Significance
We propose and test predictions of a thermodynamic and kinetic model for RNA tertiary folding that is based on separable energetic contributions of RNA elements. We define these contributions based on the principle features of RNA, and we test the basic predictions of separability by determining whether the energetic contributions of one component are affected by changes in another component. Our results support energetic separability of RNA elements and suggest that it may be possible to deconstruct RNAs into smaller parts that can be studied in isolation such that the individual folding behaviors of these parts can be used to “reconstitute” the folding of the original RNA.
Keywords: RNA folding, single-molecule FRET, RNA tertiary structure, folding kinetics, folding thermodynamics
Abstract
Decades of study of the architecture and function of structured RNAs have led to the perspective that RNA tertiary structure is modular, made of locally stable domains that retain their structure across RNAs. We formalize a hypothesis inspired by this modularity—that RNA folding thermodynamics and kinetics can be quantitatively predicted from separable energetic contributions of the individual components of a complex RNA. This reconstitution hypothesis considers RNA tertiary folding in terms of ΔGalign, the probability of aligning tertiary contact partners, and ΔGtert, the favorable energetic contribution from the formation of tertiary contacts in an aligned state. This hypothesis predicts that changes in the alignment of tertiary contacts from different connecting helices and junctions (ΔGHJH) or from changes in the electrostatic environment (ΔG+/−) will not affect the energetic perturbation from a mutation in a tertiary contact (ΔΔGtert). Consistent with these predictions, single-molecule FRET measurements of folding of model RNAs revealed constant ΔΔGtert values for mutations in a tertiary contact embedded in different structural contexts and under different electrostatic conditions. The kinetic effects of these mutations provide further support for modular behavior of RNA elements and suggest that tertiary mutations may be used to identify rate-limiting steps and dissect folding and assembly pathways for complex RNAs. Overall, our model and results are foundational for a predictive understanding of RNA folding that will allow manipulation of RNA folding thermodynamics and kinetics. Conversely, the approaches herein can identify cases where an independent, additive model cannot be applied and so require additional investigation.
Structured RNAs are integral to many biological processes, including translation, genome maintenance, and the regulation of gene expression (1, 2). These processes require RNA to fold into intricate 3D structures and to undergo a series of structural transitions (3, 4). As such, an important goal has been to characterize these states, in terms of their conformations and the rates and equilibria that describe the transitions between them (5–8). A more distant but ultimately far-reaching challenge is to quantitatively predict the kinetics and thermodynamics of RNA transitions, an accomplishment that would demonstrate fundamental understanding and effectuate engineering of these systems.
Quantitative analyses of the melting temperatures for nucleic acid duplexes led to predictive rules for RNA secondary structure stability, known as nearest neighbor or Turner rules, such that the energetics of helix formation can be predicted by considering only the identity of each base pair, the identity of its immediate neighbors, and the salt concentration (9–11). In this model, the energetic contribution of each base pair step is additive and can be summed to predict the free energy to an accuracy of 0.2–0.5 kcal/mol for simple helices (12). Although many more thermodynamic measurements will be required to predict the secondary structure stability for large RNAs and RNAs containing any possible noncanonical elements, the current model has been applied with great impact throughout biology and bioengineering (e.g., refs. 13 and 14).
An ongoing major challenge is to develop comparable quantitative and predictive algorithms for RNA tertiary structure formation. Once formulated, this knowledge may be applied to increase our understanding of biological processes that involve structured RNAs, such as RNA conformational transitions in splicing and translation, and to allow engineering of controllable RNA elements in vivo. This challenge would be greatly simplified if it were possible to model RNA tertiary structure as the collective behavior of structural modules such that the energetics of tertiary structure formation could be predicted from the sum of separable terms corresponding to each module, akin to terms used in nearest neighbor rules for predicting secondary structure.
As outlined below, considerable work by many RNA researchers over the past decades has led to a common perspective that RNA’s tertiary structure has modular properties (15–19). Based on these observations, we now ask whether this structural modularity extends to energetics, such that folding thermodynamics of complex RNAs can be described in terms of properties of components—a task that has proven elusive for proteins but may be more applicable to RNA given its structural properties. We formulate and test predictions of such a model, and our results provide initial support for energetic additivity in RNA folding thermodynamics and analogous kinetic properties. We discuss our results in terms of comparisons with proteins, noting differences between the properties of these biological macromolecules while taking advantage of the wealth of models and conceptual approaches from the more mature protein-folding field.
Background
RNA tertiary structure can be considered as a collection of helices connected by junctions (i.e., the residues connecting canonical helical elements), with sparsely distributed tertiary contacts that bring together distal parts of the structure (Fig. 1A). Several of these tertiary interactions have been described as “tertiary motifs” because their sequences are conserved in different structured RNAs and their structures have been shown to closely overlay in three dimensions, suggesting a degree of modularity (15, 16, 18, 20).
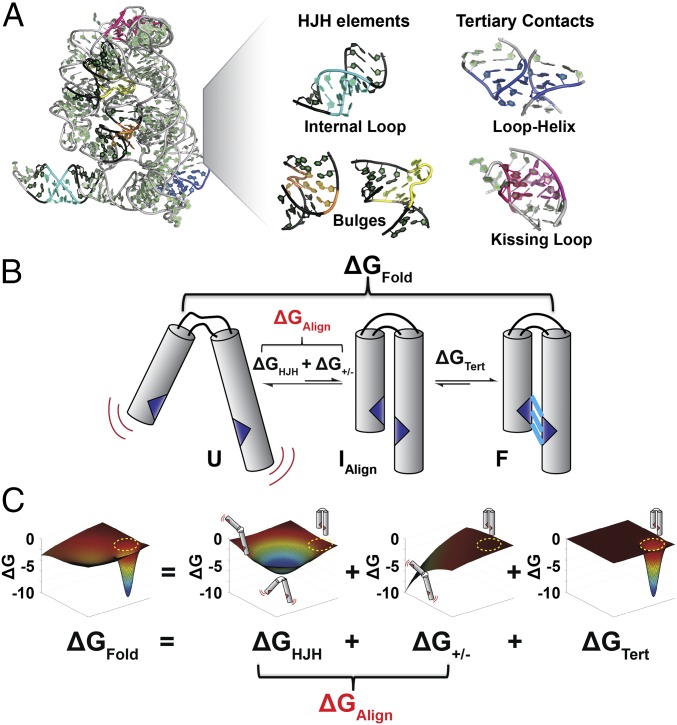
Fig. 1.
Building blocks of structured RNAs and the reconstitution hypothesis. (A) Crystal structure of the group II intron with representative RNA tertiary structure building blocks highlighted (73). Tertiary elements include HJH elements, with helices in black and junctions colored: internal loop (cyan), bulges (orange and yellow), and tertiary contacts, loop-helix (blue) and kissing-loop (magenta). (B) The overall folding (ΔGFold) of a simple HJH element with a single tertiary contact (blue) is shown to illustrate the reconstitution hypothesis. The first folding step involves a search for the conformational states in which tertiary contact partners are aligned and can form (ΔGAlign). This step can be further separated into the contributions due to the orientational preferences of the HJH elements connecting the two helices (ΔGHJH) and the electrostatic forces that bias the conformational ensemble of the HJH element to favor more extended conformations (ΔG+/−). The second step in folding involves formation of the tertiary contact (ΔGTert) that stabilizes the folded state F. (C) The reconstitution hypothesis posits that the overall folding of a HJH element (ΔGFold) is the sum of the energetic contributions from the HJH elements, electrostatics, and tertiary contact formation (ΔGFold = ΔGHJH + ΔG+/− + ΔGTert). Each energetic contribution is figuratively depicted by a 3D free energy landscape, and these landscapes are summed to give the overall folding energetics. Free energy diagrams were generated in Matlab to qualitatively depict the folding landscape. The x–y plane represents all possible states of the HJH element, and encircled in yellow is the ensemble of states in which the tertiary contact is aligned to form. The value on the z axis is free energy represented by ΔG = −RTln(Ki), where Ki is the ratio of being in the aligned state relative to all other states. Illustrative conformations for the HJH element are shown above each free energy landscape.
As the residues of these tertiary motifs typically do not directly interact with residues outside of the motif, the atomic-level process for forming interactions within a motif may be the same in different RNAs. Thus, there may be a common underlying energetic contribution of the motif to folding. Alternatively, differing connections between tertiary elements could distort the motif’s structure, alter the interactions made, and consequently lead to energetic interdependence between the motif and RNA elements that span the contacts. Initial tests of the kinetic behavior of one tertiary contact motif, the tetraloop/tetraloop receptor (TL/TLR), embedded within different structures revealed a common unfolding rate constant in different contexts, providing evidence for a modular, locally defined physical process (21).
With respect to junction elements, classic work by Lilley and coworkers (22–24) on three- and four-way junctions in DNA and RNA established rules for how the sequence and topology of these junctions determine the orientations and stacking of the helices that emanate from these junctions. Subsequent experiments demonstrated conserved orientation preferences for these and other junctions when placed in different RNAs, suggesting the transferability of junction-stacking properties (19, 25). More recently, it has been suggested that RNA substructures of two helices of defined lengths spanned by junctions of various topologies are basic building blocks of RNA tertiary structure; these substructures are referred to generically as helix-junction-helices (HJHs) (26–28).
According to this perspective, HJH elements determine the conformational ensemble explored by unfolded RNA, and ensembles have been demonstrated for highly conserved junctions such as the kink-turn and four-way junction and the biologically important trans-activation response element RNA junction (29–31). From this perspective, the probability of forming tertiary interactions depends on the fraction of time that the ensemble aligns two tertiary elements such that contacts form between them. In this way, we hypothesize that the conformational ensemble in isolation of the HJH elements that connect tertiary partners is a separable component in the determination of the probability of tertiary structure formation (27).
Given these observations, we formalized an energetic model for RNA folding consisting of separable contributors that, in principle, are predictable from the properties of isolated components and transferable between systems. We then tested quantitative predictions of this model using single-molecule FRET (smFRET) measurements of RNA folding thermodynamics and kinetics. Our results suggest that the canonical GAAA-11nt TL/TLR tertiary contact is thermodynamically and kinetically modular and that the kinetic and thermodynamic effects of mutations in this tertiary contact are transferable and thus potentially valuable tools in the dissection of folding steps for complex RNAs. Most broadly, these results provide an early step toward a quantitative and predictive understanding of RNA folding.
Results
An RNA Reconstitution Hypothesis.
As described in the Introduction, an unfolded RNA exists in an ensemble of states in which the relative locations of tertiary contact partners (multiple ones for complex RNAs) are distributed in space based on the properties of the intervening HJH elements (Fig. 1A). Once aligned, particular tertiary partners can interact and thereby provide stabilization of intermediate and folded states (25–27, 30).
The folding scheme of Fig. 1B illustrates the energetic terms that underlie an RNA folding reconstitution hypothesis, and we use this scheme to describe the hypothesis and to illustrate tests of it. Fig. 1B depicts a simple RNA with a single HJH element and a single set of tertiary contact partners, and an illustrative 2D free energy landscape for folding of this RNA is depicted in Fig. 1C, Left as a distribution of different possible states. The free energy (ΔGFold) of the overall folding process can be considered in terms of two steps and three energetic terms (Eq. 1): The first step is aligning the tertiary contact partners (ΔGAlign), and the second is formation of the tertiary contact (ΔGTert) (Fig. 1B). [As commonly discussed, the stability of RNA helices allows us to consider tertiary folding starting with secondary structure preformed. The complexity of multiple secondary structures (e.g., for riboswitches and misfolded states) (32) is accommodated in this framework by deriving a value of ΔGAlign for each underlying secondary structure.] ΔGAlign is made up of two terms, representing the properties of the junction that determine the conformational space explored by the attached helices (ΔGHJH) and the degree of charge repulsion between the helices under particular solution conditions (ΔG+/−). (In practice, ΔGHJH could be determined empirically at a particular ionic condition, with ΔG+/− providing an adjustment to ΔGAlign for a given standard state.) Fig. 1C depicts hypothetical free energies for each component’s distribution among possible conformational states, where the folded state is a small portion of the overall conformational landscape and has the helices oriented such that the tertiary contacts are aligned to form. The HJH and electrostatic terms, ΔGHJH and ΔG+/−, are both, in this example, unfavorable for folding, and ΔGTert is the sole energetic term that favors the folded state. The reconstitution hypothesis proposes that these free energy distributions can be added to obtain ΔGFold according to Eq. 1. Analogous descriptions of folding kinetics lead to parallel predictions for folding and unfolding rate constants (Testing Kinetic Predictions of the Reconstitution Hypothesis):
| [1] |
Here we test whether the energetics of tertiary contact formation () are separable from the HJH elements that connect them (), a property of the reconstitution hypothesis and one that more generally allows the most robust predictive power of RNA folding. These energetics are expected to be separable if two criteria are met: There are no additional, idiosyncratic interactions between the tertiary motif and the remainder of the RNA (i.e., the HJH elements), and the conformational ensemble of the consummated tertiary motif itself is the same (or nearly the same) when embedded in different HJH elements. Importantly, we treat tertiary motifs as distinct units and thus do not consider the energetic dependences of the interactions within the motif, as has been addressed previously (33, 34). Rather, we consider the dependences of the energetics of a tertiary motif on features outside the contact, such as electrostatic forces, the connecting HJH elements, and the formation of other tertiary contacts.
Point mutations in a tertiary contact of a complex RNA weaken folding by an amount that is the difference between the folding free energy of the wild type (WT) and the mutant RNA, ΔΔGFold,(WT–Mut). If the energetics of formation of this tertiary contact are independent of the electrostatic environment and the HJH elements that connect tertiary motif partners, as predicted by the reconstitution hypothesis, then the value of ΔΔGFold,(WT–Mut) will be the same under (i) different solution salt conditions and (ii) when the partners are embedded in different RNA contexts. Alternatively, if there is an interdependence, we will observe nonadditivity, and thus, ΔΔGFold,(WT–Mut) values will vary. In the following two sections, we test each of these predictions of the reconstitution hypothesis.
Are Electrostatic Forces (ΔG+/−) and Tertiary Contact Formation (ΔGTert) Energetically Separable?
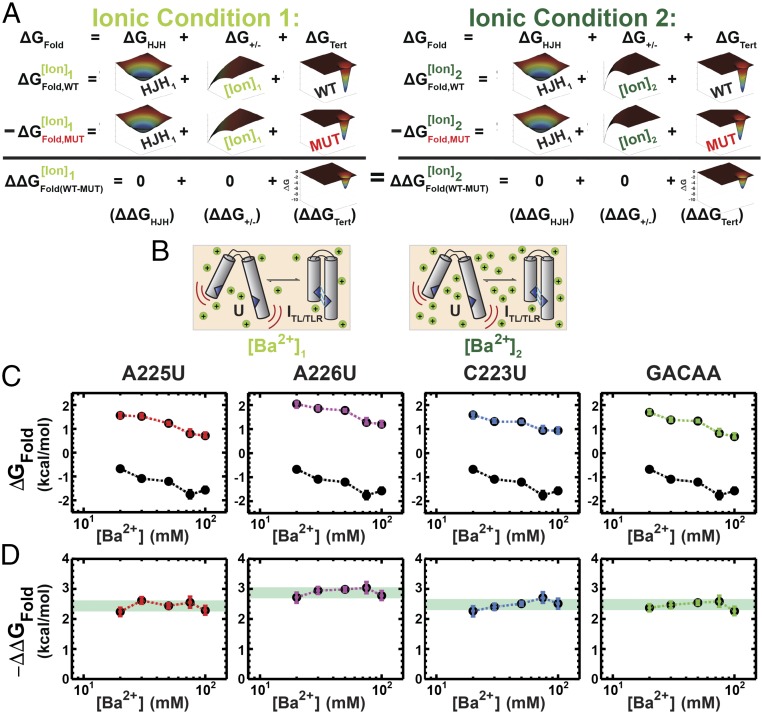
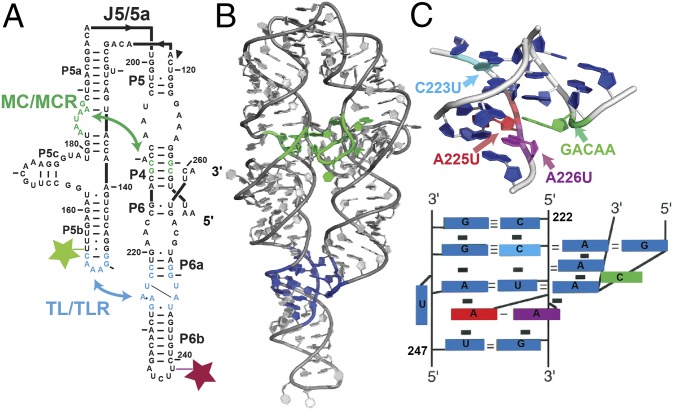
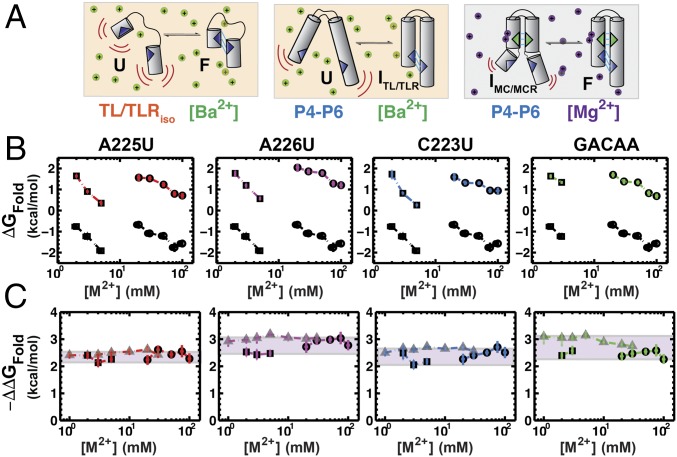
Fig. 2A illustrates the reconstitution hypothesis prediction for different ionic conditions; that is, that changes in the electrostatic term (ΔG+/−) do not alter the response of a tertiary element to mutation so that ΔΔGFold is the same under different ionic conditions (Eq. 2). To test this prediction, we measured the folding energetics of the TL/TLR tertiary contact motif of the P4–P6 RNA (Fig. 3) using smFRET. P4–P6 RNA has been widely studied as a model for RNA tertiary folding (e.g., refs. 35–39), and our recent thermodynamic and kinetic framework for its folding provides the necessary structural, thermodynamic, and kinetic context for interpretation of the results herein (21). P4–P6 has two tertiary contacts, a canonical GAAA-11nt TL/TLR and a metal core/metal core receptor (MC/MCR), and their formation bring its two helical stacks into a roughly parallel conformation (37, 40–43) (Fig. 3B).
Fig. 2.
Predicted and observed effects of changing the ion environment on the folding effects of point mutants in a tertiary contact. (A) The reconstitution hypothesis predicts that the effects of a mutation in a tertiary contact (ΔΔGFold = ΔGWT − ΔGMut) should be the same under different electrostatic conditions [e.g., [Ba2+]1 (Left) vs. [Ba2+]2 (Right)], as the different electrostatic conditions are posited to only alter ΔG+/−. This effect is depicted by a different distribution for ΔG+/− (only) in the two contexts. Mutation in a tertiary contact leads to a smaller contribution of ΔGTert to overall folding, as shown by a relatively smaller peak in the negative free energy relative to that of WT solely for ΔGTert. As depicted, ΔΔGFold (i.e., the effect of a mutation) is predicted to be the same in the two salt conditions. (B) Schematic for P4–P6 folding in Ba2+, whereby different Ba2+ concentrations modulate folding. (C) Folding across a range of Ba2+ concentrations (20–100 mM), for TL/TLR point mutants; colors designate the mutation measured and are defined in Fig. 3. WT folding at each concentration is shown in black and repeated in each panel. Errors represent the 95% confidence interval of the mean determined by bootstrapping (Methods; data in Table S1). (D) The effect of the mutation relative to WT folding, ΔΔGFold,(WT−Mut) versus Ba2+. Light green overlays designate the ΔΔGFold,(WT–Mut) range for each mutant.
Fig. 3.
P4–P6 RNA structure and point mutations. (A) Secondary structure of the P4–P6 RNA. Two tertiary contacts, the MC/MCR (green) and the TL/TLR (blue), are highlighted. (B) Tertiary structure of the P4–P6 RNA. The tertiary contacts are colored as in A [Protein Data Bank (PBD) ID code 1GID] (36). (C) Tertiary structure and schematic of the TL/TLR highlighting the residues that were mutated and studied herein: A225U (red), A226U (magenta), C223U (cyan), and GACAA (green); these colors are used throughout. A225U, A226U, and C223U are point mutations in the receptor, and GACAA is an insertion in the tetraloop.
We used Ba2+ to vary ionic conditions, rather than Mg2+, as Ba2+ does not bind to the MC of P4–P6 (44) and only the TL/TLR forms in the folded state (38) (Fig. 2B). Prior work demonstrated equivalent P4–P6 folding in Mg2+ and Ba2+ upon ablation of the MC, and independent studies demonstrated equal ion atmosphere occupancy for Mg2+ and Ba2+ surrounding a nucleic acid (21, 45). Thus, we could change the electrostatic environment without complications from site-specific metal ion binding and formation of an additional tertiary contact (Fig. 3B; see also Additional Tests of the Potential Interplay Between Structural Context and Tertiary Contact Energetics).
Fig. 2C shows the values of ΔGFold for each of four mutants in the TL/TLR of P4–P6 (colored symbols; Fig. 3C) determined using smFRET measurements of folding (21), compared with WT (black symbols, reproduced in each panel for comparison), over a series of Ba2+ concentrations. Example traces and data quality analyses for all smFRET measurements can be found in Dataset S1. The range of Ba2+ concentrations was defined by conditions that allowed observation of folding transitions for both WT and mutant P4–P6 (Methods), and we obtained measured ΔGFold values over a range of ∼4 kcal/mol.
Whereas the individual values of ΔGFold vary across Ba2+ concentrations (Fig. 2C), the differences between the WT and mutant folding free energies, ΔΔGFold, were nearly invariant (Fig. 2D). For each mutant, the variation in ΔΔGFold,(WT−Mut) was less than 0.3 kcal/mole (Fig. 2D, green swatches), which corresponds to a less than twofold difference in equilibrium constants and is similar to measurement error (Table S1). The invariance of ΔΔGFold,(WT−Mut) for each of the four mutants supports the first prediction of the reconstitution hypothesis, that tertiary contact energetics (ΔGTert) are unaffected by changes in electrostatic screening (ΔG+/−) (Fig. 1 and Eq. 2):
| [2] |
From a physical perspective, these data suggest a degree of simplicity to the conformational and energetic landscape of the TL/TLR tertiary contact, specifically that the mutations and varying electrostatic conditions do not substantially alter the interactions that make up this tertiary contact.
Table S1.
Folding parameters of P4–P6 obtained from smFRET
| Construct | [Mg2+], mM | [Ba2+], mM | kFold,* s−1 | kUnfold,* s−1 | Kfold*,† | ΔG,*,‡ kcal/mol | −ΔΔG,*,§ kcal/mol | N |
| WT | 0.0 | 20.0 | 5.68 ± 0.8 | 1.78 ± 0.15 | 3.2 ± 0.57 | −0.68 ± 0.1 | — | 106 |
| WT | 0.0 | 30.0 | 8.65 ± 0.97 | 1.34 ± 0.08 | 6.48 ± 1.1 | −1.09 ± 0.1 | — | 145 |
| WT | 0.0 | 50.0 | 8.35 ± 0.8 | 1.05 ± 0.11 | 7.94 ± 0.8 | −1.21 ± 0.06 | — | 234 |
| WT | 0.0 | 75.0 | 12.55 ± 3.94 | 0.61 ± 0.14 | 20.46 ± 7.04 | −1.76 ± 0.18 | — | 55 |
| WT | 0.0 | 100.0 | 8.37 ± 1.43 | 0.56 ± 0.1 | 14.89 ± 3.44 | −1.57 ± 0.14 | — | 104 |
| WT | 1.0 | 0.0 | 0.44 ± 0.04 | 1.51 ± 0.1 | 0.29 ± 0.04 | 0.72 ± 0.07 | — | 272 |
| WT | 2.0 | 0.0 | 1.92 ± 0.14 | 0.52 ± 0.03 | 3.71 ± 0.6 | −0.76 ± 0.1 | — | 351 |
| WT | 3.0 | 0.0 | 3.37 ± 0.89 | 0.4 ± 0.02 | 8.34 ± 2.41 | −1.23 ± 0.17 | — | 234 |
| WT | 5.0 | 0.0 | 6.3 ± 0.51 | 0.23 ± 0.02 | 26.82 ± 3.87 | −1.91 ± 0.09 | — | 368 |
| A225U | 0.0 | 20.0 | 1.19 ± 0.15 | 17.42 ± 2.78 | 0.07 ± 0.01 | 1.56 ± 0.11 | 2.24 ± 0.15 | 131 |
| A225U | 0.0 | 30.0 | 1.62 ± 0.24 | 22.28 ± 2.72 | 0.07 ± 0.01 | 1.53 ± 0.07 | 2.61 ± 0.12 | 145 |
| A225U | 0.0 | 50.0 | 2.14 ± 0.12 | 17.76 ± 2.5 | 0.12 ± 0.02 | 1.23 ± 0.07 | 2.44 ± 0.09 | 140 |
| A225U | 0.0 | 75.0 | 3.47 ± 0.42 | 13.57 ± 2.13 | 0.26 ± 0.04 | 0.79 ± 0.07 | 2.55 ± 0.19 | 115 |
| A225U | 0.0 | 100.0 | 3.77 ± 0.32 | 12.78 ± 1.7 | 0.29 ± 0.04 | 0.71 ± 0.08 | 2.28 ± 0.16 | 133 |
| A225U | 2.0 | 0.0 | 0.32 ± 0.04 | 5.36 ± 0.61 | 0.06 ± 0.01 | 1.64 ± 0.08 | 2.41 ± 0.12 | 117 |
| A225U | 3.0 | 0.0 | 0.93 ± 0.05 | 4.39 ± 0.45 | 0.21 ± 0.02 | 0.9 ± 0.06 | 2.14 ± 0.18 | 79 |
| A225U | 5.0 | 0.0 | 1.79 ± 0.12 | 3.25 ± 0.14 | 0.55 ± 0.09 | 0.35 ± 0.1 | 2.26 ± 0.13 | 96 |
| A226U | 0.0 | 20.0 | 0.48 ± 0.09 | 15.94 ± 5.73 | 0.03 ± 0.01 | 2.04 ± 0.16 | 2.72 ± 0.18 | 62 |
| A226U | 0.0 | 30.0 | 0.56 ± 0.1 | 13.73 ± 1.39 | 0.04 ± 0.01 | 1.86 ± 0.07 | 2.94 ± 0.12 | 180 |
| A226U | 0.0 | 50.0 | 0.52 ± 0.05 | 11.06 ± 1.76 | 0.05 ± 0.01 | 1.78 ± 0.06 | 2.98 ± 0.08 | 183 |
| A226U | 0.0 | 75.0 | 1.09 ± 0.13 | 9.87 ± 2.63 | 0.11 ± 0.02 | 1.28 ± 0.08 | 3.04 ± 0.2 | 92 |
| A226U | 0.0 | 100.0 | 1.15 ± 0.17 | 9.07 ± 1.33 | 0.13 ± 0.02 | 1.2 ± 0.09 | 2.77 ± 0.16 | 118 |
| A226U | 2.0 | 0.0 | 0.12 ± 0.02 | 2.57 ± 0.55 | 0.05 ± 0.01 | 1.77 ± 0.13 | 2.53 ± 0.16 | 82 |
| A226U | 3.0 | 0.0 | 0.33 ± 0.03 | 2.53 ± 0.25 | 0.13 ± 0.02 | 1.19 ± 0.1 | 2.43 ± 0.19 | 101 |
| A226U | 5.0 | 0.0 | 0.71 ± 0.04 | 1.87 ± 0.09 | 0.38 ± 0.05 | 0.56 ± 0.08 | 2.47 ± 0.12 | 107 |
| C223U | 0.0 | 20.0 | 5.43 ± 1.45 | 83.3 ± 22.1 | 0.07 ± 0.02 | 1.59 ± 0.15 | 2.27 ± 0.18 | 37 |
| C223U | 0.0 | 30.0 | 7.83 ± 0.69 | 75.05 ± 3.24 | 0.1 ± 0.01 | 1.32 ± 0.06 | 2.40 ± 0.11 | 193 |
| C223U | 0.0 | 50.0 | 5.78 ± 0.4 | 54.06 ± 4.39 | 0.11 ± 0.01 | 1.3 ± 0.03 | 2.51 ± 0.07 | 218 |
| C223U | 0.0 | 75.0 | 8.67 ± 0.6 | 44.32 ± 4.35 | 0.2 ± 0.02 | 0.95 ± 0.05 | 2.71 ± 0.19 | 126 |
| C223U | 0.0 | 100.0 | 8.5 ± 1.83 | 42.57 ± 6.99 | 0.2 ± 0.05 | 0.94 ± 0.12 | 2.51 ± 0.17 | 31 |
| C223U | 2.0 | 0.0 | 0.62 ± 0.15 | 12.02 ± 2.06 | 0.05 ± 0.02 | 1.73 ± 0.21 | 2.49 ± 0.23 | 90 |
| C223U | 3.0 | 0.0 | 2.99 ± 0.29 | 12.19 ± 0.55 | 0.25 ± 0.02 | 0.82 ± 0.05 | 2.05 ± 0.18 | 213 |
| C223U | 5.0 | 0.0 | 4.35 ± 0.41 | 6.76 ± 0.73 | 0.64 ± 0.08 | 0.26 ± 0.07 | 2.17 ± 0.11 | 77 |
| GACAA | 0.0 | 20.0 | 3.52 ± 0.91 | 64.84 ± 7.7 | 0.05 ± 0.01 | 1.69 ± 0.07 | 2.37 ± 0.12 | 102 |
| GACAA | 0.0 | 30.0 | 4.73 ± 1.00 | 50.65 ± 6.82 | 0.09 ± 0.01 | 1.38 ± 0.07 | 2.47 ± 0.11 | 153 |
| GACAA | 0.0 | 50.0 | 3.22 ± 0.16 | 31.92 ± 4.03 | 0.1 ± 0.01 | 1.33 ± 0.05 | 2.54 ± 0.07 | 165 |
| GACAA | 0.0 | 75.0 | 6.12 ± 0.71 | 25.27 ± 2.86 | 0.24 ± 0.03 | 0.83 ± 0.07 | 2.58 ± 0.19 | 94 |
| GACAA | 0.0 | 100.0 | 6.68 ± 0.6 | 21.75 ± 3.28 | 0.31 ± 0.04 | 0.69 ± 0.07 | 2.26 ± 0.15 | 158 |
| GACAA | 2.0 | 0.0 | 1.34 ± 0.16 | 22.22 ± 2.97 | 0.06 ± 0.01 | 1.64 ± 0.09 | 2.40 ± 0.13 | 140 |
| GACAA | 3.0 | 0.0 | 2.02 ± 0.23 | 20.13 ± 3.03 | 0.1 ± 0.01 | 1.34 ± 0.06 | 2.57 ± 0.18 | 90 |
N, number of molecules.
Mean and error values indicate the bootstrapped 95% confidence interval of the mean.
Kfold = kFold/kUnfold.
ΔG = −RTln(Kfold).
ΔΔG = ΔGWT − ΔGMut.
Testing the Energetics of Tertiary Contact Formation in Different RNA Contexts.
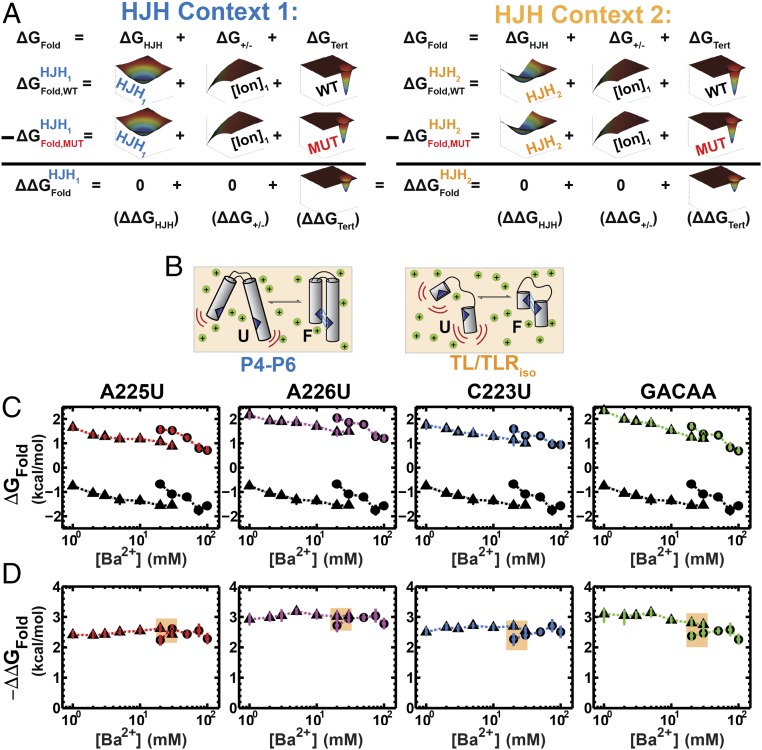
Analogous to the above prediction made for the relationship between ΔG+/− and ΔGTert (Eq. 1), the reconstitution hypothesis predicts that the effect of a mutation in a tertiary contact should be the same in different structural contexts (ΔGHJH). Fig. 4A illustrates this prediction, analogous to Fig. 2A. To test this prediction, we compared the effects of the TL/TLR point mutations in P4–P6 to the effects of these mutations in an isolated TL/TLR construct (TL/TLRiso), initially developed and studied extensively by Nesbitt and coworkers (46). In this construct, the two tertiary contact partners are connected via a flexible tether and thus lack the orientation biases imposed by P4–P6 (Fig. 4B).
Fig. 4.
Comparison of the effect of TLR mutations in P4–P6 RNA and the TL/TLRiso construct. (A) The reconstitution hypothesis predicts that the effects of a mutation in a tertiary contact (ΔΔGFold = ΔGWT − ΔGMut) will be the same in different structural contexts [HJH1 (Left) vs. HJH2 (Right)]. Mutation in a tertiary contact leads to a lower probability for formation of the tertiary contact and thus a less negative value of ΔGTert and weaker overall folding, without affecting the other free energy terms. (B) Depiction of P4–P6 folding (Left) versus the TL/TLRiso (Right) in Ba2+. (C) Folding across a range of Ba2+ concentrations (1–100 mM) for P4–P6 (circles) and TL/TLRiso (triangles); colors correspond to point mutations in the TL/TLR as in Fig. 3C, and the black points repeated in each panel are for WT P4–P6 and WT TL/TLRiso. Data are provided in Tables S1 and S2. (D) The effect of the mutation relative to WT (ΔΔGFold,(WT–Mut) = ΔGWT − ΔGMut) versus Ba2+ for P4–P6 (circles) and the TL/TLRiso (triangles). Tan overlay designates the overlapping Ba2+ concentrations where the ΔΔGFold,(WT−Mut) values can be directly compared. Errors represent the 95% confidence interval of the mean determined by bootstrapping (Methods).
Fig. 4C shows the folding free energy (ΔGFold) for each point mutation in TL/TLRiso and in P4–P6, compared with the folding free energy of the corresponding WT molecule across a range of Ba2+ concentrations (Table S2). Because of stability differences, there are only two Ba2+ concentrations where we could obtain ΔGFold values for both constructs and thus directly compare ΔΔGFold,(WT−Mut) values for TL/TLRiso and P4–P6 RNA. As shown in Fig. 4D (colored regions), the ΔΔGFold,(WT−Mut) values for each mutant at these concentrations are the same for TL/TLRiso and P4–P6. The similar ΔΔGFold,(WT−Mut) values in these starkly different structural contexts suggest that the energetic contribution of tertiary contact formation (ΔGTert) is separable from that of the HJH context (ΔGHJH) and further support the assumptions underlying the reconstitution hypothesis (Fig. 1 and Eq. 1).
Table S2.
Folding parameters of TL/TLRIso obtained from smFRET
| Construct | [Ba2+], mM | kFold,* s−1 | kUnfold,* s−1 | Kfold,*,† | ΔG,*,‡ kcal/mol | −ΔΔG,*,§ kcal/mol | N |
| WT | 1.0 | 14.72 ± 0.81 | 4.02 ± 0.35 | 3.66 ± 0.6 | −0.76 ± 0.1 | — | 108 |
| WT | 2.0 | 17.7 ± 1.59 | 2.86 ± 0.12 | 6.2 ± 0.97 | −1.06 ± 0.1 | — | 104 |
| WT | 3.0 | 20.48 ± 1.58 | 2.83 ± 0.18 | 7.22 ± 1.06 | −1.15 ± 0.09 | — | 112 |
| WT | 5.0 | 24.25 ± 1.27 | 2.48 ± 0.11 | 9.77 ± 0.94 | −1.33 ± 0.06 | — | 103 |
| WT | 10.0 | 25.52 ± 1.47 | 2.44 ± 0.15 | 10.47 ± 1.5 | −1.37 ± 0.09 | — | 126 |
| WT | 20.0 | 27.18 ± 2.55 | 1.87 ± 0.28 | 14.55 ± 2.54 | −1.56 ± 0.11 | — | 66 |
| WT | 30.0 | 30.44 ± 3.89 | 2.13 ± 0.14 | 14.26 ± 2.12 | −1.55 ± 0.09 | — | 85 |
| A225U | 1.0 | 1.85 ± 0.21 | 31.8 ± 1.43 | 0.06 ± 0.01 | 1.66 ± 0.06 | 2.41 ± 0.12 | 108 |
| A225U | 2.0 | 2.79 ± 0.14 | 27.75 ± 1.35 | 0.1 ± 0.01 | 1.34 ± 0.06 | 2.4 ± 0.11 | 114 |
| A225U | 3.0 | 3.09 ± 0.43 | 27.79 ± 1.92 | 0.11 ± 0.01 | 1.28 ± 0.05 | 2.43 ± 0.1 | 121 |
| A225U | 5.0 | 3.33 ± 0.2 | 25.17 ± 0.72 | 0.13 ± 0.02 | 1.18 ± 0.08 | 2.5 ± 0.09 | 118 |
| A225U | 10.0 | 3.34 ± 0.51 | 25.2 ± 2.47 | 0.13 ± 0.02 | 1.18 ± 0.07 | 2.54 ± 0.11 | 69 |
| A225U | 20.0 | 4.1 ± 0.29 | 25.38 ± 1.89 | 0.16 ± 0.02 | 1.06 ± 0.06 | 2.62 ± 0.12 | 129 |
| A225U | 30.0 | 5.22 ± 0.37 | 23.78 ± 0.68 | 0.22 ± 0.02 | 0.88 ± 0.05 | 2.43 ± 0.1 | 178 |
| A226U | 1.0 | 0.41 ± 0.1 | 16.93 ± 3.55 | 0.02 ± 0.01 | 2.17 ± 0.15 | 2.92 ± 0.17 | 51 |
| A226U | 2.0 | 0.6 ± 0.17 | 16.25 ± 2.88 | 0.04 ± 0.01 | 1.92 ± 0.09 | 2.98 ± 0.13 | 122 |
| A226U | 3.0 | 0.58 ± 0.08 | 15.13 ± 2.52 | 0.04 ± 0.01 | 1.9 ± 0.16 | 3.05 ± 0.17 | 69 |
| A226U | 5.0 | 0.53 ± 0.06 | 12.7 ± 1.08 | 0.04 ± 0.01 | 1.85 ± 0.07 | 3.18 ± 0.09 | 149 |
| A226U | 10.0 | 0.59 ± 0.06 | 10.85 ± 1.62 | 0.05 ± 0.01 | 1.69 ± 0.09 | 3.06 ± 0.12 | 90 |
| A226U | 20.0 | 0.82 ± 0.11 | 10.12 ± 0.74 | 0.08 ± 0.02 | 1.46 ± 0.11 | 3.02 ± 0.14 | 86 |
| A226U | 30.0 | 0.83 ± 0.09 | 10.87 ± 1.42 | 0.08 ± 0.02 | 1.49 ± 0.12 | 3.04 ± 0.14 | 73 |
| C223U | 1.0 | 11.82 ± 1.36 | 237.63 ± 17.73 | 0.05 ± 0.01 | 1.75 ± 0.08 | 2.5 ± 0.13 | 84 |
| C223U | 2.0 | 13.48 ± 1.58 | 210.38 ± 11.35 | 0.06 ± 0.01 | 1.6 ± 0.06 | 2.66 ± 0.11 | 114 |
| C223U | 3.0 | 16.14 ± 0.9 | 199.04 ± 4.55 | 0.08 ± 0.01 | 1.46 ± 0.06 | 2.61 ± 0.1 | 122 |
| C223U | 5.0 | 17.16 ± 0.97 | 187.83 ± 12.82 | 0.09 ± 0.01 | 1.39 ± 0.04 | 2.72 ± 0.06 | 162 |
| C223U | 10.0 | 19.05 ± 1.07 | 172.48 ± 10.92 | 0.11 ± 0.01 | 1.28 ± 0.04 | 2.65 ± 0.09 | 98 |
| C223U | 20.0 | 18.65 ± 1.07 | 131.21 ± 6.12 | 0.14 ± 0.01 | 1.14 ± 0.05 | 2.69 ± 0.12 | 109 |
| C223U | 30.0 | 22.47 ± 1.72 | 126.11 ± 3.04 | 0.18 ± 0.02 | 1.4 ± 0.05 | 2.55 ± 0.1 | 114 |
| GACAA | 1.0 | 1.92 ± 0.33 | 107.29 ± 14.67 | 0.02 ± 0.01 | 2.34 ± 0.25 | 3.1 ± 0.26 | 92 |
| GACAA | 2.0 | 3.11 ± 0.25 | 94.38 ± 6.8 | 0.03 ± 0.02 | 1.99 ± 0.28 | 3.05 ± 0.28 | 143 |
| GACAA | 3.0 | 4.35 ± 0.37 | 113.62 ± 3.55 | 0.04 ± 0.01 | 1.9 ± 0.07 | 3.05 ± 0.1 | 160 |
| GACAA | 5.0 | 5.21 ± 1.1 | 117.95 ± 10.19 | 0.04 ± 0.01 | 1.82 ± 0.11 | 3.14 ± 0.13 | 20 |
| GACAA | 10.0 | 7.27 ± 0.35 | 100.29 ± 4.34 | 0.07 ± 0.01 | 1.53 ± 0.05 | 2.89 ± 0.1 | 152 |
| GACAA | 20.0 | 10.35 ± 1.58 | 88.11 ± 2.74 | 0.12 ± 0.02 | 1.25 ± 0.07 | 2.8 ± 0.13 | 99 |
| GACAA | 30.0 | 11.02 ± 0.89 | 86.68 ± 4.03 | 0.13 ± 0.01 | 1.2 ± 0.06 | 2.75 ± 0.11 | 108 |
N, number of molecules.
Mean and error values indicate the bootstrapped 95% confidence interval of the mean.
Kfold = kFold/kUnfold.
ΔG = −RTln(Kfold).
ΔΔG = ΔGWT − ΔGMut.
Additional Tests of the Potential Interplay Between Structural Context and Tertiary Contact Energetics.
In the previous section, we compared the TL/TLRiso, which provides a simple flexible HJH context where the TL/TLR contact partners are able to assume a wide range of conformations in the folded state, to P4–P6, which has a more complex and conformationally restricted set of elements connecting the tertiary contact partners. Functional RNAs often contain multiple constraining tertiary contacts, and the formation of P4–P6’s other tertiary contact, the MC/MCR (Fig. 3), would further constrain the encounter angles between tertiary elements. We therefore additionally tested the relationship between the energetics of tertiary contact formation (ΔGTert) and HJH context (ΔGHJH) with the MC/MCR tertiary contact formed in P4–P6 (Figs. 3 and 5A).
Fig. 5.
Effect of HJH changes on the effect of TL/TLR point mutants. (A) Depiction of TL/TLRiso (Left) folding in Ba2+, P4–P6 folding in Ba2+ (Middle), and P4–P6 folding in Mg2+ (Right). In Ba2+ only, the TL/TLR forms in P4–P6, whereas both the MC/MCR and the TL/TLR form in Mg2+ (38). The formation of the MC/MCR changes the structural connectivity between the TL and TLR and thus the alignment term of the reconstitution hypothesis (ΔGalign; Fig. 2B and Eq. 1). (B) P4–P6 folding across a range of Ba2+ concentrations (circles; 20–100 mM) and Mg2+ concentrations (squares; 1–3 mM), for point mutations in the TL/TLR; colors are as in Fig. 3. Data are presented in Tables S1 and S2. (C) The effect of each mutation relative to WT (ΔΔGFold) for P4–P6 in Ba2+ (circles) and Mg2+ (squares) and for the TL/TLRiso in Ba2+ (triangles). Purple overlay designated the variation in the ΔΔG values for each mutant in all three HJH contexts. Errors represent the 95% confidence interval of the mean determined by bootstrapping (Methods).
The ΔGFold values for P4–P6 folding with and without MC/MCR are shown in Fig. 5B. Mg2+ was used to allow MC/MCR formation and Ba2+ to prevent it, as above (44). Also as noted above, P4–P6 MC mutants that prevent Mg2+ binding have the same folding kinetics and thermodynamics in Mg2+ and Ba2+ (21, 47), suggesting that electrostatic screening is highly similar for these divalent metal ions and supporting comparison between Mg2+ and Ba2+ to isolate and test the effects of MC/MCR formation.
Due to the large stabilization of P4–P6 folding with the MC/MCR tertiary contact formed, we could not obtain folding data with Mg2+ and Ba2+ at the same concentrations. We therefore used the TL/TLRiso construct to span the divalent metal ion concentration range for both Mg2+ and Ba2+ used in the P4–P6 measurements in Fig. 5B. In this way, the ΔΔGFold,(WT–Mut) data for TL/TLRiso serve as a common comparator for ΔΔGFold,(WT–Mut) values between conditions for P4–P6 with and without the MC/MCR, and the shallow dependence of TL/TLRiso folding on metal ion concentration further simplifies this comparison. We therefore plotted the TL/TLRiso ΔΔGFold,(WT–Mut) values from Fig. 4C with the ΔΔGFold,(WT–Mut) values for P4–P6 in Mg2+ and Ba2+ (Fig. 5C).
For three of the mutants—A225U, A226U, and C223U—there were variations of less than 0.5 kcal/mol between the ΔΔGFold,(WT–Mut) values with and without the MC/MCR, representing differences of less than twofold in equilibrium constant and providing strong support of the predictions from the reconstitution model. Intriguingly, a slightly larger variance of 0.8 kcal/mol was observed for the fourth mutant, GACAA, which is beyond our estimated experimental error (Methods, Fig. 5C, and Tables S1 and S2). Given that this mutant corresponds to an insertion into the GAAA tetraloop to give a GACAA pentaloop, the insertion of the cytosine in the loop may alter the preferred orientation of the loop such that a higher fraction of the conformational states is competent for forming the mutant TL/TLR compared with the WT tetraloop when the MC/MCR is formed; this would cause the insertion mutation to be less deleterious in the most constrained context of P4–P6 with its MC/MCR formed. In principle, comparisons such as those carried out herein can be used to assess differential conformational states of tertiary contacts.
Our results varying ionic conditions and varying the HJH context support the reconstitution hypothesis, as the constant ΔΔGFold,(WT–Mut) values meet the simplest predictions of this model. Our results also suggest the potential for more complex behavior that can arise from the presence of altered states of the tertiary interaction and provide a path to identify such altered states through outlier, nonadditive behavior.
Testing Kinetic Predictions of the Reconstitution Hypothesis.
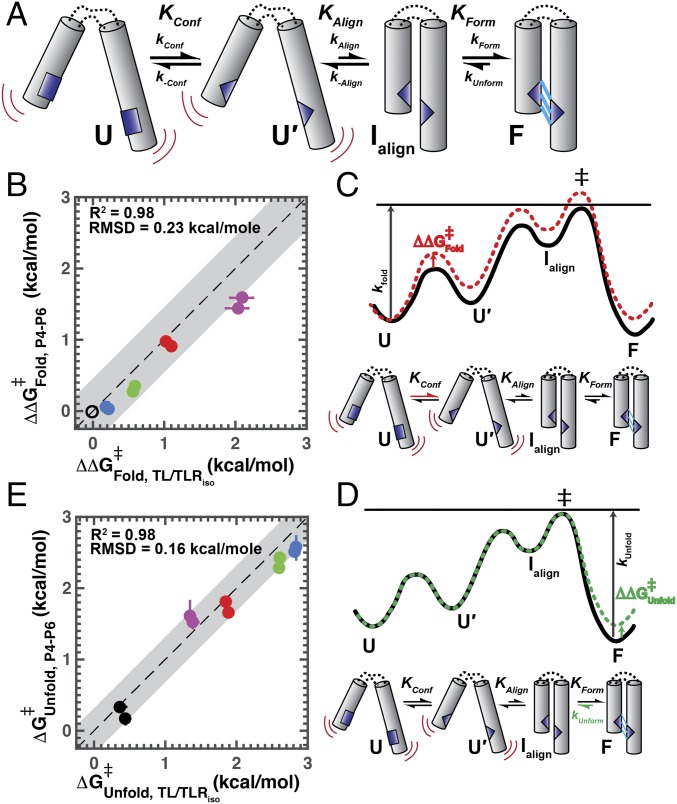
The thermodynamic modularity of the tertiary folding energetics introduces the possibility that there may also be a conservation of the underlying properties that determines the kinetics of tertiary contact formation and dissolution. We can make kinetic predictions based on the diffusion–collision model for RNA (21), whereby folding rate constants are determined by the rate constant at which tertiary elements collide weighted by the probability that the tertiary elements are in a state competent to proceed to form the interactions of the stable contact. To uncover predictions from this model, we break the overall folding process down in this model into three steps (Fig. 6A): (i) local rearrangements of the two tertiary contact elements to form a “tertiary-competent” conformation (KConf) that has a significant probability of forming the final tertiary contact from an aligned state; (ii) formation of the aligned state from which the tertiary contact can form (KAlign), which is determined by the extent to which the intervening helices and junctions favor the aligned versus all other states; and (iii) formation of the interactions, such as hydrogen bonds and stacking interactions, that make up the tertiary contact (kForm). Although in reality there will be a complex landscape of atomic-level rearrangements with many steps and pathways and a range of probabilities that they lead to the final tertiary state, we present a simplified model with three steps that we have evidence to support. In contrast, a two-step model cannot account for the data, as shown below.
Fig. 6.
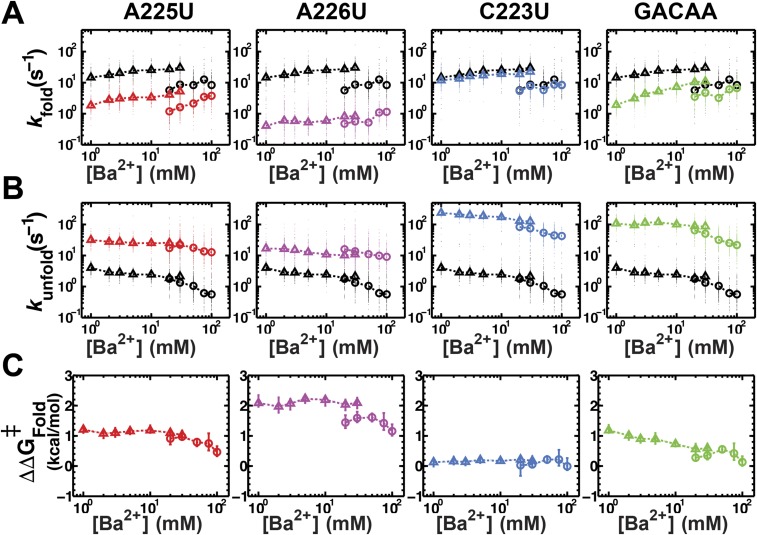
Kinetic effects of tertiary contact mutations on folding processes. (A) Simple kinetic model for tertiary contact formation based on the diffusion–collision model for RNA folding (21). Folding rates and equilibria for tertiary contact formation are composed of three steps: conformational change of tertiary contact elements to a tertiary-ready state, alignment of tertiary contact elements, and formation of the tertiary interactions between tertiary elements. Comparison of the effects on folding (B) and absolute unfolding (E) rate constants ( and ; see Methods) for TL/TLR mutations in P4–P6 versus TL/TLRiso (colors as in Fig. 1). Measurements were made at 20 and 30 mM Ba2+. Error bars represent propagation of the 95% confidence interval of the mean determined by bootstrapping. Reported is the rmsd to the fit of the data to the model y = x + b (Methods; data are presented in Tables S1 and S2) and is denoted in gray outline. Free energy diagrams depicting hypothesized effects on overall folding (C) and unfolding (D) rate constants. Effects on kFold (B) are consistent with effects on the conformational change of tertiary contact partners before formation of the tertiary contact (kConf), as depicted in the free energy diagram in C. Effects on kUnfold (D) are consistent with tertiary contact breaking as the rate-limiting transition in unfolding (kUnform), as depicted in the free energy diagram in (D).
RNA tertiary formation, including that for formation of the TL/TLR studied herein, is much slower than expected for diffusional collision; thus, there must be an additional step or steps preceding formation of the contact (21, 27, 48, 49). Although in principle there might only be a need for two steps, tertiary contact rearrangement and alignment (diffusional collision), our results provide evidence for an additional step. Specifically, the initial kConf rearrangement (Fig. 6A) is not rate limiting; if it were, then changes in the connecting helices and junctions would not alter the overall kFold for TL/TLR formation, whereas we observe that kFold is different in different RNAs (see below). Further, an alignment step involving the elements connecting the tertiary contact partners is not rate limiting (kAlign, Fig. 6A), because, if it were, mutations that disrupt the tertiary contact would not decrease the overall folding rate (kFold; see also below). Thus, a distinct, third step must be involved and rate limiting.
To build a three-step model, we posited that the initial atomic-level rearrangements leading to the “tertiary-competent” conformation (KConf) would not encompass all of the rearrangements needed to form the full interaction and that additional rearrangements would be required after alignment; these additional rearrangements are encapsulated in kForm (Fig. 6A). As kForm appears to be rate limiting, the initial two steps are expected to equilibrate before the kform step, so we discuss the kinetics below in terms of equilibria for these steps (KConf and KAlign).
The reconstitution hypothesis and kinetic model of Fig. 6A predict that the rate constants for folding vary as the HJH context is changed, since the alignment probability of tertiary contact partners (KAlign) will be different in different RNAs. In contrast, processes that are inherent to the tertiary interaction (kConf and kForm) should be the same independent of structural context (i.e., independent of the identity of the helices and junctions in the HJH that link the tertiary contact partners). While we cannot measure the rates of these individual microscopic steps to test whether they are the same, we can test instead the prediction that the effects of tertiary contact mutations on the rate constants () are the same in different contexts. That is, if these individual processes are the same, mutations will have constant effects on the pre-equilibrium of formation of the tertiary-ready state (KConf) and the rate constant for rearranging to form the tertiary interactions (kForm) and thus have the same effects on overall folding rate constants in different RNAs.
To test this model, we determined the effects on folding rate constants for each of the four tertiary mutations, and we compared the effects in P4–P6 and in TL/TLRiso. Folding rate constants were different between P4–P6 and the TL/TLRiso for all variants, as generally expected, and there were significant effects on kFold for three of the four mutants (A225U, A226U, and GACAA) of 10–30-fold (Fig. S1A and Tables S1 and S2). The effects on the folding rate constant in each structural context are plotted relative to one another in Fig. 6B in terms of for each (Methods). [These comparisons were made in Ba2+ because there are additional folding steps in Mg2+ for P4–P6, including rate-limiting formation of the MC/MCR, that would obscure information about the rate of forming the TL/TLR (21).] The congruence of the relationship between these values for TL/TLRiso and P4–P6 is excellent, with an R2 value of 0.98 and an rmsd of 0.23 kcal/mol for a linear model with the predicted slope of 1, an rmsd value that essentially matches our measurement error (error bars, Fig. 6B). Stated another way, this linear fit suggests that the effects in one context are essentially fully predictive of the effects in the other context. Free energy-reaction profiles illustrate how mutational effect on the tertiary contact conformational change (KConf; Fig. 6C) and/or the rate constant for the formation of the tertiary contact (kForm, not depicted) can lead to an overall folding rate constant effect that is independent of the alignment probability (KAlign). Fig. 6C shows the free energy diagram depicting this scenario, whereby the conformational change of the receptor (kConf) is slowed by the mutation, thereby decreasing the overall folding rate constant and adversely affecting the overall folding thermodynamics.
Fig. S1.
Folding kinetics of TL/TLR mutations in TL/TLRiso and P4–P6 RNA. Folding (A) and unfolding (B) rate constants of each mutant (colors as in Fig. 1) and WT (black; repeated in each panel) RNA across Ba2+ concentration for TL/TLRiso (open triangles) and P4–P6 (open circles). The rate constant values of each individual molecule under each condition are shown in small circles. Data are presented in Tables S1 and S2. (C) The effect of each mutant on the folding rate constant (ΔΔG‡Fold; see Methods) relative to the WT folding rate constant across [Ba2+] in the TL/TLRiso (open triangles) and P4–P6 (open circles). Errors represent the 95% confidence interval of the mean determined by bootstrapping (Methods).
With respect to unfolding rate constants, the reconstitution hypothesis predicts a conservation of the process of tertiary contact dissolution embedded in different structural contexts, as the same physical interactions are being broken regardless of the surrounding helices and junctions. This prediction will hold provided the final step in folding, kForm, is rate limiting (Fig. 6C) and, by microscopic reversibility, the first step in unfolding (Fig. 6D, kUnform). Fig. 6D shows this scenario in terms of a free energy diagram, whereby the step for dissolution of the tertiary contact (kUnform) is destabilized by the mutation, leading to an increase in the overall unfolding rate, as this step is rate-limiting in the unfolding pathway.
The unfolding rates for mutant and WT TL/TLRs in the TL/TLRiso and P4–P6 contexts are shown in Fig. S1B, and significant effects on kUnfold of 5–50-fold were observed for each of the four mutations investigated (Tables S1 and S2). As predicted, at equal Ba2+ concentrations, the unfolding rate constant for each mutant is the same within error in P4–P6 and in TL/TLRiso (Fig. 6E, kUnfold presented in terms of ). Thus, the dissociation rate constants highly correlate, with an R2 value of 0.98 and an rmsd of 0.16 kcal/mol for the model with uniform rate constants in the two contexts.
Discussion
We have formulated and tested predictions from a quantitative thermodynamic and kinetic model for RNA tertiary folding that posits that a complex RNA can be understood from the conformational and energetic behavior of its constituent elements—that is, the helices, junctions, and tertiary motifs that make up the RNA (Fig. 1). This RNA reconstitution hypothesis arises from a general perspective of RNA as modular, which in turn stems from observations that tertiary motifs have similar structures across multiple RNAs, an ability to swap these motifs as modules and maintain function, and conserved conformational properties of junctions embedded in different RNAs (15–17, 19).
Analogously, a longstanding goal within the protein-folding field has been to develop a bottom-up predictive understanding of folding kinetics and thermodynamics. Below we briefly compare and contrast the applicability of additivity models to RNA and proteins, highlighting the expectation that RNA may be more amenable to such approaches.
While hydrogen bonds and individual hydrophobic contacts in protein structures can be readily identified and counted, their energetic behavior is complex. These complexities include the typically coupled formation of secondary and tertiary structure, the differential energetic effects of solvation on buried versus exposed alpha helices and beta sheets (e.g., refs. 50 and 51), and the multiple local interactions and extended interaction networks in which most residues engage. The complications inherent to protein folding are largely absent in RNA folding, due to the simplifying features of its tertiary structure. The stability of base pairs within RNA helices results in preformed RNA secondary structure, allowing, in general, a clean energetic separation of RNA secondary and tertiary structure formation (17). [In cases such as riboswitches, there is coupling between the formation of secondary and tertiary structure, and there may be additional cases where native secondary structure is not stable in the absence of tertiary interactions (e.g., refs. 52 and 53). Nevertheless, these scenarios allow thermodynamic folding predictions to be made via a simple extension of the reconstitution model, using individual energy terms for secondary structure formation (“Turner rules”) and tertiary structure formation (Reconstitution), provided that these individual terms are known to sufficient accuracy.]
Whereas most amino acid side chains make tertiary interactions with multiple additional side chains, most RNA residues are not involved in long-range tertiary interactions. Rather, RNA bases exist in the context of helices and simple junctions that serve as connections between tertiary contacts, and these elements rarely make long-range or tertiary interactions to other parts of the structure. Thus, we might expect that the conformational properties of these helices and junctions to be the same in the structured RNA as when isolated in a much smaller RNA. In this way, the individual conformational preferences of helix and junction elements in isolation can be determined and “added” together to determine the overall conformations of a complex RNA in its unfolded states. Then, the subset of conformational states that allow the tertiary contact to be formed defines the value of ΔGAlign for an RNA (Fig. 1 B and C). While likely to hold in many cases, the proposed additivity of individual elements may find limitations in contexts where there are more extended networks of interactions such as those found near ribozyme active sites (e.g., ref. 54).
Although the goal of a complete quantitative model has not been attained for protein folding, the theories and formalisms are relevant and in some cases perhaps even more readily applied to studies of RNA folding (e.g., refs. 21, 55, and 56). These formalisms and concepts, such as the diffusion–collision model (57, 58), the contact order model (59), and hydrophobic collapse, have been proposed and tested (40, 60) and have provided useful analogies and led to evaluation of models for RNA folding (21, 55).
Analogous to the studies in RNA presented here, Pace and coworkers (61–63) compared the effects of sequence variants on alpha helix stability in small model peptide systems to the effects of the same variants in proteins with the goal to develop these peptide systems as modular pieces whose behavior could be extrapolated to proteins. Most simply, they asked whether a mutation that affects the propensity of a sequence to form an alpha helix, say decreasing this propensity, destabilizes overall folding of the protein by the same amount. This prediction bore out for mutations in exposed residues on the protein, but this simple rule is not followed for a buried side chain, as desolvation and interactions with neighboring residues affect the energetic contribution of these sides chains to folding (51, 64–66). As noted above, this approach and potential modularity may be more applicable to RNA tertiary structures given their limited packing interfaces and the sparse distribution of tertiary contacts throughout a structure.
Indeed, our results support the model that an RNA tertiary motif has the same energetic properties in different contexts. Specifically, we have shown that three out of four point mutations in the TL/TLR have the same energetic effects across different structural contexts and in different ion environments. Most directly, the kinetic and thermodynamic effects of these point mutations can be used to test whether TL/TLR formation is rate limiting in the folding or assembly of structured RNAs, as the rate of overall folding or assembly should be affected by a predictable amount if formation or dissolution of the contact is the rate-determining step in the process.
The principles of the reconstitution hypothesis are readily extendable to account for more complex behaviors such as folding cooperativity, whereby two contacts are thermodynamically coupled in their formation. When two contacts form cooperativity, like the two tertiary contacts of P4–P6 RNA (67), their successive formation can be accounted for by the reconstitution hypothesis by iteratively calculating the resultant conformational ensembles of the RNA following formation of a contact (represented by a new ΔG′HJH term). This recalculation will reflect the cooperativity between two contacts, as the conformational ensemble following the formation of a tertiary is more favorable (or less favorable in the case of negative cooperativity) for the next tertiary contact to form. For example, in the case of P4–P6, formation of the MC/MCR contact will predispose the new conformational ensemble (ΔG′HJH) to form the TL/TLR, and vice versa.
Broader characterization of more RNA modules using the approaches used herein will test whether this observed modularity applies to few or many RNA elements and can identify motifs and contexts that do not display energetic additivity. For tertiary motifs that are found to not be energetically modular, these motifs may contain multiple conformational states that could form different interactions depending on orientation or electrostatic requirements of the contact. Moreover, mutations themselves may lead to additional states to be explored, leading to nonadditivity in different contexts. Such a situation is likely for the GACAA mutant in the tetraloop studied herein, which gave a different energetic effect in one of the structural contexts consistent with the larger loop altering the conformational ensemble of the free loop and/or the conformation of the assembled tertiary motif. This result highlights the utility of these quantitative comparisons in uncovering sequence variants and motifs with different behaviors that can be targeted for further structural and energetic studies. Indeed, future work in characterizing motifs will be aided by additivity tests across a broad range of RNA contexts that can provide information about multiple orientations and conformational states. Overall, our results provide a framework to guide future work toward a quantitative and predictive understanding of RNA folding energetics and function.
Methods
Construction of Single-Molecule P4–P6 (smP4–P6) Variants.
SmP4–P6 variants were prepared using a five-piece splinted ligation protocol, as described previously (47, 67, 68). Variants were prepared with two pieces containing 5-amino-allyl-uridine modification to attach dyes for smFRET (Table S3 for sequences) and labeled using an NHS-ester reaction (69). All pieces were synthetic RNA and ordered from Integrated DNA Technologies, except for the middle section of the molecule (bases 163–234) due to its length, and were ordered as IDT Ultramer DNAs, PCR amplified, transcribed, and purified. To obtain homogenous 3′ ends following transcription, a DNAzyme complementary to the 3′ end of the transcript was used during transcription (70) with a polynucleotide kinase treatment following to remove the 2′,3′-cyclic phosphate left from DNAzyme cleavage. The ligation reaction included all five RNA pieces, added at ∼0.4 μM. The final ligated product was purified using a denaturing polyacrylamide gel [8% PAGE, 8 M urea, 1×TBE: 89 mM Tris (pH 7.4), 89 mM Boric Acid, 2 mM EDTA]. Ligated bands corresponding to the full-length ligated molecule were determined by scanning the gel with a Typhoon 9400 (GE Healthcare) at the Cy3 and Cy5 excitation wavelengths. Full-length molecules were gel purified and eluted using three freeze–thaw cycles and evaluated for homogeneity in folding and length on a native (8% acrylamide, 10 mM Mg2+) and denaturing polyacrylamide gel (8% PAGE, 8 M urea, 1×TBE).
Table S3.
DNA and RNA sequences for constructing smP4–P6 and TL/TLRiso constructs
| Construct | Variant | Name | Sequence* |
| P4–P6 | WT | 5′ piece (104–148)† | rGrGrArArUrUrGrCrGrGrGrArArArGrGrGrGrUrCrArArCrArGrCrCrGrUrUrCrArGrUrArCrCrArArGrUrCrUrCrArGrG |
| WT | Middle piece (163–234)‡ | AGTAATACGACTCACTATAGGCCTTGCAAAGGGTATGGTAATAAGCTGACGGACATGGTCCTAACCACGCAGCCAAGTCCTAAGTCAACAGGGGTGCGAG | |
| A225U | Middle piece (163–234)‡ | AGTAATACGACTCACTATAGGCCTTGCAAAGGGTATGGTAATAAGCTGACGGACATGGTCCTAACCACGCAGCCAAGTCCTTAGTCAACAGGGGTGCGAG | |
| A226U | Middle piece (163–234)‡ | AGTAATACGACTCACTATAGGCCTTGCAAAGGGTATGGTAATAAGCTGACGGACATGGTCCTAACCACGCAGCCAAGTCCTATGTCAACAGGGGTGCGAG | |
| C223U | Middle piece (163–234)‡ | AGTAATACGACTCACTATAGGCCTTGCAAAGGGTATGGTAATAAGCTGACGGACATGGTCCTAACCACGCAGCCAAGTCTTAAGTCAACAGGGGTGCGAG | |
| WT | 3′ piece (250–261 + tail)† | /5Phos/rGrGrA rUrGrC rArGrU rUrCrA rArCrC rArArA rArUrC rArArC rCrUrA rArArA rCrUrU rArCrA rCrArU | |
| WT | Cy3 oligo (149–162)§ | GGAAAC(5-N-U)UUGAGAU | |
| GACAA | Cy3 oligo (149–162)§ | GGACAAC(5-N-U)UUGAGAU | |
| WT | Cy5 oligo (235–249)§ | AUCUUC(5-N-U)GUUGAUAU | |
| WT | Surface attachment‡ | Biotin-TGTGTAAGTTTTAGGTTGATTTTGGT | |
| TL/TLRIso | WT | Loop† | 5AmMC6/rGrGrCrGrArArArGrCrCrArArArArArArArCrGrUrGrUrCrGrUrCrCrUrArArGrUrCrGrGrCrGrC |
| A225U | Loop† | 5AmMC6/rGrGrCrGrArArArGrCrCrArArArArArArArCrGrUrGrUrCrGrUrCrCrUrUrArGrUrCrGrGrCrGrC | |
| A226U | Loop† | 5AmMC6/rGrGrCrGrArArArGrCrCrArArArArArArArCrGrUrGrUrCrGrUrCrCrUrArUrGrUrCrGrGrCrGrC | |
| C223U | Loop† | 5AmMC6/rGrGrCrGrArArArGrCrCrArArArArArArArCrGrUrGrUrCrGrUrCrUrUrArArGrUrCrGrGrCrGrC | |
| GACAA | Loop† | 5AmMC6/rGrGrCrGrArCrArArGrCrCrArArArArArArArCrGrUrGrUrCrGrUrCrCrUrArArGrUrCrGrGrCrGrC | |
| WT | Base† | 5AmMC6/rGrCrGrCrCrGrArUrArUrGrGrArCrGrArCrArCrGrCrCACCAAAATCAACCTAAAACTTACACA |
Underlined residues indicate mutations.
Ordered as a synthetic RNA (IDT) and gel purified. Presented in IDT order nomenclature.
Ordered as a synthetic DNA (IDT) and transcribed.
Ordered as a synthetic RNA (Dharmacon), where (5-N-U) designates the 5-amino allyl-uridine.
Preparation of Isolated TL/TLR.
RNAs containing a 3′ primary amine were ordered from IDT for each WT and mutant TL/TLRiso (Table S3). The constructs were labeled with Cy3b/Cy5 with an NHS-ester reaction (69) and gel purified.
SmFRET Experiments.
SmFRET experiments were performed on a custom-built total internal reflection (TIRF) microscope at room temperature (23 °C). Molecules were excited with diode-pumped solid-state green laser (532 nm; Gem, laser quantum) and their fluorescence captured with an Andor Ixon Ultra Camera with an CAIRN Optosplit II to separate donor and acceptor fluorescence emissions. Before imaging, molecules were annealed to a DNA oligonucleotide containing a 5′ biotin (5′ biotin AACCAAAATCAACCTAAAACTTACACA-3′) at 50 °C for 15 min, cooled at 0.1 °C/min to room temperature, and diluted to 10–100 pM for imaging on quartz slides derivatized with biotin-BSA (1 mg/mL; Sigma-Aldrich) and streptavidin (0.1 mg/mL; Sigma-Aldrich). To stabilize dyes from bleaching and blinking, a protocatechuic acid/protocatechuate-3,4-dioxygenase (PCA/PCD) (Sigma-Aldrich) oxygen scavenging system with trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid; Sigma-Aldrich) was used. Buffers used in smFRET measurements contained 1–5 mM MgCl2 or 20–100 mM BaCl2, 50 mM potassium-Mops, pH 8.0, 100 mM KCl, 2 mM PCA, 0.01 units per mL PCD, and 1 mM trolox. SmFRET data were collected at an acquisition rate between 50 and 330 frames per second and analyzed using the SMART data analysis package (71).
Data Analysis.
Folding rate and equilibrium constants were inferred from smFRET traces by fitting donor and acceptor intensities using a HMM-based algorithm to a two-state model with a single unfolded and single folded state (21, 71). This analysis extracts transition probabilities between each state while taking into account the noise observed in each intensity channel. Criteria for trace selection were described previously (72); briefly, traces were selected based on exhibiting all of the following: (i) single-step photobleaching, (ii) anticorrelated donor and acceptor channels, (iii) total intensity consistent with a single molecule, and (iv) stable total intensity.
All reported rate and equilibrium constants are the mean of the log distribution of the values for each molecule. Reported errors are the 95% confidence intervals of the mean from bootstrapping parameters from individual molecules. The bootstrapped confidence intervals are asymmetric, so the greater of the two values is reported in the main text for simplicity (Tables S1 and S2). Both errors are reported in Dataset S1. Measurement error was determined to be the average of all confidence intervals for all measurements, which was 0.2 kcal/mol. A cutoff for signal-to-noise (SNR) of 0.75 is imposed for all P4–P6 smFRET data and 0.5 for the isolated TL/TLR, reflecting differences in the change in FRET observed for the two different molecules.
Defining ΔΔG‡Fold.
The effect of each mutation on the folding and unfolding rate constant is defined as:
The relationships between the rate constants effects between P4–P6 and TL/TLRiso in Fig. 6 were fit to a linear model where the slope is constrained to m = 1 (i.e., the model y = x + b). The goodness of these fits was determined by the rmsd of the orthogonal distance of the data to the line y = x. The coefficient of determination for a linear model (R2) was computed by the square of a Pearson correlation coefficient for two independent variables:
Supplementary Material
Acknowledgments
We thank members of the D.H. laboratory for discussion and critical advice throughout this work. This work was supported by the NIH Grant P01GM066275. N.B. was supported by the NSF graduate fellowship. M.G. was supported in part by NIH Training Grant GM008412 in biotechnology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703507114/-/DCSupplemental.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Noller HF. RNA structure: Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 5.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Rashid R, Wang K, Shan S-O. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science. 2010;328:757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parks JW, Stone MD. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun. 2014;5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner DH, Sugimoto N, Freier SM. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 10.Tinoco I, Jr, et al. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 11.Anderson CF, Record MT., Jr Salt-nucleic acid interactions. Annu Rev Phys Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 12.Xia T, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 13.Heigwer F, Kerr G, Boutros M. E-CRISP: Fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 14.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 15.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Chai D, Fraser ME, Zimmerly S. Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS One. 2012;7:e49225. doi: 10.1371/journal.pone.0049225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 18.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 19.Thomson JB, Lilley DM. The influence of junction conformation on RNA cleavage by the hairpin ribozyme in its natural junction form. RNA. 1999;5:180–187. doi: 10.1017/s1355838299981670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bisaria N, et al. Kinetic and thermodynamic framework for P4-P6 RNA reveals tertiary motif modularity and modulation of the folding preferred pathway. Proc Natl Acad Sci USA. 2016;113:E4956–E4965. doi: 10.1073/pnas.1525082113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 23.Tyagi R, Mathews DH. Predicting helical coaxial stacking in RNA multibranch loops. RNA. 2007;13:939–951. doi: 10.1261/rna.305307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murchie AI, Thomson JB, Walter F, Lilley DM. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- 25.Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 26.Chu VB, et al. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herschlag D, Allred BE, Gowrishankar S. From static to dynamic: The need for structural ensembles and a predictive model of RNA folding and function. Curr Opin Struct Biol. 2015;30:125–133. doi: 10.1016/j.sbi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hashimi HM, Walter NG. RNA dynamics: It is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohng S, et al. Conformational flexibility of four-way junctions in RNA. J Mol Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Shi X, Huang L, Lilley DMJ, Harbury PB, Herschlag D. The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol. 2016;12:146–152. doi: 10.1038/nchembio.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon L, Bascom G, Andricioaei I, Al-Hashimi HM. A general method for constructing atomic-resolution RNA ensembles using NMR residual dipolar couplings: The basis for interhelical motions revealed. J Am Chem Soc. 2013;135:5457–5466. doi: 10.1021/ja400920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klostermeier D, Millar DP. Energetics of hydrogen bond networks in RNA: Hydrogen bonds surrounding G+1 and U42 are the major determinants for the tertiary structure stability of the hairpin ribozyme. Biochemistry. 2002;41:14095–14102. doi: 10.1021/bi025551b. [DOI] [PubMed] [Google Scholar]
- 34.Silverman SK, Cech TR. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 35.Doherty EA, Doudna JA. The P4-P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry. 1997;36:3159–3169. doi: 10.1021/bi962428+. [DOI] [PubMed] [Google Scholar]
- 36.Cate JH, et al. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 37.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takamoto K, et al. Principles of RNA compaction: Insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 39.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5:1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 41.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 42.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Travers KJ, Boyd N, Herschlag D. Low specificity of metal ion binding in the metal ion core of a folded RNA. RNA. 2007;13:1205–1213. doi: 10.1261/rna.566007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebala M, Bonilla S, Bisaria N, Herschlag D. Does cation size affect occupancy and electrostatic screening of the nucleic acid ion atmosphere? J Am Chem Soc. 2016;138:10925–10934. doi: 10.1021/jacs.6b04289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc Natl Acad Sci USA. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenfeld M, Herschlag D. Measuring the energetic coupling of tertiary contacts in RNA folding using single molecule fluorescence resonance energy transfer. Methods Enzymol. 2010;472:205–220. doi: 10.1016/S0076-6879(10)72009-7. [DOI] [PubMed] [Google Scholar]
- 48.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 49.Downey CD, et al. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano L, Sancho J, Hirshberg M, Fersht AR. Alpha-helix stability in proteins. I. Empirical correlations concerning substitution of side-chains at the N and C-caps and the replacement of alanine by glycine or serine at solvent-exposed surfaces. J Mol Biol. 1992;227:544–559. doi: 10.1016/0022-2836(92)90906-z. [DOI] [PubMed] [Google Scholar]
- 51.Minor DL, Jr, Kim PS. Context is a major determinant of beta-sheet propensity. Nature. 1994;371:264–267. doi: 10.1038/371264a0. [DOI] [PubMed] [Google Scholar]
- 52.Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA. 2014;20:331–347. doi: 10.1261/rna.042747.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forconi M, Sengupta RN, Piccirilli JA, Herschlag D. A rearrangement of the guanosine-binding site establishes an extended network of functional interactions in the Tetrahymena group I ribozyme active site. Biochemistry. 2010;49:2753–2762. doi: 10.1021/bi902200n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sosnick TR, Pan T. Reduced contact order and RNA folding rates. J Mol Biol. 2004;342:1359–1365. doi: 10.1016/j.jmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Woodson SA. Folding mechanisms of group I ribozymes: Role of stability and contact order. Biochem Soc Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 57.Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 58.Karplus M, Weaver DL. Protein folding dynamics: The diffusion-collision model and experimental data. Protein Sci. 1994;3:650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 60.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 61.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Myers JK, Pace CN, Scholtz JM. A direct comparison of helix propensity in proteins and peptides. Proc Natl Acad Sci USA. 1997;94:2833–2837. doi: 10.1073/pnas.94.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Myers JK, Smith JS, Pace CN, Scholtz JM. The alpha-helix of ribonuclease T1 as an independent stability unit: Direct comparison of peptide and protein stability. J Mol Biol. 1996;263:390–395. doi: 10.1006/jmbi.1996.0583. [DOI] [PubMed] [Google Scholar]
- 64.Gao J, Bosco DA, Powers ET, Kelly JW. Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity substantially stabilizes proteins. Nat Struct Mol Biol. 2009;16:684–690. doi: 10.1038/nsmb.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albeck S, Unger R, Schreiber G. Evaluation of direct and cooperative contributions towards the strength of buried hydrogen bonds and salt bridges. J Mol Biol. 2000;298:503–520. doi: 10.1006/jmbi.2000.3656. [DOI] [PubMed] [Google Scholar]
- 66.Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19:631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solomatin SV, Greenfeld M, Herschlag D. Implications of molecular heterogeneity for the cooperativity of biological macromolecules. Nat Struct Mol Biol. 2011;18:732–734. doi: 10.1038/nsmb.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenfeld M, Herschlag D. Fluorescently labeling synthetic RNAs. Methods Enzymol. 2013;530:281–297. doi: 10.1016/B978-0-12-420037-1.00015-4. [DOI] [PubMed] [Google Scholar]
- 70.Schubert S, et al. RNA cleaving ‘10-23’ DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003;31:5982–5992. doi: 10.1093/nar/gkg791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greenfeld M, Pavlichin DS, Mabuchi H, Herschlag D. Single Molecule Analysis Research Tool (SMART): An integrated approach for analyzing single molecule data. PLoS One. 2012;7:e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi X, et al. Roles of long-range tertiary interactions in limiting dynamics of the Tetrahymena group I ribozyme. J Am Chem Soc. 2014;136:6643–6648. doi: 10.1021/ja413033d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.