Significance
AAA+ proteases are ring-shaped machines with a central narrow pore used to translocate protein substrates into a proteolytic chamber. Knotted proteins, however, may block the narrow pore by formation of an incompressible tight knot. Here, we determined how the archetypal AAA+ ClpXP protease degrades a protein substrate containing a trefoil knot. A trefoil knot is degraded quickly by ClpXP but not when it is fused to GFP placed downstream of the knot. Specifically, ClpXP releases partially degraded products when a tight knot is buttressed against GFP. Furthermore, degradation of substrates harboring linkers of different lengths between the knot and GFP allowed us to explore the dynamics of knot tightening and sliding and to postulate a new biological role for knotted proteins.
Keywords: protein degradation, knotted protein, AAA+ ATPase, translocation arrest, knot translocation
Abstract
ATP-dependent proteases translocate proteins through a narrow pore for their controlled destruction. However, how a protein substrate containing a knotted topology affects this process remains unknown. Here, we characterized the effects of the trefoil-knotted protein MJ0366 from Methanocaldococcus jannaschii on the operation of the ClpXP protease from Escherichia coli. ClpXP completely degrades MJ0366 when pulling from the C-terminal ssrA-tag. However, when a GFP moiety is appended to the N terminus of MJ0366, ClpXP releases intact GFP with a 47-residue tail. The extended length of this tail suggests that ClpXP tightens the trefoil knot against GFP, which prevents GFP unfolding. Interestingly, if the linker between the knot core of MJ0366 and GFP is longer than 36 residues, ClpXP tightens and translocates the knot before it reaches GFP, enabling the complete unfolding and degradation of the substrate. These observations suggest that a knot-induced stall during degradation of multidomain proteins by AAA proteases may constitute a novel mechanism to produce partially degraded products with potentially new functions.
Proteins with knots represent a distinctive subset of structures, in which the polypeptide chain is threaded to create complex topologies with several physical and biological consequences (1, 2). The presence of a knot slows down protein folding dynamics (3–5), confers thermodynamic stability (6), and promotes certain molecular motions required for efficient catalysis in some enzymes (7). On the other hand, knots have been proposed to impair key biological processes that involve the passage of proteins through narrow pores and channels (2, 8); however, experimental evidence supporting this hypothesis is missing.
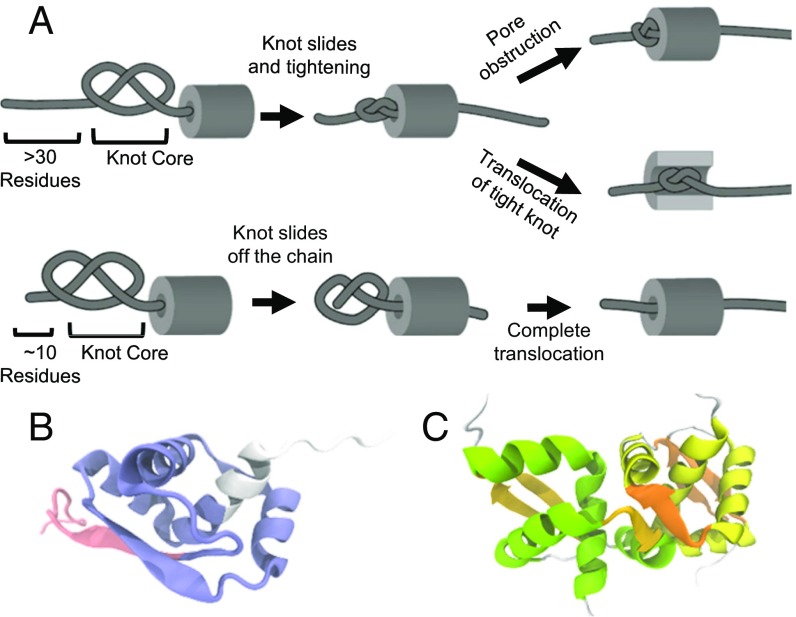
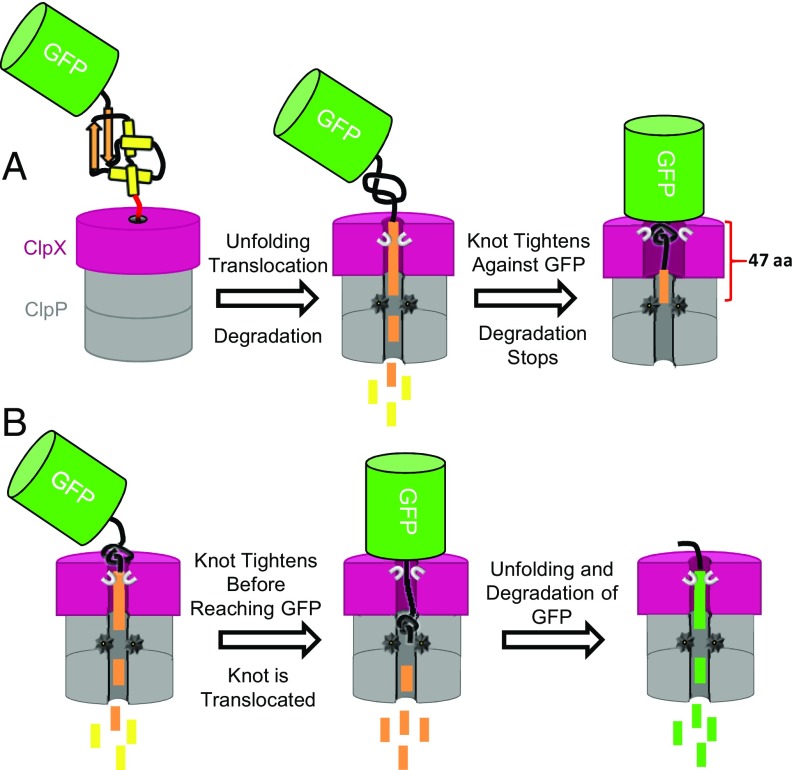
Protein translocation through narrow pores is a key process required for controlled degradation of proteins (9) and their transfer between cellular compartments (10). During these processes, proteins must be actively unraveled before translocation, because the channels are too narrow to accommodate a folded structure. Several translocation systems use ATP hydrolysis to generate the force required for unfolding and movement of proteins (9). Molecular dynamics simulations have been used to study the force-driven translocation of several knotted proteins through rigid cylinders or pores (11–15). In these simulations, the knot in the chain is seen to shrink and slide toward the free terminal end of the protein (Fig. 1A). If the knot shrinks enough before it reaches the end of the chain, it forms a tight uncompressible knot (Fig. 1A, Top). Under tension, these tight knots can no longer slide (11, 12, 14–16). If the pore’s diameter is smaller than the size of the tight knot, the pore is blocked (12–15), otherwise the tight knot is translocated through the pore, albeit slowly (11) (Fig. 1A, Top). Alternatively, the protein can be translocated if the knot slides off the chain before tightening (Fig. 1A, Bottom). The latter scenario occurs commonly in simulations of proteins whose structures contain shallow knots; that is, the segment of the chain that threads the knot core is less than 10 residues long (14–16) (Fig. 1A, Bottom).
Fig. 1.
Key events during the translocation of a knot through a pore determined by molecular dynamics. (A) The core of a knot is defined by the minimum residues that conform the knot in the structure of the protein. A deep (Top) or a shallow (Bottom) knot depends on the number of residues threading the knot core. During translocation of deep knots, a tight knot is formed more often than during the translocation of a shallow knot. A tight knot obstructs the translocation of the chain depending on the pore diameter and the knot size. Shallow knot tends to slide off the terminal, and the protein is translocated completely. (B) The backbone topology of MJ0366 creates a 31 or trefoil knot in its 3D structure (PDB ID code 2EFV). The core of the knot of MJ0366 (residues 17–78 in blue) is threaded by 14 residues at the N (red) and 12 residues at the C terminus (white). (C) Structure of the dimer of MJ0366. The dimeric interface is created by residues that conform the knot core of MJ0366. Each subunit is colored in yellow or green.
However, rigid cylinders do not necessarily represent well the particular geometry and flexibility of the pores and channels of ring-shaped ATP-dependent proteases, such as ClpXP from Escherichia coli. Due to its extensive biochemical and structural characterization, ClpXP has been used as a model system to study the mechanisms of protein unfolding and translocation by prokaryotic and eukaryotic ATP-dependent proteases (for review, see refs. 9 and 17). ClpXP is therefore also ideally suited to investigate the mechanism of knot translocation and degradation by these processing machines. Like most energy-dependent proteases, ClpXP consists of two major components: (i) the AAA+ ATPase ClpX, a homo-hexameric ring with an axial processing pore, and (ii) the barrel-shaped ClpP peptidase, which contains proteolytic active sites sequestered in an internal chamber. One of the recognition motifs mediating ClpXP degradation of protein substrates is the ssrA-tag, an 11-amino acid sequence that is appended to the C terminus of incompletely translated proteins at the ribosome (9, 17). Single-molecule force-spectroscopy experiments show that ClpXP unfolds its ssrA-tagged protein substrates by mechanically pulling from the C terminus and forcing the protein to pass through the narrow ClpX pore (18, 19). Since the proteolysis chamber is accessible only through this pore, degradation of structured proteins can occur only after unfolding and translocation of the polypeptide into the associated ClpP chamber (9, 17).
Here we explore the effect of the shallow trefoil (31) knot present in the small protein MJ0366 from Methanocaldococcus jannaschii (Fig. 1 B and C) on the operation of ClpXP. We determined that ClpXP completely degrades the MJ0366-ssrA substrate. However, when the green fluorescence protein (GFP) is appended to the N terminus of MJ0366, ClpXP is unable to degrade this fusion protein, releasing instead a partially degraded product with an intact GFP moiety. Further analysis revealed that this protection occurs because ClpXP translocation pushes the 31 knot against GFP, which prevents its unfolding and further progress of the motor. Interestingly, the insertion of unfolded linkers of different lengths between MJ0366 and GFP modifies this scenario and leads to an abrupt decrease of the GFP protection beyond certain lengths. The degree of GFP protection and the pattern of partially degraded products with an intact GFP domain observed for substrates of different linker lengths reveal additional fates for the 31 knot-containing MJ0366: (i) Once the unfolding of MJ0366 has begun, its knot does not slide for more than 36 residues before it becomes tight, and (ii) the MJ0366 knot can be translocated through the ClpXP pore if the knot is tightened before reaching GFP. Noteworthy, these observations are in agreement with molecular-dynamics simulations of knot translocation (13, 14).
Taken together, our results constitute an experimental insight into how knotted structures impair protein translocation through narrow pores. Moreover, our results indicate that a 31 knot present in multidomain protein substrates could create a barrier that ClpXP or related ATP-dependent proteases cannot overcome, leading to the release of partially degraded products that may be harmful for the cell or possess new biological activities.
Results
A Tight Knot and a Folded Domain Cooperate to Arrest Translocation by ClpXP.
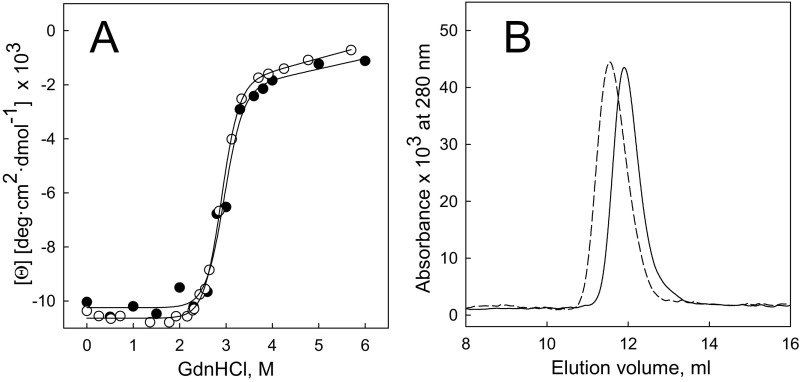
The knotted protein MJ0366 has been characterized as a stable homodimer (20) whose dimer interphase is formed by interactions of secondary structure elements that are involved in the organization of the trefoil knot in each subunit (Fig. 1C). To make MJ0366 a substrate for ClpXP, we appended a C-terminal ssrA tag, preceded by a His6 sequence for affinity purification, and tested whether the resulting MJ0366-His6-ssrA folded correctly. We found that the addition of the His6-ssrA tag to the C terminus of MJ0366 changes neither the thermodynamic stability (Fig. S1A and Table S1) nor the oligomeric state of the protein (Fig. S1B and Table S1), indicating that the knot and the native state are correctly adopted in the tagged construct.
Fig. S1.
Stability and hydrodynamics properties of MJ0366 and MJ0366-ssrA. (A) Stability curves of MJ0366 (open circle) and MJ0366-ssrA (filled circle) measured with circular dichroism at 220 nm as a function of guanidinium chloride (GdnHCl). The lines represent a fit of the experimental data to two-state model of unfolding. (B) Chromatograms of MJ0366 (solid line) and MJ0366-ssrA (dotted line) determined by size exclusion chromatography and followed at 280 nm. The apparent molecular mass and parameters of stability for each protein are indicated in Table S1.
Table S1.
Apparent molecular mass and protein stability for MJ0366 and MJ0366-ssrA
| Protein | Theoretical molecular mass for a dimer, kDa | Experimental molecular mass,* kDa | ΔGw,† kcal/mol | m value,† kcal mol−1 M−1 |
| MJ0366-ssrA | 27.8 | 24 | 9 ± 1 | 2.9 ± 0.6 |
| MJ0366 | 23 | 22 | 9 ± 0.3 | 3.1 ± 0.1 |
Show are errors obtained from the fitted procedure.
Values were determined by size exclusion chromatography.
Determinations were obtained following the circular dichroism as a function of the concentration of Guanidine hydrochloride analyzed in terms of a two-state N ↔ U transition model.
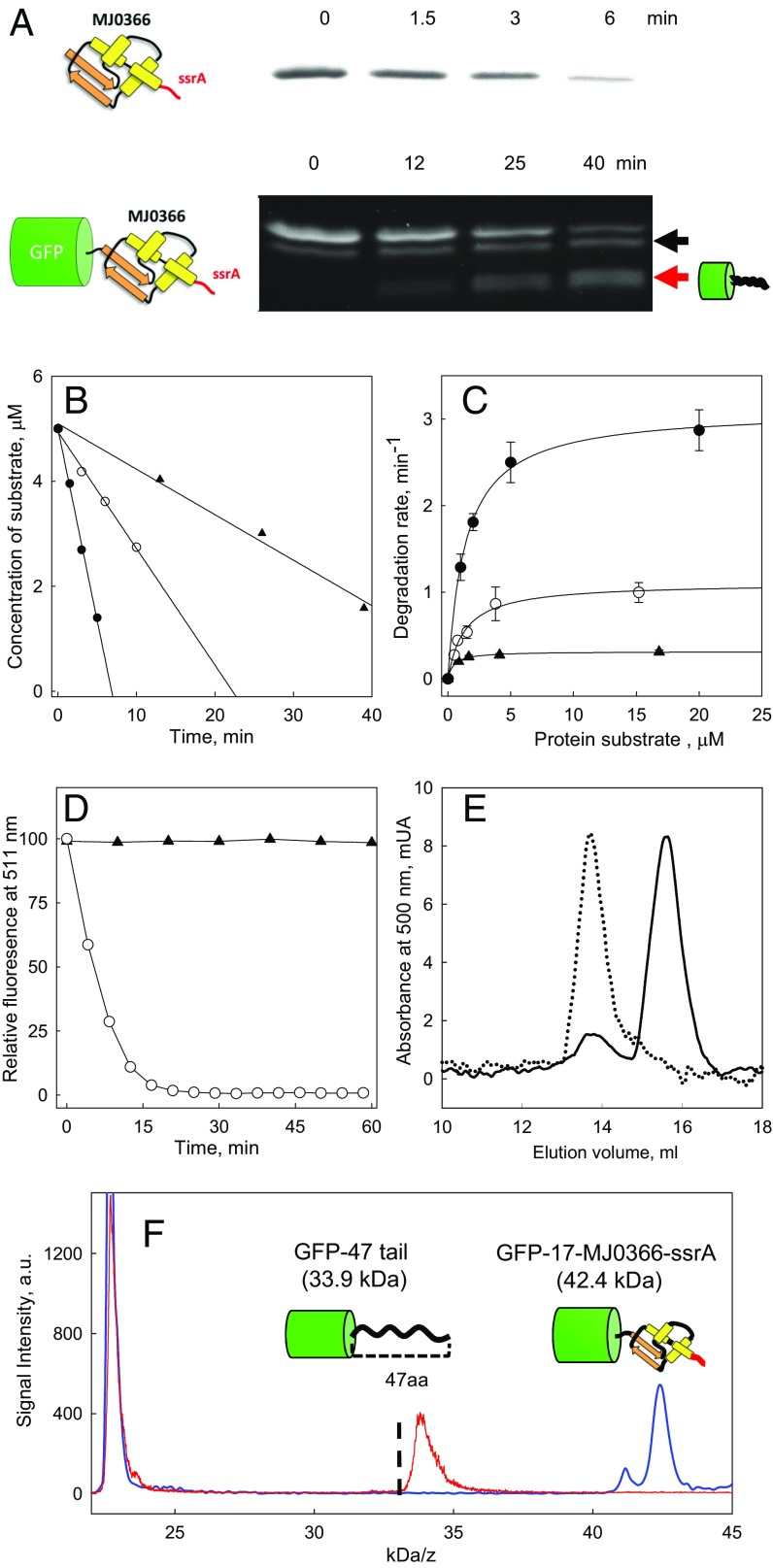
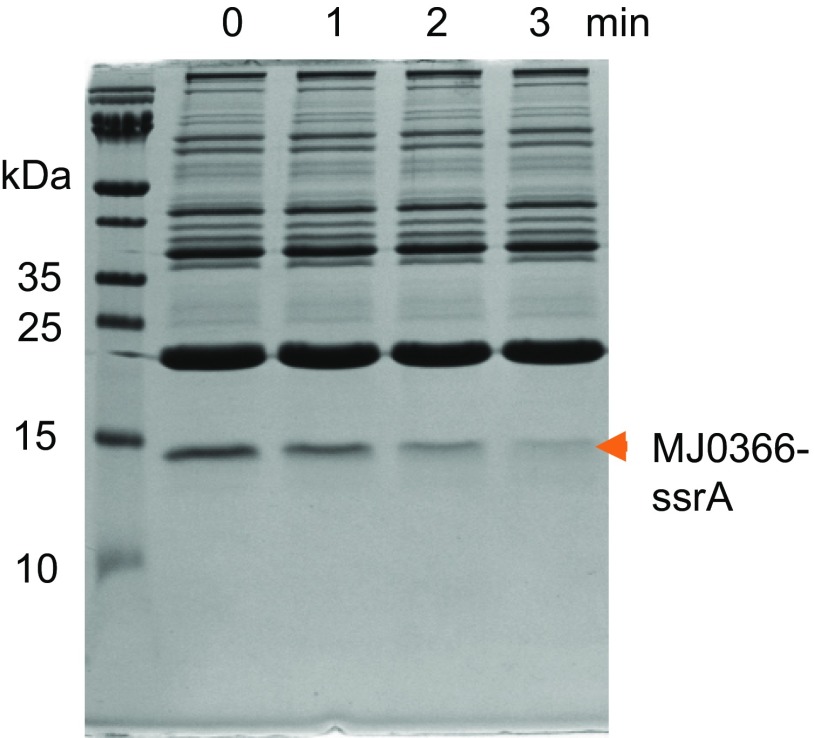
Fig. 2A depicts the ClpXP-induced degradation of MJ0366-ssrA, as followed by SDS/PAGE. The decrease in band intensity for the substrate was found to be linear during early time points (Fig. 2B), and no partially degraded products were detected (Fig. S2). Michaelis–Menten analysis of the steady-state degradation revealed kinetic parameters of KM = 1.4 ± 0.1 µM and kcat = 3.1 ± 0.05 s−1 (Fig. 2C). These values are similar to those described for other quickly degraded protein substrates of ClpXP (21), and the knot in MJ0366 hence does not confer local mechanical resistance against ClpXP-induced unfolding.
Fig. 2.
Kinetics of degradation of protein substrates by ClpXP. (A) Degradation kinetics of MJ0366-ssrA and GFP-17-MJ0366-ssrA detected by Coomasie stain and in-gel fluorescence, respectively. A red arrow indicates a fluorescent band corresponding to a partially degraded product formed during GFP-17-MJ0366-ssrA degradation. The back arrow indicates the substrate with a partially or totally proteolyzed ssrA tag. (B) Initial rates of degradation of MJ0366-ssrA (filled circle), GFP-17-MJ0366-ssrA (filled triangle), and GFP-ssrA (open circle) by quantification of the band of the intact substrate in SDS/PAGE gels. (C) Michaelis–Menten plots for ClpXP degradation of GFP-ssrA (open circle), MJ0366-ssrA (filled circle), and GFP-17-MJ0366-ssrA (filled triangle). Error bars show the SE calculated from three independent measurements. (D) Kinetics followed by total fluorescence of GFP-17-MJ0366-ssrA (filled triangle) and GFP-ssrA (open circle) at 511 nm. (E) Size exclusion chromatograms following the absorbance at 500 nm of GFP-17-MJ0366-ssrA (dotted line) and GFP-17-MJ0366-ssrA incubated by 2 h in the presence of ClpXP (continuous line). (F) MALDI-MS of GFP-17-MJ0366-ssrA and the partially degraded substrate. Samples of GFP-17-MJ0366-ssrA were incubated with ClpXP by 2 h with ATP (red spectrum) or without ATP (blue spectrum). Indicated in the graph is the size of the species identified; the partially degraded product corresponds to GFP plus a tail of 47 ± 2 residues. The dotted line represents the expected size for GFP harboring a tail of 37 residues (32.9 kD) reported for multidomain substrates without knots.
Fig. S2.
MJ0366-ssrA is degraded by ClpXP without forming partially degraded products. The kinetic of degradation of MJ0366 was followed by Coomassie staining on SDS/PAGE gel. There is not a visible partially degraded product of MJ0366-ssrA.
MJ0366 possesses a shallow knot that could easily unthread through the N terminus upon unfolding (Fig. 1B). Shallow knots in polypeptide chains often slide off through their free terminal ends during the translocation process in molecular-dynamics simulations (Fig. 1A, Bottom). Sliding off occurs because the probability that a knot becomes tight is low when the distance between the knotted core and the end of polypeptide chain is less than 30 residues (12–14). In the case of MJ0336, the free N terminus that threads the knot core comprises only 14 residues (Fig. 1B), and thus, it is likely that the knot slides off before tightening (13, 14). Accordingly, the fast degradation rate for MJ0366-ssrA described above may not provide information about how ClpXP negotiates the presence of a tight knot.
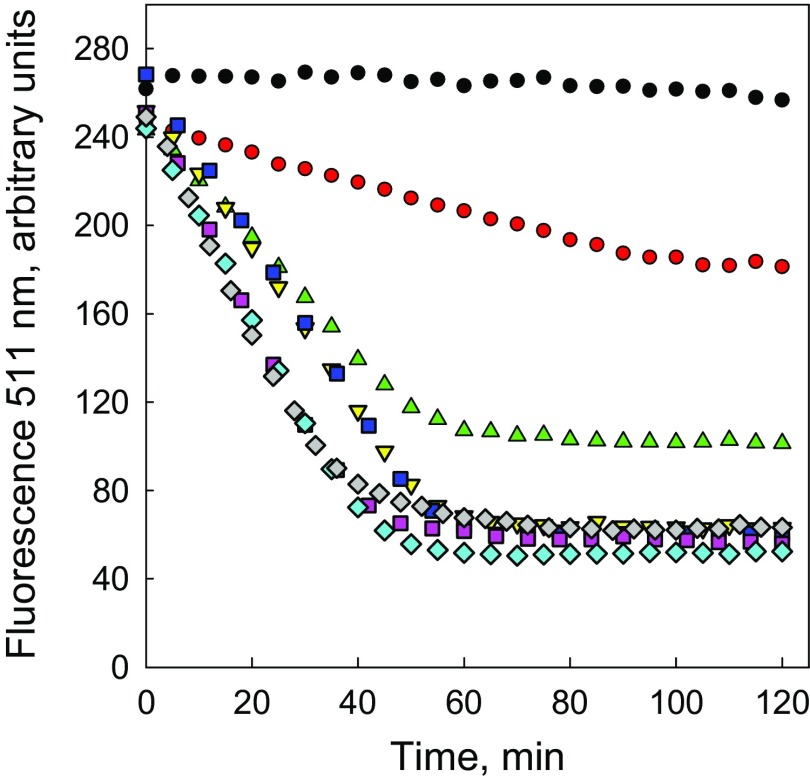
To determine the effect of a tight knot on the degradation by ClpXP, we appended GFP to the N terminus of MJ0366-ssrA, resulting in the substrate GFP-17-MJ0366-ssrA, where “17” denotes the total number of residues between the knot core of MJ0366 and GFP (14 residues provided by the N terminus of MJ0366 and 3 residues from the linker between the proteins; see substrate sequences in Dataset S1). We found that the knotted protein in the fusion construct GFP-17-MJ0366-ssrA still folds correctly, as shown by its dimerization properties tested by subunit exchange (Fig. S3). Notably, the knotted protein completely protects GFP from ClpXP degradation. In the presence of ClpXP and saturating ATP concentrations, the total fluorescence of GFP-17-MJ0366-ssrA remained invariant up to 1 h (Fig. 2D), even though ClpXP binds this substrate (Fig. S4A). In contrast, under similar experimental conditions, the fluorescence of a GFP substrate lacking the knotted protein MJ0366—that is, GFP-ssrA—rapidly decreased (Fig. 2D). To determine whether GFP protection results from a stabilization of MJ0366 by the fusion (with no substrate degradation) or involves unfolding of MJ0366 (with partial substrate degradation), we monitored substrate degradation by an in-gel fluorescence assay. Aliquots of the degradation sample were analyzed by SDS/PAGE without prior heating to preserve the fluorescence of GFP (22). As shown in Fig. 2A, the intensity of the fluorescent band corresponding to the full-length substrate GFP-17-MJ0366-ssrA decreased with time, while concomitantly a partially degraded fluorescent product appeared. The decrease in intensity for the fluorescent band of GFP-17-MJ0366-ssrA and the increase of the partially degraded fluorescent product occurs without a change in total fluorescence (Fig. 2D). We therefore conclude that although MJ0366 is unfolded and partially degraded by ClpXP, it still protects the downstream GFP from degradation. Indeed, size exclusion chromatography showed that 70–80% of the GFP-17-MJ0366-ssrA substrate is converted by ClpXP to a partially degraded product that retains GFP absorbance at 500 nm (Fig. 2E).
Fig. S3.
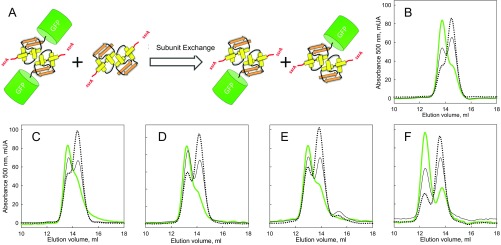
Dimeric state and subunit exchange of multidomain substrates of MJ0366. (A) Subunit exchange scheme. A homodimer of GFP-17-MJ0366-ssrA created by an interaction between domains of MJ0366 exchanges its subunits with a homodimer of MJ0366-ssrA (Left) to create two heterodimers of lower molecular mass (Right). Size exclusion chromatography of (B) GFP-17-MJ0366-ssrA, (C) GFP-23-MJ0366-ssrA, (D) GFP-58-MJ0366-ssrA, (E) GFP-78-MJ0366-ssrA, and (F) GFP-117-MJ0366-ssrA followed by GFP absorbance at 500 nm. The chromatograms in green (green line) are for the multidomain substrates, in solid line for the equimolar mix of the multidomain substrates with MJ0366-ssrA, and dotted line for the mix of the multidomain substrates with an eightfold molar excess of MJ0366-ssrA. The presence of the homodimer of MJ0366 is not observed in the chromatograms since the absorbance was followed at 500 nm.
Fig. S4.
GFP-17-MJ0366-ssrA is recognized by ClpXP from its degradation tag. (A) Degradation kinetics of GFP-ssrA in the presence of GFP-17-MJ0366-ssrA (filled circle) or absence (open circle). The inhibitory effect of GFP-17-MJ0366-ssrA disappears when cleaving the ssrA tag of GFP-17-MJ0366-ssrA with TEV protease (filled triangle). (B) The C-terminal end of GFP-17-MJ0366-ssrA has a recognition site for the tobacco etch virus protease (TEV), a hexahistidine tag followed by the ssrA degron. (C) The secondary fluorescent band of the substrate lacks the ssrA tag. Samples of GFP-17-MJ0366-ssrA display two bands that emit fluorescence: the full substrate, which can be proteolized by ClpXP, and a band of lower molecular mass (first lane), whose fluorescence remains intact in the presence of ClpXP (fourth lane). When the substrate is treated with the TEV protease to cleave the His6 and ssrA tag, observed is the formation of a single fluorescent band (second lane) unable to be degraded by ClpXP (third lane). Therefore, the secondary band of GFP-17-MJ0366-ssrA lacks part of C-terminal residues required to bind ClpXP.
The presence of partially degraded intermediates has been observed during the degradation of multidomain proteins by ClpXP (23–25). For instance, if ClpXP repeatedly fails to unfold the second domain of a multidomain substrate, the protease eventually releases a partially degraded product containing an intact second domain with a C-terminal tail (23–25). ClpXP displays a similar behavior when facing GFP-17-MJ0366-ssrA, since ClpXP generates a partially degraded product containing GFP. Importantly, GFP is not resistant to unfolding per se when fused to other moieties. GFP-TitinV15P-ssrA, for instance, is completely degraded by ClpXP (25). In contrast, MJ0366 completely protects GFP under similar ATP conditions.
A characteristic feature of the partially degraded products released by ClpXP is the presence of defined-length tails of 37–38 residues (24, 25). This number of residues is sufficient to span the distance between the pore entry of ClpX and the active sites in the ClpP peptidase (24, 25). The MALDI-TOF spectra of the partially degraded product of GFP-17-MJ0366-ssrA display a mass distribution with a median at 33.9 kDa. This mass corresponds to GFP plus a tail of 47 residues (GFP-47-tail, Fig. 2F), about 10 residues longer than the tail reported for unknotted multidomain substrates like GFP-titin-ssrA (25) or titin-titin-titin-ssrA (24). Since 13 residues are required to form an uncompressible trefoil knot (14, 16, 26), the additional 10 amino acids determined in the degradation product of GFP-17-MJ0366-ssrA can be rationalized if ClpXP translocation tightens the trefoil knot against GFP. This tightened knot may hinder the ability of the protease to unfold and degrade the fluorescent protein (Fig. 3A).
Fig. 3.
Model of degradation of knotted multidomain substrates by ClpXP. Two outcomes occur when ClpXP engages a knotted multidomain substrate. (A) The knot of MJ0366 slides and tightens against GFP, protecting GFP from the mechanical unfolding. Eventually, ClpXP releases a partially degraded product of GFP plus a tail of 47 residues. A tight knot increases the number of residues inside the channel of ClpXP from 37 to 38 to about 47: 13 residues required to create a 31 tight knot, minus the diameter of the knot itself (four residues or 1.6 nm) plus 36–37 residues to spam the distance between the pore entrance and the active site of ClpP. (B) The knot of MJ0366 gets tightened far from the GFP folded domain and is translocated through ClpXP pore, leading to GFP degradation.
Consistent with ClpXP stalling in front of GFP, we observed a significantly reduced multiple-turnover rate for GFP-17-MJ0366-ssrA. Based on the change in fluorescence intensity of the band for full-length GFP-17-MJ0366-ssrA in the gel, we determined an apparent kdeg for the MJ0366 domain of 0.31 ± 0.01 s−1 (Fig. 2C), which is 10-fold lower than the kdeg determined for the knotted protein alone (3.1 ± 0.01 s−1). As a control of our experimental procedures, the kinetic parameters for degradation of GFP, also obtained by in-gel fluorescence (Fig. 2C; KM = 1.3 ± 0.2 µM and kdeg = 1.1 ± 0.1 s−1), are very close to those reported in the literature (25). These results indicate that in the absence of GFP—that is, in the MJ0366-ssrA substrate—the knot is pushed and quickly undone, allowing ClpXP to translocate and degrade the protein at normal rates. In contrast, when GFP is placed at the N terminus—as in the GFP-17-MJ0366-ssrA substrate—the knot cannot escape through the N-terminal end but instead gets pushed against GFP and creates an unfolding barrier that ClpXP is unable to overcome. The multiple-turnover kinetics are therefore determined by the slow rate of ClpXP releasing the partially degraded substrate.
It should be noted that samples of GFP-17-MJ0366-ssrA not treated with ClpXP displayed two fluorescent bands (Fig. 2A). We established that the band of slightly lower molecular mass corresponds to a truncated GFP-17-MJ0366-ssrA substrate with an incomplete ssrA tag, which is not recognized by ClpXP and therefore not degraded (Fig. S4). In further experiments, the intensity of the smaller molecular mass band was used as a loading control to correct for variations in the concentration of added protein substrate.
Sliding and Translocation of a Knot by ClpXP.
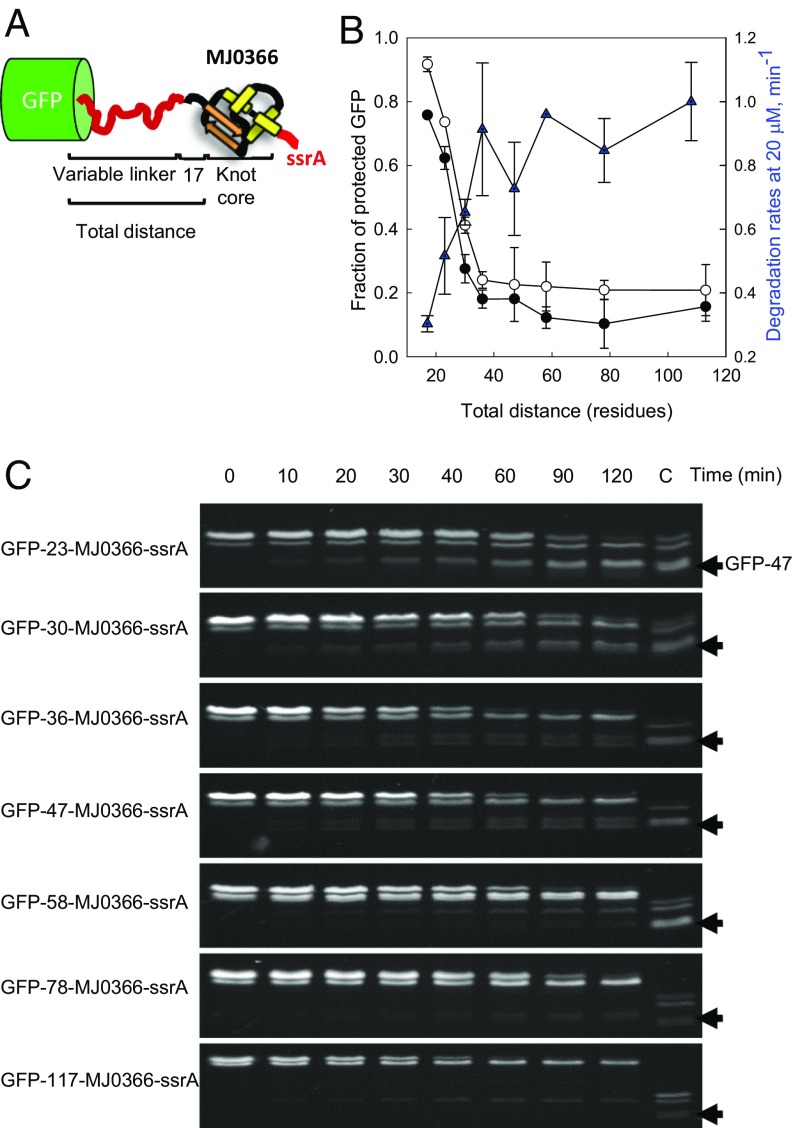
The size of the partially degraded GFP-tail product (Fig. 2F) and the kinetics of its formation by ClpXP (Fig. 2C) support the idea that GFP is protected by the presence of a knot buttressed up against its structure. During translocation of MJ0366, ClpXP slides the knot along the polypeptide until it meets the folded GFP moiety and the motor stalls (Fig. 3A). Therefore, the number of residues between GFP and the knot core of MJ3066 is too small to answer two important questions about the behavior of ClpXP in front on a knotted polypeptide: (i) How much can the knot slide along a polypeptide chain before it gets tightened, and (ii) can a knot tightened at a considerable distance from GFP be translocated through the axial pore of ClpXP? To address these questions, we increased the distance between GFP and the knot core in MJ0336, which should also increase the probability of tightening the knot before reaching GFP. Linkers of various lengths (Fig. 4A) were generated from a titin I27 mutant known to be unfolded under native conditions (27). Increasing the total distance significantly decreased the fraction of protected GFP, as determined by the residual GFP fluorescence measured after 120 min of degradation by ClpXP (Fig. 4B and Fig. S5). The residual fluorescence of GFP as a function of the total distance (Fig. 4B and Fig. S5) is well correlated with the population of the partially degraded product detected by size-exclusion chromatography (Fig. 4B and Fig. S6). Thus, as indicated in Fig. 4B, the rate of substrate degradation is the fastest and the degree of GFP protection is correspondingly the lowest when the total distance between GFP and the knot core is 36 residues (substrate GFP-36-MJ0366-ssrA).
Fig. 4.
Kinetics of degradation of substrates created with linkers of several lengths. (A) Diagram of the substrates with increasing distance between GFP and the knotted core of MJ0366. The total distance from the knot to GFP is defined by the number of residues in the linker plus 17 residues corresponding to N-terminal end of MJ0366 that threads the knot core of MJ0366. (B) The apparent degradation rate of GFP and GFP protection as a function of the total distance between GFP and the knot core of MJ3666. The protection of GFP was quantified by the kinetic amplitudes followed by total fluorescence of GFP (open circle) and by size exclusion chromatograms (filled circle). The apparent degradation rates (filled triangle) were determined by in-gel fluorescence decay of the band corresponding to the intact substrate at 20 µM of each substrate. Error bars represent the SE calculated from three independent measurements. (C) Degradation kinetics of substrates created with linkers of several lengths followed by in-gel fluorescence. The lane marked with C corresponds to a sample of GFP-17-MJ0366-ssrA incubated by 2 h with ClpXP to compare the electrophoretic mobility of the partially degraded product GFP-47-tail (black arrow). Linkers of 58 residues or smaller show a partially degraded product with similar electrophoretic mobility of the partially degraded product GFP-47-tail.
Fig. S5.
Degradation of multidomain substrates followed by GFP total fluorescence. The kinetics of degradation were followed at 511 nm (excitation 467 nm) using 0.3 µM ClpX6, 1.5 µM ClpP14, and 10 µM of the substrate GFP-17-MJ0366-ssrA (filled black circle), GFP-23-MJ0366-ssrA (filled red circle), GFP-30-MJ0366-ssrA (green triangle), GFP-36-MJ0366-ssrA (inverted yellow triangle), GFP-47-MJ0366-ssrA (purple square), GFP-58-MJ0366-ssrA (blue filled square), GFP-78-MJ0366-ssrA (blue diamond), or GFP-117-MJ0366-ssrA (green diamond). The amplitudes for each kinetic were corrected by the population of the nondegradable substrate determined by in-gel fluorescence (see Materials and Methods for calculation).
Fig. S6.
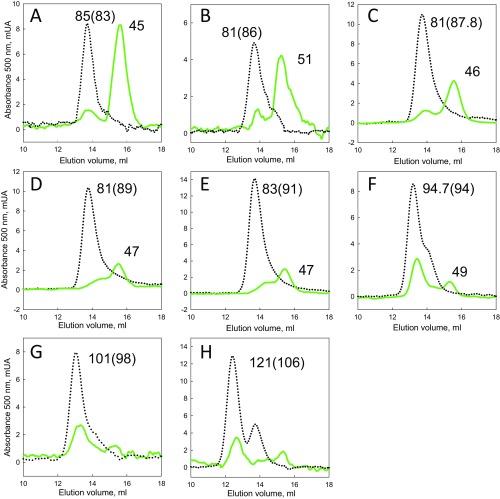
Observation of partially degraded products by size exclusion chromatography. Chromatograms determined for the substrates (A) GFP-17-MJ0366-ssrA, (B) GFP-23-MJ0366-ssrA, (C) GFP-30-MJ0366-ssrA, (D) GFP-36-MJ0366-ssrA, (E) GFP-47-MJ0366-ssrA, (F) GFP-58-MJ0366-ssrA, (G) GFP-78-MJ0366-ssrA, and (H) GFP-117-MJ0366-ssrA, incubated by 2 h with ClpXP plus ATP (green line) or in the absence of ATP (dotted line). The apparent molecular mass in kDa is shown over each peak. In parentheses is the theoretical molecular mass calculated by using the sequence for each substrate.
To determine whether the degree of GFP protection is correlated with the population of the GFP-47 intermediate, we followed the kinetics of degradation by in-gel fluorescence as a function of linker lengths, monitoring the disappearance of the full-length substrate and the appearance of the GFP-47 intermediated (Fig. 4C). When the total distance between the GFP and the knot core is extended to 36 residues (substrate GFP-36-MJ0366-ssrA), a sharp decrease of species with the electrophoretic mobility of the GFP-47 tail product is observed. This species disappears for substrates with linkers longer than 58 residues, without significant formation of other partially degraded products. Thus, when the total distance between the knot core and GFP is longer than ∼36 amino acids, substrates are degraded processively by ClpXP. These results indicate that the amino acid distance between the core of the knot and GFP plays an important role in modulating ClpXP processitvity and that a knot tightened before reaching GFP can be translocated through the ClpXP pore (Fig. 3B).
Discussion
Our results show that the knotted protein MJ0366 fused with an ssrA tag is easily unfolded by ClpXP and translocated through the machine’s pore. In close agreement, molecular-dynamics simulations of MJ0366 translocation through a rigid pore indicate that the protein is efficiently unfolded and translocated under forces of ∼30 pN (13), a magnitude that falls within the range of forces generated by ClpXP (18, 19).
Nevertheless, the structure of knots within proteins is very dynamic, since their dimensions and locations change with respect to their original position as a knotted protein is unfolded. For example, in silico simulations of a knotted protein being translocated through a pore too narrow to pass the folded polypeptide show that the knot shrinks and slides along the polypeptide toward the terminal end as translocation proceeds (refs. 11–15 and Fig. 1A, Top). This process is revealed when GFP is added as a downstream domain to the N terminus of the knotted protein, leading to a partially degraded substrate that comprises an intact GFP domain plus a tail of 47 residues. Additional support for this idea of a tightened knot in front of GFP is provided by the anomalous tail length of the GFP-47 product. It has been established that the amino acid distance between the proteolytic active site in ClpP and the pore entry in ClpX is 37 amino acids (24, 25), which is 10 amino acids shorter than the 47-residue tail observed here. Significantly, previous studies have shown that a tight knot comprises ∼13 amino acids (14, 16, 26). Therefore, we rationalize these observations with a scenario where the 47-residue tail is most likely the result of the ClpX-induced sliding of the knot along the polypeptide until it gets tightened and buttressed against GFP. Since a 47-residue tail accounts for about half of the sequence of MJ0366, clearly ClpXP is able to successfully unfold MJ0366 before the formation of the GFP-47 intermediate. These experimental observations rule out alternative explanations, such as a synergistic unfolding resistance of the GFP and MJ0366 moieties that impairs ClpXP processivity. Partial degradation by ClpXP can also occur due to reduced grip, when substrates contain slippery, Gly–Ala-rich sequences in front of a stably folded domain (28, 29). However, the constructs tested in our work do not exhibit slippery sequences adjacent to GFP (see sequences in Dataset S1), and it can be ruled out that the observed GFP protection originates from reduced grip.
How does the combination of a tight knot and a downstream GFP moiety stall the degradation by ClpXP? The strong protection of GFP suggests that the knot affects the translocation machinery either by direct contact with the translocating loops or by indirect deformation of the channel. X-ray structures of substrate-free ClpX indicate that the translocating pore loops are axially staggered with respect to each other along the translocation channel of ClpX (30). This distribution could allow interactions between pore loops near the top of the ClpX channel and the tight knot buttressed against GFP. A tight knot entering the translocation channel is certainly plausible given the flexibility of the ClpX pore (30, 31), and it would explain the 47-residue tail observed for the partially degraded GFP product: 36–37 residues normally required to span the distance between the pore entrance and the active sites of ClpP, minus 1.6 nm (or four residues in extended conformation) for the length of tight 31 knot itself, plus the 13 residues necessary to form this tight knot (14, 16, 26) (Fig. 3A).
A tight knot may prevent GFP unfolding by ClpXP, because it interferes with the translocation machinery and reduces the power of the motor—that is, the amount of work produced per unit time—by lowering the pulling force, pulling frequency, or both. It has previously been shown that to unfold GFP, ClpXP must first extract the C-terminal β-strand-11 out of the β-barrel and then quickly trap and unfold this intermediate to prevent its refolding back to native GFP (18, 25). Furthermore, it was found that increased bulkiness inside the ClpX pore lowers the pulling rate of the motor (32), whereas reduced bulkiness affects the force delivered to the substrate (32–34). The additional bulkiness of a tight knot inside the pore may thus either decrease the applied force to below the threshold necessary to overcome the mechanical barrier for β-strand extraction or render the pulling frequency too low for trapping the GFP unfolding intermediate. Further experiments are required to dissect the precise mechanism by which the structure and local dynamics of the downstream domain cooperate with a tight knot to block the translocation mediated by ClpXP.
We also explored the effect of increasing the distance between the MJ0366 and GFP on the degradation by ClpXP. Notably, the knotted domain can modulate the processivity of the protease when unfolded linkers of different lengths are inserted between MJ0366 and GFP (Fig. 4). According to molecular-dynamics simulations, a knot in folded proteins slides over short distances before it becomes tightened during translocation through a narrow pore (12–14). The sliding occurs as a stick-slip process until the knot reaches a nondiffusive tight state, in opposition to diffusible DNA knots capable of sliding smoothly over long distances (35, 36). Molecular-dynamics simulations show that from the initial event of unfolding to knot compaction, the knot core slides at most about 36 residues (refs. 13 and 14 and Fig. 1A). Our results are consistent with this behavior, since GFP protection becomes more and more apparent—the knot seems to reach GFP more often—when the number of residues between the knot’s core in MJ0366 and GFP is 36 or less (Fig. 4B). Therefore, variation of the degree of GFP protection as a function of linker length results from changes in the partitioning between two predominant molecular outcomes/pathways: (i) the sliding of the knot until it reaches GFP, leading to protection (Fig. 3A), or (ii) the tightening of the knot before it reaches GFP, in which case the knot can be successfully translocated by ClpXP (Fig. 3B). The latter situation temporally separates the less efficient translocation of the tight knot from the mechanically and/or kinetically demanding task of GFP unfolding, leading to restored ClpXP processivity and degradation of GFP (Fig. 3B).
Our observation that ClpXP completely degrades substrates with linkers longer than 36 residues indicates that a knot tightened before reaching GFP can be translocated through the ClpXP pore. Translocation of a tight knot through the ClpXP pore is plausible for three reasons: (i) Simulations indicate that once the knot is tight, its structure does not diffuse along the chain; (ii) the experimental diameter of a tight 31 knot is ∼1.6 nm (13, 14, 16), which is close to the diameter size of the expanded pore of the motor (31); and (iii) it has been shown that two fused polypeptide chains can be successfully translocated by ClpXP (31) or the proteasome (37). This is experimental evidence that a knotted topology, specifically a trefoil knot, can be successfully translocated by a AAA+ protease. However, tight knots of higher topological complexity could have more dramatic hampering effects on ATP-dependent protein degradation. Recently, Ziegler et al. (8) stretched the 51 knot present in the human ubiquitin C-terminal hydrolase isoenzyme L1 (UCH-L1) until it reached a compact size of about ∼4 nm in diameter, which is almost threefold larger than the tight knot studied here. Such a knot may completely block the pore entrance of most ATP-dependent proteases, including ClpXP. Future ClpXP degradation assays with the UCH-L1 substrate are required to test this hypothesis.
Together, our results open the route to experimentally study the elusive dynamics of a knot during the translocation of a protein through small pores and explain how the particular events of tightening and sliding of a knot alter the efficient degradation of ClpXP. A 31 knot-containing protein can be translocated and degraded by ClpXP once its knot is tightened during the process of translocation. However, the presence of a knot close to a domain whose unfolding is mechanically or kinetically more challenging can stall translocation and lead to the release of a partially degraded product, which in turn could lead to the accumulation of misfolded intermediates. Thus, even if the ClpXP pore is wide enough to translocate a tight knot, this knot may stop degradation in multidomain substrates. Our results may thus indicate an additional functional role of knots within proteins. There are several examples of proteins being incompletely degraded by the 26S proteasome (38–42), leading to the release of partially degraded fragments with new biological activities and providing a new layer of posttranslational control (43). For example, some viral proteins and transcription factors, such as NF-κB, need to be partially degraded by the proteasome to be activated (41). In these cases, a glycine/alanine-rich sequence acts as a slippery segment to stop degradation and promote the dissociation of a downstream folded domain from the proteasome (44–46). We speculate that a knot within a multidomain protein [such as the protein TTHA0275 from Thermus thermophiles; Protein Data Bank (PDB) ID code 4X3L] could work in a somewhat similar manner to the glycine/alanine-rich sequences—that is, as a stop signal for proteasomal processing. Future studies will have to address the effect of knots on the unfolding of other protein substrates by different proteases, including the 26S proteasome, to test this hypothesis.
Materials and Methods
Single-chain ClpX hexamer and ClpP were expressed and purified as previously described in refs. 25 and 47. The substrates GFP-ssrA, MJ0366-ssrA, and multidomain substrates were overexpressed in recA− E. coli strain BLR and purified by Ni2+ affinity chromatography. ClpXP degradation assays were performed with an ATP regeneration system as described in refs. 25 and 47. The size of the partially degraded product of GFP-17-MJ0366-ssrA was determined by a MALDI-TOF microflex mass spectrometrer (Bruker Daltonics Inc.). A full description of methods for protein expression, purification, and data analysis is described in SI Materials and Methods.
SI Materials and Methods
Protein Expression and Purification.
Single-chain ClpX hexamer and ClpP containing a C-terminal His6-tag (25, 47) were expressed during 3 h at 22 °C in the recA− E. coli strain BLR (DE3). ClpX single-chain hexamer was purified by Ni2+ affinity chromatography (HisTrap; GE Healthcare Life Sciences) and by size-exclusion chromatography on Superdex 200 GL as previously described in refs. 25 and 47. Clp-His6 was purified by Ni2+ affinity chromatography and by ion exchange chromatography on Bio-Rad UNO Q 1 as described previously (25, 47).
The substrates GFP-ssrA, MJ0366-ssrA and multidomain substrates were cloned in pET21d (+) (Novagen). A His6-tag was added between the protein domain and the C-terminal ssrA tag. To generate control substrates without His6 and ssrA tags, the TEV protease cleavage site (ENLYFQG) was introduced prior to His6 and ssrA of the substrates. Multidomain substrates with titin spacers were created by insertion of an unfolded double-mutant I27 domain of titin (C47E C63E, ref. 27) between GFP and MJ0366. The different lengths of the spacers were created by deleting residues from the N terminus of titin (Dataset S1). Substrates were overexpressed in recA− E. coli strain BLR (DE3) at OD600 0.7 with 1 mM IPTG for 3 h at 22 °C. Protein purification was carried out by Ni2+ affinity chromatography, applying an imidazole gradient (20–250 mM). The concentration of intact multidomain substrates was determined by GFP absorbance (ε500 55,000 M−1·cm−1) and corrected by the fraction of the intact substrate determined by quantification of the intensities of the fluorescent bands observed on SDS/PAGE. Substrates were stored at −80 °C in 25 mM Hepes (pH 7.6), 100 mM KCl, 400 mM NaCl, 10% glycerol, and 10 mM β-mercaptoethanol.
Biochemical Assays.
ClpXP degradation assays of substrates were carried out at 25 °C in 25 mM Hepes (pH 7.6), 100 mM KCl, 20 mM MgCl2, 1 mM DTT, 0.5 mM EDTA with an ATP regeneration system (5 mM ATP, 16 mM creatine phosphate, 6 mg/mL creatine phosphokinase), 0.3 µM ClpX6, and 1.5 µM ClpP14. The degradation of substrates with GFP domains was monitored by total fluorescence at 511 nm (excitation 467 nm) using an Agilent-Carry Eclypse spectrofluorimeter or by in-gel fluorescence on SDS/PAGE gels. For kinetics followed by gel fluorescence, the reaction was stopped in Laemmli buffer and loaded without heating to the SDS/PAGE gel. Samples of MJ0366-ssrA were heated and the degradation monitored by Coomasie staining. The intensity of the fluorescent and the Coomasie-stained bands were determined by GelAnalizer software (48). The size of the partially degraded product of GFP-17-MJ0366-ssrA was determined by a MALDI-TOF microflex mass spectrometrer (Bruker Daltonics Inc.).
Data Analysis.
The concentration of substrates available for ClpXP and the amplitudes of the kinetics followed by intrinsic fluorescence of GFP were corrected by the fraction of substrates harboring a damaged ssrA tag. In the case of degradation kinetics determined by total GFP fluorescence, the fraction of molecules degraded by ClpXP was determined by the quotient of the observed amplitude and the expected amplitude. The expected amplitude corresponds to the observed fluorescence of the initial concentration of substrate corrected by the fraction of molecules with damaged or lacking ssrA tag. The fraction of damaged molecules of substrate unable to be degraded by ClpXP was quantified by in gel fluorescence. In a similar way, in experiments of size exclusion chromatography, the fraction of GFP protection was calculated as the quotient between the area of the partially degraded product and the area of the peak substrate available to bind ClpXP.
Subunit Exchange of Dimers of Multidomain Substrates.
Multidomain substrates were incubated for 30 min with equimolar or eightfold excess of MJ0366-ssrA in 25 mM Hepes (pH 7.6), 100 mM KCl, 20 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA. The mixture of substrates was analyzed by size exclusion chromatography monitored by GFP absorbance at 500 nm. The formation of heterodimers by subunit exchange was determined by the apparition of a new species with GFP absorbance and smaller size than the respective multidomain substrate homodimer.
Supplementary Material
Acknowledgments
This work was supported by Fondecyt Grant 1151274 (to M.B.), Fondequip Grant EQM140151 (to M.B.), and Conicyt, Chile. A.S.M. was supported by Conicyt Master Fellowship 22151447. Travel for C.B. to and from Chile and Peru was also partially supported by the Howard Hughes Medical Institute, NIH Grant R01GM032543, and the US Department of Energy Office of Basic Energy Sciences Nanomachine Program under Contract DE-AC02-05CH11231. A.M. acknowledges support from the Howard Hughes Medical Institute and NIH Grant R01-GM094497.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705916114/-/DCSupplemental.
References
- 1.Faísca PF. Knotted proteins: A tangled tale of structural biology. Comput Struct Biotechnol J. 2015;13:459–468. doi: 10.1016/j.csbj.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Virnau P, Mirny LA, Kardar M. Intricate knots in proteins: Function and evolution. PLOS Comput Biol. 2006;2:e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sułkowska JI, Sułkowski P, Onuchic J. Dodging the crisis of folding proteins with knots. Proc Natl Acad Sci USA. 2009;106:3119–3124. doi: 10.1073/pnas.0811147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noel JK, Sułkowska JI, Onuchic JN. Slipknotting upon native-like loop formation in a trefoil knot protein. Proc Natl Acad Sci USA. 2010;107:15403–15408. doi: 10.1073/pnas.1009522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallam AL, Jackson SE. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nat Chem Biol. 2011;8:147–153. doi: 10.1038/nchembio.742. [DOI] [PubMed] [Google Scholar]
- 6.King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO. Structure and folding of a designed knotted protein. Proc Natl Acad Sci USA. 2010;107:20732–20737. doi: 10.1073/pnas.1007602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christian T, et al. Methyl transfer by substrate signaling from a knotted protein fold. Nat Struct Mol Biol. 2016;23:941–948. doi: 10.1038/nsmb.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziegler F, et al. Knotting and unknotting of a protein in single molecule experiments. Proc Natl Acad Sci USA. 2016;113:7533–7538. doi: 10.1073/pnas.1600614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 10.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Makarov DE. Translocation of a knotted polypeptide through a pore. J Chem Phys. 2008;129:121107. doi: 10.1063/1.2968554. [DOI] [PubMed] [Google Scholar]
- 12.Szymczak P. Tight knots in proteins: Can they block the mitochondrial pores? Biochem Soc Trans. 2013;41:620–624. doi: 10.1042/BST20120261. [DOI] [PubMed] [Google Scholar]
- 13.Szymczak P. Periodic forces trigger knot untying during translocation of knotted proteins. Sci Rep. 2016;6:21702. doi: 10.1038/srep21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szymczak P. Translocation of knotted proteins through a pore. Eur Phys J Spec Top. 2014;223:1805–1812. [Google Scholar]
- 15.Wojciechowski M, Gómez-Sicilia À, Carrión-Vázquez M, Cieplak M. Unfolding knots by proteasome-like systems: Simulations of the behaviour of folded and neurotoxic proteins. Mol Biosyst. 2016;12:2700–2712. doi: 10.1039/c6mb00214e. [DOI] [PubMed] [Google Scholar]
- 16.Dzubiella J. Sequence-specific size, structure, and stability of tight protein knots. Biophys J. 2009;96:831–839. doi: 10.1016/j.bpj.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang I, Chen SY, Hsu ST. Unraveling the folding mechanism of the smallest knotted protein, MJ0366. J Phys Chem B. 2015;119:4359–4370. doi: 10.1021/jp511029s. [DOI] [PubMed] [Google Scholar]
- 21.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 22.Bird LE, et al. Green fluorescent protein-based expression screening of membrane proteins in Escherichia coli. J Vis Exp. 2015;(95):52357. doi: 10.3791/52357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 24.Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc Natl Acad Sci USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat Struct Mol Biol. 2008;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- 26.He C, Lamour G, Xiao A, Gsponer J, Li H. Mechanically tightening a protein slipknot into a trefoil knot. J Am Chem Soc. 2014;136:11946–11955. doi: 10.1021/ja503997h. [DOI] [PubMed] [Google Scholar]
- 27.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyt MA, et al. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraut DA. Slippery substrates impair ATP-dependent protease function by slowing unfolding. J Biol Chem. 2013;288:34729–34735. doi: 10.1074/jbc.M113.512533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Aliaga P, Ramirez L, Kim F, Bustamante C, Martin A. Substrate-translocating loops regulate mechanochemical coupling and power production in AAA+ protease ClpXP. Nat Struct Mol Biol. 2016;23:974–981. doi: 10.1038/nsmb.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iosefson O, Olivares AO, Baker TA, Sauer RT. Dissection of axial-pore loop function during unfolding and translocation by a AAA+ proteolytic machine. Cell Rep. 2015;12:1032–1041. doi: 10.1016/j.celrep.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao XR, Lee HJ, Quake SR. Behavior of complex knots in single DNA molecules. Phys Rev Lett. 2003;91:265506. doi: 10.1103/PhysRevLett.91.265506. [DOI] [PubMed] [Google Scholar]
- 36.Vologodskii A. Brownian dynamics simulation of knot diffusion along a stretched DNA molecule. Biophys J. 2006;90:1594–1597. doi: 10.1529/biophysj.105.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–34765. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 39.Wilson MD, et al. Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell. 2013;154:983–995. doi: 10.1016/j.cell.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piwko W, Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- 41.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 42.Zanet J, et al. Pri sORF peptides induce selective proteasome-mediated protein processing. Science. 2015;349:1356–1358. doi: 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]
- 43.Rape M, Jentsch S. Taking a bite: Proteasomal protein processing. Nat Cell Biol. 2002;4:E113–E116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 44.Lin L, Ghosh S. A glycine-rich region in NF-kappaB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orian A, et al. Structural motifs involved in ubiquitin-mediated processing of the NF-kappaB precursor p105: Roles of the glycine-rich region and a downstream ubiquitination domain. Mol Cell Biol. 1999;19:3664–3673. doi: 10.1128/mcb.19.5.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 47.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 48.Skosyrev VS, et al. [Specialized software product for comparative analysis of multicomponent DNA fingerprints] Genetika. 2013;49:531–537. Russian. doi: 10.7868/s0016675813040140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.