations, while variation among evolved populations within regimes (28%) and the identity of the environmental drivers (13%) explained most of the variation in cell size. Similarly, the positive relationship between cell size and population growth rates before evolution (SI Appendix, Fig. S5A; r2 = 0.33, P < 0.0001) broke down after evolution (SI Appendix, Fig. S5B; r2 = 0.02, P = 0.006). This suggests that the minimal variation in evolved cell size is neutral or near-neutral in terms of growth.
Fig. 3.
Trait values of C. reinhardtii before and after evolution in multidriver environments. Changes in (A) cell size, (B) proportion of chlorophyll-positive cells, and (C) chlorophyll autofluorescence per cell volume (1/µm3) in populations of C. reinhardtii. In all panels, black symbols show the response (±SD) for a given number of drivers, and gray symbols show the average growth rate (±SD) for each regime. Circles represent the plastic response and triangles represent the evolved response.
In this experiment, a proportion of the population often reversibly bleached (had no detectable chlorophyll autofluorescence using a flow cytometer) as a plastic response to regimes that reduced population growth rates. Before evolution, the proportion of chlorophyll-positive cells (chlorophyll autofluorescence detectable using a flow cytometer) in populations decreased as the number of drivers in an environment increased (Fig. 3B, circles; F1, 93 = 7.945, P = 0.0058). In contrast, the number of drivers in an environment failed to explain variation in the proportion of chlorophyll-positive cells after ∼450 generations (Fig. 3B, triangles; F1, 58 = 0.800, P = 0.375). This is probably adaptive, as populations with higher proportions of chlorophyll-positive cells had higher population growth rates (SI Appendix, Fig. S6A; r2 = 0.47, P < 0.0001; SI Appendix, Fig. S6B; r2 = 0.24, P < 0.0001), and photosynthesis is essential for rapid growth in media with no carbon additions (26). While reversible cell bleaching itself requires further study, the restoration of the capacity for photosynthesis is obviously adaptive here.
Before evolution, the chlorophyll concentration in cells depended on the number of drivers in the environment (Fig. 3C, circles; F1, 93 = 24.676, P < 0.0001). This trend is absent after evolution (Fig. 3C, triangles; F1, 64 = 0.058, P = 0.811), and chlorophyll autofluorescence per cell volume in evolved populations in their own regimes did not differ from that of evolved control populations in the control environment (12.15 ± 3.07 1/µm3; mean ± SD). This is consistent with populations having adapted to their environments, as they no longer show a standard sign of stress (27–29).
Discussion
To understand how the evolutionary response of primary producers depends on the identity and number of drivers in multidriver environments, we evolved microbial populations in 96 unique single- and multidriver environments. Both the absolute growth rate of the evolved populations and the direct response to selection (the amount of evolution needed to reach that growth rate) were explained by the presence of a few dominant environmental drivers. Surprisingly, the multidriver context in which dominant drivers occurred had little effect on the growth rate of evolved populations on average. However, populations had a larger direct response to selection in environments that contained more drivers, indicating that primary producers evolve more in response to dominant drivers when they occur in multidriver environments than when they occur singly. This is largely because, on average, selection is stronger in multidriver environments. Thus, populations evolving in multidriver environments adapt more but arrive at the same final growth rates as populations evolving in single-driver environments with the same dominant drivers.
We were initially surprised by these results. We had hypothesized that pleiotropic constraints would be more important in populations evolving in environments with more drivers because more traits would be under selection (30). Instead, the response to selection increases with the number of environmental drivers. Above a threshold number of drivers (seven, in this experiment), rapid adaptation was not possible, and populations went extinct. These results suggest that the number of traits under selection does not scale with the number of environmental drivers. This makes sense given that evolutionary responses are driven by a few dominant drivers (CO2, low phosphate, temperature, and herbicide), so that the traits under selection may be more or less constant over the regimes containing a given dominant driver. As the number of drivers increases, dominant drivers are more likely to be present. The identity of dominant drivers in each regime partially explains the small differences in the response to selection in multidriver environments. The overriding effect of the dominant drivers is consistent with the acclimation response to these regimes (7), scenario-based experiments (8), and many physiological responses of microalgae to pairs of drivers (31–35). Our data are consistent with either small effects of driver interactions relative to the effects of dominant drivers or (nearly) zero-sum interactions among drivers.
Our findings highlight the importance of accurately gauging the intensity of selection for understanding the evolutionary potential of primary producers. Predicting the intensity of selection that a population is likely to experience requires knowing, first, which drivers are present locally and, second, the organismal responses to the dominant drivers. Since populations experience their local environment rather than a global average, this requires using regional rather than global patterns of multidriver change (5). The regimes in this experiment did not reflect realistic environments, nor were they intended to—the experiment was carried out using a laboratory model system and 96 different environments, with the goal of disentangling the roles of the number and identity of environmental drivers on evolution over hundreds of generations. In addition, functional groups (e.g., calcifiers, silicifiers, nitrogen fixers) of primary producers respond differently to dominant drivers (4, 36, 37). Our results also suggest that it could also be useful, when considering how primary producers may evolve under different climate change clusters (5), to group drivers based on their effects on growth (positive, negative, neutral) for different taxa when assessing whether or not we expect climate change clusters to drive evolution.
Our approach complements scenario-based studies. For example, Boyd et al. (5) modeled regional changes to multidriver regimes and used measured shifts in phytoplankton physiology to make qualitative predictions about shifts in biogeography. They detail the responses of coccolithophores (calcifiers) and diatoms (silicifiers) in two high latitude ocean provinces to shifts in temperature, CO2, photosynthetically active radiation (PAR), iron, silicate, nitrate, and phosphate, as well as interactions between driver pairs. Based on plastic responses, they suggest that elevated temperature is likely to cause a poleward shift in coccolithophores, and high PAR, low phosphate, and low silicate are likely to favor coccolithophores over diatoms. A decrease in calcification is predicted for coccolithophores and a decrease in silicification for diatoms. Our study suggests that, in addition, selection to increase both calcification and silicification could drive evolution in multidriver environments. We also expect that evolution reverses some of the trait change predicted based on Boyd et al. (5). Adaptation could also result in (positive and negative) changes in growth rates eventually attenuating, based on data from single-driver evolution experiments (1, 2). This illustrates how we can use an understanding of how natural selection acts to refine predictions of trait change in multidriver environments.
Despite some populations having a large direct response to selection, evolution reversed plastic changes in several traits. This suggests that as populations adapt to multidriver environments, the function of evolving groups may change less than expected based on plastic responses to multidriver environments (plastic responses are circles in Fig. 3). Population growth rates, however, are not restored to control values in all regimes. Since the populations in this experiment are propagated by batch culture and were not allowed to reach carrying capacity, overall selection was to increase cell division rates (20). The lower growth rates in some regimes may be the result of physiological constraints, since resources can limit growth rates even after adaptation. For example, growth rates are lower in low-phosphate regimes than in phosphate-replete ones regardless of driver number (SI Appendix, Fig. S4). Despite this, the proportion of cells showing signs of stress (chlorophyll-negative and/or small cells) in evolved populations in the low-phosphate and low-nutrient environments returned to control values (Fig. 3). In contrast, populations evolved in the control environment produce small cells and high proportions of chlorophyll-negative cells when grown in low-nutrient environments. Both the proportion of chlorophyll-positive cells and cell size are correlated with population growth rates, but this correlation is much weaker in evolved populations, largely because those traits have converged. This indicates that cells adapted to tolerate the low nutrient multidriver environments, even if they lack the resources to increase growth rates. Similarly, in environments with seven drivers, populations have low growth rates but normal chlorophyll content and cell sizes. This is in line with other studies that demonstrated that in poor-quality environments populations invest in maintaining cell condition (38, 39). In C. reinhardtii, cell size is related to population growth through its effect on cell division, as a critical size must be reached before cells divide (40–42). This trend of phenotypic reversion during adaptive evolution has also been seen in high-CO2 environments (43), indicating that some phenotypic reversion may be a common outcome of evolution (44).
We did not examine the molecular basis of trait reversion here but offer two nonexclusive explanations. First, our results are consistent with compensatory mutations affecting trait evolution in multidriver environments. Following a large drop in population fitness, the first beneficial mutation fixed often has a large effect (45) and can change several traits simultaneously. This may be followed by compensatory mutations that increase fitness by reversing the effect of the initial mutation on traits where change was not adaptive (46–48). Second, heritable epigenetic mutations or transgenerational plasticity can contribute to early adaptation (7) but eventually be replaced by genetic mutations (49–52). Understanding how trait reversion is linked to adaptation presents an opportunity to improve our predictions of functional trait values in primary producers in aquatic environments.
Laboratory evolution experiments use simplified environments and populations to gain insights into the fundamental action of natural selection. Applying the results of this experiment to natural phytoplankton population requires taking into account how population size and diversity (among other factors, such as recombination rates) affect adaptation (53). Previous work has shown that higher standing genetic variation can allow adapting populations to evolve faster (54–57). Similarly, recombination and plasticity (1, 58) can both speed up adaptation (59). The power of simplified laboratory experiments lies in providing insight into how natural selection works; applying these insights requires accounting for other processes that can modify evolution, and for the specific biology of wild populations.
Conclusions
We show that populations adapt more in response to dominant drivers when those drivers occur in a multidriver context, until environments deteriorate enough to cause extinctions. Alongside this, adaptation can result in the reversion of the initial changes to trait values in multidriver environments. While we expect the result of evolution being driven primarily by a few dominant drivers in multidriver environments to be general, the identity of dominant drivers will be organism- and context-dependent, such that a variety of approaches (large factorial experiments like this one, scenario-based models and experiments, taxa-specific physiological, and evolution studies) are needed to understand how primary producers will respond to multiple environmental drivers. In addition, the evolutionary potential of populations will depend on demography, existing genetic variation, and the rate at which new variation can be generated by recombination, migration, and the availability of spatial and temporal refugia. Our results emphasize the importance of gauging the intensity of selection on populations under global change (60) by linking complex environmental change to organismal fitness. This informs our understanding of the extent to which primary producers evolve in multidriver environments. A second challenge is understanding the evolutionary reversion of plastic responses in functional traits, as this will determine the function of primary producers under complex environmental change.
Methods and Materials
Selection Experiment.
All populations were founded from one colony originating from a single cell of C. reinhardtii (CC-2931, mt-; Chlamydomonas Resource Center, University of Minnesota), grown in sterile Sueoka’s high salt medium with Tris⋅HCl (HSMT; ref. 61) (SI Appendix, Tables S2 and S3). The ancestral population was split into 576 populations (Fig. 1 and SI Appendix, Table S1). A single founder colony ensures that population evolution uses de novo variation. Environmental changes occurred in one step at the beginning of the experiment (control level in brackets): increased CO2 to 2,000 ppm (420 ppm), temperature to 26 °C (25 °C), decreased pH to 6.5 (7.2 pH), light levels to 18 μmol⋅m−2⋅s−1 (32 μmol⋅m−2⋅s−1), reduced phosphate to 1.69 mM (13.56 mM), nutrient depletion to 25% (100% nutrients SI Appendix, Table S3), and 0.5 µM of atrazine (no herbicide). Regimes with UV were dosed with 8.1 KJ⋅m−2 UVB twice a week (SI Appendix, Table S4). Populations evolved in environments with at least one driver are multidriver-evolved populations. Populations evolved in the control environment are control populations. All populations were propagated by batch culture. See SI Appendix for details.
Assays of Population Growth Rates.
An acclimation period of one transfer cycle was used for all assays (SI Appendix).
The average rate of cell division per day was calculated over a single batch culture-length time using Eq. 1 (7). See SI Appendix for details.
| [1] |
Nt is the cell density (cells per mL) at time t (hours) and N0 is the cell density at time t0.
The direct response to selection was measured by comparing the growth of a multidriver-evolved population and a control population in the same multidriver regime (Fig. 1). The direct response to selection was calculated using Eq. 2 and scaled to the number of divisions (d−1) of the control population in the relevant regime. See SI Appendix for details.
| [2] |
E is the number of divisions (d−1) of multidriver-evolved populations in their regimes, and C is the number of divisions (d−1) of evolved control populations in the same regimes.
Flow Cytometric Analysis of Physiological Parameters.
An acclimation period of one transfer cycle was used for all assays, as above. A FACS CANTO was used to determine red autofluorescence (chlorophyll a and b), event number (cell density), and forward scatter (cell size) (1, 62). See SI Appendix.
Statistical Analysis.
The effects of (i) driver identity and (ii) the plastic response on the response to selection were analyzed with a mixed model in R (63), using the packages lme4 and lmerTest. The plastic response and the presence/absence of drivers are fixed factors. The effects of (i) the number of drivers and the (ii) identity of the drivers on evolved growth rate were also analyzed using a mixed model. The number of drivers (0–7) and driver identity (e.g., CO2) are fixed factors. Regime identity (SI Appendix, Table S1), batch number, and replicate populations within regime are random factors (see SI Appendix for details).
The contribution of fixed factors was estimated using Eq. 3 (as described in ref. 7):
| [3] |
where is the variance of the fixed effect, b is the slope of the fixed effect, se is the SE of the fixed effect, and is the variance of the response variable.
Supplementary Material
Acknowledgments
We thank A. Phillimore and J. Hadfield for discussion on statistics and H. Kuehne and M. Waterfall for technical and flow cytometry assistance. This work was supported by the European Research Council (FP7 Grant 260266) and a Royal Society (UK) University Research Fellowship (to S.C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The R codes and data reported in this paper have been deposited in the PANGAEA database, www.pangaea.de (doi: PANGAEA.879517).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703375114/-/DCSupplemental.
References
- 1.Schaum E, Collins S. Plasticity predicts evolution in marine alga. Proc Biol Sci. 2014;281:20141486. doi: 10.1098/rspb.2014.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lohbeck K, Riebesell U, Reusch T. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat Geosci. 2012;5:346–351. [Google Scholar]
- 3.Hutchins DA, et al. Irreversibly increased nitrogen fixation in Trichodesmium experimentally adapted to elevated carbon dioxide. Nat Commun. 2015;6:8155. doi: 10.1038/ncomms9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins S, Rost B, Rynearson TA. Evolutionary potential of marine phytoplankton under ocean acidification. Evol Appl. 2014;7:140–155. doi: 10.1111/eva.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd PW, Lennartz ST, Glover DM, Doney SC. Biological ramifications of climate-change-mediated oceanic multi-stressors. Nat Clim Change. 2015;5:71–79. [Google Scholar]
- 6.Dutkiewicz S, et al. Impact of ocean acidification on the structure of future phytoplankton communities. Nat Clim Change. 2015;5:1002–1006. [Google Scholar]
- 7.Brennan G, Collins S. Growth responses of a green alga to multiple environmental drivers. Nat Clim Change. 2015;5:892–897. [Google Scholar]
- 8.Boyd PW, et al. Physiological responses of a Southern Ocean diatom to complex future ocean conditions. Nat Clim Change. 2015;6:207–213. [Google Scholar]
- 9.Gao K, Helbling EW, Häder DP, Hutchins DA. Responses of marine primary producers to interactions between ocean acidification, solar radiation, and warming. Mar Ecol Prog Ser. 2012;470:167–189. [Google Scholar]
- 10.Tatters AO, et al. Short- versus long-term responses to changing CO2 in a coastal dinoflagellate bloom: Implications for interspecific competitive interactions and community structure. Evolution. 2013;67:1879–1891. doi: 10.1111/evo.12029. [DOI] [PubMed] [Google Scholar]
- 11.Schlüter L, et al. Adaptation of a globally important coccolithophore to ocean warming and acidification. Nat Clim Change. 2014;4:1024–1030. [Google Scholar]
- 12.Taucher J, et al. Combined effects of CO2 and temperature on carbon uptake and partitioning by the marine diatoms Thalassiosira weissflogii and Dactyliosolen fragilissimus. Limnol Oceanogr. 2015;60:901–919. [Google Scholar]
- 13.Folt C, Chen C. Synergism and antagonism among multiple stressors. Limnol Oceanogr. 1999;44:864–877. [Google Scholar]
- 14.Rynearson TA, Armbrust EV. Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol Ecol. 2005;14:1631–1640. doi: 10.1111/j.1365-294X.2005.02526.x. [DOI] [PubMed] [Google Scholar]
- 15.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: The structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 16.Orr HA. Adaptation and the cost of complexity. Evolution. 2000;54:13–20. doi: 10.1111/j.0014-3820.2000.tb00002.x. [DOI] [PubMed] [Google Scholar]
- 17.Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. The role of functional traits and trade-offs in structuring phytoplankton communities: Scaling from cellular to ecosystem level. Ecol Lett. 2007;10:1170–1181. doi: 10.1111/j.1461-0248.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 18.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 19.Scheinin M, Riebesell U, Rynearson TA, Lohbeck KT, Collins S. Experimental evolution gone wild. J R Soc Interface. 2015;12:1–5. doi: 10.1098/rsif.2015.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 21.Litchman E, Klausmeier CA. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst. 2008;39:615–639. [Google Scholar]
- 22.Behrenfeld M, Falkowski PG. A consumer’s guide to phytoplankton primary productivity models. Limnol Oceanogr. 1997;42:1479–1491. [Google Scholar]
- 23.Jacox MG, Edwards CA, Kahru M, Rudnick DL, Kudela RM. The potential for improving remote primary productivity estimates through subsurface chlorophyll and irradiance measurement. Deep Sea Res Part II Top Stud Oceanogr. 2015;112:107–116. [Google Scholar]
- 24.Finkel ZV, et al. Phytoplankton in a changing world: Cell size and elemental stoichiometry. J Plankton Res. 2010;32:119–137. [Google Scholar]
- 25.Hansen PJ, Bjørnsen PK, Hansen BW. Zooplankton grazing and growth: Scaling within the 2–2,000-µm body size range. Limnol Oceanogr. 1997;42:687–704. [Google Scholar]
- 26.Blifernez-Klassen O, et al. Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii. Nat Commun. 2012;3:1214. doi: 10.1038/ncomms2210. [DOI] [PubMed] [Google Scholar]
- 27.Fischer BB, Wiesendanger M, Eggen RIL. Growth condition-dependent sensitivity, photodamage and stress response of Chlamydomonas reinhardtii exposed to high light conditions. Plant Cell Physiol. 2006;47:1135–1145. doi: 10.1093/pcp/pcj085. [DOI] [PubMed] [Google Scholar]
- 28.Müller P, Li XP, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado R, Rioboo C, Herrero C, Cid A. Characterization of cell response in Chlamydomonas moewusii cultures exposed to the herbicide paraquat: Induction of chlorosis. Aquat Toxicol. 2011;102:10–17. doi: 10.1016/j.aquatox.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Fisher R. The Fundamental Theorum of Natural Selection. Oxford Univ Press; Oxford: 1930. [Google Scholar]
- 31.Jackson MC, Loewen CJG, Vinebrooke RD, Chimimba CT. Net effects of multiple stressors in freshwater ecosystems: A meta-analysis. Glob Change Biol. 2016;22:180–189. doi: 10.1111/gcb.13028. [DOI] [PubMed] [Google Scholar]
- 32.Crain CM, Kroeker K, Halpern BS. Interactive and cumulative effects of multiple human stressors in marine systems. Ecol Lett. 2008;11:1304–1315. doi: 10.1111/j.1461-0248.2008.01253.x. [DOI] [PubMed] [Google Scholar]
- 33.Schafer RB, Kuhn B, Malaj E, Konig A, Gergs R. Contribution of organic toxicants to multiple stress in river ecosystems. Freshw Biol. 2016;61:2116–2128. [Google Scholar]
- 34.Aristi I, et al. Nutrients versus emerging contaminants-Or a dynamic match between subsidy and stress effects on stream biofilms. Environ Pollut. 2016;212:208–215. doi: 10.1016/j.envpol.2016.01.067. [DOI] [PubMed] [Google Scholar]
- 35.Noyes PD, et al. The toxicology of climate change: Environmental contaminants in a warming world. Environ Int. 2009;35:971–986. doi: 10.1016/j.envint.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Vinebrooke RD, et al. Impacts of multiple stressors on biodiversity and ecosystem functioning: The role of species co-tolerance. Oikos. 2004;104:451–457. [Google Scholar]
- 37.Litchman E, Edwards KF, Klausmeier CA, Thomas MK. Phytoplankton niches, traits and eco-evolutionary responses to global environmental change. Mar Ecol Prog Ser. 2012;470:235–248. [Google Scholar]
- 38.Zhang X-X, Rainey PB. Bet hedging in the underworld. Genome Biol. 2010;11:137. doi: 10.1186/gb-2010-11-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rioboo C, González O, Herrero C, Cid A. Physiological response of freshwater microalga (Chlorella vulgaris) to triazine and phenylurea herbicides. Aquat Toxicol. 2002;59:225–235. doi: 10.1016/s0166-445x(01)00255-7. [DOI] [PubMed] [Google Scholar]
- 40.Machado MD, Soares EV. Modification of cell volume and proliferative capacity of Pseudokirchneriella subcapitata cells exposed to metal stress. Aquat Toxicol. 2014;147:1–6. doi: 10.1016/j.aquatox.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Matsumura K, Yagi T, Yasuda K. Role of timer and sizer in regulation of Chlamydomonas cell cycle. Biochem Biophys Res Commun. 2003;306:1042–1049. doi: 10.1016/s0006-291x(03)01089-1. [DOI] [PubMed] [Google Scholar]
- 42.Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 43.Schaum C-E, Rost B, Collins S. Environmental stability affects phenotypic evolution in a globally distributed marine picoplankton. ISME J. 2016;10:75–84. doi: 10.1038/ismej.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins S. Growth rate evolution in improved environments under Prodigal Son dynamics. Evol Appl. 2016;9:1179–1188. doi: 10.1111/eva.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Moore FB-G, Rozen DE, Lenski RE. Pervasive compensatory adaptation in Escherichia coli. Proc Biol Sci. 2000;267:515–522. doi: 10.1098/rspb.2000.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kronholm I, Collins S. Epigenetic mutations can both help and hinder adaptive evolution. Mol Ecol. 2016;25:1856–1868. doi: 10.1111/mec.13296. [DOI] [PubMed] [Google Scholar]
- 50.Klironomos FD, Berg J, Collins S. How epigenetic mutations can affect genetic evolution: Model and mechanism. BioEssays. 2013;35:571–578. doi: 10.1002/bies.201200169. [DOI] [PubMed] [Google Scholar]
- 51.Burggren W. Epigenetic inheritance and its role in evolutionary biology: Re-evaluation and new perspectives. Biology (Basel) 2016;5:1–22. doi: 10.3390/biology5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Dea RE, Noble DWA, Johnson SL, Hesselson D, Nakagawa S. The role of non-genetic inheritance in evolutionary rescue: Epigenetic buffering, heritable bet hedging and epigenetic traps. Environ Epigenetics. 2016;2:1–12. doi: 10.1093/eep/dvv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29:33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 55.Rigato E, Fusco G. Enhancing effect of phenotype mutational robustness on adaptation in Escherichia coli. J Exp Zoolog B Mol Dev Evol. 2016;326:31–37. doi: 10.1002/jez.b.22662. [DOI] [PubMed] [Google Scholar]
- 56.Wünsche A, et al. Diminishing-returns epistasis decreases adaptability along an evolutionary trajectory. Nat Ecol Evol. 2017;1:1–6. doi: 10.1038/s41559-016-0061. [DOI] [PubMed] [Google Scholar]
- 57.Vogwill T, Kojadinovic M, MacLean RC. Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc Biol Sci. 2016;283:20160151. doi: 10.1098/rspb.2016.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghalambor CK, et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525:372–375. doi: 10.1038/nature15256. [DOI] [PubMed] [Google Scholar]
- 59.McDonald MJ, Rice DP, Desai MM. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–236. doi: 10.1038/nature17143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyd PW, et al. Biological responses to environmental heterogeneity under future ocean conditions. Glob Change Biol. 2016;22:2633–2650. doi: 10.1111/gcb.13287. [DOI] [PubMed] [Google Scholar]
- 61.Harris E. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic; San Diego: 1989. [DOI] [PubMed] [Google Scholar]
- 62.Prado R, Rioboo C, Herrero C, Suárez-Bregua P, Cid A. Flow cytometric analysis to evaluate physiological alterations in herbicide-exposed Chlamydomonas moewusii cells. Ecotoxicology. 2012;21:409–420. doi: 10.1007/s10646-011-0801-3. [DOI] [PubMed] [Google Scholar]
- 63.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.