Significance
Invasion of human intestinal epithelial cells resulting from the delivery of bacterial effectors into the host cell cytoplasm via a type III secretion system (T3SS) is the hallmark of Shigella, the causal agent of bacillary dysentery. Here, we provide evidence that human lymphocytes are mainly targeted by injection of T3SS effectors not resulting in cell invasion. These findings highlight the diversity of mechanisms triggered by Shigella to enlarge the panel of targeted cells and thus counteract host immunity, including adaptive immune responses. The potential impact of these data on vaccine design, in particular for live-attenuated vaccine candidates, deserves further investigation.
Keywords: Shigella, T3SS, bacterial effectors, human lymphocytes, host–pathogen cross talk
Abstract
The enteroinvasive bacterium Shigella is a facultative intracellular bacterium known, in vitro, to invade a large diversity of cells through the delivery of virulence effectors into the cell cytoplasm via a type III secretion system (T3SS). Here, we provide evidence that the injection of T3SS effectors does not necessarily result in cell invasion. Indeed, we demonstrate through optimization of a T3SS injection reporter that effector injection without subsequent cell invasion, termed the injection-only mechanism, is the main strategy used by Shigella to target human immune cells. We show that in vitro-activated human peripheral blood B, CD4+ T, and CD8+ T lymphocytes as well as switched memory B cells are mostly targeted by the injection-only mechanism. B and T lymphocytes residing in the human colonic lamina propria, encountered by Shigella upon its crossing of the mucosal barrier, are also mainly targeted by injection-only. These findings reveal that cells refractory to invasion can still be injected, thus extending the panel of host cells manipulated to the benefit of the pathogen. Future analysis of the functional consequences of the injection-only mechanism toward immune cells will contribute to the understanding of the priming of adaptive immunity, which is known to be altered during the course of natural Shigella infection.
The Gram-negative enteroinvasive bacteria Shigella spp. are responsible for bacillary dysentery, an acute rectocolitis causing high levels of morbidity and mortality predominantly in children under 5 y old in low- and middle-income countries (1). Shigella pathogenicity relies on a type III secretion system (T3SS), a device delivering bacterial effectors into the host cell cytoplasm that interfere with intracellular signaling pathways. In intestinal epithelial cells, these effectors manipulate the host cell cytoskeleton to promote bacterial internalization and reprogram cell gene expression to modulate the host innate inflammatory response, which is key for efficient establishment of Shigella infection (2). To understand why several rounds of Shigella infection induce only a short-term protective humoral immunity and thus overcome the challenges of vaccine development (3, 4), a better knowledge of Shigella–immune cell interactions is necessary. Over the last years, Shigella has been shown to not only target innate immune cells (5) but also to directly impact adaptive immune cells through mechanisms dependent upon its T3SS (6–8). Our previous studies of Shigella–lymphocytes cross talks suggested that, besides invasion of these cells, injection of Shigella T3SS effectors not resulting in invasion might also occur (6). Direct visualization of T3SS effector injection can be assessed by taking advantage of a fluorescence resonance energy transfer (FRET)-based β-lactamase assay. This translocation assay originally reported to monitor enteropathogenic Escherichia coli effector translocation (9, 10) is based on reporter bacteria expressing an effector translationally fused to the TEM-1 β-lactamase. When injected into cells loaded with the FRET substrate CCF2-AM (11), the β-lactamase cleaves the cephalosporin core of CCF2-AM, inducing a detectable shift in fluorescence from green to blue (Fig. S1). By using this assay, we showed injection of the Shigella T3SS effector IpgD into human T lymphocytes while using a high multiplicity of infection (MOI) and preventing invasion with the actin polymerization inhibitor cytochalasin D (6). These findings suggested that injection of T3SS effectors might indeed be uncoupled from cellular invasion. The purpose of the current study was thus to assess the different targeting mechanisms used by Shigella while interacting with human lymphocytes, including the formal demonstration of injection occurring in the absence of invasion. To document these events in a more physiological setting (use of a lower MOI and no inhibition of bacterial invasion), we improved both parts of the injection reporter, identifying a better-secreted T3SS effector and a more efficient β-lactamase enzyme. In addition, direct distinction between invaded vs. “injected-only” cells was implemented by using imaging flow cytometry analysis combined with Shigella expressing the DsRed fluorescent protein. This strategy successfully demonstrated that the predominant Shigella-mediated mechanism to target primary human lymphocytes is indeed “injection-only.”
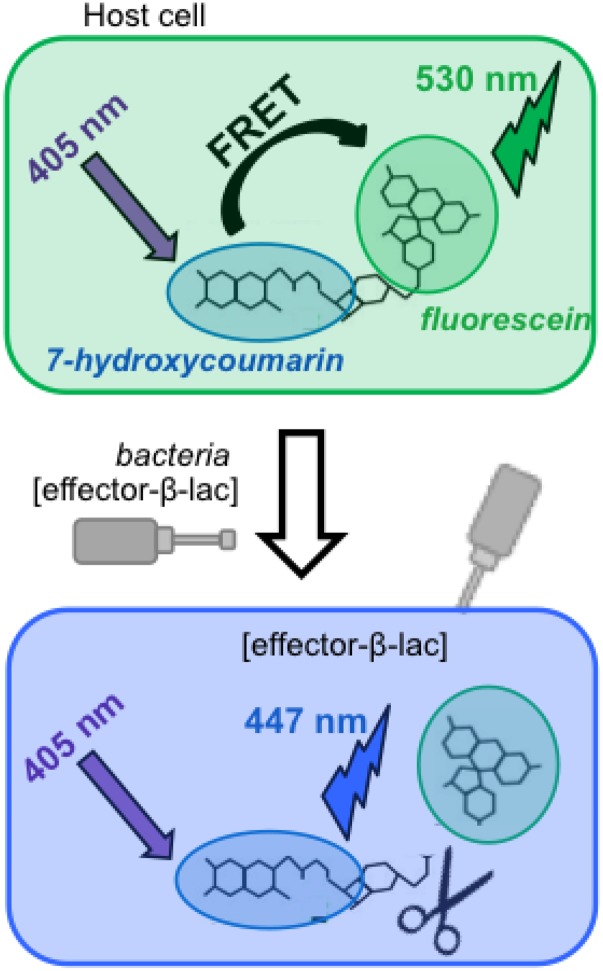
Fig. S1.
FRET-based translocation assay principle. Eukaryotic cells loaded with the fluorescent substrate CCF2-AM display a green fluorescence (emission at 530 nm) when excited at 405 nm. Upon translocation of a β-lactamase–based chimeric protein into the host cell cytoplasm, the cephalosporin core of CCF2-AM is cleaved, resulting in blue fluorescence (emission at 447 nm) upon the same excitation (11).
Results
Optimization of the Injection Reporter.
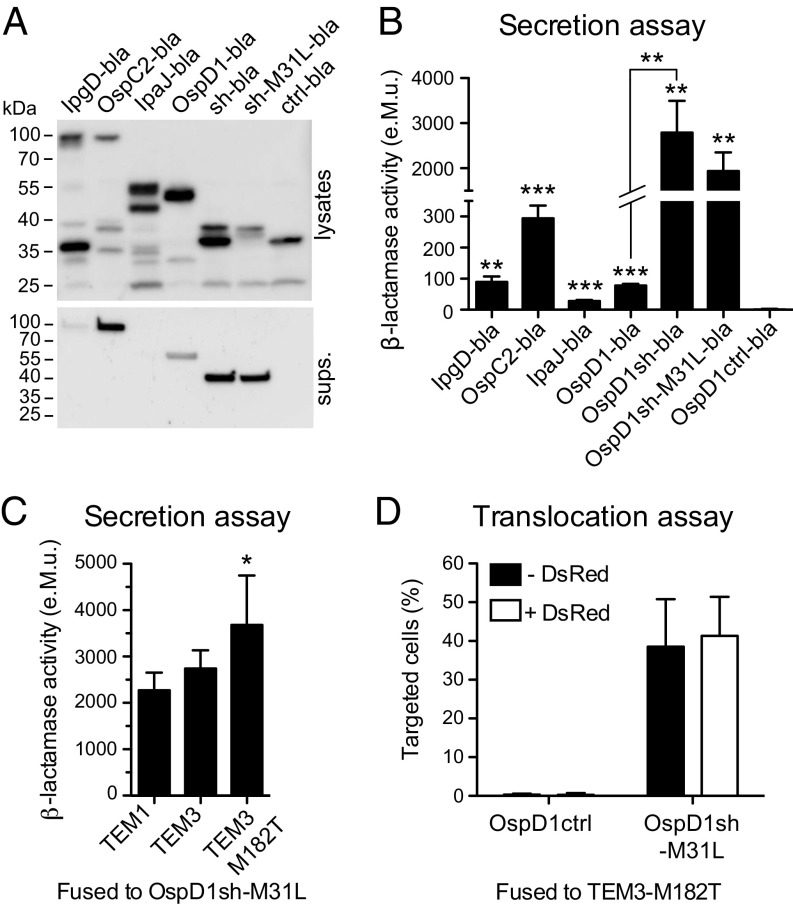
Both parts of the injection reporter were optimized by identifying a more efficiently secreted T3SS effector and a more active β-lactamase enzyme. First, a panel of T3SS effectors was expressed as chimeric proteins fused to the β-lactamase TEM-1 (bla) in the constitutively secreting ipaD Shigella mutant strain (12), and secretion of most of the full-length chimeric proteins was confirmed in culture supernatants (Fig. 1A). The β-lactamase activity measured in culture supernatants, using the colorimetric substrate nitrocefin, was shown to be optimal with the enzyme fused to the first 80 aa of the T3SS effector OspD1, named OspD1sh (Fig. 1B). Interestingly, the enzymatic activity was 35 times higher in the supernatants of ipaD strains expressing OspD1sh-bla compared with full-length OspD1-bla (Fig. 1B). Exchange of the methionine-31 residue to leucine (M31L) abolished an alternative translation start site in the OspD1sh-bla construct, resulting in only the full-length fusion protein being expressed (Fig. 1A), with no impact on the enzymatic activity detected in ipaD supernatants compared with strains expressing OspD1sh-bla (Fig. 1B). As a negative control, the sequence coding for residues 2–30 of the OspD1sh-M31L-bla construct was deleted, abolishing secretion of the resulting OspD1ctrl-bla chimeric protein into ipaD supernatants (13) (Fig. 1 A and B). Noteworthy, the enzymatic activity measured in the supernatant of ipaD bacteria expressing OspD1sh-M31L-bla was 40 times higher compared with the previously used IpgD-bla reporter construct (6) (Fig. S2A). Moreover, detection of T3SS-mediated effector translocation into CCF2-AM–loaded Jurkat T lymphocytes was significantly improved upon infection with WT Shigella expressing OspD1sh-M31L-bla compared with IpgD-bla (Fig. S2 B–E).
Fig. 1.
Optimization of Shigella T3SS injection reporter. (A) Immunoblotting with anti–β-lactamase antibody of lysates (Top) and supernatants (sups.; Bottom) of ipaD strains expressing TEM-1 chimeric proteins. The expected molecular weights of chimeric proteins were as follows (in kDa): IpgD-bla (89), OspC2-bla (85), IpaJ-bla (58), OspD1-bla (54), OspD1sh-bla (sh-bla) (38), OspD1sh-M31L-bla (sh-M31L-bla) (38), and OspD1ctrl-bla (ctrl-bla) (35). (B) Measure of β-lactamase activity in the ipaD supernatants after incubation with nitrocefin. Data are from three independent experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (Student’s t test compared with OspD1ctrl-bla condition, unless depicted otherwise). (C) Measure of β-lactamase activity in the supernatants of ipaD expressing OspD1sh-M31L fused to TEM variants after incubation with CCF2-FA. Data are from four independent experiments. *P ≤ 0.05 (Student’s t test compared with TEM1 condition). (D) Flow cytometry analysis of CCF2-AM–loaded Jurkat T lymphocytes infected for 1 h at an MOI of 50 with WT Shigella expressing the optimized T3SS injection reporter with or without DsRed. Data are from five independent experiments.
Fig. S2.
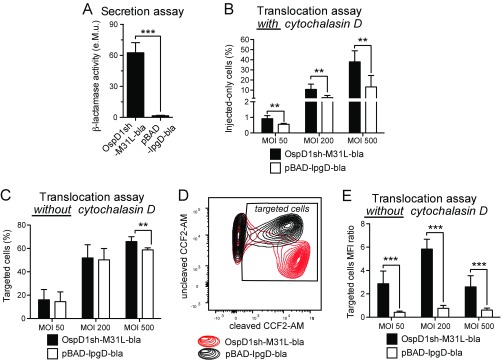
The optimized T3SS injection reporter is more sensitive at detecting T3SS translocation than the previously described IpgD-based protein. (A) Supernatants of ipaD strains expressing TEM1 (bla) fused to the optimized T3SS injection reporter or to IpgD [pBAD-IpgD-bla (6)] were incubated with nitrocefin. Enzymatic activity was calculated based on measurement of A486nm. Data are from three independent experiments. (B–E) CCF2-AM–loaded Jurkat T lymphocytes were infected for 30 min at different MOIs with WT Shigella expressing OspD1sh-M31L-bla or pBAD-IpgD-bla, followed by 2 h of incubation in medium supplemented with gentamycin to kill extracellular bacteria. Upon treatment with cytochalasin D to prevent invasion, injected-only cells were detected by flow cytometry (B). Without cytochalasin D treatment, all targeted cells were detected by flow cytometry (C–E). Under this condition, the same proportion of cells was detected as targeted at low MOIs with both reporters (C). However, fluorescence intensity of the targeted cells indicated that they were significantly more injected by the optimized reporter (D, red) compared with pBAD-IpgD-bla (D, black), which was confirmed by quantification of the ratio of mean fluorescence intensity (MFI) of cleaved CCF2-AM over uncleaved CCF2-AM (E). Data are from three independent experiments. **P ≤ 0.01; ***P ≤ 0.001 (Student’s t test compared with pBAD-IpgD-bla).
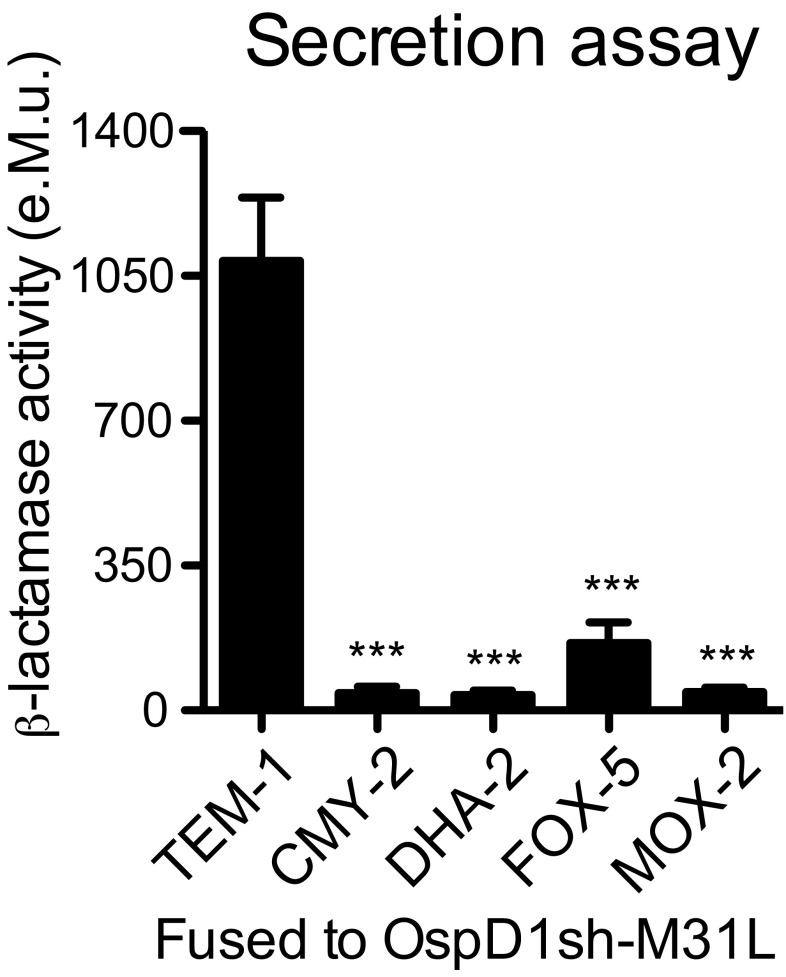
Since the core of CCF2 is composed of a cephalosporin, a class of antibiotics most efficiently processed by class C β-lactamases (14), members of this group were selected and their coding sequence fused in 3′ of the sequence coding for OspD1sh-M31L. The enzymatic activity measured upon incubation of CCF2 with supernatants of ipaD strains expressing these class C β-lactamases–based chimeric proteins was only 4–15% of that measured in supernatants of strains expressing TEM-1–based chimeric proteins (Fig. S3). OspD1sh-M31L was then translationally fused to the TEM-1 natural variant TEM-3 (also known as CTX-1) displaying increased resistance to cephalosporins (15). In addition, the point mutation M182T known to stabilize TEM β-lactamases was introduced (16). The resulting construct displayed a 60% increase in activity toward CCF2 in ipaD supernatants compared with strains expressing the TEM-1–based chimeric protein, and was thus selected (Fig. 1C). Last, to discriminate between T3SS effector translocation coupled to invasion as opposed to injection-only, the sequence coding for the DsRed fluorescent protein was inserted into the reporter plasmid under the control of a constitutive promoter (Fig. S4A). No impact on the detection of reporter translocation into Jurkat T lymphocytes was observed (Fig. 1D). Hereafter, plasmids encoding DsRed and either the T3SS injection reporter OspD1sh-M31L-TEM3-M182T or its control counterpart OspD1ctrl-TEM3-M182T are referred to as Rep-bla and Ctrl-bla, respectively. WT strains expressing Rep-bla or Ctrl-bla were able to invade, replicate, and spread within epithelial cell monolayers to a similar extent compared with the WT strain expressing DsRed only (Fig. S4B).
Fig. S3.
Class C β-lactamases are less active than TEM-1 for CCF2-FA cleavage. Supernatants of ipaD strains expressing OspD1sh-M31L fused to class C β-lactamases were incubated with CCF2-FA. Enzymatic activity was calculated based on measurement of fluorescence emission at 450 nm upon 405-nm excitation. Data are from three independent experiments. ***P ≤ 0.001 (Student’s t test compared with the TEM-1–based chimeric protein).
Fig. S4.
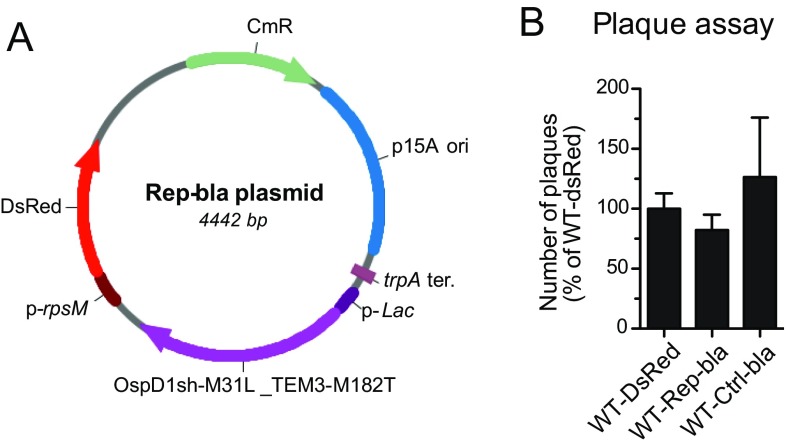
WT Shigella expressing Rep-bla or Ctrl-bla invade, replicate, and spread within epithelial cell monolayers. (A) Map of the final reporter plasmid Rep-bla. OspD1sh-M31L translationally fused to TEM-3 M182T is expressed under the control of the lac promoter (p-Lac). The DsRed fluorescent protein is expressed under the control of the constitutive rpsM promoter (p-rpsM). CmR, chloramphenicol resistance gene; p15A ori, p15A origin of replication; trpA ter., transcription terminator of trpA. (B) Confluent TC7 cells were infected for 3 d with WT Shigella expressing DsRed (WT-DsRed), Rep-bla, or Ctrl-bla. Plaques were visualized by Giemsa staining and counted. Data are from three independent experiments. No statistical difference between the different conditions was found (Student’s t test).
T3SS Injection Occurs Independently from Invasion in Jurkat T Lymphocytes.
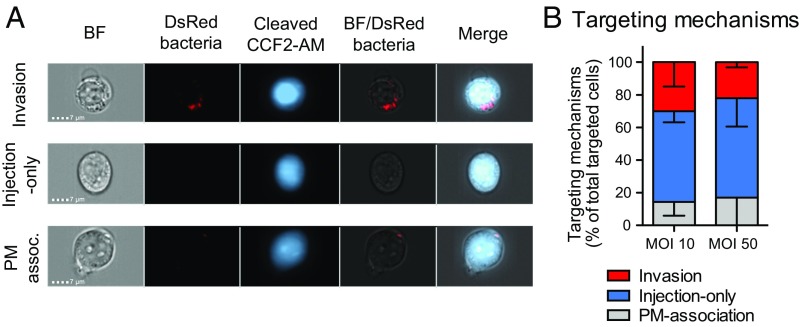
Occurrence of invasion vs. injection-only events was assessed by using imaging cytometry, a technology combining flow cytometry with single-cell epifluorescence microscopy. By applying the analysis strategy described in Fig. S5 to infection of the Jurkat human T-lymphocyte cell line, three types of targeting mechanisms were identified within WT-Rep-bla–infected cells (Fig. 2A) and their respective proportion quantified (Fig. 2B). While confirming invasion (6), injection-only appeared to be a major targeting strategy toward these cells. Among targeted cells, those with a bacterium associated to their plasma membrane (PM-association) were considered a separated category of events since the outcome of bacterial attachment to cell surface in terms of invasion vs. injection-only is unpredictable. Similar results were obtained using multiplicity of infections (MOIs) of 50 and 10, demonstrating that the number of targeted cells had no impact on targeting mechanism proportions. As previously checked for epithelial cells, the invasiveness of WT-Rep-bla was compared with that of the control strains. Its invasion rate was about twofold lower than that of WT-Ctrl-bla or WT-DsRed (Fig. S6A). Similar results were obtained with different sources of human primary cells, that is, blood lymphocytes (Fig. S6B) and lamina propria cells from colonic explants (Fig. S6C). Consequently, quantification of injection-only vs. invasion mechanisms was considered to be relevant only when the proportion of targeted cells exceeded at least twice that of invaded cells.
Fig. S5.
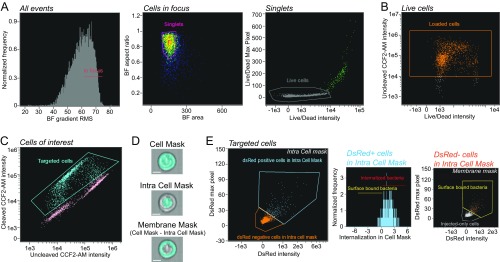
Imaging flow cytometry analysis strategy. (A) Unfocused cells were excluded before restricting the analysis to single cells (singlets) and live cells. (B) Analysis was limited to CCF2-AM–loaded cells, which are the ones with high-intensity values in the uncleaved CCF2-AM fluorescence channel, compared with nonloaded cells. (C) Among the loaded cells, the Shigella-targeted ones were identified in WT-Rep-bla–infected samples as those with high-intensity values for the cleaved CCF2-AM fluorescence. The gate to select those Shigella-targeted cells was set using the WT-Ctrl-bla–infected samples as the negative control, which did not display any targeted cells. (D) The different targeting mechanisms occurring among the targeted cells were identified by defining three masks: (i) a Cell Mask covering the entire cell, (ii) an Intra Cell Mask excluding the plasma membrane (PM), and (iii) a Membrane Mask delineating the PM by subtracting the Intra Cell Mask from the Cell Mask. The noninvasive Shigella mxiD strain was used as a control to design the mask allowing the distinction between intracellular vs. extracellular bacteria. (Scale bar: 5 μm.) (E) Among the WT-Rep bla–targeted cells, Shigella-associated cells were selected as those with a positive DsRed signal within the Intra Cell Mask (Left). Those cells were further categorized into Shigella-invaded cells (internalized bacteria) and cells with PM-associated bacteria (surface-bound bacteria) based on the DsRed intensity ratio inside the cell over the entire cell with a threshold set at 0.3 (Middle). Additional cells with surface-bound Shigella were identified among the cells displaying a negative DsRed signal in the Intra Cell Mask but a positive DsRed signal in the Membrane Mask (Right). The WT-Rep-bla–targeted cells nonclassified either as displaying internalized bacteria or PM-associated bacteria were identified as injected-only cells. Details of the features and masks used for the described gating strategy: Gradient RMS (root-mean-square for image sharpness) Area: number of microns squared in a mask; Aspect ratio: minor axis intensity divided by major axis intensity; Intensity: sum of the background subtracted pixels values; Max Pixel Intensity: the largest background-subtracted pixels; Internalization: ratio of the DsRed intensity inside the cell to the intensity of the entire cell, mapped to a log scale; Cell Mask [Erode(M01,4)]; Intra Cell Mask [Erode(Object(M01, BF, Tight), 6)].
Fig. 2.
T3SS injection occurs independently from invasion in Jurkat T lymphocytes. Imaging cytometry analysis of CCF2-AM–loaded Jurkat T lymphocytes infected with WT-Rep-bla, WT-Ctrl-bla, and WT-DsRed at an MOI of 10 or 50 for 1 h (strategy detailed in Fig. S5). (A) Identification of three events: invasion, injection-only, and association of a bacterium with the plasma membrane (PM-association). (B) Quantification of the targeting mechanism proportions among WT-Rep-bla targeted cells. Data are from two independent experiments (analysis of 403 and 1,133 targeted cells upon infection at MOIs of 10 and 50, respectively).
Fig. S6.
Invasion rates of WT-Rep-bla and control strains compared with total targeting of cells. Invasion rates were calculated based on imaging flow cytometry data analysis, by identifying invaded cells through the previously described strategy on focused, single, live, CCF2-AM–loaded cells (Fig. S5). In Jurkat T cells (A), blood-derived primary lymphocytes (B), and colonic lamina propria lymphocytes (C), WT-Rep-bla (black bars) displayed an invasion rate twofold to threefold lower than the control strains WT-Ctrl-bla and WT-DsRed (gray bars). Targeting mechanisms among WT-Rep-bla–targeted cells were analyzed in lymphocyte subsets for which the proportion of targeted cells (blue bars) exceeded by at least twofold that of cells invaded with the control strains (gray bars). Data are from two (Jurkat T cells and blood switched memory B cells) or three (activated blood lymphocytes and colonic lamina propria cells) independent experiments.
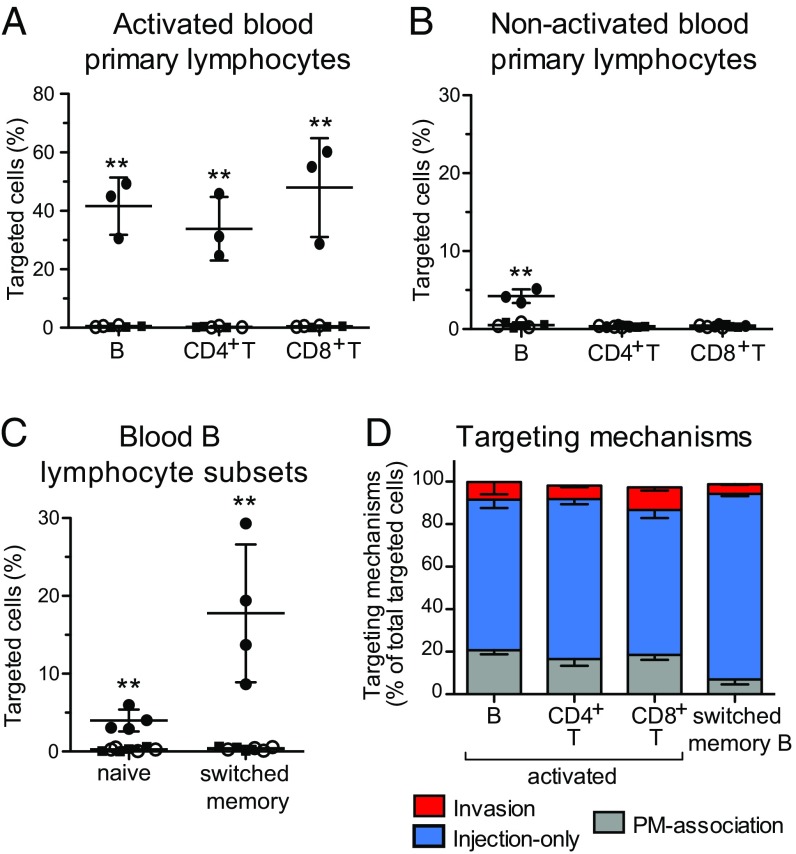
In Vitro-Activated Primary Blood Lymphocytes and Switched Memory B Cells Are Largely Targeted by Injection-Only.
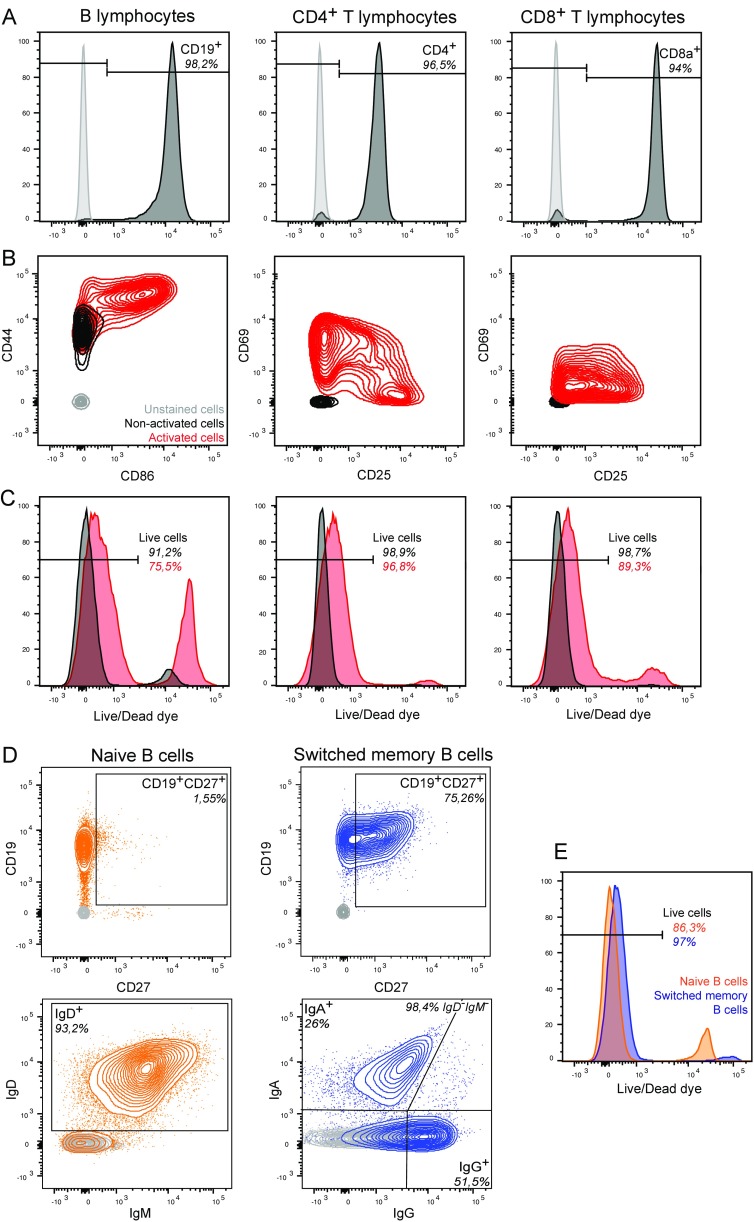
To investigate Shigella targeting of primary lymphocytes, peripheral blood mononuclear cells (PBMCs) and, subsequently, B, CD4+ T, and CD8+ T lymphocytes were isolated from different donors. Lymphocyte viability and purity were systematically assessed (Fig. S7 A–C). For each donor, some cells were kept nonactivated while others were activated in vitro using interleukin-2 (IL-2) and protein A from Staphylococcus aureus for B lymphocytes (SAC) or phorbol 12-myristate 13-acetate (PMA) for T lymphocytes. Activation efficiency and cell viability were verified before infection (Fig. S7 B and C).
Fig. S7.
Characterization of lymphocytes isolated from human peripheral blood. Lymphocyte subsets were purified from peripheral blood of healthy donors. Following isolation, viability, purification efficiency, and activation status were assessed by flow cytometry using Live/Dead fluorescent dye and surface antibody staining. Purity (A), activation status (B), and viability (C) of B, CD4+ T, and CD8+ T cells were assessed with anti-human CD19, CD4, CD8a, CD44/CD86, CD69/CD25 fluorophore-conjugated antibodies and Live/Dead dye (black, nonactivated cells; red, activated cells; gray, unstained cells). Purity (D) and viability (E) of B-lymphocyte subsets was assessed with anti-human CD19/CD27, IgD/IgM/IgA/IgG fluorophore-conjugated antibodies and Live/Dead dye (gray: unstained cells; orange, blue: stained cells). Data are from one donor representative of three.
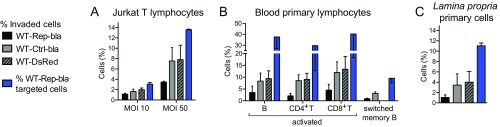
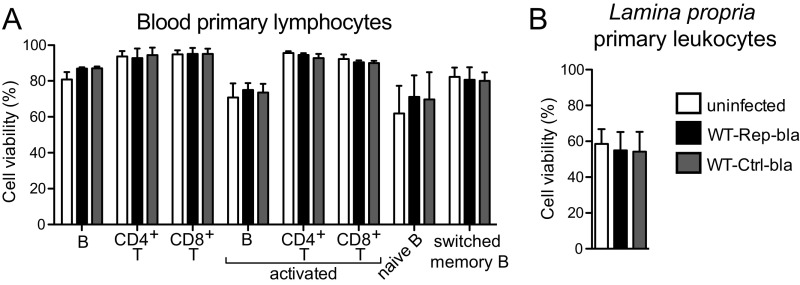
By implementing an analysis similar to that applied to the Jurkat cell line (Fig. S5), we showed that 30–60% of in vitro-activated primary B, CD4+ T, and CD8+ T lymphocytes were targeted upon infection with WT-Rep-bla with a MOI of 10 (Fig. 3A). Regarding nonactivated lymphocytes that were infected the day following isolation, only B cells were significantly targeted by Shigella (Fig. 3B), but to a lesser extent than their activated counterparts. Among these, switched memory B cells were preferential targets as opposed to naive B cells (Fig. 3C and Fig. S7 D and E). Overall, incubation with Shigella had no impact on blood primary lymphocyte viability during the time frame of the experiment (Fig. S8A). Regarding the absence of detection of nonactivated blood T lymphocytes targeted by Shigella, we cannot rule out that minor subsets, such as regulatory CD4+ T cells, might have been missed due to the analysis performed on all CD4+ T lymphocytes. Among the blood-derived activated B and T lymphocytes and the switched memory B cells, we further demonstrated that only a minor proportion of targeted cells were invaded, while injection-only constituted the main targeting mechanism (Fig. 3D).
Fig. 3.
Injection-only is a mechanism used by Shigella to target human blood primary B and T lymphocytes in vitro. Analysis of CCF2-AM–loaded human blood primary lymphocytes infected for 1 h at an MOI of 10 with Shigella WT-Rep-bla and WT-Ctrl-bla. (A–C) Flow cytometry detection of targeted cells within activated (A) and nonactivated (B) B, CD4+ T, and CD8+ T lymphocytes from three independent donors, naive and switched memory B lymphocytes from four independent donors (C). Each dot represents one donor. Filled circles: WT-Rep-bla–infected cells; open circles: WT-Ctrl-bla–infected cells; filled squares: uninfected cells. **P ≤ 0.01 (Student’s t test compared with WT-Ctrl-bla–infected cells). (D) Imaging cytometry quantification of invasion and injection-only events among targeted cells (strategy detailed in Fig. S5). Data are from three (activated lymphocytes) and two (switched memory B cells) donors (analysis of ≈5,000 and 800 targeted cells for activated lymphocytes and switched memory B cells, respectively).
Fig. S8.
Cell viability upon infection. Viability of infected primary cells was assessed by flow cytometry using Live/Dead fluorescent dye. Infection with Shigella WT-Rep-bla or WT-Ctrl-bla had no impact on the viability of cells isolated from peripheral blood (A) or colonic lamina propria (B). Data are from four independent blood and colonic tissue donors.
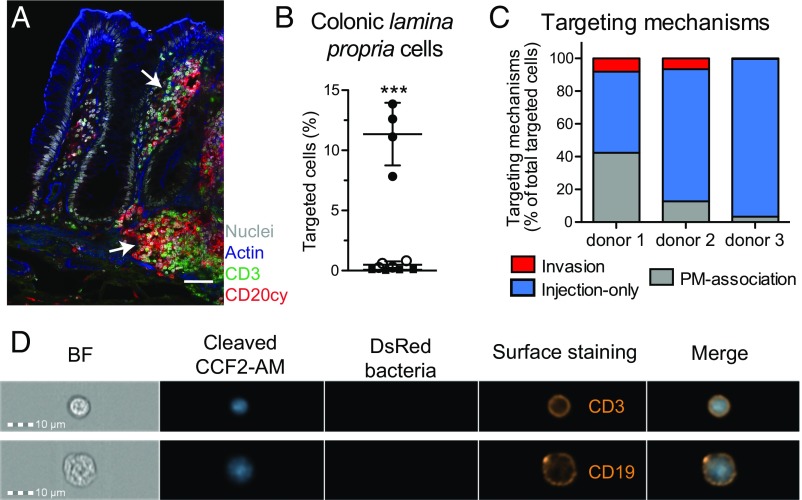
Human Colonic Lamina Propria B and T Lymphocytes Are Almost Exclusively Targeted by Injection-Only.
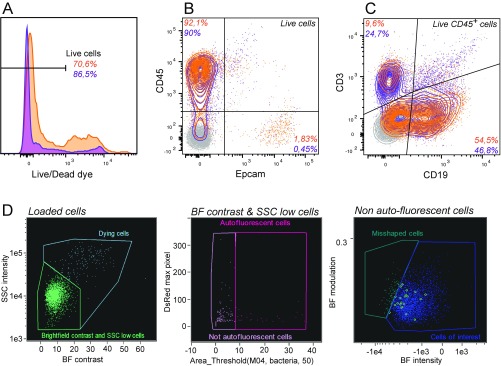
Immune cells isolated from the lamina propria of human colonic explants (Fig. 4A), which are the first cells encountered by Shigella upon crossing the colonic epithelium barrier, were investigated. Briefly, the mucosa was dissected away from the muscle layer and epithelial cells were removed through successive EDTA incubations. Lamina propria cells were released upon collagenase treatments before CCF2-AM loading. Cells isolated through this procedure displayed variable viability from donor to donor (usually around 75%; Fig. S9A). Only a minor contamination of epithelial cells (below 2%) was observed while more than 90% of isolated cells were CD45+ (Fig. S9B), among which the majority was either CD19+ or CD3+ (Fig. S9C). Following infection with WT-Rep-bla and WT-Ctrl-bla for 1 h at a MOI of 50, 60% of cells were still alive (Fig. S8B) and 8–14% of live loaded cells were detected as targeted by Shigella WT-Rep-bla (Fig. 4B).
Fig. 4.
Human colonic lamina propria lymphocytes are preferentially targeted via injection-only upon in vitro infection with Shigella. (A) Confocal single slices of human colon specimen. B (CD20cy, red) and T lymphocytes (CD3, green); DAPI-stained nuclei (gray); phalloidin-associated actin (blue). Arrows denote B- and T-lymphocyte aggregates. (Scale bar: 50 μm.) (B–D) Analysis of CCF2-AM–loaded cells isolated from human colonic lamina propria and infected for 1 h at an MOI of 50 with Shigella WT-Rep-bla or WT-Ctrl-bla. (B) Flow cytometry analysis of targeted cells. Each dot represents one donor. Filled circles: WT-Rep-bla–infected cells; open circles: WT-Ctrl-bla–infected cells; filled squares: uninfected cells. ***P ≤ 0.001 (Student’s t test compared with WT-Ctrl-bla–infected cells). Data are from four independent donors. (C) Imaging cytometry quantification of invaded, injected-only, and PM-associated event proportions among targeted cells. Data from three independent donors are represented (analysis of ≈250 targeted cells per donor). (D) Representative images of imaging cytometry acquisition displaying lamina propria injected-only targeted cells with anti-human CD3 or CD19 surface staining.
Fig. S9.
Characterization of human colonic specimen and derived cells. Viability (A) and purity (B and C) of cells isolated from human colonic lamina propria were systematically assessed by flow cytometry before infection, using Live/Dead fluorescent dye and anti-human CD45, Epcam, CD3, and CD19 fluorophore-conjugated antibodies, respectively. Data from two independent donors are represented in orange and purple. Unstained cells are depicted in gray. (D) For imaging cytometry analysis, apoptotic, autofluorescent, and misshapen cells were excluded after selection of focused, single, live loaded cells (Fig. S5 A and B) and before quantification of targeting mechanisms (Fig. S5 C–E). Apoptotic cells (dying cells) were excluded from the analysis as those with high values in the Contrast (measuring the sharpness quality of the BF image) and the Side-Scatter intensity features of the BF image (Left). Cells with high autofluorescence in the DsRed image were removed by creating a threshold mask delineating the top 50% intensity pixels in this channel (Middle). Remaining misshapen cells were removed using the “find the best feature” procedure (41) that identified the Intensity and Modulation (measures the intensity range of an image, normalized between 0 and 1) features calculated on the BF channel (Right).
The imaging flow cytometry procedure to quantify invaded vs. injected-only events was then implemented with some additional steps to exclude apoptotic, autofluorescent, and misshapen cells after gating on live loaded cells (Fig. S9D). About 50–96% of the Shigella-targeted lamina propria lymphocytes were shown to be injected-only (Fig. 4C), including CD19+ and CD3+ lymphocytes (Fig. 4D). Altogether, these findings revealed a thus-far-undetected mechanism used by Shigella to target host cells.
Discussion
In this study, we developed a Shigella T3SS-injection reporter tool adapted to the investigation of targeting mechanisms toward immune cells, using imaging flow cytometry to directly visualize invasion vs. injection-only events at the single-cell level. In the field of host–pathogen interactions, imaging flow cytometry has been used so far to study the intracellular fate of bacteria or parasites (17–20). This report describes an experimental strategy to simultaneously distinguish and quantify different Shigella targeting mechanisms among a given population of cells. Using this approach, we demonstrated that injection of Shigella T3SS effectors into some B- and T-lymphocytes subsets, including those relevant during natural infection of the human colon, does not necessarily result in cell invasion. We propose that this injection-only mechanism is at the core of interactions with cells refractory to invasion, thus extending the panel of host cells manipulated to the benefit of the pathogen. We hypothesize that susceptibility to Shigella T3SS effector injection, followed or not by invasion, depends on host cell morphology. This concept is based on our observations using the optimized translocation reporter of a preferential targeting of activated lymphocytes over their nonactivated counterparts, as well as switched memory B cells over naive B cells. Indeed, the amount of cytoskeleton components and the size of the cytoplasmic compartment, which are required for bacterial invasion and replication, are largely reduced in lymphocytes compared with epithelial cells but increase upon their activation (21, 22). In particular, considering the critical role of host cytoskeleton rearrangements for Shigella-induced invasion of epithelial cells (23), we speculate that the amount of cytoskeleton components that can be hijacked by injected bacterial effectors is critical for the outcome of T3SS injection. Above the threshold required to trigger the full process of bacterial internalization, T3SS injection results in cellular invasion, while below this threshold injection-only occurs, thus permitting an increased diversity in the nature of targeted cells, beyond those sensitive to invasion.
Thus far, targeting of human colonic lymphocytes by enteropathogenic bacteria has been poorly investigated. Indeed, modulation of B- and T-lymphocyte functions upon infection with Yersinia, Salmonella, EPEC, and EHEC has been studied using in vitro human cell lines or in vivo mouse models of infection (24–28). In contrast, our findings constitute a description of primary human colonic lymphocyte interactions with enteropathogenic bacteria.
We consider that the current findings along with our previous report on B lymphocytes (8) leaves the door open to reexamining the current understanding of T3SS-mediated Shigella pathogenicity. We previously reported a Shigella targeting mechanism toward some B-lymphocyte subsets that is independent from both invasion and T3SS effector injection, demonstrating that these cells undergo apoptosis after activation of a TLR2-dependent signaling pathway triggered by the T3SS apparatus needle tip protein IpaD (8). Therefore, we propose a paradigm for Shigella pathogenicity termed the “kiss-and-run” strategy. This depicts the ability of Shigella to manipulate targeted cells via a T3SS-dependent contact, which results in either no T3SS effector delivery into the host cell cytoplasm or their delivery not followed by cell invasion. Long studied for its life cycle as an intracellular pathogen based on its ability to invade almost any cell type in vitro (29, 30), these findings highlight an overlooked aspect of Shigella’s extracellular lifestyle. Considering the large panel of T3SS effectors reported to alter host cell processes (2, 31–33), the impact of injection-only on lymphocyte functions is likely to contribute to the observed impaired priming of host-specific immunity. It is likely that the lack of CD8+ T-cell priming reported using a mouse model of Shigella infection (suggested to be due to a T3SS effector-mediated mechanism actively interfering with antigen presentation in Shigella-infected cells) (34) might result from T3SS injection-only occurring in these cells, as reported in this work. The injection-only mechanism may also explain the Shigella-induced modulation of antimicrobial peptide production observed in noninvaded differentiated intestinal epithelial cells (35). Future studies at the single-cell level will probe the impact of the injection-only outcome on immune cells targeted by Shigella within the human colonic mucosa and might significantly impact efforts to develop a Shigella vaccine. Indeed, in particular, the design of live, rationally attenuated, orally administered vaccine candidates will have to take into consideration the so-far-underestimated T3SS activity toward B and T lymphocytes that are key for the priming of efficient protective immunity.
Materials and Methods
Bacterial Strains and Cloning Reagents.
The Shigella flexneri 5a (referred to as Shigella or S. flexneri) M90T WT strain (36) and its derivative mutant strain ipaD (37) were grown in tryptic soy broth (TSB), supplemented when appropriate with 10 μg/mL chloramphenicol. Chemically competent Escherichia coli DH5α bacteria (Invitrogen) were used for all cloning steps and grown at 37 °C in yeast tryptone medium (2xYT), supplemented when appropriate with 30 μg/mL chloramphenicol. NucleoSpin Plasmid miniprep, PCR clean-up, and gel extraction kits (Macherey-Nagel), and PureLink HiPure midiprep kit (Invitrogen) were used for DNA preparation. Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase, Taq DNA polymerase, and Phusion polymerase were purchased from Thermo Fisher Scientific.
Plasmids.
The pBAD-IpgD-TEM1 plasmid was previously described (6). Details of plasmids construction for the obtention of WT-Rep-bla are described in Supporting Information.
Bacterial Lysates and Immunoblotting.
Bacterial overnight cultures were subcultured 1/50 in TSB at 37 °C without antibiotics. Subcultures were spun at 9,000 × g for 1 min, and an equivalent amount of bacteria or supernatants was resuspended in Laemmli buffer (Bio-Rad). Boiled lysates were analyzed by SDS/PAGE, and proteins of interest were detected with mouse anti–β-lactamase antibody (Abcam, ab12251; or Novus Biologicals, NB120-12251; 1/1,000) and anti–mouse-HRP antibody (GE Healthcare; NXA931; 1/5,000), using film detection or Amersham Imager 600 (GE Healthcare).
Secretion Assay.
Overnight cultures of ipaD bacteria were grown at 30 °C and subcultured 1/50 in TSB at 37 °C for 4 h without antibiotics. Subcultures were spun at 9,000 × g for 1 min, and 100 μL of supernatants at different dilutions were incubated with β-lactamase substrates: 20 μL of nitrocefin (500 μg/mL stock; EMD Millipore) or 20 μL of CCF2-FA (25 μM stock; Thermo Fisher Scientific). Hydrolysis of the colorimetric substrate nitrocefin [absorbance, 486 nm (abs486)] and cleavage of the fluorescent substrate CCF2-FA [excitation, 405 nm; emission, 460 nm (em460)] were measured over time using TECAN Infinite M200 Pro (LifeSciences). β-Lactamase activity was calculated in equivalent Miller units (e.M.u.) as follows: Activity [e.M.u.] = 1,000 × (abs486 or em460[sample] − abs486 or em460[TSB])/(time [min] × volume [mL] × OD600[subculture] × sample dilution).
Human Cell Lines.
Jurkat T cells (clone E6-1; ATCC TIB-152) were cultured in RPMI 1640 (Gibco) supplemented with 10% heat-inactivated FCS (HI-FCS) at 37 °C in 5% CO2 incubator. TC7 cells (a clone of Caco-2) were cultured in DMEM, 1 g/L glucose, supplemented with nonessential amino acids, glutamine (all from Gibco), and 20% HI-FCS at 37 °C in 10% CO2 incubator.
Isolation of Primary B and T Lymphocytes from Human Blood.
Samples were obtained following patients’ informed consent from Etablissement Français du Sang (study approved by the Institut Pasteur’s ethical and medical committee, agreement HS2015-24009). PBMCs were purified by density separation on Ficoll Paque Plus (GE Healthcare) upon centrifugation at room temperature (RT) for 30 min at 800 × g without brake. Remaining red blood cells (RBCs) were lysed using RBC lysis buffer (Sigma). B, CD4+ T, CD8+ T, naive B, and switched memory B lymphocytes were isolated using Miltenyi MicroBeads kits (B-cell kit II, CD4+ T-cell kit, CD8+ T-cell kit, naive B-cell kit II, and switched memory B-cell kit, respectively). Cells were cultured in RPMI 1640 supplemented with 10% HI-FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco) in 37 °C and 5% CO2 incubator. When indicated, activation of lymphocytes was performed for 3 d with 100 U/mL recombinant human IL-2 (Peprotech), supplemented with 0.001% (wt/vol) SAC for B lymphocytes (protein A from Staphylococcus aureus; Sigma) or 5 ng/mL PMA for T lymphocytes (PMA; Sigma). Antibodies used for purity and activation status are listed in Table S1.
Table S1.
List of antibodies
| Antibody | Source | Method |
| CD3-APC | Beckman, IOTest IM2467 | FC |
| CD3-Biotin | eBioscience, 13-0038-82 | FC |
| CD3-FITC | eBioscience, 11-0037-42 | FC |
| CD3 | Dako, A0452 | IHC |
| CD4-PerCPCy5.5 | eBioscience, 45-0048-73 | FC |
| CD8a-PE | eBioscience, 12-0088-73 | FC |
| CD19-APC | eBioscience, 17-0199-73 | FC |
| CD19-Biotin | eBioscience, 13-0199-82 | FC |
| CD19-PacificBlue | BioLegend, 302232 | FC |
| CD19-PE | eBioscience, 12-0199-42 | FC |
| CD20cy | Dako, M0755 | IHC |
| CD25-PE | BD Pharmingen, 555432 | FC |
| CD27-PECy7 | eBioscience, 25-0279-42 | FC |
| CD44-APC | BD Pharmingen, 559942 | FC |
| CD45-PE-Vio770 | Miltenyi, 130-098-148 | FC |
| CD45-APC | BioLegend, BLE304011 | FC |
| CD69-APC | BD Pharmingen, 555533 | FC |
| CD86-PE | eBioscience, 12-0869-73 | FC |
| Epcam-PE | Miltenyi, 130-098-115 | FC |
| IgA-FITC | Miltenyi, 130-093-071 | FC |
| IgD-PerCPCy5 | BD Pharmingen, 561315 | FC |
| IgG-PE | BD Pharmingen, 555787 | FC |
| IgM-APCCy7 | BioLegend, 314520 | FC |
| Streptavidin-Qdot705 | Invitrogen, Q10163MP | FC; 2 μL |
| Mouse Ig-AF568 | Thermo Fisher, A11021 | IHC; 1/500 |
| Rabbit Ig-AF647 | Thermo Fisher, A21245 | IHC; 1/500 |
All anti-human antibodies were used for surface staining at 1/100 dilution, unless stated otherwise. FC, flow cytometry; IHC, immunohistochemistry.
Isolation of Primary Cells from Human Colonic Specimens.
Tissue specimens were obtained following patients’ informed consent from Hôpital Mondor, Gastroenterology Department, Creteil, France (collaboration with A. Amiot and I. Sobhani; study approved by the Institut Pasteur’s ethical and medical committee, agreement 2012-37). Samples were taken at a distance from the tumor in patients undergoing surgery for colonic cancer. Details of cell isolation are described in Supporting Information. Antibodies used for purity and targeted cell phenotyping are listed in Table S1.
Immunofluorescence Staining of Colon Specimens.
Immunohistochemistry procedures are presented in Supporting Information.
Shigella Infection and Translocation Assay.
Bacterial overnight cultures grown at 30 °C were subcultured 1/50 in TSB at 37 °C without antibiotics until early exponential phase was reached. Cells were seeded 1 h before infection in 96-well plates (round bottom) at 3–5 × 105 cells per well in translocation assay medium [RPMI 1640 supplemented with 2.5 mM probenecid (VWR) and CCF2-AM (Thermo Fisher Scientific); final concentration of 1 μM was used for Jurkat cells, 2 μM for primary cells]. When indicated, cytochalasin D was used at a final concentration of 1 μg/mL (Sigma-Aldrich). Bacterial subcultures were adjusted in RPMI 1640–probenecid to the indicated MOI, and centrifuged onto the cells at 300 × g and 37 °C for 5 min. Infection was allowed to proceed for indicated times at 37 °C in a 5% CO2 incubator. When indicated, gentamycin was used at 50 μg/mL to kill extracellular bacteria. Following infection, probenecid was kept in all buffers, including during washing steps. To assess viability, cells were washed in cold PBS–probenecid and stained for 30 min on ice with Live/Dead fixable near-infrared dead cell stain kit (Thermo Fisher Scientific). Infected cells were kept on ice in cold PBS–probenecid supplemented with 0.1% BSA for analysis. For imaging cytometry, cells were fixed in 1% PFA–probenecid for 10 min at RT before acquisition.
Flow Cytometry and Imaging Flow Cytometry Data Analysis.
Flow cytometry data acquisition was achieved using a FACSCantoII flow cytometer (BD Biosciences) equipped with 405-, 488-, and 633-nm lasers. FlowJo, version 10.0.8, software was used to analyze the data. Dead cells were excluded based on Live/Dead staining (Thermo Fisher Scientific) and/or forward- and side-scatters profile. A minimum of 10,000 live cells was analyzed per sample. Imaging flow cytometry data acquisition was achieved using an ImageStream X Mk I (Amnis, part of EMD Millipore-Merck) equipped with dual camera and 405-, 488-, 561-, and 633-nm excitation lasers. Samples were acquired at 40× (N.A., 0.75) magnification with the 488-nm laser switched off. A minimum of 5,000 events corresponding to focused, single cells was collected for each sample. Channels used were channels 1 and 9 [brightfield (BF)], channel 3/4 (Shigella-DsRed fluorescence), channels 7 and 8 (cleaved and uncleaved CCF2-AM fluorescence, respectively), channel 11 (staining with Qdot705 fluorophore-conjugated antibodies), and channel 12 (Live/Dead staining fluorescence). Data analysis was performed using the IDEAS, version 6.2, software (Amnis). Masks (areas of interest) and features (calculations made from masks) were generated to give quantitative measurement of the collected images.
Plaque Assay.
Plaque assays were performed as described previously (38), using TC7 cells cultured to confluency for 2 wk. Following 2-h infection with 106 bacteria at 37 °C in 10% CO2 incubator, an overlay of 0.5% agarose containing 50 μg/mL gentamycin in culture medium was poured into each well after extensive washing of the cell monolayers. Three days later, cells were fixed for 5 min in 100% ethanol and stained for 10 min in Giemsa R solution (RAL Diagnostics; diluted 1/20 in water), and plaques were enumerated.
Data Presentation and Statistical Analysis.
Prism 6.0 (GraphPad Software) was used for graphs and statistical analyses. Means and SDs are represented. Unpaired two-tailed Student’s t test was used to compare two groups. The Illustrator CS5 software (Adobe) was used to assemble figures.
Construction of the WT Rep-Bla Reporter Tool
The β-lactamase reporter plasmid is derived from pSU2718 (GenBank accession no. M64731) in which the transcription terminator of trpA has been added upstream of the lac promoter, resulting in pSU2.1tt plasmid. The sequence coding for the β-lactamase TEM-1 without its periplasm-addressing peptide was amplified from pUC18, with 5′ insertion of the Shine and Dalgarno (SD) sequence carried by pQE-60 (39), a start codon and a sequence coding for a 6-aa linker PGGGGS. This sequence was inserted downstream of the lac promoter via KpnI/XbaI sites, resulting in pSU2.1tt_TEM1 plasmid. The sequences coding for Shigella effectors and the first 80 aa of OspD1 (OspD1sh) were amplified from M90T virulence plasmid DNA and inserted in-frame via EcoRI/SmaI sites downstream of the lac promoter. Exchange of methionine-31 for a leucine into OspD1sh and the subsequent removal of the sequence coding for the N-terminal residues 2–30 (OspD1ctrl) were obtained by site-directed mutagenesis PCR. The sequences coding for the β-lactamases TEM-3, MOX-2, and DHA-2 were amplified without their periplasm-addressing peptide from Klebsiella pneumoniae strains and CMY-2 and FOX-5 from Escherichia coli strains (strains kindly provided by Marc Galimand, Antibacterial Agents Unit, Institut Pasteur, Paris and Delphine Girlich, Bacteriology-Hygiene Unit, Université Paris-Sud, Université Paris-Saclay, Paris and Hôpital Bicêtre, Assistance Publique Hôpitaux de Paris, Le Kremlin Bicêtre, France). These sequences were inserted via BamHI/XbaI restriction sites to replace TEM-1 into pSU2.1tt_OspD1sh-M31L-TEM1 plasmid. Point mutations into TEM-1 and TEM-3 were obtained by site-directed mutagenesis PCR. The coding sequence for the DsRed protein was amplified from pMW211 (40) and subcloned under the control of the rpsM promoter. This sequence was inserted downstream of the effector–β-lactamase coding sequence via XbaI/HindIII restriction sites. The final optimized plasmids pSU2.1tt_OspD1sh-M31L-TEM3-M182T_DsRed and pSU2.1tt_OspD1ctrl-TEM3-M182T_DsRed were termed Rep-bla and Ctrl-bla, respectively. The control M90T-DsRed strain expressing only DsRed was constructed by PCR removal of the reporter from the Rep-bla plasmid.
Immunofluorescence Staining of Colon Specimens
Human colon mucosa specimens were pinned flat onto 4% agarose dishes and fixed overnight in 4% paraformaldehyde (PFA). Tissues were cryoprotected by successive immersions in 15% and 30% sucrose (Sigma) in PBS at 4 °C overnight. Samples were embedded in Tissue-Tek OCT compound (Sakura) using a flash-freeze protocol and frozen at −80 °C. Twenty-micrometer-cut frozen sections were permeabilized in PBS supplemented with 0.1% Triton X-114 (Sigma) for 10 min at RT. Unspecific antibody binding was prevented by incubation in PBS supplemented with 1.5% BSA (staining buffer) for 30 min at RT. Antibodies were diluted in staining buffer supplemented with 0.1% Triton. Primary antibodies incubation were performed overnight at 4 °C in a humidified chamber. Slides were washed three times in PBS supplemented with 0.05% Tween 20 (Sigma). Secondary antibodies incubation was performed for 1.5 h at RT together with Alexa Fluor 488 phalloidin (1/500; Thermo Fisher Scientific; A12379). Antibodies used for these stainings are listed in Table S1. Slides were washed in PBS, and nuclei were stained with 125 ng/mL DAPI (Sigma; D9542) in PBS for 5 min at RT. Prolong Gold (Invitrogen) was used to mount the slides. Acquisitions were performed using an Axio Observer.Z1 microscope (Zeiss) equipped with a swept field confocal Opterra system (Bruker) and a Evolve 512 Delta EMCCD camera (Photometrics), using a 63× Plan-Apochromat oil-immersion/1.4 N.A. objective (Zeiss) and 0.8-μm Z stacks.
Isolation of Primary Cells from Human Colonic Specimens
Upon reception, specimens were washed and stored at 4 °C in PBS supplemented with 0.1% BSA. The following day, mucosa was dissected from the muscle layer and cut in small pieces. Tissue pieces were incubated in chelation buffer [calcium- and magnesium-free PBS (CMF-PBS) (Gibco) supplemented with 2 mM EDTA (Thermo Fisher Scientific)] for 30 min on ice with gentle and regular shaking before trituration in fresh chelation buffer using 25-mL pipet. These steps were repeated until no more epithelial cells were released from the tissue. Remaining colonic tissue pieces were rinsed in PBS containing calcium and magnesium (PBS+/+) (Gibco) before incubation in PBS+/+ supplemented with 10 mM Hepes, 1 mg/mL collagenase VIII, and 800 U/mL DNaseI (both from Sigma-Aldrich) at 37 °C with 400 rpm agitation for 35 min. Remaining tissue pieces were cut smaller, and the collagenase incubation step was repeated in fresh medium. Released lamina propria cells from both incubations were pooled, serial filtered on 100-, 70-, and 40-μm cell strainers, and washed several times in CMF-PBS. If necessary, RBC lysis buffer was used (Sigma). Lamina propria cells were resuspended in RPMI 1640, counted, and further processed for Shigella infection.
Acknowledgments
We thank Aurélien Amiot for providing human colonic tissue specimen and Céline Mulet for its characterization by microscopy; Marc Galimand and Delphine Girlich for supplying bacteria strains expressing class C β-lactamases; Giulia Nigro, Daniel Scott-Algara, Asmaa Tazi, and Nicolas Doucet for discussion and advice; Mark Anderson for reviewing the manuscript; the Centre for Human Immunology for the use of their technical facilities; and Florence Videau for her expertise in imaging cytometry. This study was supported by the French Ministry of Higher Education and Research (L.P. and F.S.); French Medical Research Foundation Grants FDT20150532160 and SPF20121226366 (to L.P., M.L.F., and F.-X.C.-V.); European Research Council Grants 232798 and 339579 (to M.L.F. and I.B.); the Marie Curie Fellowship Program (to F.-X.C.-V.); the National Science and Engineering Research Council of Canada and the Faculty of Science of the University of Ottawa (F.-X.C.-V.); and the French Government’s Investissement d’Avenir Program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (Grant ANR-10-LABX-62-IBEID).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707098114/-/DCSupplemental.
References
- 1.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Phalipon A, Sansonetti PJ. Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: A tool box for survival? Immunol Cell Biol. 2007;85:119–129. doi: 10.1038/sj.icb7100025. [DOI] [PubMed] [Google Scholar]
- 3.Phalipon A, Mulard LA, Sansonetti PJ. Vaccination against shigellosis: Is it the path that is difficult or is it the difficult that is the path? Microbes Infect. 2008;10:1057–1062. doi: 10.1016/j.micinf.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 4.Barry EM, et al. Progress and pitfalls in Shigella vaccine research. Nat Rev Gastroenterol Hepatol. 2013;10:245–255. doi: 10.1038/nrgastro.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashida H, Ogawa M, Mimuro H, Sasakawa C. Shigella infection of intestinal epithelium and circumvention of the host innate defense system. Curr Top Microbiol Immunol. 2009;337:231–255. doi: 10.1007/978-3-642-01846-6_8. [DOI] [PubMed] [Google Scholar]
- 6.Konradt C, et al. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe. 2011;9:263–272. doi: 10.1016/j.chom.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Salgado-Pabón W, et al. Shigella impairs T lymphocyte dynamics in vivo. Proc Natl Acad Sci USA. 2013;110:4458–4463. doi: 10.1073/pnas.1300981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nothelfer K, et al. B lymphocytes undergo TLR2-dependent apoptosis upon Shigella infection. J Exp Med. 2014;211:1215–1229. doi: 10.1084/jem.20130914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe. 2008;3:104–113. doi: 10.1016/j.chom.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Zlokarnik G, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 12.Ménard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsot C, et al. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- 14.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirot D, et al. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: Identification of CTX-1, a novel beta-lactamase. J Antimicrob Chemother. 1987;20:323–334. doi: 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- 16.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson J, Karlsson A, Bylund J, Welin A. Phagocyte interactions with Mycobacterium tuberculosis—simultaneous analysis of phagocytosis, phagosome maturation and intracellular replication by imaging flow cytometry. J Immunol Methods. 2015;427:73–84. doi: 10.1016/j.jim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Bonnefois T, et al. Development of fluorescence expression tools to study host-mycoplasma interactions and validation in two distant mycoplasma clades. J Biotechnol. 2016;236:35–44. doi: 10.1016/j.jbiotec.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Terrazas C, et al. Uncovering Leishmania-macrophage interplay using imaging flow cytometry. J Immunol Methods. 2015;423:93–98. doi: 10.1016/j.jim.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drechsler-Hake D, et al. Mononuclear phagocytes contribute to intestinal invasion and dissemination of Yersinia enterocolitica. Int J Med Microbiol. 2016;306:357–366. doi: 10.1016/j.ijmm.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Pulvertaft RJ, Pulvertaft I. Activation of lymphocytes. J Clin Pathol. 1967;20:795–805. doi: 10.1136/jcp.20.6.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuschieri A, Mughal S. Surface morphology of mitogen-activated human lymphocytes and their derivatives in vitro. J Anat. 1985;140:93–104. [PMC free article] [PubMed] [Google Scholar]
- 23.Clerc P, Sansonetti PJ. Entry of Shigella flexneri into HeLa cells: Evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55:2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köberle M, et al. Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS Pathog. 2009;5:e1000551. doi: 10.1371/journal.ppat.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geddes K, Cruz F, III, Heffron F. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 2007;3:e196. doi: 10.1371/journal.ppat.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerke C, Falkow S, Chien Y-H. The adaptor molecules LAT and SLP-76 are specifically targeted by Yersinia to inhibit T cell activation. J Exp Med. 2005;201:361–371. doi: 10.1084/jem.20041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trülzsch K, et al. Yersinia outer protein P inhibits CD8 T cell priming in the mouse infection model. J Immunol. 2005;174:4244–4251. doi: 10.4049/jimmunol.174.7.4244. [DOI] [PubMed] [Google Scholar]
- 28.Torres A, et al. Asparagine deprivation mediated by Salmonella asparaginase causes suppression of activation-induced T cell metabolic reprogramming. J Leukoc Biol. 2016;99:387–398. doi: 10.1189/jlb.4A0615-252R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansonetti PJ, Ryter A, Clerc P, Maurelli AT, Mounier J. Multiplication of Shigella flexneri within HeLa cells: Lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardini ML, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsot C. Shigella type III secretion effectors: How, where, when, for what purposes? Curr Opin Microbiol. 2009;12:110–116. doi: 10.1016/j.mib.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Campbell-Valois FX, Pontier SM. Implications of spatiotemporal regulation of Shigella flexneri type three secretion activity on effector functions: Think globally, act locally. Front Cell Infect Microbiol. 2016;6:28. doi: 10.3389/fcimb.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashida H, Mimuro H, Sasakawa C. Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol. 2015;6:219. doi: 10.3389/fimmu.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jehl SP, et al. Antigen-specific CD8+ T cells fail to respond to Shigella flexneri. Infect Immun. 2011;79:2021–2030. doi: 10.1128/IAI.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio B, et al. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J Exp Med. 2008;205:1121–1132. doi: 10.1084/jem.20071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sansonetti PJ, Kopecko DJ, Formal SB. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ménard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oaks EV, Wingfield ME, Formal SB. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell-Valois F-X, et al. A fluorescent reporter reveals on/off regulation of the Shigella type III secretion apparatus during entry and cell-to-cell spread. Cell Host Microbe. 2014;15:177–189. doi: 10.1016/j.chom.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Sörensen M, et al. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 2003;552:110–114. doi: 10.1016/s0014-5793(03)00856-1. [DOI] [PubMed] [Google Scholar]
- 41.de la Calle C, Joubert PE, Law HK, Hasan M, Albert ML. Simultaneous assessment of autophagy and apoptosis using multispectral imaging cytometry. Autophagy. 2011;7:1045–1051. doi: 10.4161/auto.7.9.16252. [DOI] [PubMed] [Google Scholar]