Significance
Salmonella enterica serovar Typhi (S. Typhi) causes ∼20 million cases of typhoid fever every year. We carried out a genome-wide association study to identify genetic differences that correlate with the susceptibility of cells from hundreds of individuals to S. Typhi invasion. A SNP in VAC14 was associated with susceptibility to S. Typhi invasion and VAC14 expression. Cells mutated for VAC14 displayed increased S. Typhi docking due to increased plasma membrane cholesterol levels. The same SNP was associated with risk of typhoid fever in a Vietnamese population. Furthermore, treating zebrafish with a cholesterol-lowering drug reduced their susceptibility to S. Typhi infection. Therefore, this work demonstrates the power of coupling multiple genetic association studies with mechanistic dissection for understanding infectious disease susceptibility.
Keywords: single nucleotide polymorphism, lymphoblastoid cell line, phosphoinositide, ezetimibe, Salmonella pathogenicity island 1
Abstract
Risk, severity, and outcome of infection depend on the interplay of pathogen virulence and host susceptibility. Systematic identification of genetic susceptibility to infection is being undertaken through genome-wide association studies, but how to expeditiously move from genetic differences to functional mechanisms is unclear. Here, we use genetic association of molecular, cellular, and human disease traits and experimental validation to demonstrate that genetic variation affects expression of VAC14, a phosphoinositide-regulating protein, to influence susceptibility to Salmonella enterica serovar Typhi (S. Typhi) infection. Decreased VAC14 expression increased plasma membrane cholesterol, facilitating Salmonella docking and invasion. This increased susceptibility at the cellular level manifests as increased susceptibility to typhoid fever in a Vietnamese population. Furthermore, treating zebrafish with a cholesterol-lowering agent, ezetimibe, reduced susceptibility to S. Typhi. Thus, coupling multiple genetic association studies with mechanistic dissection revealed how VAC14 regulates Salmonella invasion and typhoid fever susceptibility and may open doors to new prophylactic/therapeutic approaches.
The etiologic agent of typhoid fever, Salmonella enterica serovar Typhi (S. Typhi), causes ≈20 million infections worldwide every year (1). Susceptibility, symptom presentation, and disease progression of typhoid fever are variable among people. Approximately 1–5% of infected individuals become carriers of the disease (2), most famously in the case of Typhoid Mary, who was forcibly isolated to prevent further transmission (3, 4). Recently, the first genome-wide association study (GWAS) of typhoid fever identified one locus in the MHC region as associated with typhoid fever risk (5), but undoubtedly other typhoid susceptibility loci have yet to be discovered. Furthermore, while a GWAS of disease can successfully identify associated genetic variants, mechanisms of how these SNPs affect disease are usually not apparent. Elucidating these mechanisms could reveal unexpected biomarkers and therapeutic strategies.
As a complementary approach to GWAS of disease, GWAS of molecular and cellular phenotypes can help elucidate how genetic differences impact genes and cellular phenotypes to affect disease physiology. GWAS of gene-expression quantitative trait loci (eQTLs) (6, 7) and protein QTLs (pQTLS) (8) can reveal what genes are being affected by genetic variation. Recent work has focused on eQTLs that are induced upon immune stimulation (9, 10). Our laboratory has developed and validated a cellular GWAS approach called “Hi-HOST” (high-throughput human in vitro susceptibility testing), using live pathogens as probes to bridge human genetic variation, host cell biology, and disease (11–13). Using hundreds of genotyped lymphoblastoid cell lines (LCLs), we previously identified SNPs that regulate caspase-1–mediated cell death (pyroptosis) and are associated with sepsis in humans (11, 12, 14, 15). Here, we report a cellular GWAS of susceptibility to bacterial cell entry.
We have applied the Hi-HOST approach to the phenotype of S. Typhi invasion. In mouse models, Salmonella mutants that do not invade efficiently have severely decreased virulence in oral infections (16–18), demonstrating the importance of this cellular phenotype to disease. Salmonella injects effector proteins into the host cell to induce its own uptake through macropinocytosis. The effectors SopB and SopE induce actin ruffling and facilitate macropinocytosis at the site of invasion (19). SopE activates CDC42 and RAC1 by acting as a guanine nucleotide exchange factor (GEF) (20). SopB is a phosphatidylinositol phosphatase with several functions, primarily activating Rho GTPases to mediate actin assembly and modulating the phosphoinositide composition on the Salmonella-containing vacuole (SCV) (21, 22). Thus, while phosphoinositide changes induced by Salmonella during invasion have been characterized, the host’s role in phosphoinositide metabolism in the context of invasion is still poorly understood. Furthermore, although approaches using bacterial genetics to identify and characterize key mechanisms of virulence have been very successful, GWAS of human variation provides a complementary approach that can reveal the more subtle complexities that occur when multiple pathways contribute to quantitative cellular traits and human disease susceptibility.
In this study we show that natural genetic variation influencing VAC14, a gene encoding a scaffolding protein involved in phosphoinositide metabolism, affects Salmonella invasion. We determined that a SNP (rs8060947) associated with decreased VAC14 expression was also associated with increased Salmonella invasion. Increased invasion was also observed by experimentally decreasing VAC14 expression. The mechanism underlying this increase in invasion is due to elevated plasma membrane cholesterol that enhances docking of Salmonella to the host cell. Remarkably, the same SNP identified through Hi-HOST showed an association with increased risk of typhoid fever in a Vietnamese population. Finally, depleting cholesterol in zebrafish through the Food and Drug Administration (FDA)-approved drug ezetimibe increased clearance of S. Typhi. This multidisciplinary approach to understanding how human genetic variation affects a cellular trait relevant for disease has revealed a role for a phosphoinositide scaffolding protein in the regulation of plasma membrane cholesterol that could be exploited to reduce the risk of typhoid fever.
Results
S. typhi Invasion into LCLs Requires SPI1 Effectors and Is Regulated by Heritable Genetic Variation.
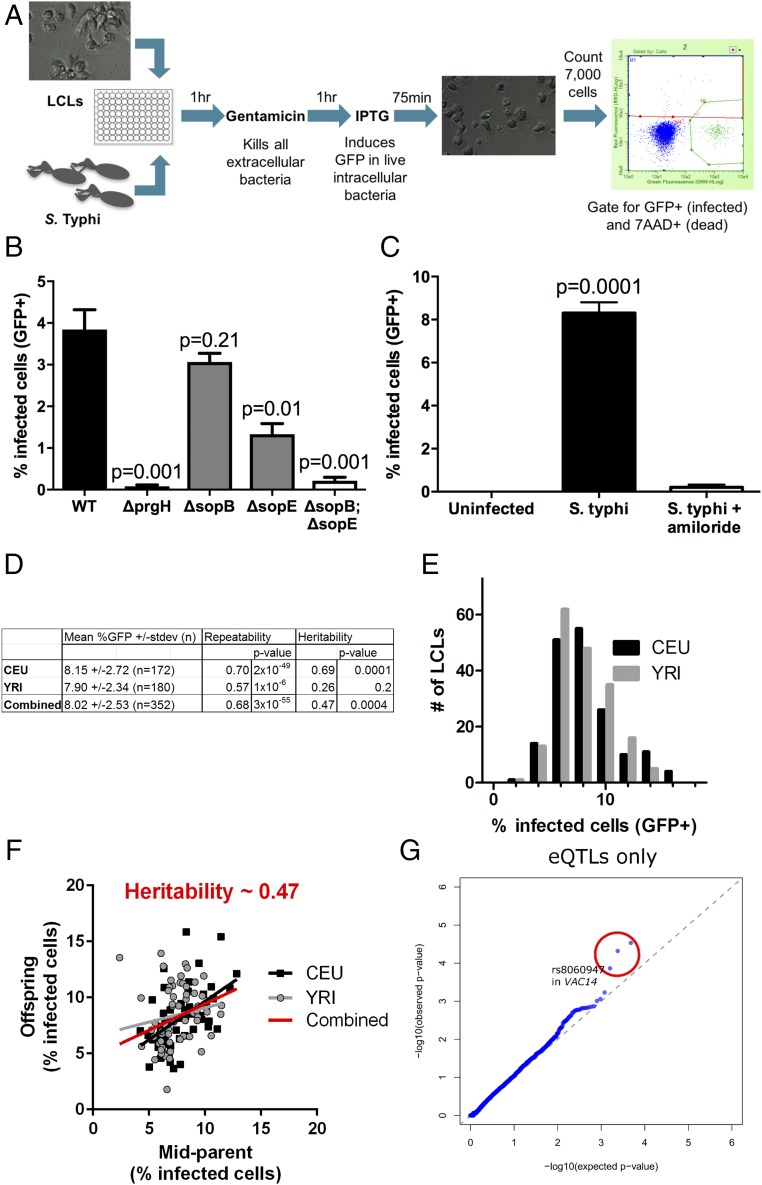
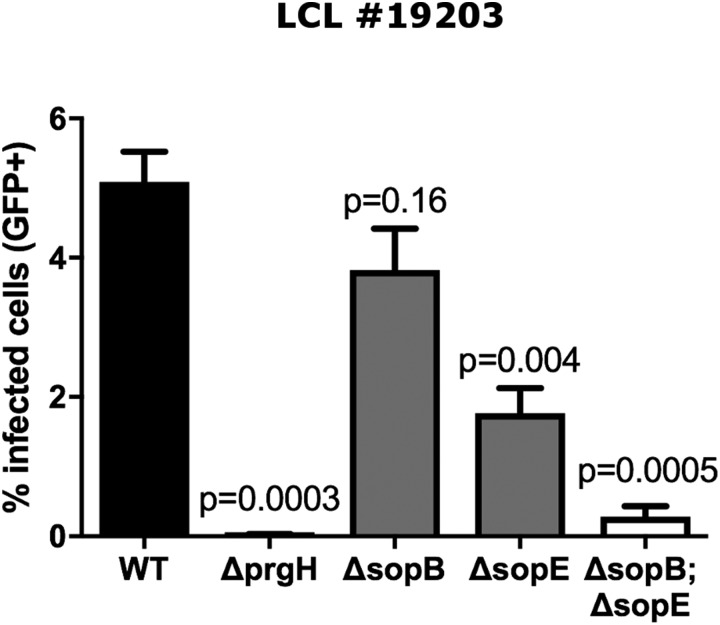
As invasion of S. Typhi into LCLs had not been previously characterized, we determined whether the process was similar to that observed in other human cell types. LCLs are immortalized B cells, and, notably, B cells have been shown to be in vivo targets of Salmonella infection (23–25). To quantitatively measure Salmonella invasion into cells, we used a modified gentamicin protection assay where cell entry and early intracellular survival into cells was assessed with flow cytometric measurement of GFP (Fig. 1A). Salmonella invasion into HeLa cells has been extensively characterized (26–28) and is known to occur through macropinocytosis (29), mediated by the Salmonella pathogenicity island-1 (SPI-1) effectors sopB and sopE/E2 (21). Consistent with this, sopB and sopE are necessary for invasion in LCLs, as seen by the decrease in infected cells when each gene is deleted individually and the nearly complete loss of invasion when both are deleted (sopE2 is not present in S. Typhi Ty2). See Fig. 1B for data from an LCL from the CEU population [Utah residents with Northern and Western European ancestry from the Centre d’Étude du Polymorphisme Humain (CEPH) collection] and Fig. S1 for data from an LCL from the Yoruba in Ibadan, Nigeria (YRI) population. Similarly, deletion of prgH, encoding a component of the type three secretion system (TTSS) and necessary for the translocation of sopB and sopE into the host cell, also abrogates invasion. Furthermore, an inhibitor of macropinocytosis, amiloride (30), also blocked the ability of S. Typhi to invade (Fig. 1C).
Fig. 1.
Invasion of S. Typhi into LCLs occurs via SPI-1–dependent macropinocytosis. (A) Schematic of the flow cytometric assay of S. Typhi invasion into LCLs. Following 1 h incubation with S. Typhi, gentamicin was added to kill extracellular bacteria. IPTG was added to induce expression of GFP in living, intracellular bacteria, and the percentage of GFP+ infected cells was quantified by flow cytometry. (B) Invasion of S. Typhi into LCLs requires SPI-1 TTSS and SPI-1 effectors. The percentage of infected cells identified by flow cytometry is dramatically reduced with deletion of the gene encoding the SPI-1 TTSS component prgH or by deletion of the genes encoding the secreted effectors sopE and sopB. Data presented are the mean ± SEM from three independent experiments. P values in B and C are from t tests. Data are from LCL 7056 from the CEU population. Similar data were observed with LCL 19203 from the YRI population (Fig. S1). (C) Invasion of S. Typhi into LCLs is blocked by amiloride, an inhibitor of macropinocytosis. Cells were pretreated for 30 min with 1 mM amiloride before infection with S. Typhi. Data presented are the mean ± SEM from four biological replicates. (D) Highly reproducible and heritable variation in S. Typhi invasion into LCLs. Data presented are the mean ± SD from independent measurements from three serial passages of LCLs from CEU and YRI populations. Repeatability of the measurement was calculated as the interindividual component of variance from ANOVA. Heritability was calculated by parent–offspring regression, and P values are significance of nonzero slope. For D–G, n = 352 LCLs. (E) Histogram of distribution of S. Typhi invasion (percentage GFP+ at 3.5 h) into LCLs. (F) Invasion of S. Typhi into LCLs is heritable. Parent–offspring regression from CEU (black squares) and YRI (gray circles) trios gives a slope of 0.47, estimating that 47% of the variance for the trait is heritable. (G) A Q–Q plot of P values for only cis-eQTLs reveals P values lower than expected by chance for P < 0.001. In ref. 7, 4787 cis-eQTLs were identified. rs8060947 in VAC14 has the third lowest P value in the Q–Q plot (1.4 × 10−4). Characteristics of the cis-eQTLs within the red circle are given in Table S1. A Q–Q plot of all SNPs is shown in Fig. S2.
Fig. S1.
Specificity of the invasion phenotype demonstrated in LCL 19203 from the YRI population. The percentage of infected cells quantified by flow cytometry is dramatically reduced with deletion of the gene encoding the SPI-1 TTSS component prgH or by deletion of the genes encoding the secreted effectors sopE and sopB. Data shown are the mean ± SEM from three independent experiments.
The similarity of S. Typhi invasion into LCLs to epithelial cell invasion prompted us to assess the feasibility of carrying out a Hi-HOST cellular GWAS screen of invasion. We observed repeatable invasion measurements with the between-individual component of variance at ≈68% (the remainder of the variance being intraindividual variation for the same LCL measured on different days) (Fig. 1D and invasion measurements from Hi-HOST screen in Dataset S1). Similar means (P = 0.35 by t test) and distributions (Fig. 1E) were observed in 352 LCLs from two different human populations from the HapMap collection (31, 32), CEU and YRI (Fig. 1D), indicating no evidence of population differentiation for this trait. Furthermore, a substantial portion of the variation (47%) was heritable, based on parent–offspring regression of these LCL trios (Fig. 1F). Encouraged by the high repeatability and heritability of this trait, we carried out genome-wide association.
Hi-HOST Cellular GWAS Reveals a SNP in VAC14 Associated with S. typhi Invasion.
Genome-wide association was carried out on data from 352 LCLs using family-based association analysis in PLINK (33) on HapMap phase 3 (32) genotypes with imputation based on 1,000 Genomes phase 1 haplotypes. With this relatively modest sample size, we focused on SNPs associated with the expression level of nearby genes (cis-eQTLs). GWAS signals are enriched for cis-eQTLs (34), and we previously demonstrated that focusing on cis-eQTLs in a Hi-HOST screen of pyroptosis revealed true-positive hits based on subsequent experimental validation and genotyping in clinical samples (11). We used cis-eQTLs found in the same LCLs used in our Hi-HOST screening (7) to limit our search space to SNPs that have a robust association between SNP and gene expression in the cell type used in our screen. We found that cis-eQTLs deviated toward lower P values than expected by chance in a quantile–quantile (Q–Q) plot of S. Typhi invasion (Fig. 1G and Fig. S2 show a Q–Q plot of all SNPs). Of the three cis-eQTLs that deviated from neutrality (Table S1), rs8060947, located within an intron of VAC14, was particularly intriguing because of the known function of VAC14. VAC14 encodes a phosphoinositide scaffolding protein that regulates levels of the phosphatidylinositol PtdIns(3,5)P2 through its binding partners PIKfyve and FIG4 (35–37). While the Salmonella-secreted effector sopB has been demonstrated to regulate phosphoinositides (38–40), how host regulation of phosphoinositides impacts invasion is poorly characterized, and the role of human genetic variation in this process is unknown. The remaining text focuses on the association of VAC14 with S. Typhi invasion, its mechanism of action, and how variation in VAC14 impacts Salmonella infection.
Fig. S2.
A Q–Q plot of P values for all SNPs for the invasion association screen demonstrates that most SNPs follow the expected neutral distribution.
Table S1.
eQTLs associated with susceptibility to S. Typhi invasion
| SNP | Human gene whose expression is associated with SNP | P value |
| rs3781626 | SPI1 | 0.000029 |
| rs755553 | SPI1 | 0.000048 |
| rs8060947 | VAC14 | 0.00014 |
P values are given for family-based association (QFAM-parents) of 352 LCLs from the CEU and YRI populations. All eQTLs with P < 0.0005 are shown. rs8060947 (bold) is described in detail in this article.
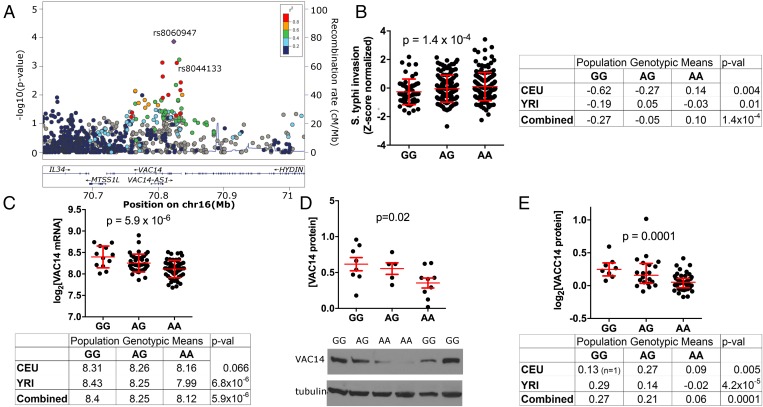
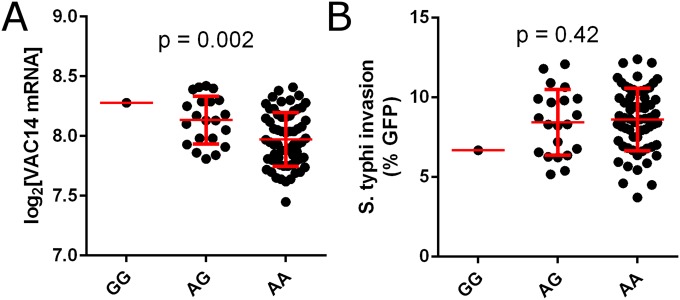
rs8060947 is associated with both the expression of VAC14 and the level of S. Typhi invasion. A regional association plot demonstrates that rs8060947 is within the first intron of VAC14 and is the SNP in the region showing the strongest association with S. Typhi invasion (Fig. 2A). Although family-based genome-wide association was conducted on both CEU and YRI families combined to detect this association (P = 1.4 × 10−4), we also observed the association when CEU and YRI populations were analyzed separately (P = 0.004 in CEU, P = 0.02 in YRI) (Fig. 2B). In both populations, the “A” allele was associated with higher levels of invasion (Fig. 2B) as well as with lower levels of VAC14 mRNA (P = 5.9 × 10−6 based on microarray data of LCLs) (Fig. 2C) (7), protein levels by Western blot (P = 0.02 from 22 LCLs randomly selected with representation of each genotype) (Fig. 2D), and protein levels by proteomics (P = 0.0001 based on mass spectrometry LCL data in ref. 8) (Fig. 2E). Thus, the association of rs8060947 with VAC14 expression was observed with three different methods. While not part of our initial screen, we also tested the association of rs8060947 with VAC14 expression and invasion in HapMap Asian LCLs in the Chinese from Beijing (CHB) and Japanese from Tokyo (JPT) populations. Similar to the data from CEU and YRI, the A allele was associated with lower levels of VAC14 mRNA (P = 0.002) (Fig. S3A). For invasion, the predicted trend of AA > AG > GG was observed, but there was no significant association (P = 0.42); this result is not entirely surprising given the limited power from 85 Asian LCLs with only one of the GG genotype.
Fig. 2.
A SNP in VAC14 is associated with VAC14 expression and S. Typhi invasion. (A) Regional plot around the VAC14 gene demonstrates an association of rs8060947 with S. Typhi invasion. SNPs are plotted by position on chromosome 16 and by −log(P value) and are color-coded by r2 value to rs8060947 from 1,000 Genomes European data. rs8060947 is located within the first intron of VAC14. A second labeled SNP in high LD, rs8044133, is described in the text. (B) rs8060947 is associated with susceptibility of LCLs to S. Typhi invasion. The derived allele A is associated with increased levels of invasion in CEU and YRI populations. For genotypic means, percent invasion for each individual has been normalized into a Z-score to minimize a batch effect due to measurement of LCLs at two different times. P values are from family-based association analysis using QFAM-parents in PLINK. (C) rs8060947 is associated with the expression of VAC14 mRNA. The derived allele A is associated with lower levels of VAC14 mRNA in CEU and YRI populations (n = 60 unrelated individuals in each population). Gene expression values for each LCL are from ref. 7. Genotypic means are given for each population and for individuals from both populations combined. P values in C–E are from linear regression. (D) rs8060947 is associated with the expression of VAC14 protein levels. The derived allele A is associated with lower VAC14 protein. VAC14 protein was quantified by immunoblotting of 22 LCLs with β-tubulin as a loading control. The intensity of the VAC14 band normalized to β-tubulin was averaged from two separate scanned blots. (E) Confirmation of the association of rs8060947 with VAC14 protein. VAC14 protein levels were obtained from a mass spectrometry dataset with CEU (n = 47) and YRI (n = 28) LCLs (8).
Fig. S3.
(A) rs8060947 is associated with the expression of VAC14 mRNA. The derived allele A is associated with lower levels of VAC14 mRNA in Asian (CHB and JPT) populations (P = 0.002; 85 unrelated individuals). Gene expression values for each LCL are from ref. 7. The P value is from linear regression. (B) rs8060947 is not associated with susceptibility of LCLs to S. Typhi invasion. As in CEU and YRI populations, the AA > AG > GG pattern is observed, but the association is not significant (P = 0.42). The P value is from linear regression using with PCA correction in PLINK.
The A allele, which is the derived allele based on the chimpanzee genome, is found at a higher frequency in the CEU (80%) than in the YRI (46%) population. rs8060947 shows the strongest association in the region, but linkage disequilibrium (LD) extends for ∼100 kb with numerous other SNPs showing an association; any of these SNPs could be the true causal variant(s) in the region regulating VAC14 expression. One plausible causal variant is rs8044133, which shows the fourth strongest association in the region (P = 7.6 × 10−4) and is on a DNA segment bound by the serum response factor (SRF) transcription factor in ENCODE ChIP-sequencing (ChIP-seq) data (41).
VAC14 Is an Inhibitor of S. typhi Invasion.
The association data demonstrated strong correlations between rs8060947 and VAC14 expression and between rs8060947 and S. Typhi invasion. The direction of effects for the different alleles led to the hypothesis that reducing VAC14 expression would increase S. Typhi invasion. This hypothesis was tested through both RNAi and CRISPR/Cas9 knockout of the VAC14 gene.
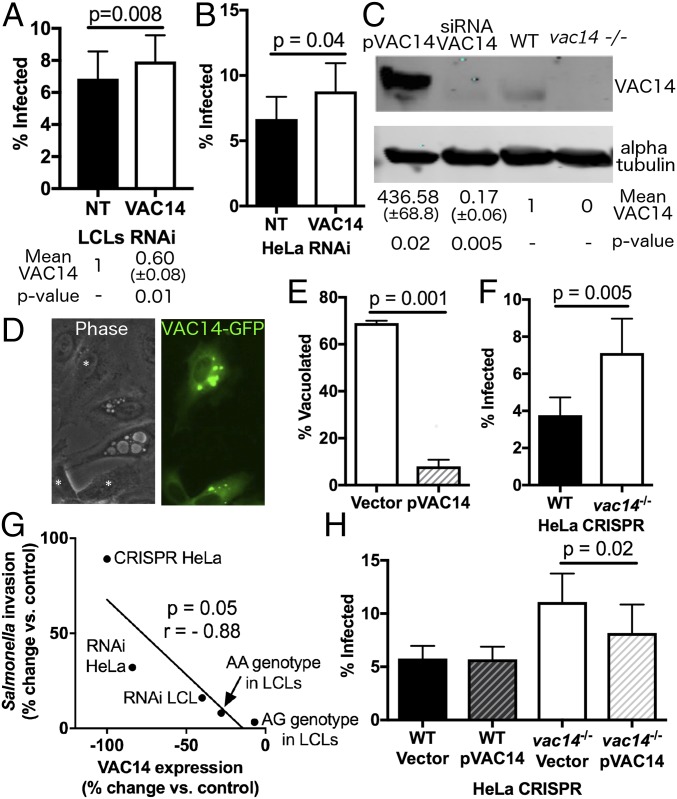
In LCLs, RNAi decreased VAC14 protein expression by 40% and increased S. Typhi invasion by 16% (P = 0.008) (Fig. 3A). This effect was mirrored in HeLa cells (P = 0.04) (Fig. 3B). The inhibitory activity of VAC14 on invasion was confirmed using CRISPR knockout. Effective targeting of VAC14 was demonstrated by Western blot (Fig. 3C) and through sequencing of a characterized clone of the targeted region (Fig. S4). Consistent with the phenotype of vac14−/− mouse embryonic fibroblasts (42), these vac14−/− mutant HeLa cells have abnormally enlarged vacuoles (Fig. 3D). Plasmid complementation of the vacuolation phenotype demonstrated that the phenotype was indeed attributable to VAC14 (P = 0.01) (Fig. 3 D and E). Furthermore, the vac14−/− mutant HeLa cells had an even larger increase in invasion than vac14 RNAi (P = 0.005) (Fig. 3F). Indeed, the relative decrease in VAC14 expression seen with natural variation (AA vs. GG genotype; 28% decrease) or with RNAi in LCLs (40% decrease) or HeLa cells (84% decrease) or with vac14 knockout (100% decrease) was inversely correlated with the relative increases in S. Typhi invasion (8% AA/GG, 16% by RNAi in LCLs, 32% by RNAi in HeLa cells, and 89% by CRISPR knockout in HeLa cells). Thus, decreased VAC14 expression resulted in higher levels of invasion, and the magnitude of the increase in invasion was larger the more VAC14 expression was diminished (Fig. 3G).
Fig. 3.
Loss-of-function studies and complementation indicate that VAC14 limits Salmonella invasion. (A) Reduction of VAC14 expression in LCLs by RNAi increases S. Typhi invasion. Percentages of S. Typhi invasion of 18,507 LCLs (YRI population) treated with either nontargeting (NT) or VAC14 siRNA demonstrated increased invasion with VAC14 depletion (P = 0.008). Data shown are the mean ± SEM of three experiments. Quantification of three Western blots of VAC14 knockdown showed 40% reduction in VAC14 protein levels (P = 0.01). (B) Reduction of VAC14 expression in HeLa cells by RNAi increased S. Typhi invasion. Shown are percentages of S. Typhi invasion in HeLa cells treated with either NT or VAC14 siRNA (P = 0.02). Data shown are the mean ± SEM from four experiments. (C) Representative Western blot of VAC14 protein demonstrated endogenous protein levels (WT), effective RNAi (siRNA VAC14), CRISPR knockout (vac14−/−), and plasmid overexpression (pVAC14) in HeLa cells. Protein extracted from each lane was collected from 300,000 cells, and α-tubulin was used as a loading control. Values below the blots show the mean ± SEM of three Western blots. (D) vac14−/− HeLa cells contain enlarged vacuoles, and transfection of pVAC14 rescued the vacuolated phenotype. Asterisks in the phase image denote cells that are transiently transfected with pVAC14-GFP. (E) Quantified (n = 100) vacuole-containing vac14−/− HeLa cells transfected with pVAC14-GFP demonstrated complementation (P = 0.001). (F) Complete loss of VAC14 protein expression in HeLa cells by CRISPR/Cas9 mutation increased S. Typhi invasion. S. Typhi invasion percentages demonstrated increased invasion in vac14−/− compared with WT cells (P = 0.005). Data shown are the mean ± SEM from four experiments. (G) Increase in Salmonella invasion is inversely correlated with VAC14 depletion (P = 0.05, r = −0.88). Increases in invasion percentage and the percentage of VAC14 protein depletion are calculated relative to Salmonella invasion with the GG allele in LCLs, NT siRNA controls, or WT HeLa cell controls. (H) Transient transfection of pVAC14 in vac14−/− cells complements invasion phenotype (P = 0.02). Data shown are the mean ± SEM from five experiments. All P values are calculated from paired t tests.
Fig. S4.
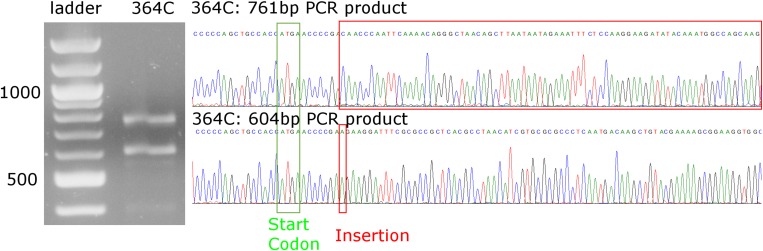
Characterization of the genetic lesion in VAC14 CRISPR knockout. PCR amplification of a 603-bp region in the VAC14 gene of genomic DNA from the 364C mutant cell line revealed two bands of equal intensity, one around 600 bp and the other around 750 bp. DNA sequencing of the individually cloned mutant alleles revealed two mutant alleles with insertions (red boxes): a 761-bp PCR product found to have a 158-bp insertion and a 604-bp PCR product containing a single additional adenine located 12 bp downstream of the start codon (green box). Both insertions are predicted to cause premature stop codons.
While RNAi and CRISPR can have off-target effects, we were able to complement the invasion phenotype. Transient transfection of VAC14 plasmid into WT HeLa cells had no effect, while VAC14 plasmid transfection into vac14−/− mutant cells reduced invasion toward WT levels (P = 0.02) (Fig. 3H). A lack of effect for overexpression of VAC14 in the presence of endogenous VAC14 is consistent with VAC14 being part of a protein complex, where overexpression of single components have been observed to have no phenotype in yeast (43). Thus, using RNAi, CRISPR knockout, and complementation, we have demonstrated that VAC14 negatively regulates S. Typhi invasion.
VAC14 Inhibits Invasion at the Step of SPI-1 TTSS Docking to the Plasma Membrane.
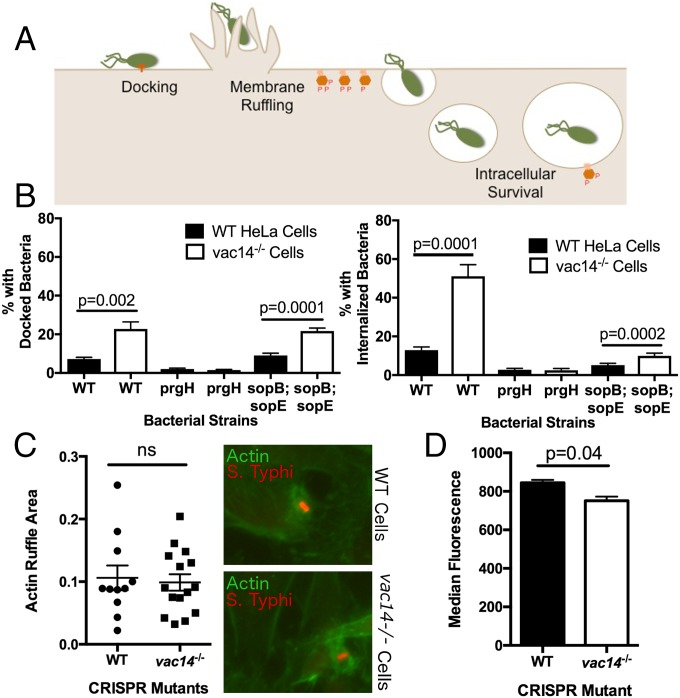
We systematically determined which step in invasion (docking, membrane ruffling, or early intracellular survival) was being affected by VAC14 (Fig. 4A). Increased S. Typhi invasion could be due to greater docking of the bacteria to the host cell plasma membrane. Adhesion to host cells by bacterial flagellin and LPS is reversible, but once a bacterium has injected its TTSS into a host cell, it is docked and attached firmly to the plasma membrane (27). Selective staining of intracellular vs. extracellular bacteria was used to compare levels of invasion vs. docking (Fig. 4B). The ΔprgH bacterial mutant, lacking the needle complex, exhibited very low invasion and docking in both WT and vac14−/− cells. The ΔsopBΔsopE double mutant, lacking the effectors necessary to induce macropinocytosis, could effectively dock but had dramatically reduced invasion. Importantly, WT and ΔsopBΔsopE bacteria demonstrated higher levels of docking in vac14−/− mutant cells, indicating that the increase in invasion could be explained by an increase in this early step.
Fig. 4.
Loss of VAC14 enhances S. Typhi docking. (A) Schematic of cellular processes where VAC14 could affect Salmonella invasion. Phosphoinositides (orange hexagons) are known to be involved in macropinocytosis and SCV maturation. (B) Loss of VAC14 increases S. Typhi docking. WT and vac14−/− HeLa cells were infected with three different S. Typhi bacterial strains (WT, ΔprgH, and a ΔsopB;ΔsopE double mutant). Cells were infected with fluorescently green S. Typhi for 1 h, washed, and fixed. External, adhered bacteria were stained with anti-Salmonella LPS (red). Bacteria were counted as either green only (internalized) or green and red (adhered). Cell counts were obtained by counting DAPI-stained nuclei. (C) Loss of VAC14 has no effect on membrane ruffling. WT and vac14−/− cells were infected with fluorescently labeled S. Typhi (pseudocolored red) for 15 min at a MOI of 50, washed, fixed, stained with Phalloidin-647 (pseudocolored green) for 20 min, and imaged. The area of actin ruffle was measured using Fiji (103); no difference between WT and vac14−/− cells was detected (P = 0.749). (D) Loss of VAC14 does not increase S. Typhi intracellular survival. Early intracellular survival was measured by quantifying median fluorescence of each cell 8 h post invasion. GFP fluorescence was induced 75 min before measurement; after a 1-h gentamicin treatment the green fluorescence represents living intracellular bacteria. Therefore, higher median fluorescence reflects an increased number of living GFP fluorescent bacteria inside each cell. A slightly significant decrease was detected (P = 0.04) and therefore cannot account for the increase in invasion. In all panels the mean ± SEM for three independent experiments or a minimum of 100 imaged cells are shown. P values are calculated from a paired t test.
In contrast, there were no gross defects of the vac14−/− mutant in the area of the plasma membrane ruffles engulfing the bacteria (Fig. 4C). Furthermore, no increase in intracellular survival was detected by median fluorescence of GFP Salmonella. (Fig. 4D). Therefore, our data demonstrate VAC14 regulates the level of S. Typhi invasion at the early step of docking to the host cell plasma membrane.
VAC14 Inhibits Docking by Reducing the Cholesterol Content of the Plasma Membrane.
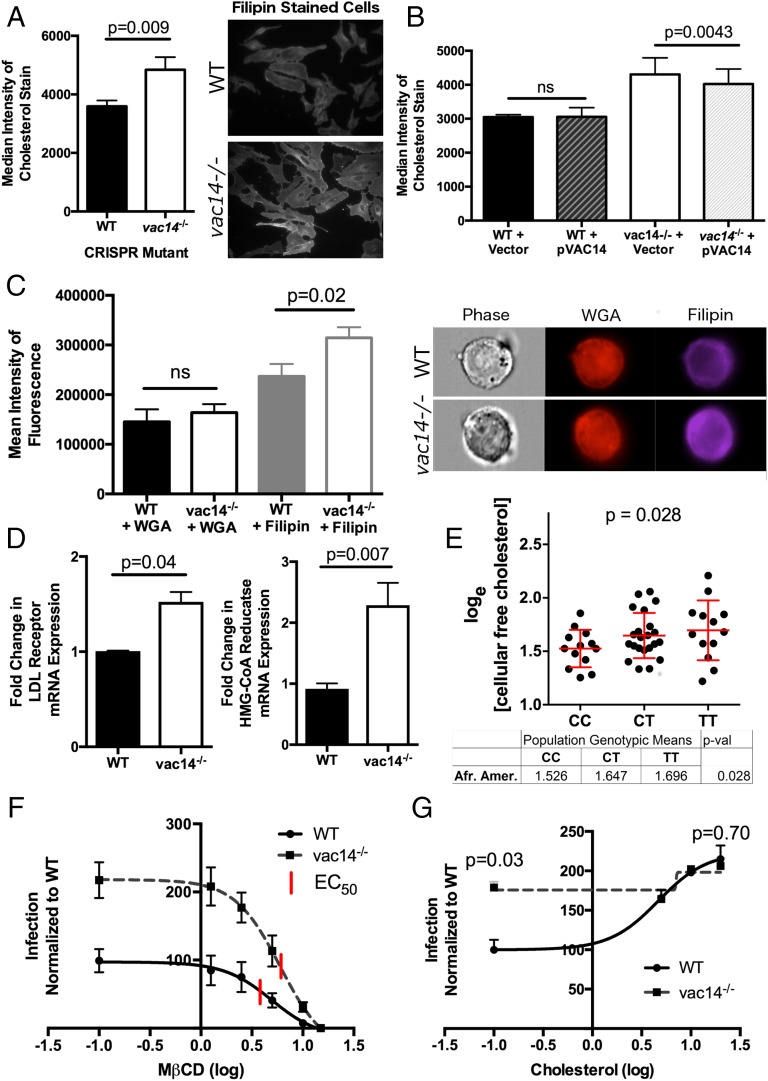
A role for VAC14 in plasma membrane attachment was unexpected, as VAC14 and the signaling lipids it regulates are cytosolic. We hypothesized that VAC14-mediated effects on phosphoinositide localization/abundance could alter abundance of plasma membrane constituents regulating docking. Specifically, attachment of the SPI-1 TTSS is partially mediated by direct binding of sipB to plasma membrane cholesterol (44). Therefore, we compared cholesterol localization and levels in WT vs. vac14−/− mutant cells by staining with the fluorescent cholesterol-binding molecule filipin (45, 46). Disruption of vac14 increased cellular cholesterol content measured by both flow cytometry and microscopy (P = 0.009) (Fig. 5A). Transfection of VAC14 plasmid decreased cholesterol in vac14−/− cells but had no effect on WT cells (Fig. 5B), mirroring the effect on S. Typhi invasion (Fig. 3H).
Fig. 5.
Loss of VAC14 increases cholesterol at the plasma membrane. (A) vac14−/− cells have increased total cholesterol. WT and vac14−/− cells were fixed and stained with filipin, and fluorescence was measured by flow cytometry (P = 0.009). Fluorescent microscopy of WT and vac14−/− cells also shows increased filipin staining in the vac14−/− cells. Data shown are the mean ± SEM from three independent experiments. (B) Transient transfection of pVAC14 partially rescues cholesterol phenotype. Decreased filipin staining by flow cytometry was measured in vac14−/− cells transfected with pVAC14, while no difference was detected in transfected WT cells (P = 0.004). Data shown are the mean ± SEM from four independent experiments. ns, not significant. (C) vac14−/− cells have increased cholesterol at the plasma membrane. Imaging flow cytometry was used to image and measure WGA (cell membrane staining) and filipin staining in WT and vac14−/− cells. No difference is seen in WGA staining, while filipin staining at the plasma membrane is significantly increased in vac14−/− cells (P = 0.02). Data shown are the mean ± SEM from three independent experiments. (D) Expression of LDLR and HMG-Co-A Reductase mRNA are increased in vac14−/− cells. qPCR analysis of LDLR and HMGCR was done on WT and vac14−/− cells using 18S rRNA to normalize. Data shown are the mean ± SEM from three independent experiments for LDLR and four independent experiments for HMGCR. (E) rs8044133 is associated with free cholesterol levels in 48 CAP African American LCLs (P = 0.028). Cellular free cholesterol was measured using the Amplex Red Cholesterol Assay Kit, and rs8044133 genotypes were imputed. One heterozygous outlier was removed based on Grubbs’ test (P < 0.01). One-tailed P values are from linear regression. African American data without the outlier removed (P = 0.05) and European American data (P = 0.47) are shown in Fig. S5. (F) Cholesterol depletion with MβCD reduces S. Typhi invasion. The EC50 was significantly higher in vac14−/− cells than in WT cells (P = 0.02) indicating that greater amounts of MβCD are needed to overcome the higher cellular cholesterol in vac14−/− cells. Data shown are the mean ± SEM from nine independent experiments. (G) Repletion of cholesterol increases S. Typhi invasion. Exogenous cholesterol increases invasion in WT cells to levels similar to vac14−/− cells. Data shown are the mean ± SEM from four independent experiments. The P value is calculated from a paired t test.
The increased cholesterol localized to the plasma membrane based on imaging flow cytometry. We saw no difference in the intensity of a nonspecific plasma membrane stain (wheat germ agglutinin, WGA) (47) at the plasma membrane, but vac14−/− cells had significantly higher filipin intensity when intracellular fluorescence in images was masked (Fig. 5C).
The increase in plasma membrane cholesterol was accompanied by increased expression of genes involved in both cholesterol uptake and synthesis. qPCR of the LDL receptor (LDLR) showed a 40% increase, and HMG-CoA reductase, the rate-limiting enzyme in cholesterol synthesis (48), showed a 125% increase (Fig. 5D). Therefore, vac14−/− cells have elevated plasma membrane cholesterol and increased expression of genes regulating uptake and synthesis.
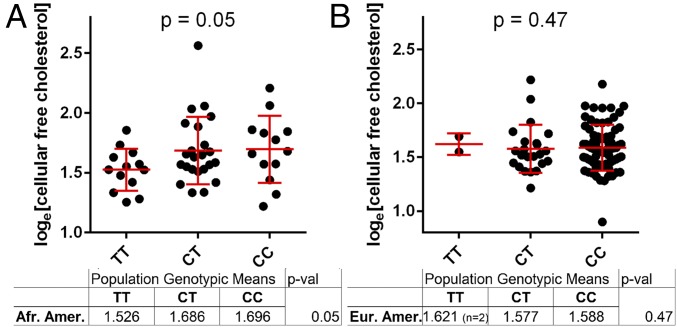
To determine whether cellular cholesterol levels were also associated with natural variation influencing VAC14, we used LCLs from the Cholesterol and Pharmacogenetics (CAP) simvastatin clinical trial. In 49 LCLs from African American participants, rs8060947 showed a trend toward significance in the predicted direction: the A allele was associated with higher free cholesterol (P = 0.08). rs8044133, a SNP in high LD with rs8060947 (r2 = 0.74 in CEU; r2 = 0.94 in YRI) that demonstrated an association with S. Typhi invasion nearly as strong as rs8060947 (Fig. 2A), was weakly associated with free cholesterol levels (P = 0.028) (Fig. 5E and Fig. S5). No association was noted in 98 European American LCLs (Fig. S5); however, due to the reduced minor allele frequency of this SNP in Europeans, only two LCLs homozygous for the minor allele were present in the dataset, and therefore power was limited. Thus, elevated cholesterol was observed not only in the vac14−/− cells but also with natural genetic variation that reduces VAC14 levels, although the association was observed only in the African American LCLs.
Fig. S5.
(A) rs8044133 is associated with free cholesterol levels in 49 CAP African American LCLs (P = 0.05). Cellular free cholesterol was measured using the Amplex Red Cholesterol Assay Kit, and rs8044133 genotypes were imputed. One-tailed P values are from linear regression. (B) rs8044133 is not associated with free cholesterol levels in 98 CAP European American LCLs. Cellular free cholesterol was measured using the Amplex Red Cholesterol Assay Kit, and rs8044133 genotypes were imputed. One-tailed P values are from linear regression.
To confirm the role of cholesterol in S. Typhi invasion, we depleted cholesterol at the plasma membrane using methyl-β-cyclodextrin (MβCD), a compound that sequesters cholesterol (49, 50). Increasing doses of MβCD in WT cells caused a decrease in invasion, consistent with S. Typhi binding to cholesterol to facilitate invasion. vac14−/− cells were modestly resistant to the effects of MβCD. The EC50 increased from 4.0 to 5.9 (P = 0.02) (Fig. 5F), consistent with an overabundance of cholesterol requiring a higher dose of MβCD to see an effect on invasion. Furthermore, exogenous cholesterol increased S. Typhi invasion and at higher doses resulted in equivalent levels of invasion between WT and vac14−/− cells (Fig. 5G). The vac14−/− cells appear already to have reached a nearly maximal level of invasion that is minimally enhanced with exogenous cholesterol. Based on the elevated plasma membrane cholesterol in the vac14−/− cells and the dependence of S. Typhi docking on plasma membrane cholesterol, we conclude that VAC14 inhibits S. Typhi invasion by modulating cholesterol at the plasma membrane.
VAC14 Is Associated with Susceptibility to Typhoid Fever in People.
While our cellular studies demonstrated that rs8060947 is associated with S. Typhi invasion into cells and that VAC14 inhibits invasion through the regulation of cellular cholesterol, we turned to human genotyping to assess the relevance of VAC14 to the risk of typhoid fever. We genotyped rs8060947 and compared the allele frequencies in 496 typhoid fever cases and 500 population controls. rs8060947 was in Hardy–Weinberg equilibrium in controls (P = 0.97). Table 1 shows that rs8060947 was associated with typhoid fever in this Vietnamese cohort. Remarkably, people who carry the rs8060947 A allele, which results in more invasion in Hi-HOST, had increased susceptibility to typhoid fever [P = 0.01; allelic odds ratio (OR) = 1.38; recessive OR = 3.60]. Although the effect of rs8060947 is not as large as, for example, the protection afforded by the sickle cell allele against malaria (OR = 10) (51), the effect is comparable to other infectious disease susceptibility loci, such as ABO and malaria (52, 53).
Table 1.
Association of rs8060947 with typhoid fever
| Alleles (%) | Genotypes (%) | ||||||||
| Group | G | A | GG | GA | AA | P value* | Allelic, G vs. A, OR (95% CI)† | Recessive, GG+GA vs. AA, OR (95% CI)‡ | |
| Control (n = 496) | 166 (17) | 826 (83) | 14 (2.8) | 138 (27.8) | 344 (69.4) | 0.01 | 1.38 (1.08–1.77) | 3.60 (1.18–11.02) | |
| Typhoid (n = 500) | 127 (13) | 873 (87) | 4 (0.8) | 119 (23.8) | 377 (75.4) | ||||
Genotypes are relative to the positive strand of chromosome 16. The VAC14 coding sequence is on the negative strand.
P value is from Pearson’s χ2 test.
ORs are for an additive model with 95% CU in parentheses. The A allele associated with higher S. typhi invasion in Hi-HOST is associated with increased odds of typhoid fever.
Odds ratios are for a homozygous model with 95% CI in parentheses. The A allele associated with higher S. typhi invasion in Hi-HOST is associated with increased odds of typhoid fever.
Pharmacologic Reduction of Cholesterol Is Protective Against S. Typhi in Zebrafish.
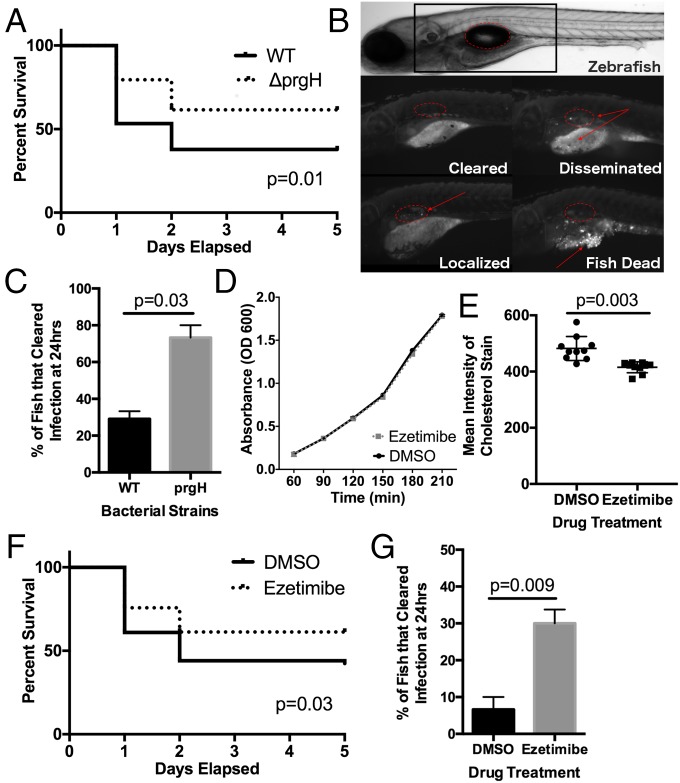
To experimentally assess the importance of cholesterol modulation during Salmonella infection in vivo, we developed a zebrafish S. Typhi infection model. Previous work has described zebrafish as a useful model to study pathogenesis, transmission, and vaccine efficacy of Salmonella Typhimurium (54, 55), but infection models using S. Typhi, a human-specific pathogen, had not been previously reported. The zebrafish larva is optically transparent and contains canonically organized epithelial surfaces and a functional innate immune system by 48 h postfertilization. To assess the invasion phenotype in whole animals, we made use of a previously developed epithelial infection model using the zebrafish swim bladder, an organ with a well-defined and visually accessible epithelial layer (56). A SPI-1 mutant (ΔprgH) was used to demonstrate the relevance of this model for studying invasion and infection outcomes. Survival curves demonstrated improved survival of animals infected with ΔprgH compared with animals infected with WT bacteria (P = 0.01) (Fig. 6A). Appreciating that zebrafish larvae are optically transparent, we imaged the swim bladders 24 h post infection to better assess the infection process. Infections were categorized as cleared, localized, disseminated, or dead (Fig. 6B). Swim bladder infections with the ΔprgH mutant bacterial strain were more effectively cleared than WT infections (P = 0.03) (Fig. 6C).
Fig. 6.
Ezetimibe is protective in a zebrafish model of S. Typhi infection. (A) Fish infected with S. Typhi prgH had increased survival compared with fish infected with WT S. Typhi (P = 0.01). The survival curve was carried out for 5 d; fish were checked once each day. (B) Zebrafish were scored 24 h post S. Typhi infection as cleared (no bacteria), localized (bacteria only in the swim bladder), disseminated (bacteria found outside the swim bladder), or dead (fish dead due to bacterial burden). The swim bladders are denoted by red circles; bacteria are denoted by the red arrows. (C) Fish infected with the S. Typhi prgH mutant had increased clearance of bacteria at 24 h (P = 0.03). (D) Ezetimibe had no effect on S. Typhi bacterial growth. Bacteria were diluted from an overnight stock and grown with DMSO or 10 µM ezetimibe. The OD600 was taken every 30 min for 3.5 h. Data points are the mean from two separate experiments. (E) Ezetimibe decreased filipin staining in fish. Twenty-four–hour pretreatment with 10 µM ezetimibe reduced filipin (0.05 mg/mL) staining (P = 0.003) in zebrafish; n = 20 fish from two separate experiments; P value from an unpaired t test. (F) Fish pretreated with ezetimibe had increased survival from S. Typhi infection compared with DMSO-pretreated controls (P = 0.03). (G) Ezetimibe treatment increased bacterial clearance in fish. Twenty-four–hour pretreatment with 10 µM ezetimibe increased the percentage of fish that cleared the bacteria 24 h postinfection from 8 to 30% (P = 0.009). Infection data for each survival curve and clearance comparisons are from three independent experiments with a minimum of n = 60 fish. P values from survival curves are from the Mantel–Cox test; P values for other comparisons are from unpaired t tests.
We hypothesized that pharmacologic reduction of cholesterol could reduce the amount of invasion during S. Typhi infection. Therefore, we treated the fish with ezetimibe, a drug that has been demonstrated to reduce cholesterol levels in zebrafish larvae (57, 58). Ezetimibe did not cause any morphological abnormalities in the fish, even at higher doses, and had no effect on bacterial growth (Fig. 6D). Filipin staining of whole fish embryos showed a significant decrease in fluorescence consistent with ezetimibe inhibiting cholesterol transport (Fig. 6E). Ezetimibe-treated fish showed both improved survival (P = 0.03) (Fig. 6F) and increased bacterial clearance (P = 0.009) (Fig. 6G) compared with DMSO-treated fish.
Discussion
Through a multidisciplinary approach, we have discovered that modulation of plasma membrane cholesterol, through natural genetic variation in VAC14 or pharmacological manipulation, decreases risk of S. Typhi infection. While cellular GWAS with Hi-HOST identified the association of the SNP in VAC14 with Salmonella invasion, association studies of different phenotypic scales (molecular, cellular, and organismal) were necessary to fully delineate the chain of causality leading from SNP to typhoid fever. Furthermore, mechanistic studies were necessary to uncover the unexpected role of VAC14 in limiting bacterial docking through cholesterol regulation at the host cell plasma membrane. Previous work has demonstrated a role for cholesterol metabolism and localization in bacterial invasion and survival (44, 59–61). This study provides host genetic evidence supporting this role and leads to the hypothesis that repurposing FDA-approved cholesterol-lowering drugs, such as ezetimibe, could have potential prophylactic or therapeutic use against typhoid fever.
This study provides evidence for a role for VAC14 in cholesterol metabolism. Previous work has emphasized the role of VAC14 in late endosomal trafficking based on localization of VAC14 and its binding partners to late endosomes (62, 63). Additionally, the inhibition of the VAC14 complex causes large endosomal vacuoles to form, and VAC14 has been reported to physically interact with regulators of the endolysosomal pathway, including Rab proteins (37, 64). As LDL-derived cholesteryl esters are delivered to late endosomes for hydrolysis before being trafficked to subsequent cellular sites, including the plasma membrane (65), it is perhaps not surprising that VAC14 and the phosphoinositide it regulates, PtdIns(3,5)P2, could play an important role in cellular cholesterol trafficking and homeostasis. Deletion of VAC14 increased intracellular cholesterol levels, including at the plasma membrane. We speculate that the effect size of common genetic variation influencing VAC14 and Salmonella invasion is constrained by the crucial role of plasma membrane cholesterol in cellular functions that operate within an optimal range (66). Nevertheless, our findings in cells, zebrafish, and humans demonstrate that moderate changes in cellular host–pathogen phenotypes can significantly alter infectious disease risk and severity.
Future studies will be necessary to determine the exact mechanism by which VAC14 is altering cholesterol. Determining localization of PtdIns(3,5)P2 in WT and vac14−/− cells will be important, and a PtdIns(3,5)P2 fluorescent probe has been described (67). However, recent work calls into question the specificity of this probe (68), and therefore new tools will likely be required before PtdIns(3,5)P2 localization can be accurately assessed. Recently, a key mechanism of nonvesicular lipid trafficking has been demonstrated to involve sterol-binding proteins facilitating countercurrent exchange of cholesterol and phospholipids at membrane contact sites (MCS) (69, 70). This has been most extensively characterized for OshP4 mediating the exchange of PtdIns(4)P and cholesterol between the endoplasmic reticulum (ER) and the Golgi (71, 72). We speculate a similar mechanism may be at work to regulate plasma membrane cholesterol levels. Indeed, our data with increased expression levels of LDL receptor (LDLR) and HMG-CoA reductase could support a model where impaired trafficking from the plasma membrane to the ER leads to a depletion of ER cholesterol and up-regulation of SREBP2 targets despite high total cholesterol levels.
While our studies provide a demonstration of the role of VAC14 in infectious disease, most studies on VAC14 have focused on its role in neurodegenerative disorders (42, 73). Neurons appear highly sensitive to loss of vac14 as seen by the lethal neurodegeneration of the CNS and peripheral nervous system in vac14−/− mice. Additionally, recessive mutations of vac14 in humans also lead to progressive neurodegeneration (74). Recently, it was also found that the VAC14 protein complex binds to amyloid precursor protein (APP), a protein essential to the development of Alzheimer’s disease (75–77). There have been several studies of the link between cholesterol and Alzheimer’s disease (78–80) and even trials of cholesterol-reducing drugs as potential therapeutics for patients with the disease (81, 82). Our elucidation of the functional links between VAC14, cholesterol, and disease could inform not only our understanding of Salmonella infection but also neurodegenerative disease. For example, the clinical presentation of Charcot–Marie–Tooth type 1 caused by mutation in the phosphatase FIG4 can be quite variable, onset can range from childhood to late adulthood, and severity can vary from loss of mobility to death (73). As VAC14 stabilizes the FIG4 protein from degradation, we hypothesize the genotype of rs8060947 and subsequent levels of VAC14 could help determine the severity of this disease.
While vaccines for S. Typhi exist, they are only moderately effective (55–60%) (83), have limited protection duration, and are not suitable in young children, the population most at risk. Therefore, the current vaccines are not widely deployed to populations in need. The new generation of conjugate vaccines for typhoid fever becoming available is showing considerably greater efficacy and applicability to younger children. However, no effective vaccines exist against enteric fever caused by S. Paratyphi pathovars, and unfortunately the incidence of S. Paratyphi A is increasing in many at-risk nations (84, 85). There is therefore a need for further development of typhoid vaccines, and our work suggests that the use of cholesterol-reducing drugs in combination with vaccines may potentially improve protection against typhoid fever. Metabolomic approaches have recently reported significantly higher plasma cholesterol levels in afebrile controls compared with enteric fever patients during infection (86). Although these data provide evidence for a role for cholesterol in typhoid fever, it is unclear how these levels measured during infection would correlate with cellular cholesterol levels before infection.
Finally, it is important to note that Salmonella spp. are not the only pathogens that use cholesterol to facilitate invasion. Ebola (87), Chlamydia (88), HCV (89), and malaria (90) are part of a growing list of pathogens whose entry into cells is regulated by cholesterol. In this regard, our work leads to two predictions that will be important to test in future work: (i) rs8060947 may be predictive of risk of other infectious diseases regulated by plasma membrane cholesterol, and (ii) ezetimibe might prove useful as a potential adjunctive therapy to prevent/treat not only typhoid fever but other infectious diseases as well.
Materials and Methods
Requests for data and reagents should be directed to Dennis Ko (dennis.ko@duke.edu). More detailed materials and methods are in Supporting Information.
Cell Biology.
HapMap LCLs (31, 32) were purchased from the Coriell Institute. LCLs used in cholesterol measurement were established from the CAP simvastatin clinical trial (91). Growth and assaying LCLs and HeLa cells for Salmonella spp/ infection was conducted as previously described (12) and as further detailed in SI Materials and Methods. S. typhi Ty2 was tagged with an inducible GFP plasmid [pMMB67GFP from ref. 92]. Ty2 deletion mutants were constructed with lambda red (93).
For RNAi, LCLs were treated for 3 d in Accell medium (Dharmacon) with either nontargeting Accell siRNA #1 or an Accell SmartPool directed against human vac14 (1 μM total siRNA) (Dharmacon). HeLa cells were treated for 2 d in 100 μL of DMEM medium (Invitrogen) with either nontargeting siGENOME siRNA #5 or a siGENOME SMARTpool directed against human vac14 (0.33 μM total siRNA) (Dharmacon).
For CRISPR/Cas9 knockouts, targeting vectors for the mutation of VAC14 were constructed following the protocol described in ref. 94. For plasmid complementation, HeLa cells were transfected using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher).
The bacterial docking assay was performed by infecting both WT HeLa and vac14−/− cells with isopropyl β-d-1-thiogalactopyranoside (IPTG)-induced S. Typhi at multiplicity of infection (MOI) of 50 for 1 h. Cells were washed twice with PBS, fixed with 3% paraformaldehyde (PFA) at RT for 30 min, washed twice again, and blocked with 5% normal donkey serum (NDS). Cells were treated with rabbit anti-Salmonella (MA183451; Fisher), 1:500 dilution, overnight at 4 °C. The secondary antibody used was donkey anti-rabbit 568 for 1 h at RT. Cells were stained for 20 min with Hoechst dye before imaging.
Filipin staining was quantified using the BD FACSCanto II from the Duke Flow Cytometry Shared Resource. Imaging flow cytometry was performed on 1 million cells per sample, which were washed with PBS, fixed with 3.7% PFA for 20 min at RT, and stained with WGA Alexa Fluor 680 (Thermo Fisher) for 10 min, 0.05 mg/mL Filipin (Sigma) for 2 h, or both stains and were run through the ImageStream X Flow Cytometer (Amnis Corp.). Analysis was performed using the ISX software to determine colocalization and intensity of WGA and filipin at the plasma membrane. Expression levels of genes that regulate cholesterol synthesis (HMGCR) and uptake (LDLR) were determined by TaqMan qPCR assays (Thermo Fisher) on a StepOnePlus Real-Time PCR machine (Thermo Fisher). Cholesterol depletion was performed by pretreating the cells with MβCD (Sigma) at the indicated concentrations for 1 h. After treatment, cell medium was changed and cells were subsequently infected with S. Typhi. Cholesterol repletion in cells was done by pretreating cells with water-soluble cholesterol (Sigma) at the indicated concentrations for 1 h, changing medium, and infection with S. Typhi. For the CAP LCLs, free cholesterol was quantified using the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific).
Typhoid Fever Association Study.
DNA samples from typhoid patients (n = 500) were collected as part of larger epidemiological or clinical studies that were undertaken in Viet Nam between 1992 and 2002. These clinical studies, which have been described previously (95–98), took place at the Hospital for Tropical Diseases in Ho Chi Minh City and Dong Thap Provincial Hospital, Vietnam, and the samples have been previously used in human genetic studies (5). Patients were defined as children or adults with clinical signs and symptoms of typhoid fever with culture-confirmed S. Typhi in their blood or bone marrow. The population control group comprised 496 DNA samples extracted from the umbilical cord blood of newborn babies born in 2003 at Hung Vuong Obstetric Hospital in Ho Chi Minh City, Vietnam. Written informed consent for clinical study participation was obtained by the treating physician from the patient or the patient’s parent/guardian and from the baby’s mother for population controls. All protocols were approved by the scientific and ethical committees at the Hospital for Tropical Diseases, the Dong Thap Provincial Hospital, and the Health Services of Dong Thap Province in Vietnam. Ethical approval was also granted by the Oxford Tropical Research Ethics Committee, Oxford University, Oxford, United Kingdom, the Duke University IRB, and the Human Research Ethics Committee of the University of Melbourne.
rs8060947 was genotyped by Taqman using a predesigned assay kit (Applied Biosystems). Pearson’s χ2 test was performed to assess an association between disease phenotype, allele frequencies, and recessive genotype (AA). The OR and 95% CI were used to measure the risk of disease associated with a specific allele (A).
The typhoid fever case and control samples used in this candidate gene study are primarily a subset of a larger Vietnamese sample set that has undergone genome-wide genotyping (5). Principal component analysis of this GWAS dataset found that all enteric fever cases had sufficiently matched controls (figures S2 and S3 in ref. 5), which provides strong evidence that no significant population stratification is present in our candidate gene study population.
Zebrafish Infections.
Studies involving zebrafish (strain AB*) were approved by the Duke University Institutional Animal Care and Use Committee and were performed in accordance with national and institutional guidelines. WT zebrafish embryos (strain AB*) were injected in the swim bladder at 4 d postfertilization (dpf) with ≈100 cfu of S. Typhi. Ezetimibe (10 µM) dissolved in DMSO was added to the fish water 24 h before S. Typhi injections. Filipin staining was performed on whole fixed fish at 5 dpf.
Statistical Analysis.
Descriptive statistics, parent–offspring regression, and Q–Q plots were performed with GraphPad Prism 6 (GraphPad Software) and with R (99). Genome-wide association analysis was conducted with PLINK v1.07 (33). Analysis was carried out with QFAM-parents with adaptive permutation under default settings. For CHB+JPT LCLs, which are all unrelated, GWAS was conducted in PLINK using a linear model with the top two principal components as covariates (using default parameters). Genotypes were from HapMap phase 3 release 2 (1,439,782 SNPs). The imputation of autosomal genotypes included two steps: a prephasing step using SHAPEIT2 (100) and an imputation step using IMPUTE2 (101) with 1,000 Genomes phase 1 haplotypes (201312 version provided by IMPUTE2 software). A regional Manhattan plot of the VAC14 region was made using LocusZoom (102). P values for eQTL analysis were calculated using linear regression in GraphPad Prism using unrelated individuals (parents only in trio data).
SI Materials and Methods
Cells.
HapMap LCLs (352) from CEU and YRI HapMap populations (31, 32) were purchased from the Coriell Institute. LCLs used in cholesterol measurement were established from the CAP simvastatin clinical trial as previously described (91). HeLa cells were purchased from ATCC. Cells were maintained at 37 °C in a 5% CO2 atmosphere. LCLs were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, 2 mM glutamine, 100 U/mL penicillin-G, and 100 mg/mL streptomycin. HeLa cells were grown in DMEM supplemented with 10% FBS, 1 mM glutamine, 100 U/mL penicillin-G, and 100 mg/mL streptomycin.
Bacterial Infection.
S. typhi Ty2 was tagged with an inducible GFP plasmid [pMMB67GFP from ref. 92]. prgH, sopB, sopE, and sopB; sopE deletion mutants were constructed with lambda red (93) and verified by PCR. Assaying LCLs and HeLa cells for Salmonella spp. infection was conducted as previously described (12). Briefly, overnight bacterial cultures were subcultured with a 1:33 dilution and grown for 2 h 40 min at 37 °C. For Hi-HOST screening of LCLs, invasion was conducted for 1 h at a MOI of 10, followed by the addition of gentamicin (50 μg/mL) for 1 h and then IPTG (1.5 mM) for 75 min. For HeLa cells, bacterial invasion was conducted for 30 min at an MOI of 30 (or 50 where indicated), followed by gentamicin (50 μg/mL) treatment for 1 h and IPTG (1.5 mM) for 75 min. Cellular invasion and cell death were assessed by staining with 7-AAD 7-aminoactinomycin D (7-AAD) (Enzo Life Sciences) and measuring green and red fluorescence on a Guava easyCyte Plus flow cytometer (Millipore). To measure intracellular survival, the infection assay was kept the same, but cells were read 8 h postinfection. After infection for 30 min at an MOI of 30, cells were incubated with gentamicin (50 μg/mL) for 6.25 h followed by IPTG (1.5 mM) for 75 min. In amiloride-treatment experiments cells were pretreated with 1 mM amiloride for 1 h before Salmonella infection.
RNAi Experiments.
LCLs (2 × 105 cells) were treated for 3 d in 500 μL of Accell medium (Dharmacon) with either nontargeting Accell siRNA #1 or an Accell SMARTPool directed against human vac14 (1 μM total siRNA; Dharmacon). Before infection, cells were plated at 1 × 105 in 100 μL RPMI complete medium (without antibiotics) in 96-well plates. Salmonella infections were conducted as described above. Cell invasion by S. Typhi was assessed after 3.5 h.
HeLa cells (1.5 × 105 cells) were treated for 2 d in 100 μL of DMEM medium (Invitrogen) with either nontargeting siGENOME siRNA #5 or a siGENOME SMARTpool directed against human vac14 (0.33 μM total siRNA; Dharmacon). Salmonella infections were conducted as described above.
Western Blotting.
For LCLs, 1,000,000 cells were harvested after 3 d of culture in 30 μL RIPA buffer and were rocked at 4 °C for 15 min; then insoluble material was removed by centrifugation at 8,000 × g for 5 min. Protein was transferred to a PVDF membrane using TE77X Semi-Dry transfer units (Hoefer) at 60 mA for ≈1 h. Blots were probed overnight at 4 °C (antibody dilutions are specific to each antibody and are indicated in figure legends) followed by 1:10,000 HRP- or LI-COR IRDye-conjugated secondary antibodies for 1 h at RT. Membranes were developed using ECL substrate (Pierce) with X-ray film or the LI-COR Odyssey Infrared Imager. X-ray film was scanned and quantified using ImageJ. IRDye Western blots were quantified using LI-COR Odyssey Software. For measuring the association of VAC14 protein and genotype in LCLs, 22 LCLs were randomly selected to provide several individuals of each genotype. More homozygous individuals were included (8GG, 5AG, 9 AA), as we reasoned the difference between homozygous individuals would be greatest.
For HeLa cells, 300,000 cells were harvested after 48 h of RNAi treatment and processed as above using LI-COR.
Generation of VAC14 CRISPR Mutant Cells.
Targeting vectors for the mutation of VAC14 were constructed following the protocol described in ref. 94. pSpCas9(BB)-2A-Puro gRNAs were selected from ref. 104. Sequence-confirmed constructs were transfected into HeLa cells using the Fugene6 transfection reagent (Promega) according to the manufacturer’s instructions. Cells were selected using puromycin at 1 μg/mL and subsequently split into 96-well plates to grow individual clones. To confirm that some cells had been mutated by the CRISPR constructs, a SURVEYOR assay (Transgenomic) was performed on the cells after selection to detect heteroduplexes that result from successful targeting. Clone 364C displayed increased invasion that could be complemented by VAC14 plasmid expression. DNA was extracted from clone 364C by QuickExtract (Epicentre), and the region was PCR amplified with Herculase II DNA polymerase (Agilent Technologies) and cloned using the Zero-Blunt TOPP PCR Cloning kit (Invitrogen). Plasmid was transformed into One Shot Top10 Competent Escherichia coli, and six individual colonies were chosen for sequencing. Three clones had a 1-bp insertion, and three clones had a 158-bp insertion, both resulting in a premature stop codon early in the VAC14 ORF.
HeLa Transfection Experiments.
HeLa cells were plated at 1.5 × 105 cells per well in 96-well plates and transfected using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher). Plasmids used included human VAC14mycDDK (OriGene clone #RC214195) and VAC14-gfp. BglII and XhoI were used to move VAC14-mycDDK into the BglII and SalI sites of pEGFP-N3 (Clontech).
For assessing inhibition of Salmonella spp. invasion, each well contained 100 ng of plasmid, 0.2 µL p3000, and 0.1 µL of Lipofectamine 3000. At 24 h cells were imaged to determine rescue of the vacuolated phenotype by scoring 100 cells as either “vacuoles” or “no vacuoles.” At 48 h the invasion assay was performed. Expression levels were assessed by Western blot for human vac14 (1:500) (SAB4200074; Sigma).
Docking Assay.
The bacterial docking assay was performed by infecting both WT HeLa and vac14−/− cells with IPTG-induced S. Typhi at an MOI of 50 for 1 h. Cells were washed twice with PBS, fixed with 3% PFA at RT for 30 min, washed twice again, and blocked with 5% NDS. Cells were treated with rabbit anti-Salmonella (1:500 dilution) (MA183451; Fisher) overnight at 4 °C. The secondary antibody used was donkey anti-rabbit 568 for 1 h at RT. Cells were stained for 20 min with Hoechst dye. Cells were washed, mounted, and imaged. Ten images were taken per well at 40×. Nuclear staining was quantified using Fiji (threshold, particle analysis), and green- and red-stained bacteria were counted manually.
Cholesterol Quantification and Modulation.
Filipin staining was quantified using the BD FACSCanto II system (Becton Dickinson) from the Duke Flow Cytometry Shared Resource. Both WT HeLa and vac14−/− cells were trypsinized and fixed with 3% PFA for 30 min at RT. They were then stained with 0.05 mg/mL filipin diluted in PBS (Sigma F9765) for 2 h at RT. Ten thousand cells were counted, and the mean intensity was used to determine the relative amount of cholesterol in the cells. Imaging flow cytometry was performed on 1 million cells per sample. Cells were washed with PBS, fixed with 3.7% PFA for 20 min at RT, and stained with WGA Alexa Fluor 680 (Thermo) for 10 min, 0.05 mg/mL filipin (Sigma) for 2 h, or both and were run through the ImageStream X Flow Cytometer (Amnis Corp). Analysis was performed using ISX software to determine the colocalization and intensity of WGA and filipin at the plasma membrane. Expression levels of genes that regulate cholesterol synthesis (HMGCR) and uptake (LDLR) were determined by TaqMan qPCR assays (Thermo) on a StepOnePlus Real-Time PCR machine (Thermo).
Cholesterol depletion was performed by pretreating the cells with MβCD (Sigma) at the indicated concentrations for 1 h. After treatment, cell medium was changed, and cells were subsequently infected with S. Typhi. Cholesterol repletion in cells was done by pretreating cells with water-soluble cholesterol (Sigma) at the indicated concentrations for 1 h, changing medium, and infecting cells with S. Typhi.
For the CAP LCLs, free cholesterol was quantified using the Amplex Red Cholesterol Assay Kit (Thermo Fisher Scientific) in 17 batches of ≤12 LCLs per batch and was normalized to total cellular protein content. rs8060947 and rs8044133 were imputed from Omni2.5+Exome and Cardio-MetaboChip genotypes using MaCH-Admix (CAP African Americans) or as previously described for the CAP European Americans (105).
Microscopy.
HeLa cells were fixed at the indicated times with 3.7% PFA for 20 min at RT. Blocking and permeabilization were carried out for 30 min with 0.2% saponin, 5% NDS in PBS. Primary antibody staining was carried out against anti-VAC14 (1:500 dilution) (Abnova H00055697-B01P) or anti-Salmonella (1:500 dilution) (Sigma). Secondary antibodies were conjugated to Alexa Fluor 568 and Alexa Fluor 647 (Molecular Probes). Actin filaments were stained with Alexa Fluor 647 Phalloidin (1:500 dilution) (Molecular Probes). The area of ruffling around Salmonella bacteria was calculated using Fiji by drawing a circle around the ruffles and calculating the area. Cholesterol staining was performed using 0.05 mg/mL filipin (Sigma) for 2 h at RT. Imaging was done with a Zeiss 710 inverted fluorescence microscope and an EVOS fluorescent microscope.
Typhoid Fever Association Study.
DNA samples from typhoid patients (n = 500) were collected as part of larger epidemiological or clinical studies that were undertaken in Viet Nam between 1992 and 2002. These clinical studies, which have been described previously (95–98), took place at the Hospital for Tropical Diseases in Ho Chi Minh City and Dong Thap Provincial Hospital, Vietnam, and the samples have been previously used in human genetic studies (5). Patients were defined as children or adults with clinical signs and symptoms of typhoid fever with culture-confirmed S. Typhi in their blood or bone marrow. Blood samples for DNA extraction were collected at the time of patient recruitment to the clinical study, and case report forms were used to record demographic and clinical information.
The population control group comprised 496 DNA samples extracted from the umbilical cord blood of newborn babies born in 2003 at Hung Vuong Obstetric Hospital in Ho Chi Minh City, Vietnam. All patients and controls were unrelated, and the majority of individuals were ethnic Vietnamese Kinh (>98%) as assessed by questionnaire.
Written informed consent for clinical study participation was obtained by the treating physician from the patients or the patient’s\\ir parent/guardian and from the baby’s mother for population controls. All protocols were approved by the scientific and ethical committees at the Hospital for Tropical Diseases, the Dong Thap Provincial Hospital and the Health Services of Dong Thap Province in Vietnam. Ethical approval was also granted by the Oxford Tropical Research Ethics Committee, Oxford University, Oxford, United Kingdom, the Duke IRB, and the Human Research Ethics committee of the University of Melbourne.
Blood was collected in EDTA anticoagulant, and genomic DNA was extracted from the blood pellet using Qiagen blood midi or maxi kits (Qiagen) or the Nucleon BACC genomic DNA extraction kits (GE Healthcare). DNA was quantified by PicoGreen (Molecular Probes, Invitrogen).
rs8060947 was genotyped by Taqman using a predesigned assay kit (Applied Biosystems). This was performed using a LightCycler 480 Probes Master kit on the LightCycler 480 real-time PCR system (Roche).
Genotypic deviations from Hardy–Weinberg equilibrium were assessed using a χ2 statistical test. Pearson’s χ2 test was performed to assess an association between disease phenotype, allele frequencies, and recessive genotype (AA). The OR and 95% CI were used to measure the risk of disease associated with a specific allele (A).
The typhoid fever case and control samples used in this candidate gene study are primarily a subset of a larger Vietnamese sample set that has undergone genome-wide genotyping (5). Principal component analysis of this GWAS dataset found that all enteric fever cases had sufficiently matched controls (Figs. S2 and S3 and ref. 5), which provides strong evidence no significant population stratification is present in our candidate gene study population.
Zebrafish Infections.
Studies involving zebrafish (strain AB*) were approved by Duke Institutional Animal Care and Use Committee and were performed in accordance with national and institutional guidelines. WT zebrafish embryos (strain AB*) were injected in the swim bladder at 4 dpf with ≈100 cfu of S. Typhi. Images were taken 24 h postinfection at 2× and 10× magnification. Fish were scored based on bacterial localization as cleared, localized to the swim bladder, disseminated out of the swim bladder, or dead due to bacterial burden. Approximately 25 fish were injected per bacterial strain for each experiment. Ezetimibe (10 μM) dissolved in DMSO was added to the fish water 24 h before S. Typhi injections. Filipin staining was performed on whole fixed fish at 5 dpf. Fish were initially fixed with 4% PFA overnight at 4 °C, washed three times with PBS, and then stained with filipin (0.05 mg/mL) overnight at 4 °C and subsequently imaged.
Statistical Analysis.
Descriptive statistics, parent–offspring regression, and Q–Q plots were performed with GraphPad Prism 6 (GraphPad Software) and with R (99). Genome-wide association analysis was conducted with PLINK v1.07 (33). Analysis was carried out with QFAM-parents with adaptive permutation under default settings. The QFAM procedures implemented in PLINK use linear regression to test for association while using permutation of within- and between-family components separately to control for family structure (106). Genotypes were from HapMap phase 3 release 2 (1,439,782 SNPs). The imputation of autosomal genotypes included two steps: a prephasing step using SHAPEIT2 (100) and an imputation step using IMPUTE2 (101). After filtering out imputation accuracy scores (IMPUTE’s INFO) <0.9 and minor allele frequency <0.01, we obtained 17,759,517 SNPs (6,967,393 SNPs have minor allele frequency >0.05). Q–Q plots were plotted using the quantile–quantile function in R. A regional Manhattan plot of the VAC14 region was made using LocusZoom (102). P values for eQTL analysis were calculated using linear regression in GraphPad Prism using unrelated individuals (parents only in trio data).
Supplementary Material
Acknowledgments
We thank Samuel I. Miller for early support of this work; T. Y. Chang for thoughtful discussions on cholesterol distribution and trafficking; the clinical staff from the Hospital of Tropical Diseases, Ho Chi Minh City, and Dong Thap Provincial Hospital, Vietnam, who initially diagnosed and studied the patients with typhoid fever; and Dr. Nguyen Thi Hieu from Hung Vuong Obstetric Hospital for the collection of the cord blood controls. M.I.A., L.C.G., P.L., L.W., D.M.T., S.J.D., and D.C.K. were supported by NIH Grants R01AI118903 and K22AI093595. D.C.K. was also supported by a Duke University Whitehead Scholarship and the Butler Pioneer Award. M.I.A. is supported by a National Science Foundation Predoctoral Fellowship. L.C.G. was supported by a Duke Molecular Genetics and Microbiology Summer Undergraduate Research Engagement Fellowship. S.H.O. was supported by an Australian National Health and Medical Research Council CJ Martin Early Career Fellowship, Grant 1053407. S.J.D. and T.T.B.T. were supported by the Wellcome Trust Major Overseas Program in Viet Nam, Grant 089276/Z/09/Z. E.T., Y.-L.K., and M.W.M. were supported by NIH Grants U19 HL069757 and P50 GM115318. J.I.R. was supported by NIH National Center for Advancing Translational Science University of California, Los Angeles Clinical and Translational Science Institute Grant UL1TR001881. C.M.M. was supported by National Institute of Allergy and Infectious Diseases/NIH Grant F30 AI126693. The graphical abstract was generated by itFigures Consulting. Research reported in this publication was supported by the Duke Light Microscopy Core and Flow Cytometry Shared Resource. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding sources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706070114/-/DCSupplemental.
References
- 1.Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 2014;68:317–336. doi: 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- 2.Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet. 2005;366:749–762. doi: 10.1016/S0140-6736(05)67181-4. [DOI] [PubMed] [Google Scholar]
- 3.Marineli F, Tsoucalas G, Karamanou M, Androutsos G. Mary Mallon (1869–1938) and the history of typhoid fever. Ann Gastroenterol. 2013;26:132–134. [PMC free article] [PubMed] [Google Scholar]
- 4.Mason WP. Typhoid Mary. Science. 1909;30:117–118. doi: 10.1126/science.30.760.117-a. [DOI] [PubMed] [Google Scholar]
- 5.Dunstan SJ, et al. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat Genet. 2014;46:1333–1336. doi: 10.1038/ng.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu L, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko DC, et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc Natl Acad Sci USA. 2012;109:E2343–E2352. doi: 10.1073/pnas.1206701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko DC, et al. A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease. Am J Hum Genet. 2009;85:214–227. doi: 10.1016/j.ajhg.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko DC, Urban TJ. Understanding human variation in infectious disease susceptibility through clinical and cellular GWAS. PLoS Pathog. 2013;9:e1003424. doi: 10.1371/journal.ppat.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas RE, et al. A cellular genome-wide association study reveals human variation in microtubule stability and a role in inflammatory cell death. Mol Biol Cell. 2014;25:76–86. doi: 10.1091/mbc.E13-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, et al. Human genetic and metabolite variation reveal methylthioadenosine is a prognostic biomarker and inflammatory regulator in sepsis. Sci Adv. 2017;3:e1602096. doi: 10.1126/sciadv.1602096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan JE. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 17.Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson PR, Paulin SM, Bland AP, Jones PW, Wallis TS. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect Immun. 1995;63:2743–2754. doi: 10.1128/iai.63.7.2743-2754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int J Med Microbiol. 2002;291:479–485. doi: 10.1078/1438-4221-00156. [DOI] [PubMed] [Google Scholar]
- 20.Friebel A, et al. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem. 2001;276:34035–34040. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- 21.Patel JC, Galan JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol. 2006;175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakowski MA, et al. The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe. 2010;7:453–462. doi: 10.1016/j.chom.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Rosales-Reyes R, et al. Salmonella infects B cells by macropinocytosis and formation of spacious phagosomes but does not induce pyroptosis in favor of its survival. Microb Pathog. 2012;52:367–374. doi: 10.1016/j.micpath.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Souwer Y, et al. Selective infection of antigen-specific B lymphocytes by Salmonella mediates bacterial survival and systemic spreading of infection. PLoS One. 2012;7:e50667. doi: 10.1371/journal.pone.0050667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro-Eguiluz D, et al. B cell precursors are targets for Salmonella infection. Microb Pathog. 2009;47:52–56. doi: 10.1016/j.micpath.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Giannella RA, Washington O, Gemski P, Formal SB. Invasion of HeLa cells by Salmonella typhimurium: A model for study of invasiveness of Salmonella. J Infect Dis. 1973;128:69–75. doi: 10.1093/infdis/128.1.69. [DOI] [PubMed] [Google Scholar]
- 27.Misselwitz B, et al. Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect Immun. 2011;79:330–341. doi: 10.1128/IAI.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mroczenski-Wildey MJ, Di Fabio JL, Cabello FC. Invasion and lysis of HeLa cell monolayers by Salmonella typhi: The role of lipopolysaccharide. Microb Pathog. 1989;6:143–152. doi: 10.1016/0882-4010(89)90017-x. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-del Portillo F, Finlay BB. Salmonella invasion of nonphagocytic cells induces formation of macropinosomes in the host cell. Infect Immun. 1994;62:4641–4645. doi: 10.1128/iai.62.10.4641-4645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koivusalo M, et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010;188:547–563. doi: 10.1083/jcb.200908086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium IH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International HapMap Consortium et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alghamdi TA, et al. Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J Biol Chem. 2013;288:9363–9372. doi: 10.1074/jbc.M113.453712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dove SK, et al. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- 37.Schulze U, et al. The Vac14-interaction network is linked to regulators of the endolysosomal and autophagic pathway. Mol Cell Proteomics. 2014;13:1397–1411. doi: 10.1074/mcp.M113.034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernandez LD, Hueffer K, Wenk MR, Galan JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- 39.Mallo GV, et al. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J Cell Biol. 2008;182:741–752. doi: 10.1083/jcb.200804131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roppenser B, Grinstein S, Brumell JH. Modulation of host phosphoinositide metabolism during Salmonella invasion by the type III secreted effector SopB. Methods Cell Biol. 2012;108:173–186. doi: 10.1016/B978-0-12-386487-1.00009-2. [DOI] [PubMed] [Google Scholar]
- 41.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayward RD, et al. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- 45.Cadigan KM, Spillane DM, Chang TY. Isolation and characterization of Chinese hamster ovary cell mutants defective in intracellular low density lipoprotein-cholesterol trafficking. J Cell Biol. 1990;110:295–308. doi: 10.1083/jcb.110.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sewda K, et al. Cell-surface markers for colon adenoma and adenocarcinoma. Oncotarget. 2016;7:17773–17789. doi: 10.18632/oncotarget.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endo A. The discovery and development of HMG-CoA reductase inhibitors. J Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 49.Ilangumaran S, Hoessli DC. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahammad S, Parmryd I. Cholesterol depletion using methyl-beta-cyclodextrin. Methods Mol Biol. 2015;1232:91–102. doi: 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 51.Ackerman H, et al. A comparison of case-control and family-based association methods: The example of sickle-cell and malaria. Ann Hum Genet. 2005;69:559–565. doi: 10.1111/j.1529-8817.2005.00180.x. [DOI] [PubMed] [Google Scholar]
- 52.Band G, et al. Imputation-based meta-analysis of severe malaria in three African populations. PLoS Genet. 2013;9:e1003509. doi: 10.1371/journal.pgen.1003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timmann C, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- 54.Howlader DR, et al. Zebrafish as a novel model for non-typhoidal Salmonella pathogenesis, transmission and vaccine efficacy. Vaccine. 2016;34:5099–5106. doi: 10.1016/j.vaccine.2016.08.077. [DOI] [PubMed] [Google Scholar]
- 55.van der Sar AM, et al. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 2003;5:601–611. doi: 10.1046/j.1462-5822.2003.00303.x. [DOI] [PubMed] [Google Scholar]
- 56.Gratacap RL, Rawls JF, Wheeler RT. Mucosal candidiasis elicits NF-kappaB activation, proinflammatory gene expression and localized neutrophilia in zebrafish. Dis Model Mech. 2013;6:1260–1270. doi: 10.1242/dmm.012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baek JS, Fang L, Li AC, Miller YI. Ezetimibe and simvastatin reduce cholesterol levels in zebrafish larvae fed a high-cholesterol diet. Cholesterol. 2012;2012:564705. doi: 10.1155/2012/564705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clifton JD, et al. Identification of novel inhibitors of dietary lipid absorption using zebrafish. PLoS One. 2010;5:e12386. doi: 10.1371/journal.pone.0012386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Catron DM, et al. Salmonella enterica serovar Typhimurium requires nonsterol precursors of the cholesterol biosynthetic pathway for intracellular proliferation. Infect Immun. 2004;72:1036–1042. doi: 10.1128/IAI.72.2.1036-1042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai CH, et al. Cholesterol depletion reduces Helicobacter pylori CagA translocation and CagA-induced responses in AGS cells. Infect Immun. 2008;76:3293–3303. doi: 10.1128/IAI.00365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nawabi P, Catron DM, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol Microbiol. 2008;68:173–185. doi: 10.1111/j.1365-2958.2008.06142.x. [DOI] [PubMed] [Google Scholar]
- 62.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sbrissa D, et al. A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol Cell Biol. 2004;24:10437–10447. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ikonomov OC, Sbrissa D, Shisheva A. Localized PtdIns 3,5-P2 synthesis to regulate early endosome dynamics and fusion. Am J Physiol Cell Physiol. 2006;291:C393–C404. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]
- 65.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 66.Goluszko P, Nowicki B. Membrane cholesterol: A crucial molecule affecting interactions of microbial pathogens with mammalian cells. Infect Immun. 2005;73:7791–7796. doi: 10.1128/IAI.73.12.7791-7796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, et al. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci USA. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammond GR, Takasuga S, Sasaki T, Balla T. The ML1Nx2 phosphatidylinositol 3,5-bisphosphate probe shows poor selectivity in cells. PLoS One. 2015;10:e0139957. doi: 10.1371/journal.pone.0139957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 70.Henne WM. Organelle remodeling at membrane contact sites. J Struct Biol. 2016;196:15–19. doi: 10.1016/j.jsb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Saint-Jean M, et al. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du X, Brown AJ, Yang H. Novel mechanisms of intracellular cholesterol transport: Oxysterol-binding proteins and membrane contact sites. Curr Opin Cell Biol. 2015;35:37–42. doi: 10.1016/j.ceb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Lenk GM, et al. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenk GM, et al. Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am J Hum Genet. 2016;99:188–194. doi: 10.1016/j.ajhg.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Currinn H, Wassmer T. The amyloid precursor protein (APP) binds the PIKfyve complex and modulates its function. Biochem Soc Trans. 2016;44:185–190. doi: 10.1042/BST20150179. [DOI] [PubMed] [Google Scholar]
- 76.Currinn H, Guscott B, Balklava Z, Rothnie A, Wassmer T. APP controls the formation of PI(3,5)P(2) vesicles through its binding of the PIKfyve complex. Cell Mol Life Sci. 2016;73:393–408. doi: 10.1007/s00018-015-1993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balklava Z, et al. The amyloid precursor protein controls PIKfyve function. PLoS One. 2015;10:e0130485. doi: 10.1371/journal.pone.0130485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casserly I, Topol E. Convergence of atherosclerosis and Alzheimer’s disease: Inflammation, cholesterol, and misfolded proteins. Lancet. 2004;363:1139–1146. doi: 10.1016/S0140-6736(04)15900-X. [DOI] [PubMed] [Google Scholar]
- 79.Di Paolo G, Kim TW. Linking lipids to Alzheimer’s disease: Cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puglielli L, Tanzi RE, Kovacs DM. Alzheimer’s disease: The cholesterol connection. Nat Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- 81.Schneider LS, et al. Clinical trials and late-stage drug development for Alzheimer’s disease: An appraisal from 1984 to 2014. J Intern Med. 2014;275:251–283. doi: 10.1111/joim.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mangialasche F, Solomon A, Winblad B, Mecocci P, Kivipelto M. Alzheimer’s disease: Clinical trials and drug development. Lancet Neurol. 2010;9:702–716. doi: 10.1016/S1474-4422(10)70119-8. [DOI] [PubMed] [Google Scholar]
- 83.Sur D, et al. A cluster-randomized effectiveness trial of Vi typhoid vaccine in India. N Engl J Med. 2009;361:335–344. doi: 10.1056/NEJMoa0807521. [DOI] [PubMed] [Google Scholar]
- 84.Karki S, Shakya P, Cheng AC, Dumre SP, Leder K. Trends of etiology and drug resistance in enteric fever in the last two decades in Nepal: A systematic review and meta-analysis. Nephrol Dial Transplant. 2013;57:e167–e176. doi: 10.1093/cid/cit563. [DOI] [PubMed] [Google Scholar]
- 85.McGregor AC, Waddington CS, Pollard AJ. Prospects for prevention of Salmonella infection in children through vaccination. Curr Opin Infect Dis. 2013;26:254–262. doi: 10.1097/QCO.0b013e32835fb829. [DOI] [PubMed] [Google Scholar]
- 86.Näsström E, et al. Salmonella Typhi and Salmonella Paratyphi A elaborate distinct systemic metabolite signatures during enteric fever. eLife. 2014;3:e03100. doi: 10.7554/eLife.03100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hacke M, et al. Inhibition of Ebola virus glycoprotein-mediated cytotoxicity by targeting its transmembrane domain and cholesterol. Nat Commun. 2015;6:7688. doi: 10.1038/ncomms8688. [DOI] [PubMed] [Google Scholar]
- 88.Jutras I, Abrami L, Dautry-Varsat A. Entry of the lymphogranuloma venereum strain of Chlamydia trachomatis into host cells involves cholesterol-rich membrane domains. Infect Immun. 2003;71:260–266. doi: 10.1128/IAI.71.1.260-266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Voisset C, et al. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793–7799. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 90.Samuel BU, et al. The role of cholesterol and glycosylphosphatidylinositol-anchored proteins of erythrocyte rafts in regulating raft protein content and malarial infection. J Biol Chem. 2001;276:29319–29329. doi: 10.1074/jbc.M101268200. [DOI] [PubMed] [Google Scholar]
- 91.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pujol C, Bliska JB. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun. 2003;71:5892–5899. doi: 10.1128/IAI.71.10.5892-5899.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chinh NT, et al. A randomized controlled comparison of azithromycin and ofloxacin for treatment of multidrug-resistant or nalidixic acid-resistant enteric fever. Antimicrob Agents Chemother. 2000;44:1855–1859. doi: 10.1128/aac.44.7.1855-1859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.House D, et al. Cytokine release by lipopolysaccharide-stimulated whole blood from patients with typhoid fever. J Infect Dis. 2002;186:240–245. doi: 10.1086/341298. [DOI] [PubMed] [Google Scholar]
- 97.Luxemburger C, et al. Risk factors for typhoid fever in the Mekong delta, southern Viet Nam: A case-control study. Trans R Soc Trop Med Hyg. 2001;95:19–23. doi: 10.1016/s0035-9203(01)90318-9. [DOI] [PubMed] [Google Scholar]
- 98.Vinh H, et al. Double blind comparison of ibuprofen and paracetamol for adjunctive treatment of uncomplicated typhoid fever. Pediatr Infect Dis J. 2004;23:226–230. doi: 10.1097/01.inf.0000114905.87426.c2. [DOI] [PubMed] [Google Scholar]
- 99.Team RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2016. Available at https://www.r-project.org/. Accessed January 2017. [Google Scholar]
- 100.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 101.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Humphreys D, Davidson A, Hume PJ, Koronakis V. Salmonella virulence effector SopE and Host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe. 2012;11:129–139. doi: 10.1016/j.chom.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Theusch E, et al. Statin-induced expression change of INSIG1 in lymphoblastoid cell lines correlates with plasma triglyceride statin response in a sex-specific manner. Pharmacogenomics J. 2017;17:222–229. doi: 10.1038/tpj.2016.12. and erratum (2016) 16:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Purcell S, Sham P, Daly MJ. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76:249–259. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.