Significance
Enzymes responsible for the clipping of histone tails and removal of arginine-methylated histone tails still remain elusive. The underlying mechanism of high histone turnover rate in nonproliferated cells is still a mystery. How RNA polymerase II overcomes nucleosome barriers during transcription is unknown. This article described the discovery of a JmjC domain containing subfamily members JMJD5 and JMJD7, which could be responsible for these unsolved puzzles in epigenetics and transcription fields.
Keywords: histone tail, arginine methylation, clipping, JMJD5/7
Abstract
Two of the unsolved, important questions about epigenetics are: do histone arginine demethylases exist, and is the removal of histone tails by proteolysis a major epigenetic modification process? Here, we report that two orphan Jumonji C domain (JmjC)-containing proteins, JMJD5 and JMJD7, have divalent cation-dependent protease activities that preferentially cleave the tails of histones 2, 3, or 4 containing methylated arginines. After the initial specific cleavage, JMJD5 and JMJD7, acting as aminopeptidases, progressively digest the C-terminal products. JMJD5-deficient fibroblasts exhibit dramatically increased levels of methylated arginines and histones. Furthermore, depletion of JMJD7 in breast cancer cells greatly decreases cell proliferation. The protease activities of JMJD5 and JMJD7 represent a mechanism for removal of histone tails bearing methylated arginine residues and define a potential mechanism of transcription regulation.
Regulation of gene transcription by epigenetic mechanisms is one of the most important areas of inquiry in modern molecular biology (1). Much epigenetic regulation occurs via posttranslational modifications (PTMs) of histone tails and a key assumption is that such modifications should be reversible via enzymes (“erasers”) that remove the marks. However, despite the confirmed importance of methylation of histone arginines in transcriptional regulation (2), the question of whether arginine methylation is reversible is unsolved and controversial. On the other hand, the phenomenon of clipping of histone tails has been reported and has received significant attention in recent years (3–5). However, unlike other epigenetic modifications, no enzyme family has been determined to be responsible for histone clipping, although potential individual enzyme candidates have been implicated in this mechanism (3–5).
Interestingly, proteins of the JmjC domain family have been reported to possess diverse enzymatic activities (6–8). Generally, these functions are related to the JmjC/cupin-like dioxygenase domain that is the hallmark of this protein family. Published activities of various JmjC proteins include demethylation of lysines in histones, demethylation of RNA and DNA, and hydroxylation of various amino acids (6, 8, 9). While many members of the JmjC protein family have been characterized biochemically, the functions of several members, including JMJD4–JMJD8 and HSPBAP1 remain uncharacterized or controversial. For example, JMJD6 has been reported variously to be an arginine demethylase (10, 11), a hydroxylase (12, 13), a single-stranded RNA-binding protein (14), or RNA demethylase (11). Although JMJD6 has been reported to possess histone arginine demethylase activity (10, 11), we and others have been unable to replicate this result (12, 14–17). Thus, the identities of proteins that reverse methylation of arginine residues, either through conventional demethylation or others, have not been resolved.
Here, we report a mechanism by which mammalian cells remove methylated arginine residues on histone tails: JMJD5 and JMJD7 proteins are proteases that recognize and cleave histone tails with methylated arginine residues. Furthermore, after the initial cleavage, JMJD5 and JMJD7 function as aminopeptidases that progressively digest the C-terminal products. Knockouts of JMJD5 and JMJD7 dramatically increase the content of arginine-methylated histone tails as well as the overall content of histone subunits in cells. Deletion of JMJD7 genes in human breast cancer cells drastically reduces their growth in soft agar. We conclude that JMJD5 and JMJD7 specifically cleave histone tails containing methylated arginine and progressively digest the cleaved histone product, which might regulate nucleosome stability, replacement, or turnover. Our identification of functions of these JmjC proteins sheds light on regulatory mechanisms that may impact transcription regulation and human diseases in the field of epigenetic gene regulation and tumorigenesis.
Results
c-JMJD5 and c-JMJD7 Remove Methyl Arginine from Histones.
Although at least nine arginine methyltransferases have been characterized, no specific enzyme has been conclusively proven to be responsible for removing methyl groups on methylated arginine residues within histone tails, although a report showed that some lysine demethylases also have activities against methylated arginines (18). In the hunt for histone arginine demethylase candidates, we hypothesized that one or more orphan JmjC domain-containing proteins, including JMJD4, JMJD5, JMJD7, and JMJD8, may be such a candidate. JMJD5 has been reported to play a critical role in embryonic development (19, 20). The exact functions of JMJD5 remain controversial. JMJD5 has been suggested to possess H3K36 lysine demethylase (21) and/or a hydroxylase activities (22, 23). The possibility remains that JMJD5 and some other “orphan” JmjC proteins target methyl arginines in histones and/or other proteins via different mechanisms.
The overall structural fold and catalytic cores of all members of the JmjC family are well defined (24). Therefore, to investigate the biochemical and biological function of orphan JmjC proteins, the catalytic cores of JMJD5 (residues 183–416, c-JMJD5) and JMJD7 (residues 26–318, c-JMJD7) (both mouse and human cDNA clones, Thermo Scientific) were overexpressed in Escherichia coli to produce recombinant JmjC proteins (Fig. S1).
Fig. S1.
The final purification results of c-JMJD5 and c-JMJD7. (A) c-JMJD5 and c-JMJD7 are used for all activity assays and crystallization. (B) GST fusion proteins of c-JMJD5 and c-JMJD7. The second major bands are GST proteins copurified with fusion proteins.
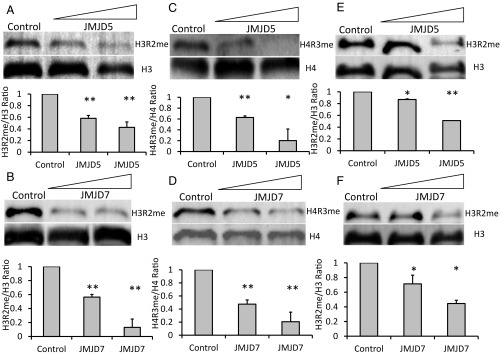
Following expression and purification of c-JMJD5 and c-JMJD7, we tested the proteins’ abilities to demethylate arginine substrates bulk histones from calf thymus, which are rich in posttranslation modifications, including methylated arginine as substrates. Initially, reaction conditions were similar to those reported previously for histone lysine demethylase assays (24–26), but were refined to new conditions later on. Following incubation of recombinant c-JMJD5 or c-JMJD7 and substrates, the reaction products were resolved using SDS/PAGE and Western blotted for detection of methylated arginines or control epitopes using commercial antibodies. c-JMJD5 and c-JMJD7 each decreased detection of histone H3 arginine 2 with dimethylation [H3R2(me2)] (Fig. 1 A and B) and histone H4 arginine 3 with symmetric dimethylation [H4R3(me2)] (Fig. 1 C and D) in a dose-dependent manner.
Fig. 1.
The reduction of methylated arginine of bulk histones and chromatin by c-JMJD5 and c-JMJD7. Each section contains two parts: (Top) Western blotting; (Bottom) quantified data of the Western blot above. A–D are results of c-JMJD5 and c-JMJD7 on bulk histone. E and F are results of c-JMJD5 and c-JMJD7 on chromatin. (A) c-JMJD5 on H3R2me2; lane 1, control; 2–1 μg, 3–2 μg. (B) c-JMJD7 on H3R2me2; lane 1, control; 2–1 μg, 3–2 μg. (C) c-JMJD5 on H4R3me2, lane 1, control; 2–1 μg, 3–2 μg. (D) c-JMJD7 on H4R3me2; lane 1, control; 2–1 μg, 3–2 μg. (E) c-JMJD5 on H3R2me2 of chromatin; lane 1, control; 2–1 μg, 3–2 μg. (F) c-JMJD7 on H3R2me2 of chromatin; lane 1, control; 2–1 μg, 3–2 μg. *P < 0.05 compared with control, **P < 0.01 compared with control.
Bulk histones differ from histones in vivo, which are assembled as octamer cores and wrapped with DNA in nucleosomes, the basic subunit of chromatin. To test whether c-JMJD5 and c-JMJD7 act similarly on isolated histones and on histones associated with chromatin, chromatin purified from calf thymus was treated with c-JMJD5 or c-JMJD7 in the same way as the bulk histones as described above. Similar to their activities on isolated histones, c-JMJD5 and c-JMJD7 decreased the content of methylated arginine H3R2(me2) within chromatin (Fig. 1 E and F).
Several mechanisms could account for the loss of the methylated arginines on histone tails. First, methyl groups may be removed directly from arginine by demethylation, similar to mechanisms used by JmjC proteins to demethylate methyl lysine (27). Second, methylated arginine may be converted to citrulline, similar to the products of peptidylarginine deiminase (PADI) enzymes (27–30). Third, methyl arginine may be removed by more extensive cleavage of histone tails by proteolysis, thus using a clipping process (3). The following experiments support the third mechanism, in which proteolytic clipping of histone tails removes methylated arginines, and, potentially, the entire tail from targeted histones.
Protease Activities of c-JMJD5 and c-JMJD7 on Bulk Histones.
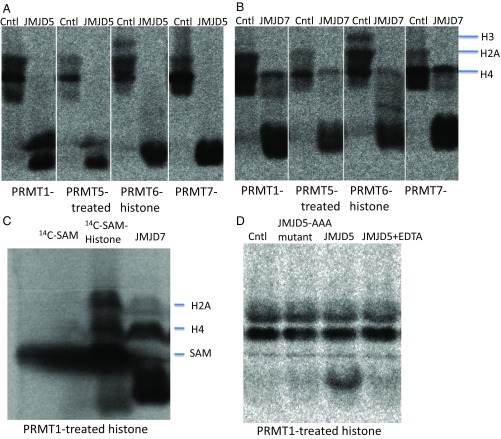
A major challenge in the study of epigenetic modifications of histone tails is the lack of specificity of some commercially available antibodies (31). Thus, instead of using antibodies, we used a method in which S-[14C-methyl]-adenosyl-l-methionine (14C-SAM) and recombinant arginine methyl transferases (PRMT1 and PRMT5–7) (32–39) were used to introduce 14C-labeled methyl moieties on bulk histones at specific sites. Methylated products were then assessed for modifications by the c-JMJD proteins. After methylation, the radioactive intensities of the 14C-methylated bulk histones were compared with or without incubation with recombinant c-JMJD5 or c-JMJD7. Changes of 14C signal were quantitated using phosphorimaging.
We assessed the specificities of the recombinant PRMT enzymes. PRMT1 has been reported to methylate H4, while PRMT5 targets H2A, H3, and H4, and PRMT6 is specific for H3. However, in our hands PRMT1 transferred 14C-methyl groups to both H2A and H4, PRMT5 modified H2A and H4 but not H3 and PRMT6 methylated H2A, H3, and H4 (Fig. 2 A and B, control lanes). Incubation of 14C-containing bulk histones with c-JMJD5 dramatically reduced the amounts of labeled histones, regardless of the PRMT used to add methyl groups (Fig. 2A, lanes c-JMJD5). c-JMJD7 exhibited similar activity, but demethylated H4 less efficiently (Fig. 2B, lanes c-JMJD7). These results occurred no matter whether the histones were labeled with PRMTs that generate symmetric dimethyl- (PRMT5) or asymmetric dimethyl-labeled (PRMT1 and PRMT6) or monomethylated (PRMT1 and PRMT7) arginines.
Fig. 2.
14C labeled bulk histone reaction with c-JMJD5 and c-JMJD7. Each section is labeled both on top and bottom. (Top) Controls and enzymes used; (Bottom) 14C-labeled bulk histone generated with individual PRMT proteins. (A) The activities of c-JMJD5 on bulk histone treated with PRMT1, PRMT5, PRMT6, and PRMT7, respectively. (B) The activities of c-JMJD7 on bulk histone treated with PRMT1, PRMT5, PRMT6, and PRMT7, respectively. (C) The 14C-SAM and digested bulk histone. (D) The activities of c-JMJD5, c-JMJD5 with three point of mutations on –HD/E(X)nH– metal chelating site (HDH → AAA), and c-JMJD5 in the presence of EDTA on bulk histone generated with PRMT1. Cntl, control; H2A, histone subunit 2A; H3, histone subunit H3; H4, histone subunit H4.
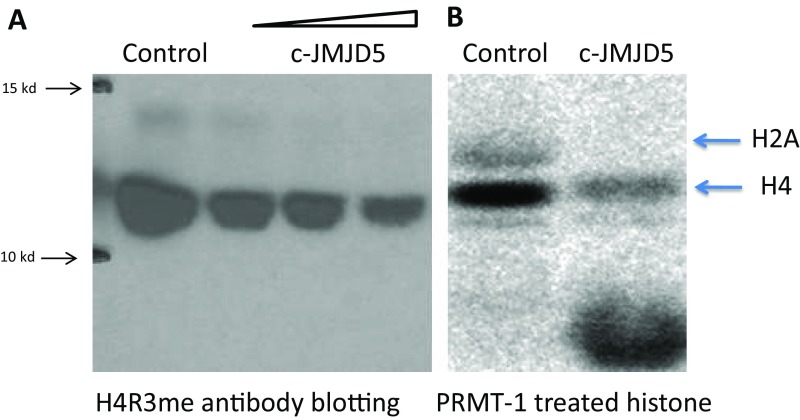
Incubation with either of the c-JMJD5/7 proteins generated additional bands that migrated faster on SDS/PAGE, suggesting conversion of the methylated moieties to products with reduced mass (Fig. 2 A and B). However, we did not observe the faster migrating bands in the Western blotting experiments shown in Fig. S2. To account for this paradox, we posit that the fragments released from these histone tails were too small to be detected using standard SDS/PAGE gels and/or because small fragments were below the resolution of Western blotting. Based on the relative location of the products (Fig. 2C), we suspected that the smaller fragments of the histone tails were too small for antibody recognition in our previous assays (Fig. S2).
Fig. S2.
(A) Western blotting of bulk histone with anti-H4R3me antibodies after reaction with c-JMJD5. (B) Bulk histone labeled with C14-SAM by PRMT1 after reaction with c-JMJD5.
Our data suggest that c-JMJD5 and c-JMJD7 are both proteases, because they cleave off histone tails with all possible variations of methyl arginine, including monomethyl-, asymmetric dimethyl-, and symmetric dimethyl arginine. To estimate the approximate size of the cleavage products, we electrophoresed 14C-SAM (molecular weight of 398.44 Da) by itself or in a mixture of 14C-SAM and bulk histones as a control with and without c-JMJD7 (Fig. 2C). The bands generated by c-JMJD7 migrated faster than SAM alone (Fig. 2C). This suggests that these small radioactive bands probably contain only one, two, or three amino acid residues.
Overall, these results suggest that c-JMJD5 and c-JMJD7 both remove methylated arginines, whether they are monomethylated (products of PRMT1 and PRMT7), symmetrically (products of PRMT5) or asymmetrically (products of PRMT1 or PRMT6) dimethylated, from histones by proteolytic cleavage.
To further characterize and confirm the proteolytic activities of c-JMJD5 and c-JMJD7, we ruled out protease contamination of the proteins. Several approaches were used. First, a mixture of protease inhibitors was added to all reactions. Second, if c-JMJD5 and c-JMJD7 were the responsible enzymes, they should be metalloproteases, given the dependence of other members of this family on divalent cations. To find out whether the proteolytic activity we observed was from metalloproteases, we added EDTA to the assays. The activities of both the c-JMJD5 and c-JMJD7 preparations were dependent on divalent cations (Fig. 2D). Third, to determine whether the iron chelating motif –HXD/E(X)nH–, which is conserved in all JmjC proteins, is required for activity, the two His residues and the Asp residue of the motif were mutated to Ala in c-JMJD5 (c-JMJD5-AAA, triple mutation). The activity of c-JMJD5-AAA was almost completely abolished (Fig. 2D).
Cleavage Sites of c-JMJD5 and c-JMJD7 on Histone Tails.
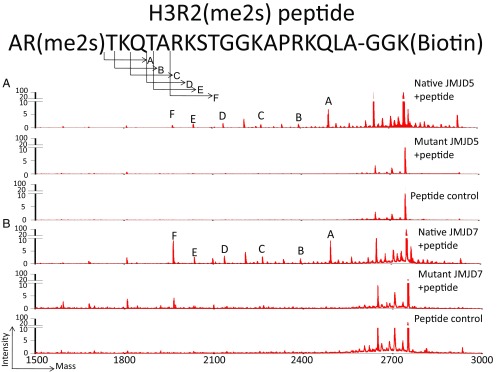
Our detection of the proteolytic activities of c-JMJD5 and c-JMJD7 on methylated histone tails begs the question as to which sites are recognized specifically by these proteins. Due to the nonspecific reactions (or cross-reactions) of antibodies and the multiple potential methylation sites of PRMT1, PRMT5, PRMT6, and PRMT7, neither approach described above identified the specific sites recognized by c-JMJD5 or c-JMJD7. It was unclear whether c-JMJD5 and c-JMJD7 recognize methylated H4R3(me2), H3R2(me2), and/or other sites. To define the specificities of c-JMJD5 and c-JMJD7, synthetic histone peptides marked with pH3R2(me2s) were used as substrates in cleavage assays. After the reactions, the mixtures were subjected to mass spectrometry analysis on a MALDI-TOF/TOF analyzer. Both c-JMJD5 and c-JMJD7 generated a series of peaks from pH3R2(me2s)-containing H3 tail peptide (Fig. 3). Of particular interest, we detected a peak with molecular weight of 2,494.30 Da, which is 255.17 less than the original peptide (MW2749.47 Da) (peak A, Fig. 3A). This cleavage product is predicted to result from cleavage of the first two N-terminal residues of pH3R2(me2s). We found a similar result of c-JMJD7 (peak A, Fig. 3B). This suggests that both c-JMJD5 and c-JMJD7 cleave the H3 tail just after the dimethylated arginine residue (Fig. 3). This cleavage product is consistent with results of the 14C-labeled bulk histone assays, which generated peptide fragments smaller than the 14C-labeled SAM itself after digestion with c-JMJD5 or c-JMJD7 (Fig. 2D).
Fig. 3.
The endopeptidase and exopeptidase activities of c-JMJD5 and c-JMJD7 on synthesized histone tails. (A) c-JMJD5 and mutant c-JMJD5 cleavage on pH3R2me2. The Top portion is the sequence of the pH3R2me2 peptide with symmetric dimethylation on R2 with molecular weight of 2,749.47 Da. After cleavage, a major band of molecular weight of 2,494.3 (peak A) is the product of peptide with the first two residues missing. (B) c-JMJD7 generated a similar profile.
In addition to the 2,494.30 peak, we also observed a series of peaks corresponding to the C-terminal fragments generated by cleavage at T3(peak B), K4(peak C), Q5(peak D), T6(peak E), and A7(peak F) (Fig. 3). Together, these peaks indicate aminopeptidase activities of c-JMJD5 and c-JMJD7, with removal of one residue at a time progressively from the N termini of the peptides following their initial cleavage after R2. One possible explanation is that, after removal of the first two residues, both c-JMJD5 and c-JMJD7 continue to digest the remaining C-terminal products of the peptides via aminopeptidase activity. This raises the possibility that these enzymes possess aminopeptidase activity for the original unmethylated peptide. However, we did not observe a product resulting from the loss of Ala1 (MW 2,678.4 Da). The data confirm that both c-JMJD5 and c-JMJD7 cleave the peptides preferentially following the methylated arginine residues, followed by progressive digestion of C-terminal products. To confirm that both endopeptidase and exopeptidase activities is originally from c-JMJD5/JMJD7 proteins, point mutations of three ion chelating residues within the catalytic center of JMJD5 (c-JMJD5-AAA)/JMJD7 (JMJD7-AAA) do not show obvious cleavage activity (Fig. 3).
Specificity of c-JMJD5 Toward pH3 Peptide with Different Modifications.
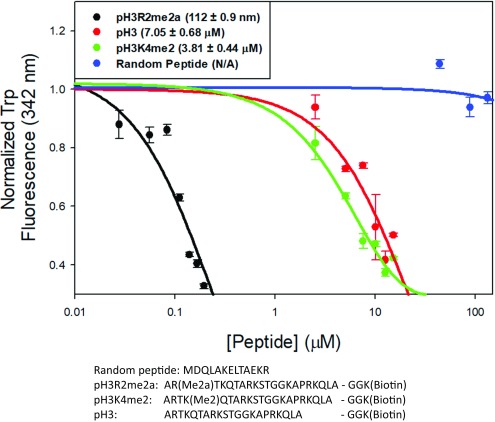
Interestingly, c-JMJD5 also showed weak activity toward pH3 peptide without modification (Fig. S3), although its activity was undetectable on pH3K4me2 (Fig. S4). Although c-JMJD5 and c-JMJD7 showed both endopeptidase and exopeptidase activities toward arginine-methylated pH3 peptide, the question here is whether c-JMJD5 or c-JMJD7 has different binding affinity, depending on the methylation status of arginine. In this regard, fluorescence polarization experiments were performed to characterize the specific binding between c-JMJD5 and pH3 peptides with different modification. In a c-JMJD5-containing mixture, a random peptide does not show any change (Fig. 4), native form pH3 peptide yields measurable binding affinity ∼7 μM (Fig. 4), while pH3R2me2a binds with surprisingly high affinity ∼0.112 μM (Fig. 4), or ∼70 times that of pH3. This binding affinity is similar to those between PHD domains and methylated lysines (0.16–30 μM; ref. 40), but is more than one order of magnitude higher than affinities between Tudor domains and methylated arginines (∼4 μM; ref. 41). This is also true compared with pH3K4me2, where the binding affinity between c-JMJD5 and pH3K4me2 is ∼4 μM (Fig. 4), or ∼40 times lower than that of pH3R2me2a (Fig. 4). These results showed that c-JMJD5 discriminates pH3R2me2a from pH3 or pH3K4me2 by exhibiting a much higher binding affinity.
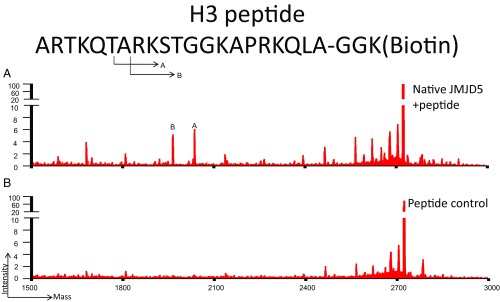
Fig. S3.
Mass spectrometry results of (A) pH3 peptide [ARTKQTARKSTGGKAPRKQLAGGK (biotin)] without incubation, and (B) pH3 peptide after 3 h incubation with c-JMJD5. These results suggest that c-JMJD5 exhibits weak activity toward the unmodified H3 peptide.
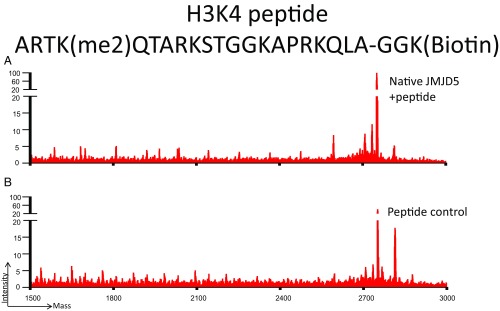
Fig. S4.
Mass spectrometry results of (A) pH3K4me2 peptide [ARTK(Me2)QTARKSTGGKAPRKQLAGGK(biotin)] without incubation, and (B) pH3K4me2 peptide after 3 h of incubation with c-JMJD5. These results suggest that c-JMJD5 exhibits weak activity toward the lysine-methylated H3 peptide, pH3K4me2.
Fig. 4.
The specific binding between c-JMJD5 and synthetic peptides. The mixture of c-JMJD5 to a random peptide showed no binding. The mixture of c-JMJD5 to pH3 peptide results in binding constant of ∼7 μM. The mixture of c-JMJD5 to pH3K4me2 peptide results in binding constant of ∼4 μM. The mixture of c-JMJD5 to pH3R2me2a peptide results in binding constant of ∼0.1 μM or ∼100 nM. The sequences of four peptides are shown.
Activities of JMJD5 and JMJD7 in Vivo.
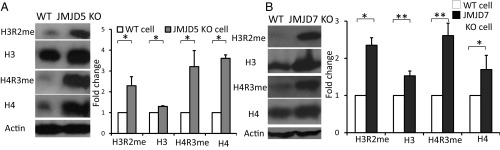
It was reported that JMJD5 is a negative regulator of cell cycle repressors, including p21 and p53, which control the transition from G1 phase to S phase (20, 42). However, how JMJD5 regulates the expression of p21 and p53 remains elusive. The cleavage activity of c-JMJD5 on methylated histone tails in vitro indicates that JMJD5 may modify histones similarly in vivo, which in turn controls the transcription of genes encoding repressors of the cell cycle. To confirm this hypothesis, we generated mouse fibroblast cells with conditionally deleted JMJD5 alleles. Levels of arginine methylation in fibroblasts with knockout of JMJD5 were compared with levels in normal fibroblasts. We found that the arginine methylation of H3R2 was dramatically increased in the JMJD5-deficient cells (Fig. 5A). This was also true for H4R3. However, it is surprising that the overall content of both H3 and H4 was also dramatically increased (Fig. 5A). This result may indicate that cleavage of methylated histone tails by JMJD5 renders the remaining nucleosomes prone to degradation, either through the aminopeptidase function of JMJD5 or by other proteases. This is consistent with the aminopeptidase functions of c-JMJD5 and c-JMJD7, which continue to clip off histone tails after the first cleavage at the methylated arginine site.
Fig. 5.
JMJD5 knockout dramatically increased the content of both arginine methylated and total H3 and H4 in mouse embryonic fibroblast (MEF) cells. JMJD7 knockout has similar effect. A and B contain two parts: (Left) Western blot of interested target and (Right) quantified results of corresponding Western blot. (A) The arginine methylation of H3 and H4 is increased in JMJD5 knockout MEFs. However, the overall content of H3 and H4 increases as well, suggesting degradation of H3 and H4 in the presence of JMJD5. (B) The knockout of JMJD7 in breast cancer cell lines leads to the increase in arginine-methylated H3 and H4, as well as the total H3 and H4. *P < 0.05 compared with WT cell, **P < 0.01 compared with WT cells.
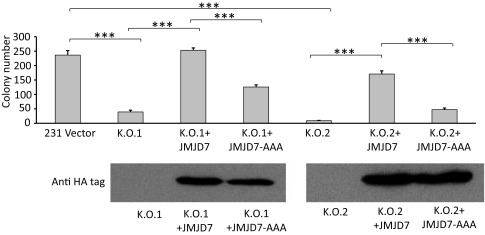
The exact functions of JMJD7 have not been determined in vivo. One report suggests a critical role of JMJD7 in the development of autism (43). A particularly puzzling aspect of JMJD7 is its readthrough transcription into the Phospholipase A2 gene, which generates a putative JMJD7–PLA2G4B fusion with predominant localization within the cytoplasm (44). We hypothesize that JMJD7 may function similarly to JMJD5, but may recognize different forms of histone tails and/or nonhistone proteins. In this regard, we used the CRISPR/Cas9 system to knock out JMJD7 in a breast cancer cell line, MDA-MB231. The content of arginine methylation of both H3R2/H4R3 and overall H3/H4 was increased with the knockout of JMJD7 (Fig. 5B), although not as dramatically as JMJD5 (Fig. 5A). Most importantly, the colony-forming abilities of JMJD7-deficient MDA-MB231 cells were greatly decreased (Fig. 6), which is similar to the inhibition of breast cancer cell growth upon down-regulation of JMJD5 (21). To avoid potential errors introduced by CRISPR/CAS9 and to cross-confirm whether the phenotype of JMJD7-deficient breast cancer cells is indeed caused by reducing JMJD7, either the native form of JMJD7, or an enzymatically inactive version of JMJD7 (JMJD7-AAA) defective in divalent cation binding, were introduced back into JMJD7-deficient cells. The native form of JMJD7 rescued normal proliferation in JMJD7-deficient cells, while JMJD7-AAA did not in two independent knockout cell lines (Fig. 6). It will be of great interest to determine the identities of genes regulated by JMJD7 as well as how JMJD7 is regulated and localized to the nucleus.
Fig. 6.
(Top) Knockout of JMJD7 in MDA-MB231 cells significantly attenuate the soft agar forming abilities of two independent JMJD7-knockout populations (K.O.1 and K.O.2). Introduction of wild-type JMJD7 rescues the phenotypes of both knockout cell lines (K.O.1 + JMJD7 and K.O.2 + JMJD7) but not when enzymatic-inactive JMJD7 was introduced (K.O.1 + JMJD7-AAA and K.O.2 + JMJD7-AAA). (Bottom) Western blot analysis of JMJD7 or JMJD7-AAA expression. ***P < 0.001.
Discussion
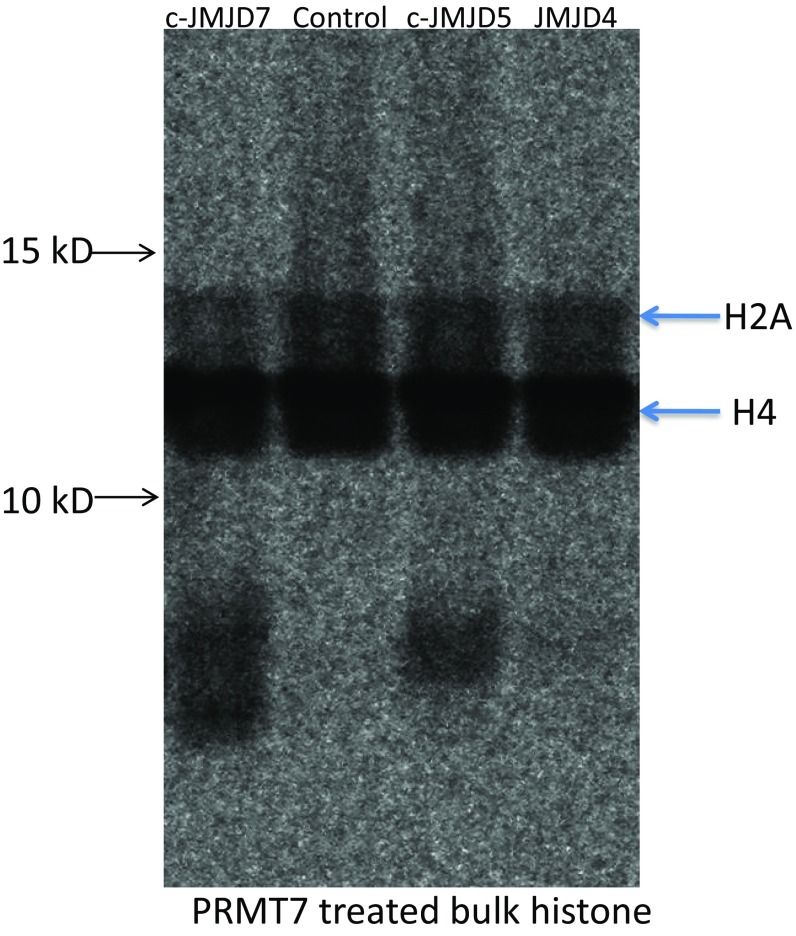
Over three decades ago, Allis et al. reported the cleavage of H3 histone tails and its potential critical role in transcriptional regulation (45). Furthermore, cathepsin L was identified as an enzyme that cleaves histone tails (46). Kouzarides and colleagues also reported that an endopeptidase in Saccharomyces cerevisiae cleaves the first 21 residues from H3 (47). A number of groups have reported different candidate enzymes responsible for the cleavage of histone tails (4, 5). However, systematic characterization of the enzyme family(ies) responsible for this activity has been lacking. Most importantly, the biological significance of this modification is not well understood. It has been suggested that the proteolytic cleavage of histone tails plays critical roles in a variety of processes, including control of the cell cycle, development, response to infection, embryonic stem cell differentiation, aging, spermatogenesis, and sporulation (4, 5). Due to the diversity of histone tail modifications, a critical question is whether a family of proteases specifically recognizes individual modified histone tails in different contexts, similar to other histone-modifying enzyme families. Our experiments suggest that JMJD5 and JMJD7 and the JmjC family of proteins meet these criteria. Biological significance of this mechanism is further supported by requirements for JMJD5 (and potentially JMJD7) in cell cycle control. Our data suggest that the clipping of histone tails is essential for epigenetic regulation. Furthermore, our results indicate the potential existence of a histone-modifying family that includes JMJD5, JMJD7, and other closely related JmjC proteins (but not JMJD4, Fig. S5).
Fig. S5.
Activities of JMJD4, c-JMJD5, and c-JMJD7. Both c-JMJD5 and c-JMJD7 show activities toward PRMT7-treated bulk histone, while JMJD4 does not.
With the identification of lysine demethylase I (LSD1) in 2004 (48), lysine demethylation was confirmed as a significant eraser of epigenetic marks. Additional studies demonstrated that this reaction is carried out by multiple oxidases, including proteins of the JmjC domain family (27). However, similar progress has not been made concerning demethylation of arginine, despite the extensive characterization of nine protein arginine methyltransferase (PRMT1–9) family members (2). A recent report claimed that some lysine demethylases have arginine demethylation activities too (18); however, nonspecific hydroxylase activities are common for JmjC protein family members. In contrast, our data showed that c-JMJD5 specifically recognizes arginine-methylated H3 (Fig. 4). In our search for arginine demethylase candidates among orphan JmjC family members, we found that JMJD5 and JMJD7 each dramatically reduced detection of methyl arginine in H3R2 and H4R3. Surprisingly, the reduction of methyl arginine is not due to simple demethylation of arginine residues, but is due instead to direct cleavage and loss of the peptide backbone adjacent to methyl arginine residues on histone tails by c-JMJD5 and c-JMJD7. These results are further confirmed by our mass spectrum data on synthetic methylated peptides. We also determined that c-JMJD5 and c-JMJD7 function as both endopeptidases and aminopeptidases (exopeptidases) to further trim histone tails, where we detected removal of individual amino acids, including Ala, Gly, Ser, Thr, Pro, Arg, Lys, and Gln residues from the H3 and H4 tails (Fig. 3). The unique nonspecific activities of these enzymes represent a mechanism of catalysis. It is worthwhile to characterize this activity further, including the full range and mechanism of amino acid recognition. These discoveries suggest that the methylated arginines removed from histone tails are neither recycled nor reversed, but rather are completely destroyed through degradation. This result is consistent with the turnover rate of histones in nonproliferating cells (49).
JMJD5 has been reported to be critical for the cell cycle (20, 21, 42). Epigenetic modifications are closely related to various cancers and are promising drug targets, notably, JMJD5 is highly expressed in cancer patients and is critical for proliferation of cancer cells (21, 50). Our knockout of JMJD7 in breast cancer cells dramatically impacts soft agar growth of MDA-MB231 cells. These results suggest that JMJD5, JMJD7, and possibly other orphan JmjC proteins contribute to tumorigenesis.
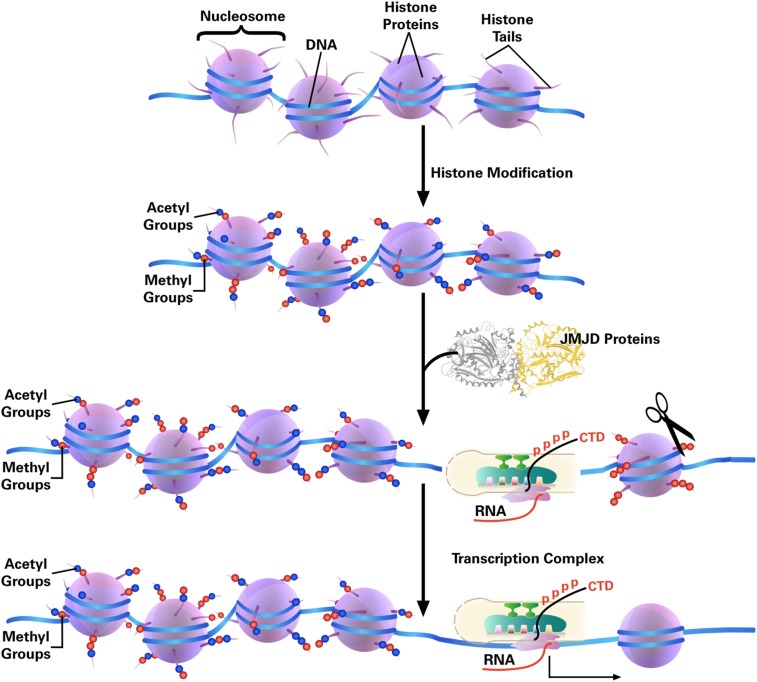
The clipping phenomenon of histone tails has been known for several decades but is largely understudied, possibly because the permanent removal of histone tails is not in line with the notion that the reversibility of epigenetic marks is critical for epigenetic modification. What could be the effect of removing the highly positively charged histone tail, in conjunction with the variety of modifications present on the transcription machinery Pol II (RNA polymerase II)? For a relatively simple bacterial system, recruitment of RNA polymerase (RNAP) to the promoter region of a target gene by transcription factors triggers transcription initiation (51, 52). However, for a eukaryotic system, the nucleosome, which contains eight subunits of histone proteins, provides another layer of structural complexity, which prevents Pol II from accessing promoters, transcription initiation, and elongation. There is abundant evidence showing that ATP-dependent chromatin remodelers, such as switch/sucrose nonfermentable (SWI/SNF), act to alter nucleosome or chromatin architecture via histone subunit exchange, ejection, and spatial reorganization (i.e., by sliding) (53–55). Based on our findings, we propose a possible mechanism in which the cleavage of the highly charged histone tails lead to weaker association between DNA and histone subunits so as to the final depletion of histone subunits from nucleosome and creation of nucleosome-free regions (Fig. 7).
Fig. 7.
Transcription elongation model. Compared with a prokaryotic system, eukaryotic DNA is wrapped around histones and not accessible for Pol II. The highly positively charged histone tails play critical roles in the packing of chromatin. Acetylation of the tails could loosen up the tight packing while cleavage of histone tails by JMJD5, JMJD7, and possibly others could lead to complete depletion of nucleosomes for the accessibility of Pol II, or Pol II is able to overcome the tailless nucleosomes as that in Archaea.
The aminopeptidase activities of both JMJD5 and JMJD7 suggest that both enzymes will continue to digest histones after the first specific cleavage at the methylated arginine residues. A scenario may be the following: Arginine methylations of histone tails act as a mark of histone degradation beyond ubiquitination. Nucleosomes or chromatin, upon truncation of histone tails, which dramatically decrease the stability of nucleosomes, will eventually be degraded. This is evidenced by our observation of the dramatic increase in H3 and H4 levels in the JMJD5 knockout mouse fibroblast cells and JMJD7 knockout human breast cancer cells (Fig. 5) and confirmed by rescue experiments (Fig. S6). The degradation of histone tails could be a part of the larger regulatory mechanism. Interestingly, it was well known that Archaea contain histone-like proteins and form nucleosome-like structures but without tails or chromatin remodeling complex (56–58). It seems that RNA polymerases alone in Archaea could overcome tailless nucleosomes during transcription since there is no known chromatin remodeling complex in Archaea (58). It may be true for Pol II to overcome nucleosomes after tail cleavage by JMJD5 and JMJD7 (Fig. 7). However, we cannot rule out if it needs the help of chromatin remodeling complexes.
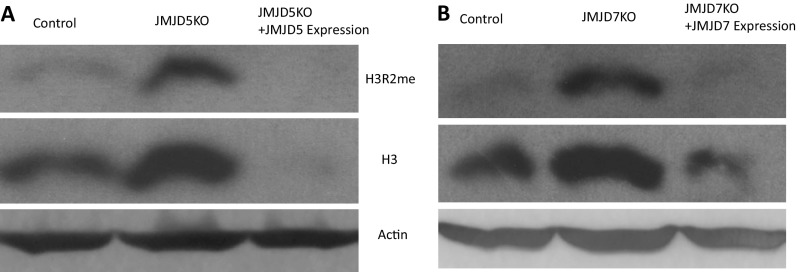
Fig. S6.
JMJD5/7 reexpression in the JMJD5/7 knockout cell lines reverses histone 3 content. (A) Blotting of H3R2me and H3 in the JMJD5 knockout cells and JMJD5 reexpression cells. (B) Blotting of H3R2me and H3 in the JMJD7 knockout cells and JMJD7 reexpression cells.
Materials and Methods
Cloning, Expression, and Purification.
The N-terminal 26-residue truncated mouse JMJD7 was cloned into the pGEX 4T-2 plasmid, which was transformed into Rosetta competent cells. A tobacco etch virus (TEV) protease cut site was engineered between the GST tag and c-JMJD7. When the cell density reached OD600 = 0.8, protein expression was induced by 1 mM IPTG at 18 °C overnight. GST–c-JMJD7 protein was purified using glutathione beads, and c-JMJD7 was excised from the fusion protein using the TEV protease. c-JMJD7 protein was further purified using a sizing column. Cloning, expression, and purification of human c-JMJD5 and mouse c-JMJD5 followed a similar procedure as that of JMJD7 (Fig. S1). Only the catalytic core of human JMJD5 (183–416) is used for activity assays.
Detection of Arginine-Methylated Histone Assay Using Calf Bulk Histone.
A total of 2 μg calf bulk histone (Sigma) was dissolved in buffer [20 mM Tris-Bis (pH ∼ 6.5–7.0), 150 mM NaCl, 50 mM (NH4)2Fe(SO4)2/ZnCl2, 1 mM α-ketoglutarate, and 2 mM ascorbic acid]. Different amounts of c-JMJD5 and c-JMJD7 were added to the tubes in the presence of protease inhibitor mixture (Roche). The reaction was carried out at 37 °C for 3–4 h. The sample was separated by SDS/PAGE, transferred to nitrocellulose membrane, and detected with H3R2me2 (Millipore) and H4R3me2a antibodies (Active Motif). After the first blotting, the bound antibody was washed away using Restore Western Blot Stripping Buffer (Thermo Scientific). H3 and H4 total antibody (Active Motif) was then probed as the loading control.
Calf Thymus Chromatin Preparation.
Calf thymus was kindly provided by L. Wysocki’s laboratory. In brief, the thymus was minced using a razor blade and homogenized with a Dounce homogenizer in sucrose/Tris/PMSF buffer (10 mM Tris, 5 mM MgCl2 and 0.3 M sucrose pH7.2 with 1 mM PMSF added before use). The homogenate was then passed through cheesecloth and pelleted by centrifugation. Nuclei were released from cells in TKM buffer (50 mM Tris, 50 mM KCl and 15 mM MgCl2) and resuspended in 1/2 TKM buffer with 1% Triton X-100. The mixture was aliquoted into 15-mL conical tubes and the cell suspension was underlaid with 60% sucrose with 1/2 TKM buffer, and centrifuged at 1,000 × g for 10 min with no break. Nuclei were collected at the interface and resuspended in 1 mM EDTA for the lysis of nuclei. The chromatin was collected by centrifugation.
Radiolabeling of Bulk Histone by PRMTs Using 14C-SAM.
A total of 100 μg calf bulk histone was dissolved in methyl transfer buffer [50 mM Tris⋅HCl (pH 8.0), 2 mM EDTA, 1 mM DTT]. Ten micrograms of PRMT1, PRMT5, PRMT6, or PRMT7 was added to separate tubes in the presence of protease inhibitor mixture. As a radioactive methyl donor, 1μCi adenosyl-l-methionine, S-[methyl-14C] (PerkinElmer) was used. The reaction was carried out at 30 °C for 2 h. The excess 14C-SAM was removed by centrifugation using Amicon centrifugation tubes with molecular weight cutoff (MWCO) of 10 kDa. The c-JMJD5 and c-JMJD7 reaction was performed as described above, differing only in that the duration time was 4 h at 37 °C. After the reaction, the sample was separated by SDS/PAGE gel, which was then dried using a vacuum pump. The 14C radioactive signal was detected using a Typhoon 9500 imager (GE Healthcare).
Mass Spectrum Analysis of Reaction Mixture.
Briefly, 1–5 μg of purified c-JMJD5 or c-JMJD7 was incubated with 1–5 μg of dimethyl pH3R2me2s (symmetric and asymmetric) in a reaction buffer [20 mM Tris-Bis (pH ∼ 6.6–7), 20 mM NaCl, 50 µM (NH4)2Fe(SO4)2, 1 mM a-ketoglutarate, and 2 mM ascorbic acid] for 1–2 h at 37 °C or room temperature. The reaction was stopped by adding 0.1% trifluoroacetic acid (TFA). Then the reaction mixture was diluted 10 times with water before adding 10 mg/mL a-cyano-4-hydroxycinnamic acid MALDI matrix in 70% acetonitrile/0.1% TFA. The mixed material was spotted on a plate to evaporate the solvent and cocrystallize the peptide/matrix. Samples were analyzed by a MALDI-TOF/TOF analyzer (4800 Plus; SCIEX) at the University of Colorado Health Sciences Center. Data were processed using Data Explorer software.
Fluorescence Polarization Experiment.
All of the regular and methylated peptides were synthesized by AnaSpec. A total of 10 μM c-JMJD5 was titrated and equilibrated with fixed concentrations of each peptide, respectively (pH3, pH3R2me2a, pH3K4me2, and random peptide), incubated at 25 °C for 30 min between each titration interval, and subject to fluorescence measurement. The buffer used in the fluorescence quenching assay was 20 mM Tris⋅HCl pH 6.5. The excitation wavelength of 280 nm and the emission wavelength of 342 nm were used for data collection and recorded with a Fluoromax-3 spectrometer. Data were normalized and the dissociation constant (Kd) for each peptide was calculated by fitting to a four-parameter sigmoidal dose–response curve with SigmaPlot v11.0.
SI Materials and Methods
Peptides.
All peptides used in this manuscript are from Anaspec: pH3R2(me2s) (AS-64631): AR(Me2s) TKQTARKSTGGKAPRKQLA–GGK(biotin)–NH2; pH3 (AS-61702): ARTKQTARKSTGGKAPRKQLA–GGK(biotin)–NH2; and pH3K4(me2) (AS-64192): ARTK(Me2)QTARKSTGGKAPRKQLA–GGK(biotin)–NH2.
The random control peptide sequence for the fluorenscence polarization experiment is MDQLAKELTAEKR.
Antibodies.
The dimethyl histone H3R2 (me2) antibody is Millipore 07585. The histone H4R3(me2a) asymetry antibody is Active Motif 39705. The histone H3 antibody is Abcam ab10158. The histone H4 antibody is Abcam ab1791. The actin antibody is Abcam ab3280.
Generation of Cells with JMJD5 Knockouts and Knockins.
Primary JMJD5 genetrap and conditional knockout MEFs were isolated at embryonic day 12.5∼13.5. For conditional JMJD5 knockout MEFs, JMJD5F/F MEFs were immortalized with SV40T (hygromycinR) and selected with 1:10 low dilution seeding for at least 10 passages, then infected with lenti-Puro-IRES-GFP or lenti-Puro-IRES-CRE:GFP and selected with 1 μg/mL puromycin. Reexpression of JMJD5 in the JMJD5 knockout cell lines was carried out by retrovirus transduction carrying the full length of mouse JMJD5.
JMJD7 Cloning, Mutagenesis, and Reexpression.
Human JMJD7 was cloned from MCF10A cell cDNA. HA tag was fused at the C-terminal of JMJD7. Then the JMJD7-HA fragment was cloned into PCDNA3.1 vector for additional expression experiments.
To avoid cutting by sgRNA that targets JMJD7, PAM- and sgRNA-targeted sequences of JMJD7 were mutated by the following primers: F2, CTGCCCAAATCGACCATGTATAATAAGGAACGCTCTGCAGCACTGGCCGGCCCTCCAG and R2, GCGTTCCTTATTATACATGGTCGATTTGGGCAGACCCAGTCCCGGTAGAAGTGGAGCGG.
To generate JMJD7 functional mutant, two pairs of primers were designed for the two-step mutagenesis. The first pair of primers was used for generating H178A and D180A. The second pair of primers was used for generating H277A: F1, CTTTGGCCAAGGCCCACTATGAGAACCTCTACTGCGTGGTCTCAGGAG and
R1, CATAGTGGGCCTTGGCCAAAGAAGTCACTGCAGCCGCCTCCCCCAGCC; and F2, GGTTCGCCCACGTCCAGCAGTCCCAGGGCTGCATC and R2, GACGTGGGCGAACCACAGAGCCGGCAGATAGAGC.
sgRNA Cloning and Lentivirus Production.
sgRNA targeting human JMJD7 was cloned into lentiCrispr vector and then transduced into 293T cells together with second generation packaging plasmids from the D. Trono laboratory (59) (psPAX2, pMD2.G) following a published, calcium phosphate-based D. Trono laboratory protocol (tronolab.epfl.ch). Supernatant was collected at 24–48 h.
The sgRNA primers sequences are: F, CACCGGCGGATAATGCACGGCCTGT and
R, AAACACAGGCCGTGCATTATCCGCC.
Soft-Agar Assay.
About 500 MDA-MB231 cells in growth medium were plated into six-well plates with 1.5 mL 0.3% (wt/vol) low-melting agar (BD), which was overlaid onto a 1.5-mL 0.6% (wt/vol) bottom agar layer. Soft-agar cultures were maintained at 37 °C and fed twice a week by dropwise addition of growth medium for colony formation. After 3 to 4 wk in culture, the colonies were counted by staining with 0.005% crystal violet.
Acknowledgments
We thank Nicole Dufour, Monika Dzieciatkowska, Frances Crawford, Kara Lukin, Desiree Straign, Gina Gill, Ryan Walsh, Abbey Esbenson, Peter Henson, James Crapo, and other researchers at National Jewish Health for their kind support; Dr. Thiago Detanico and Lawrence J. Wysocki for providing calf chromatin; Dr. Joanie Hevel for PRMT1 cDNA; Drs. Ruimin Xu and Na Yang for PRMT5 cDNA; Dr. Mark T. Bedford for PRMT6 cDNA; and Dr. Yunyu Shi for PRMT7 cDNA. H.L. is partially supported by Howard Hughes Medical Institute (HHMI), NIH Grant AI109219 (to G.Z.), and NIH Training Grant 5T32AI074491-07. S.L. is supported by Training Grant T32AI007405-26 (to P.M.). A.M.J. was supported by R35GM119575 from NIH/National Institute of General Medical Sciences. J.H. was supported by NIH Grants AI081878 and AI115696, and the Wendy Siegel Fund for Leukemia and Cancer Research. P.M. and J.W.K. are supported by HHMI. C.D. was supported by a Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology fellowship. C.W. and G.Z. were partially supported by NIH AI22295 (to P.M.), AI115696 (to J.H.), and AI109219 (to G.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706831114/-/DCSupplemental.
References
- 1.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 2.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan EM, Allis CD. Errors in erasure: Links between histone lysine methylation removal and disease. Prog Drug Res. 2011;67:69–90. doi: 10.1007/978-3-7643-8989-5_4. [DOI] [PubMed] [Google Scholar]
- 4.Zhou P, Wu E, Alam HB, Li Y. Histone cleavage as a mechanism for epigenetic regulation: Current insights and perspectives. Curr Mol Med. 2014;14:1164–1172. doi: 10.2174/1566524014666141015155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhaenens M, Glibert P, Meert P, Vossaert L, Deforce D. Histone proteolysis: A proposal for categorization into ‘clipping’ and ‘degradation’. BioEssays. 2015;37:70–79. doi: 10.1002/bies.201400118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson C, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics. 2014;6:89–120. doi: 10.2217/epi.13.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmons JM, Müller TA, Hausinger RP. Fe(II)/alpha-ketoglutarate hydroxylases involved in nucleobase, nucleoside, nucleotide, and chromatin metabolism. Dalton Trans. 2008:5132–5142. doi: 10.1039/b803512a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez S, Hausinger RP. Catalytic mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases. J Biol Chem. 2015;290:20702–20711. doi: 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 13.Han G, et al. The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J Cell Biochem. 2012;113:1663–1670. doi: 10.1002/jcb.24035. [DOI] [PubMed] [Google Scholar]
- 14.Hong X, et al. Interaction of JMJD6 with single-stranded RNA. Proc Natl Acad Sci USA. 2010;107:14568–14572. doi: 10.1073/pnas.1008832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unoki M, et al. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6) J Biol Chem. 2013;288:6053–6062. doi: 10.1074/jbc.M112.433284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014;12:e1001819. doi: 10.1371/journal.pbio.1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckel JN, et al. Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc Natl Acad Sci USA. 2011;108:3276–3281. doi: 10.1073/pnas.1008098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walport LJ, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun. 2016;7:11974. doi: 10.1038/ncomms11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HJ, et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc Natl Acad Sci USA. 2014;111:279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh S, Janknecht R. Histone demethylase JMJD5 is essential for embryonic development. Biochem Biophys Res Commun. 2012;420:61–65. doi: 10.1016/j.bbrc.2012.02.115. [DOI] [PubMed] [Google Scholar]
- 21.Hsia DA, et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Rizzo PA, Krishnan S, Trievel RC. Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol Cell Biol. 2012;32:4044–4052. doi: 10.1128/MCB.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, et al. Structure of the JmjC-domain-containing protein JMJD5. Acta Crystallogr D Biol Crystallogr. 2013;69:1911–1920. doi: 10.1107/S0907444913016600. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci USA. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 27.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc Natl Acad Sci USA. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 30.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Rothbart SB, et al. An interactive database for the assessment of histone antibody specificity. Mol Cell. 2015;59:502–511. doi: 10.1016/j.molcel.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Branscombe TL, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 33.Pollack BP, et al. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 35.Strahl BD, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 36.Guccione E, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 37.Hyllus D, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iberg AN, et al. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, et al. Structural determinants for the strict monomethylation activity by trypanosoma brucei protein arginine methyltransferase 7. Structure. 2014;22:756–768. doi: 10.1016/j.str.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA. 2010;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu H, Hu S, Baker J. JMJD5 regulates cell cycle and pluripotency in human embryonic stem cells. Stem Cells. 2014;32:2098–2110. doi: 10.1002/stem.1724. [DOI] [PubMed] [Google Scholar]
- 43.Matsunami N, et al. Identification of rare DNA sequence variants in high-risk autism families and their prevalence in a large case/control population. Mol Autism. 2014;5:5. doi: 10.1186/2040-2392-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leslie CC. Cytosolic phospholipase A2: Physiological function and role in disease. J Lipid Res. 2015;56:1386–1402. doi: 10.1194/jlr.R057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: A physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 46.Duncan EM, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos-Rosa H, et al. Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc Natl Acad Sci USA. 1982;79:1163–1165. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Z, et al. Overexpression of histone demethylase JMJD5 promotes metastasis and indicates a poor prognosis in breast cancer. Int J Clin Exp Pathol. 2015;8:10325–10334. [PMC free article] [PubMed] [Google Scholar]
- 51.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 52.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 53.Kadoch C, Copeland RA, Keilhack H. PRC2 and SWI/SNF chromatin remodeling complexes in health and disease. Biochemistry. 2016;55:1600–1614. doi: 10.1021/acs.biochem.5b01191. [DOI] [PubMed] [Google Scholar]
- 54.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 55.Henikoff S. Mechanisms of nucleosome dynamics in vivo. Cold Spring Harb Perspect Med. 2016;6:a026666. doi: 10.1101/cshperspect.a026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pereira SL, Reeve JN. Histones and nucleosomes in Archaea and Eukarya: A comparative analysis. Extremophiles. 1998;2:141–148. doi: 10.1007/s007920050053. [DOI] [PubMed] [Google Scholar]
- 57.Ammar R, et al. Chromatin is an ancient innovation conserved between Archaea and Eukarya. Elife. 2012;1:e00078. doi: 10.7554/eLife.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koster MJ, Snel B, Timmers HT. Genesis of chromatin and transcription dynamics in the origin of species. Cell. 2015;161:724–736. doi: 10.1016/j.cell.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 59.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]