Significance
A core tenet in genetics is that genotype influences phenotype. In an individual, the same genome can be expressed in substantially different ways, depending on the tissue. Expression quantitative trait locus (eQTL) analysis, which associates genetic variants at millions of locations across the genome with the expression levels of each gene, can provide insight into genetic regulation of phenotype. In each of 13 tissues we performed an eQTL analysis, represented significant associations as edges in a network, and explored the structure of those networks. We found clusters of eQTL linked to shared functions across tissues and tissue-specific clusters linked to tissue-specific functions, driven by genetic variants with tissue-specific regulatory potential. Our findings provide unique insight into the genotype–phenotype relationship.
Keywords: GTEx, expression quantitative trait locus, eQTL, bipartite networks, GWAS
Abstract
Characterizing the collective regulatory impact of genetic variants on complex phenotypes is a major challenge in developing a genotype to phenotype map. Using expression quantitative trait locus (eQTL) analyses, we constructed bipartite networks in which edges represent significant associations between genetic variants and gene expression levels and found that the network structure informs regulatory function. We show, in 13 tissues, that these eQTL networks are organized into dense, highly modular communities grouping genes often involved in coherent biological processes. We find communities representing shared processes across tissues, as well as communities associated with tissue-specific processes that coalesce around variants in tissue-specific active chromatin regions. Node centrality is also highly informative, with the global and community hubs differing in regulatory potential and likelihood of being disease associated.
More than a decade after the sequencing of the human genome, our understanding of the relationship between genetic variation and complex traits remains limited. Genome-wide association studies (GWASs), which look for association between common genetic variants and phenotypic traits, have resoundingly shown that complex phenotypes are influenced by many variants of relatively small effect size (1, 2), the overwhelming majority of which () lie in noncoding regions of the genome (3, 4). Those single-nucleotide polymorphisms (SNPs) associated with complex traits are enriched for variants likely to affect gene expression, as measured by expression quantitative trait locus (eQTL) analysis (5), suggesting that they influence phenotypes through changes in gene regulation (6, 7). Identifying the regulatory role of these variants likely also depends on the tissues relevant to the phenotype. For example, eQTL identified in skeletal muscle and adipose tissues for type 2 diabetes (T2D) have been shown to explain a greater proportion of the disease heritability than those identified across tissues (8). Furthermore, variants far from the transcriptional start site (TSS) of a gene, trans-eQTL, explain more of the heritability of T2D than those near the gene, cis-eQTL, and there is mounting evidence for the importance of these variants in a variety of phenotypes (9–11). That trans-eQTL, which can influence hundreds of genes in humans (12), might have an impact on the phenotype is consistent with similar observations in model organisms (13). However, large-scale detection of trans-eQTL across populations and tissues (14) has only recently become feasible in humans, and our understanding of how multiple cis- and trans-eQTL influence gene expression and cellular functions in different tissues is incomplete.
We performed a systems genetics analysis of the regulatory effects of common [minor allele frequency (MAF) 5%] variants in 13 tissues collected by the Genotype–Tissue Expression (GTEx) consortium. By constructing tissue-level eQTL networks from cis- and trans-eQTL, where each significant SNP–gene association within a tissue is cast as an edge, we find that these networks provide insight into the shared and tissue-specific regulatory roles of common variants and their collective impact on biological pathways. In particular, we find three aspects of the eQTL network topology that inform tissue-level regulatory biology: (i) Communities—which are composed of SNPs and genes with a high density of within-group edges—are enriched for pathways, functionally related genes, and SNPs in tissue-specific active chromatin regions (actively transcribed and open regulatory regions); (ii) community hubs (core SNPs)—which are SNPs highly connected to genes in their community—are enriched for active chromatin regions close to the transcriptional start site and for GWAS association; and (iii) global hubs—which are connected to many genes throughout the network—are enriched for distal elements such as nongenic enhancers and devoid of GWAS association. The picture that emerges from analysis of the eQTL networks is a complex web of associations that reflects the polygenic architecture across tissues and that provides a natural framework for understanding both the shared and tissue-specific effects of genetic variants. These networks, along with SNP and gene network properties across all 13 tissues, are available at networkmedicine.org:3838/eqtl/.
Results and Discussion
eQTL Networks Are Highly Modular.
We downloaded RNA-Seq data and imputed genotypes obtained from postmortem samples coming from 50 tissue types and two cell lines from the GTEx version 6.0 dataset (phs000424.v6.p1, 2015-10-05 release) from the database of Genotypes and Phenotypes (dbGaP) (approved protocol no. 9112). After quality control and preprocessing (see SI Appendix, Fig. S1 and Materials and Methods for details of RNA-Seq and genotyping data preprocessing and tissue-specific gene filtering), we retained 29,242 genes (15, 16) and 5,096,867 SNPs across all tissues. For statistical power purposes, we considered only tissues for which we had both RNA-Seq and imputed genotyping data for at least 200 individuals (SI Appendix, Fig. S2). Twelve tissues and one cell line met all criteria (SI Appendix, Fig. S1 and Table S1).
For each of the 13 tissues, we tested for association between SNP genotypes and gene expression levels both in cis and in trans, correcting for reported sex, age, ethnic background, and the top three principal components obtained using genotyping data (SI Appendix, Figs. S1 and S3 and Materials and Methods). Including SNPs within 1 Mb of each gene, we found between 285,283 and 691,333 significant cis-eQTL (5,301–11,035 genes) and between 7,151 and 15,183 significant trans-eQTL (326–955 genes) at a false discovery rate (FDR) of 5% in each tissue. Despite differences in normalization, number and type of covariates, and eQTL P-value calculation, on average 73% of our cis-eQTL were also detected by the GTEx project in each tissue (minimum 70% in artery aorta and maximum 76% in thyroid; SI Appendix, Table S2). Consistent with previous reports (17–19), we find most cis-eQTL are located around TSSs, with 50% of SNPs located within 16,000 bp of the nearest TSS (14,767 bp for whole blood and 17,109 bp for thyroid). We also observed that cis-eQTL were highly replicable across tissues (70–88% were replicated in at least one other tissue), while trans-eQTL replicability across tissues was more varied (37–88% were replicated in at least one other tissue).
If our trans-eQTL reflect actual associations, we would expect that they should be preferentially located in regulatory genomic regions. To test for this, we used the Roadmap Epigenomics Project core 15-states model, which classifies genomic regions into 15 chromatin states based on epigenetic marks measured in a specific tissue or cell line; these classifications were available for eight tissues (adipose subcutaneous, artery aorta, fibroblast cell line, esophagus mucosa, heart left ventricle, lung, skeletal muscle, and whole blood) (20). Correcting for local linkage disequilibrium, we found that trans-eQTL are significantly more likely to be located in regulatory and actively transcribed regions (TSSs and their flanking regions, enhancers) in at least seven of the eight tissues and significantly less likely to fall into quiescent regions and constitutive heterochromatin (Dataset S1). This is confirmed by P values combined across tissues obtained using conditional logistic regression (stratified by tissue; Dataset S1). In addition, we found that a majority (50–66%) of trans-eQTL were also associated with genes in cis.
For each of the 13 tissues, we represented the significant eQTL as a bipartite network, with nodes representing either SNPs or genes and edges representing significant SNP–gene associations (for example, heart left ventricle tissue; Fig. 1A). Each network was composed of a “giant connected component” (GCC) plus additional small connected components. To increase the size of the GCC and because network centrality measures are more sensitive to false negative than to false positive edges (21, 22), we relaxed the FDR cutoff and included all eQTL with FDR q values under 0.2. At this threshold, about 11% of SNPs are associated with at least one gene in cis (333,225–751,418 SNPs) and 0.3% in trans (10,814–22,570 SNPs; Dataset S2). For all subsequent analyses, we focused on the networks defined by these GCCs.
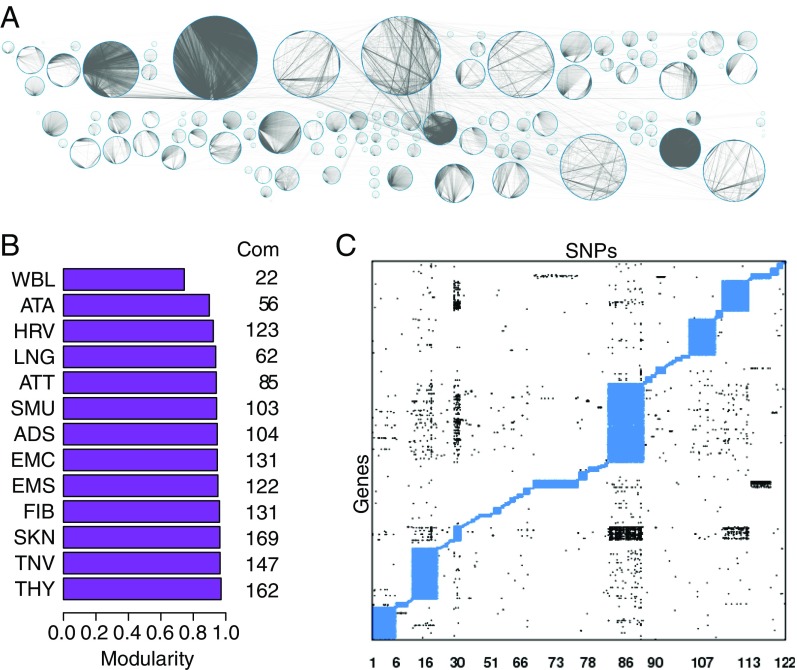
Fig. 1.
Structure of eQTL networks. (A) The eQTL network from heart left ventricle. Each circle represents a community. The nodes, both SNPs and genes, are located around each circle. Gray lines represent network edges (significant cis- and trans-eQTL associations). (B) Modularity of the eQTL network from each of the 13 tissues. Modularity assesses the strength of division of the network in communities and corresponds to the fraction of edges observed within each community minus the expected fraction if edges were randomly distributed. B, Right shows the number of communities (Com) in each network. (C) Structure of communities within the eQTL heart left ventricle network. The heart left ventricle network is represented as a matrix with SNPs in columns and genes in rows. Each network edge is represented by a point. Intracommunity edges are plotted in blue and intercommunity edges in black. Community structure for the other 12 networks is presented in SI Appendix, Fig. S4.
We used the R condor package (23) to identify communities in each of the 13 eQTL networks (SI Appendix, Fig. S1). We found between 22 (whole blood) and 169 (skin) communities in each tissue-specific network. The modularities of the eQTL networks ranged from 0.74 (whole blood) to 0.97 (thyroid, Fig. 1B). This suggests that within each network, the communities were tightly clustered, with the density of edges linking nodes from the same community being much higher than the density of edges linking nodes from different communities (example in Fig. 1C and SI Appendix, Fig. S4). The size of the communities ranged broadly, between 2 and 1,220 genes and between 3 and 26,056 SNPs.
To determine how much correlated gene expression determined community structure, we computed the Pearson correlation coefficient (Pearson ) for each pair of genes within each community and each network. The distributions of pairwise coefficients by tissue and community ranged from −0.2 to 0.2 for most of the communities (SI Appendix, Fig. S5A). The median absolute correlation was lower than 0.2 in 86% of communities, with about 2% of communities presenting a median correlation higher than 0.4. Moreover, small-size communities (fewer than 10 genes) were overrepresented among those with median correlation higher than 0.4 (SI Appendix, Fig. S5B, odds ratio of 29, P value of ). This shows that our communities are not driven by gene expression correlation, except for a tiny fraction that contain few genes, and thus not likely to be found using coexpression network methods.
Finally, we recognize that genetic recombination effects, including local linkage disequilibrium, might lead to a clustering of SNPs and genes from the same chromosomal region to cluster together. To test this, we examined the chromosomal distribution of genes and SNPs in each community. Across all 13 tissues, between 71% (fibroblast cell line) and 94% (adipose subcutaneous) of communities included genes and SNPs from two or more chromosomes (SI Appendix, Fig. S6). Overall, our results suggest that the structure of eQTL network communities is not based exclusively on genomic correlations such as linkage disequilibrium or gene expression correlation.
eQTL Community Structure Reflects Shared and Tissue-Specific Biological Functions.
To investigate the biological relevance of these communities, we tested all communities in each tissue for overrepresentation of Gene Ontology (GO) biological processes (24). Across all tissues, we found 208 communities enriched for at least one biological process at a FDR of 5% (Dataset S3). We compared the observed biological processes across communities and tissues and found that some communities were enriched for genes involved in tissue-specific functions while others included genes with biological functions relevant to all tissues (Figs. 2 and 3).
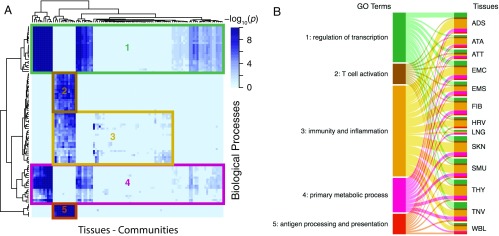
Fig. 2.
Some network communities are enriched for biological functions shared across tissues. A complete list of the significantly overrepresented biological processes in each community and each network can be found in Dataset S3. (A) Heatmap clustering the similarity of GO biological processes in communities from all tissues. Only GO terms that were significant in at least 12 tissues are included. (B) Sankey diagram linking clusters from the heatmap to the tissues that contain at least one community enriched for genes involved in the clustered functions. A ribbon’s thickness is proportional to the number of communities enriched for each cluster of GO terms in each TS network.
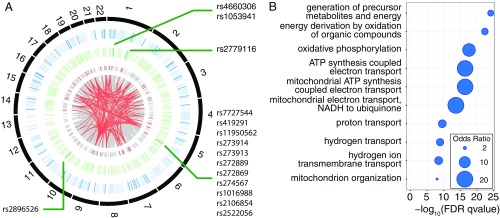
Fig. 3.
Close-up of a community enriched for TS GO biological processes: the heart ventricle community 86. (A) Circos diagram for heart left ventricle eQTL in community 86. From the outside to the inside: chromosomes (in black), genes from community 86 (in blue, dark blue bars represent genes associated with cellular metabolism), SNPs (in green, with dark green bars indicating an association with metabolic traits), and SNP–gene assocations (in gray: all associations, in red: eQTL linked to genes involved in cellular respiration or SNPs involved in metabolism). Details about GWAS annotation of the SNPs and genes are given in Dataset S4. (B) Heart left ventricle community 86 is enriched for genes involved in cellular respiration.
We first identified communities enriched for ubiquitous biological functions, which we defined as GO terms that were enriched in a community from at least 12 of the 13 tissue-specific eQTL networks. Using the Fisher combined probability test (FCPT), we showed that these shared GO terms have significant combined P values across all tissues (P values shown in Dataset S3). To further ensure that this enrichment in shared GO terms was not a spurious signal due to the number of tests run, we computed a null distribution of 1,000 P-values for each community enriched for a shared GO term in each tissue by rerunning the enrichment test using a randomly selected set of genes equivalent in size to the community in the original test (Dataset S3). An example of a null distribution of P values for enrichment in the shared GO term GO:0010468 “regulation of gene expression” is shown in SI Appendix, Fig. S8. For all shared GO terms across all tissues and communities, the P value associated with the observed enrichment was in the bottom 4% of the null distributions, indicating that the observed enrichment reflects shared biological processes rather than random enrichment in common GO terms.
To analyze these results, we clustered communities using their enrichment P values in these ubiquitous biological functions only (Fig. 2A). We identified five groups of GO terms related to regulation of transcription and RNA metabolism and immunity that defined five groups of communities (Fig. 2 and Dataset S3). For example, we found that genes involved in functions linked to pathogen recognition, innate and adaptive immune response triggering, T-cell activation, and inflammation (groups 2, 3, and 5) are overrepresented in at least one community in all tissue-specific eQTL networks except lung. These communities include many genes from the major histocompatibility complex (MHC) class II and may reflect the presence of infiltrated macrophages in all tissues. Similarly, 12 of the 13 tissue-specific networks present communities strongly enriched for genes related to the control of transcription and RNA metabolism (groups 1 and 4).
Communities that are enriched for the same GO biological process tend to share many of the same SNPs, genes, and edges, with the average pairwise Jaccard index between tissues for each GO biological process ranging from 0.16 to 0.69 for SNPs, 0.17 to 0.47 for genes, and 0.10 to 0.46 for edges. The content of these communities with shared biological functions can vary slightly from tissue to tissue (1–5%, 1–10%, and 9–32% for SNPs, genes, and edges, respectively). However, these differences are small and likely due to slight variations in sample size between tissues, affecting the significance of the eQTL associations, network clustering, and the small differences in the populations analyzed for each tissue (average proportion of samples shared with other tissues varies from 64% to 95%).
We also used the GO enrichment analysis to identify tissue-specific functions, which we defined as a community-enriched GO term appearing in no more than two tissues. An interesting example is community 86 from the heart left ventricle network (Fig. 3). This community is enriched for genes involved in cellular respiration and the mitochondrial respiratory chain (Fig. 3B and Dataset S3). These functions, despite being ubiquitous, are particularly important in heart due to its high metabolic requirements, and studies have shown that dysfunction of the respiratory chain is involved in many heart diseases (25). This heart community contains 13,143 SNPs (1,834 linkage disequilibrium blocks) linked to 1,182 genes (gray links, Fig. 3A). Of these, there is a subset involved in cellular respiration (red links) that includes 547 SNPs [100 linkage disequilibrium (LD) blocks] linked to 52 genes, located on 20 autosomes. GWASs have reported 5 of these 52 genes to be associated with cardiovascular-related traits and metabolism such as conotruncal heart defects, obesity, and blood metabolite levels, a significant enrichment [resampling , Fig. 3A and Dataset S4 (26)]. This community also contains SNPs from three genomic regions that have been linked to metabolic traits and blood metabolite levels; these include association with the metabolite carnitine, a molecule involved in the transport of fatty acid from cytoplasm to the mitochondrial matrix where those fatty acids are metabolized (Fig. 3A and Dataset S4).
Other examples of tissue-specific overrepresentation of biological functions include transmission of nerve impulse and myelination in community 4 from tibial nerve and muscle development and contraction in muscular tissues. Community 30 from the heart left ventricle network is enriched for genes related to ventricular cardiac muscle tissue morphogenesis and contraction, community 75 from skeletal muscle for striated muscle contraction and cell differentiation, and community 83 from esophagus muscularis for smooth muscle contraction (SI Appendix, Fig. S7 and Dataset S3).
This suggests that eQTL network community structure reflects many SNPs working together to influence groups of functionally related genes and that some of the communities capture unique features of tissues and phenotypic states. Our analysis of these eQTL communities paints an empirically robust picture of communities that involve functions shared across tissues as well as those that are highly specialized to individual tissues.
Biological Characteristics of Tissue-Specific Communities.
Given that genetic variants are present in all tissue types, it may be that tissue-specific epigenetic activation influences their regulatory effect on gene expression. Consequently, we searched for chromatin state changes associated with the tissue-specific communities we observed. We defined tissue-specific and shared communities, using the enrichment in GO biological processes. Tissue-specific (TS) communities were defined as those that showed a higher than expected proportion of GO biological processes that were present in no more than two tissues, while shared communities showed a higher than expected proportion of GO biological processes present in at least 12 of the 13 tissues. We compared the proportion of TS SNPs, genes, and edges (that is, SNPs, genes, and edges that are present in no more than 2 tissues) between TS and shared communities. As expected, we observed that in all tissues, TS communities tend to have a higher proportion of TS genes than shared communities (average proportions range from 5% to 37% in TS communities and from 2% to 23% in shared communities, depending on the tissue). This difference is significant in 9 of the 13 tissues (Fig. 4A), with a combined P value across tissues using the FCPT of . We also observed a significantly higher proportion of TS SNPs and edges in TS communities, which can represent up to 100% of the content of the communities (P values corrected for LD are presented in Fig. 4A, and across-tissue FCPT P values are and , respectively).
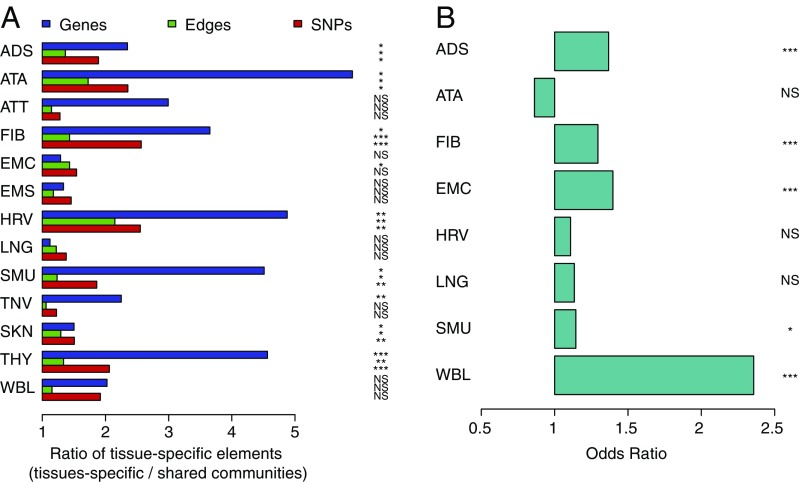
Fig. 4.
Characteristics of TS communities. (A) Enrichment in TS genes, SNPs, and edges among communities with genes involved in TS GO biological processes compared with communities with genes involved in shared pathways. Barplots represent the ratio of mean proportion of unique elements in TS vs. shared communities. A, Right shows P values obtained using the Mann–Whitney U test and correcting for LD. (B) TS SNPs are more likely to be located in TS activated chromatin regions among TS communities compared with shared communities. P values were obtained using the Fisher test and correcting for LD. (A and B) ADS, adipose subcutaneous; ATA, aorta; ATT, artery tibial; EMC, esophagus mucosa; EMS, esophagus muscularis; FIB, fibroblast; HRV, heart left ventricle; LNG, lung; NS, nonsignificant; SKN, skin; SMU, skeletal muscle; TNV, tibial nerve; THY, thyroid; WBL, whole blood (WBL); *, **, ***.
These TS SNPs and edges (eQTL associations) in TS communities may result from TS activation of genomic regions. To test for this, we first extracted the chromatin state for all TS SNPs in the eight tissues for which chromatin-state maps were available in the Roadmap Epigenomics Project data. For each network, we assigned the TS SNPs to one of the two following categories: those that were located in a chromatin region that was specifically activated in the tissue of interest and those that were located in chromatin regions that presented a repressed, similar, or exactly identical chromatin state in the tissue of interest compared with the seven other tissues. By comparing TS and shared communities, we found that in five of the eight tissues, TS SNPs in TS communities are significantly more likely to show TS chromatin activation than TS SNPs in shared communities (odds ratios and P values corrected for LD are presented in Fig. 4B; the combined odds ratio and P value using the Cochran–Mantel–Haenszel test were 1.21 and , respectively).
Global Hubs and Community Cores Reflect Different SNP Functional Roles.
A number of studies have shown that eQTL SNPs are overrepresented in regulatory regions (3, 14, 27), but not all eQTL SNPs have obvious regulatory roles. One natural hypothesis that emerges from our structural analysis of eQTL networks is that there may be network properties that are associated with the various regulatory roles of SNPs. Specifically, one might expect that SNPs that are more highly connected, either globally or locally, would have different regulatory roles than SNPs at the periphery of the network.
We investigated the relationship between eQTL network structure and regulatory role, using two measures of centrality: the degree—the total number of genes to which each SNP is linked—and the modularity contributed by each SNP or “core score” (Eq. 2 in Materials and Methods). A SNP’s degree reflects the global SNP centrality within the entire network, while the core score reflects the local centrality of each SNP within its community. The core score is also normalized to account for global hubs, which may have more within-community edges.
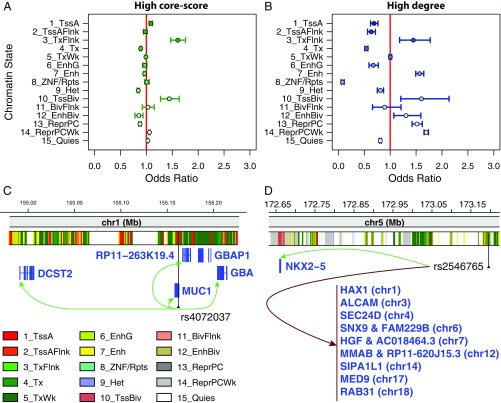
For each TS network, we studied the enrichment of central SNPs (either high–core-score or high-degree SNPs) across 15 chromatin states in a TS manner, using the Roadmap Epigenomics Project data. We calculated enrichment of central SNPs (Materials and Methods) in the eight tissues for which chromatin-state maps were available, using all SNPs in the GCC as background and correcting for gene density (Materials and Methods). Using a conditional logistic regression, which allows us to obtain combined odds ratio and P value across all eight tissues, we found that core SNPs are enriched for promoters (states 1 and 10, Fig. 5A and Dataset S5). Tissue-specific odds ratios can be found in Dataset S5. For example, rs4072037, a SNP located in MUC1 on chromosome 1 that has been associated with esophageal and gastric cancer in a GWAS (28), is a core SNP of community 31 in the esophagus mucosa network and located in an active TSS region in this tissue according to the core 15-states model from the Roadmap Epigenomics Project (20) (Fig. 5C). It is a cis-eQTL of GBAP1, DCST2, GBA, RP11-263K19.4, and MUC1, a gene coding for a protein of the mucin family involved in forming mucous barriers on epithelial surfaces including the esophagus, and associated with the proliferation, migration, and invasion capacities of esophageal adenocarcinomatous cells (29).
Fig. 5.
Network hub SNPs are more likely to lie in active chromatin regions than nonhub SNPs. (A and B) Symbols shows the odds ratios across all eight tissues with matching chromatin states from the Roadmap Epigenomics Project. Odds ratios are combined across tissues, using conditional logistic regression. Bars represent the 95% confidence interval. Each odds ratio measures the enrichment of central SNPs in a particular functional category, corrected for number of genes within 1 Mb of the SNP. (A) Enrichment in each chromatin state for SNPs with a core score in the top 25%. (B) Enrichment in each chromatin state for SNPs with network degree greater than 10. Odds ratios and P values for each TS network are listed in Dataset S5. Enrichment in each chromatin state for all trans-eQTL are presented in Dataset S1. (C) Example of a core SNP located in an active TSS region: rs4072037, associated with esophageal and gastric cancer. (D) Example of a high-degree SNP located in an active enhancer: rs2546765. The chromatin-state tracks are based on results from the Roadmap Epigenomics Project core 15-states model for esophagus mucosa (E079) for C and heart left ventricle (E095) for D.
Global hubs are generally significantly more likely to lie within nongenic enhancers and Polycomb repressed regions (states 7, 12, and 13–14; Fig. 5B and Dataset S5). A good example is rs2546765, which is an intergenic SNP on chromosome 5, a high-degree SNP in community 86 in the heart left ventricle network, connected to 13 genes across 10 chromosomes and 3 communities and located in an enhancer region in this tissue (Fig. 5D). It is a cis-eQTL of NKX2-5, a gene involved in heart development, and a trans-eQTL of among others ALCAM, involved in heart development in vertebrates; HAX-1, a regulator of contractility and calcium cycling in the heart; SIPA1L1, associated in the GWAS with QRS intervals; and SEC24D, HGF, and MMAB, associated in the GWAS with cardiovascular diseases and/or dyslipidemia. As expected, both global and core SNPs are depleted in constitutive heterochromatin (state 9).
Overall, we find that global hubs and community cores each have different associations with active regulatory regions: Global hubs are preferentially located in distal regulatory regions (enhancers) while core SNPs are significantly overrepresented in proximal regulatory elements (promoters). This is another case where the eQTL network structure provides insight into biological processes, an insight that is reproducible across tissues.
SNPs in Community Cores Are Associated with Trait and Disease Phenotypes.
We tested whether the SNPs with high centrality were more likely to be associated with complex traits in the GWAS by mapping trait-associated SNPs in the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) GWAS catalog (26) to each TS eQTL network, considering only SNPs with GWAS P values less than (GWAS SNPs). In each tissue, we compared the distribution of degrees (number of edges per node) for all SNPs and GWAS SNPs. We found that SNPs of low degree (1–2) were significantly depleted in GWAS SNPs while SNPs of intermediate degree (5–10) showed consistent significant enrichment for GWAS SNPs across all 13 tissues and SNPs of degree greater than 15 were devoid of GWAS associations (SI Appendix, Fig. S9).
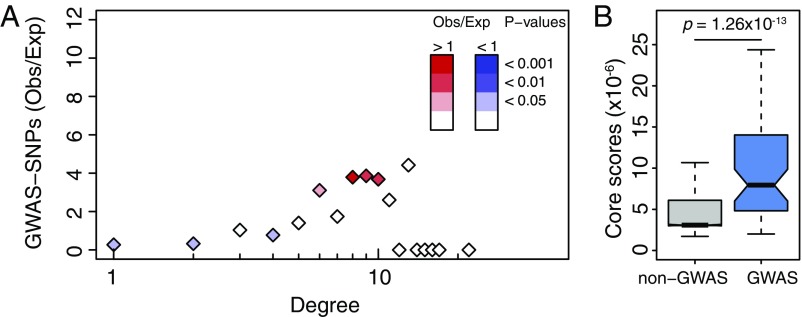
These results are even more striking when we consider only GWAS SNPs associated with phenotypes related to the tissue of interest. For example, when we mapped SNPs associated with autoimmune diseases to the whole-blood eQTL network, we found that they were enriched for intermediate degree (Fig. 6A). This overrepresentation of intermediate-degree SNPs led us to hypothesize that GWAS SNPs tend to be highly central not on a global scale, but instead within the eQTL communities. We tested this by comparing the core scores for GWAS SNPs from the NHGRI-EBI GWAS catalog to those for non-GWAS SNPs. Using a likelihood-ratio test and controlling for LD, we found that GWAS and non-GWAS SNPs had significantly different core-score distributions, with the median core scores for GWAS SNPs higher in all 13 tissues (SI Appendix, Fig. S10, FCPT P value ). This enrichment for high core scores was also present when considering only SNPs associated with autoimmune diseases (shown for whole blood in Fig. 6B, ).
Fig. 6.
Network properties of GWAS SNPs associated with autoimmune diseases in whole blood. (A) Ratio of observed vs. expected number of SNPs associated with autoimmune diseases by GWASs depending on their network degree. P values were obtained using 1,000 resamplings, taking into account gene density around each SNP. Ratios including all GWAS traits and diseases in each TS network are in SI Appendix, Fig. S9. (B) Distribution of core scores for SNPs associated (in blue) or not (in gray) to autoimmune diseases by GWAS. P values were obtained using a LRT and pruning for SNPs in LD. Distributions for all TS networks including all GWAS traits and diseases are in SI Appendix, Fig. S10.
The consistency of these results across tissues supports the idea that GWAS SNPs have a unique pattern of connectivity within eQTL networks that differs widely from those of non-GWAS SNPs. In each tissue, we find that GWAS SNPs cluster within a relatively small number of communities and within those communities, they are significantly overrepresented in the local, but not the global, hubs within the network. These findings are in agreement with what one might expect from genotype associations in complex disease. Disease-linked SNPs with effect sizes large enough to be identified through the GWAS map to a relatively small number of relevant processes in each tissue, and the likelihood of being disease associated is related to the likelihood that a variant is at the center of its functional community—and therefore more likely to perturb function.
Discussion
We have carried out a systematic analysis of eQTL networks constructed from cis- and trans-eQTL in 13 different tissues, using data available from the GTEx project. We have found that the structural properties of these networks provide functional insight into the regulatory roles of genetic variants across and within tissues. Using a community detection algorithm (30) to search for communities of densely connected SNPs and genes, we found that the eQTL network in each of the 13 tissues was organized into highly modular communities. When we examine the genes represented in each community, we find an enrichment for genes, located on multiple chromosomes, that share similar functions or are associated with coherent biological processes. While the FDR may not be well controlled for the large number of GO terms tested, our resampling analysis for GO terms shared across tissues suggests that the observed enrichment is unlikely to be due to the large number of tests. However, the possibility of post hoc plausibility explanations cannot be completely ruled out. Contrary to what one might expect, these communities are not driven by coexpression (excepting communities with very few genes), suggesting that it is the genetic influence of multiple cis- and trans-eQTL SNPs on functionally related groups of genes that drives the organization and structure of these communities.
When comparing communities between tissues, we find many communities with common patterns of functional enrichment across tissues, reinforcing the pleiotropic role of the SNPs in these communities. We do, however, also find TS communities that contain genes involved in TS functional processes such as cellular respiration in heart left ventricle or smooth muscle contraction in esophagus muscularis. There is a plausible mechanistic explanation for the tissue specificity of some of these communities: Using data in eight tissues from the Roadmap Epigenome Project we find that TS eQTL SNPs in these TS communities are enriched for active chromatin regions that are unique to that tissue. This suggests that the organization of these communities is driven by the epigenetic activation of chromatin regions surrounding specific SNPs and that these SNPs act in cis and trans to exert genetic effects on the expression of functionally related genes, genes with important roles in their respective tissue-level processes. In addition, these communities are not only enriched for specific tissue-relevant gene function; they are also enriched for tissue-specific edges (eQTL), SNPs, and genes. This is relevant to the ongoing discussion of the tissue specificity of eQTL. Although most eQTL appear to be shared, TS eQTL emerge in concert with TS epigenetic changes and not only influence single genes, but also help coalesce TS gene expression into regulatory communities.
We find these 13 eQTL networks possess two informative types of hubs: community hubs or “cores,” which are SNPs highly connected to genes in their community, and global hubs, which are connected to many genes throughout the network. These two types of hubs have different biological properties across tissues: Community hubs are enriched for active chromatin regions close to the transcriptional start site, but not enhancers, while global hubs are enriched for distal elements such as nongenic enhancers. Moreover, community hubs are enriched for GWAS-associated variants, while global hubs are not. The degree distribution for trait-associated variants from the GWAS is also highly consistent across the 13 tissues: GWAS SNPs are enriched for intermediate network degree, depleted for low degree, and absent from global hubs. The significant overrepresentation of GWAS SNPs among the community cores provides another important insight. Across tissues, disease SNPs are those most likely to perturb groups of genes and, in doing so, may disturb important biological processes.
While the observed relationships between eQTL network properties and SNP/gene function are consistent across tissue types, we cannot rule out the possibility that the large number of statistical tests performed in the cis- and trans-eQTL analysis could lead to identification of some individual eQTL associations as significant when they are not. Although we cannot conclude, based on our analysis, that any individual SNP–gene association is correct, the consistency of our findings regarding the structure of the networks across multiple tissues and the consistent functional enrichment we observe for global and local hubs indicate that the higher-order structural organization of the networks likely provides a robust model of SNP regulatory effects. While one could imagine that the observed network patterns might be driven by unwanted systematic variations in the genotype and gene expression data, our identification of similar structural properties in an eQTL network derived using an independent chronic obstructive pulmonary disease (COPD) dataset (23) further supports the network structural associations we have described. Nevertheless, this possibility, along with more detailed analysis of specific network-prioritized SNPs, should be further investigated as additional TS gene expression and genotype data become available through the next release of GTEx and other large-cohort studies.
Our analysis of bipartite networks built from both cis- and trans-eQTL in 13 tissues provides important evidence about the collective role of eQTL in TS function and disease. The network communities reveal biological processes under the shared genetic influence of many variants, including both processes shared across tissues and those that are TS. The TS genetic regulation we observe is driven in part by SNPs that lie in TS active chromatin regions. This suggests that epigenetic profile analysis, applied to both genic and nongenic elements, will be essential for understanding the processes responsible for TS function. The eQTL networks also group together functionally related sets of variants, including GWAS SNPs, and the structure of the network provides a model of how multiple cis- and trans-acting variants can work together to influence function and phenotype. While the network models we describe do not fully resolve the question of how weak-effect variants determine complex traits and disease, this network approach provides a framework with distinct explanatory power that can serve as a basis for further exploration of the link between genotype and phenotype.
Materials and Methods
GTEx Data Preprocessing, Filtering, and Merging.
We downloaded NHGRI GTEx version 6 imputed genotyping data and RNA-Seq data from the dbGaP database. The RNA-Seq data were preprocessed using the Bioconductor R YARN package (15, 16) and normalized using the Bioconductor R qsmooth package (31). We excluded five sex-specific tissues (prostate, testis, uterus, vagina, and ovary) and merged the skin samples from the suprapubic and lower leg sites. We limited our eQTL analysis to the 13 tissues for which there were available data for at least 200 individuals. The RNA-Seq and genotyping data were mapped by the GTEx Consortium to GENCODE version 19, which was based on human genome build GRCh37.p13 (Dec 2013). We accounted for RNA extraction kit effects, using the removeBatchEffect function in the R limma package (32).
eQTL Mapping and Bipartite Network Construction.
For eQTL analysis, we excluded SNPs from all analyses if they had a call rate under 0.9 or an allele frequency lower than 5% in any tissue. A gene was considered expressed in a sample if its read count was greater than or equal to 6. Genes that were expressed in fewer than 10 of the samples in a tissue were removed for the eQTL analysis in that tissue. To correct for varying degrees of admixture in the African-American subjects, we used the first three principal components of the genotyping data provided by the GTEx consortium and included these in our eQTL model. We used the R MatrixEQTL package (33) to calculate eQTL with an additive linear function that models gene expression levels () in function of genotypes () and included age, sex, and ethnic background (), as well as the first three genotype principal components (), as covariates:
We tested for association between gene expression levels and SNPs both in cis and in trans, where we defined cis-SNPs as those within 1 Mb of the TSS of the gene based on mapping using the Bioconductor R biomaRt package (34). P values were adjusted for multiple testing using Benjamini–Hochberg correction for cis- and trans-eQTL separately and only those with adjusted P values less than 0.2 were used in subsequent analyses.
To compare our results with those reported by the GTEx consortium, we downloaded the single-tissue cis-eQTL from the GTEx portal (www.gtexportal.org/).
Community Identification.
For each tissue, we represented the significant eQTL as edges of a bipartite network linking SNPs and gene nodes. To identify highly connected communities of SNPs and genes in the eQTL networks, we used the R condor package (23), which maximizes the bipartite modularity (30). As recursive cluster identification and optimization can be computationally slow, we calculated an initial community structure assignment on the weighted, gene-space projection, using the multilevel.community function in the R igraph package (35). This function is an implementation of a fast unipartite modularity maximization algorithm (36). Using this initial assignment, we then iteratively converged on a community structure corresponding to a maximum bipartite modularity.
The bipartite modularity is defined in Eq. 1, where is the number of links in the network, is the upper right block of the network adjacency matrix (a binary matrix where a 1 represents a connection between a SNP and a gene and 0 otherwise), is the degree of SNP , is the degree of gene , and , are the community indexes of SNP and gene , respectively:
| [1] |
SNP Core-Score Calculation.
We defined a SNP’s core score as the SNP’s contribution to the modularity of its community. Specifically, For SNP in community , its core score, , is defined by Eq. 2. To normalize SNPs across communities, we accounted for community membership in our downstream testing (Eqs. 3 and 4), which better accounts for community variation compared with the normalization method used in ref. 23. Indeed, is dependent on community size (SI Appendix, Fig. S11):
| [2] |
Gene Ontology Functional Category Enrichment.
We extracted the list of genes within each community in each TS network and then used the R GOstat package (37) to perform a tissue-by-tissue analysis of the overrepresentation of Gene Ontology biological processes within each community. Our reference set consisted of all of the genes present in the corresponding tissue-specific network. Communities were considered significantly enriched in a given category if the FDR-adjusted P value was .
TS SNP Enrichment.
We defined TS communities and shared communities based on their enrichment in GO biological process (BP) terms. We first calculated for each community the proportion of TS BP terms (defined as the BP terms that were significant in no more than 2 of the 13 networks) and the proportion of shared BP terms (defined as the BP terms that were significant in at least 12 of the 13 networks). In each network, we then extracted communities that had a higher than average proportion of either TS BP terms (TS communities) or shared BP terms (shared communities). In each of these communities, we computed the proportion of TS elements (SNPs, genes, or edges), defined as the proportion of elements present in no more than 2 of the 13 networks. To control for LD in the case of SNPs and edges, we generated lists of SNPs falling into the same LD block using the plink2 –blocks option, a 5-Mb maximum block size, and an of 0.8. In each community, we collapsed SNPs by LD blocks and number of network in which they were present and, in the case of edges, by genes to which they were associated. We then compared the distribution of proportion of unique elements between TS and shared communities, using a Mann–Whitney U test.
Enrichment in TS Activated Regions Among Unique SNPs.
We first determined the chromatin state at each TS SNP (as defined above) in eight tissues, using the core 15-states model of the Roadmap Epigenomics Project (below). We then compared the chromatin state at each TS SNP in the tissue in which it was present to each of the seven others. To control for LD, we randomly selected TS SNPs by LD blocks (definition above) and chromatin state in the tissue corresponding to the network in which they were present. We split chromatin states into three classes: active chromatin (1_TssA, 2_TssAFlnk, 3_TxFlnk, 4_Tx, 5_TxWk, 6_EnhG, 7_Enh, 8_ZNF/Rpts), bivalent chromatin (10_TssBiv, 11_BivFlnk, 12_EnhBiv), and silent chromatin (9_Het, 13_ReprPC, 14_ReprPCWk, 15_Quies). For each pairwise comparison, we classified TS SNPs in four categories: same (chromatin state is exactly the same), similar (chromatin-state change from one to another chromatin state of the same class, for example 4_Tx 5_TxWk), repressed (from an active to any other class or from a bivalent to a silent class), and activated (from a silent to any other class or from a bivalent to an active class). We then calculated the number of TS SNPs in the activated and in the three other categories for each pairwise comparison. Summing over all seven comparisons for each network, we calculated enrichment in activated category among TS SNPs in TS communities, compared with TS SNPs in shared communities (definition above). P values were calculated using a Fisher test.
GWAS Analysis.
We downloaded the NHGRI-EBI GWAS catalog (accessed December 8, 2015, version v1.0) from the EBI website (https://www.ebi.ac.uk/gwas). We filtered out associations with P values greater than . We then compared the distribution of SNP core scores between GWAS-associated SNPs from the NHGRI-EBI catalog and those not associated with traits or diseases for each TS network, using a likelihood-ratio test (LRT). In our setting, the LRT assesses whether a linear model that includes GWAS status (Eq. 4) fits the observed data better than a linear model that does not include this variable (Eq. 3). As the distribution of SNP core scores () is not uniform across communities, we added community identity as a covariate in the linear regression. In Eqs. 3 and 4, is the core score of SNP in community , the number of communities in the tissue, an indicator function equal to 1 if the SNP is associated with a trait or disease in the GWAS and equal to 0 otherwise, and an indicator function equal to 1 if the SNP belongs to community and equal to 0 otherwise:
| [3] |
| [4] |
To control for LD between SNPs, we generated lists of SNPs falling into the same LD block, using the plink2 –blocks option, a 5-Mb maximum block size, and an of 0.8. In each community, for each LD block, we extracted the median of for GWAS SNPs and non-GWAS SNPs separately and used these values as input in the linear regressions.
Chromatin-State Category Definition.
We downloaded the genome-wide core 15-state model chromatin-state data from the Roadmap Epigenomics Project website (www.roadmapepigenomics.org/) for the eight tissues for which data are available: adipose subcutaneous, artery aorta, fibroblast cell line, esophagus mucosa, heart left ventricle, lung, skeletal muscle, and whole blood (20). The 15 chromatin states are active TSS (TssA), flanking active TSS (TssAFlnk), transcribed at a gene’s 5 and 3 ends (TxFlnk), strong transcription (Tx), weak transcription (TxWk), genic enhancers (EnhG), enhancers (Enh), ZNF genes and repeated regions (ZNF/Rpts), constitutive heterochromatin (Het), bivalent/poised TSS (TssBiv), flanking bivalent TSS/enhancers (BivFlnk), bivalent enhancers (EnhBiv), repressed Polycomb (ReprPC), weak repressed Polycomb (ReprPCWk), and quiescent (Quies).
Chromatin-State Enrichment Analyses.
For chromatin-state analyses, we calculated enrichment in each functional category among either global or local hubs, using a logistic regression model (38) which allows for covariates,
where is an indicator function equal to 1 if the SNP belongs to the functional category and equal to 0 otherwise, and is an indicator function equal to 1 if the SNP is central (in the top quartile of core scores or for degree) and equal to 0 otherwise. The odds ratios are estimated by . Similarly, we used a conditional logistic regression to calculate the combined odds ratio across tissues, using the model
where strata(Tissue) allows us to stratify the analysis by tissue. We used all SNPs in each TS network as background.
Similar to the calculation of enrichment in GWAS SNPs, we included indicators of community as a covariate when computing enrichment in Roadmap Epigenomics Project categories among high–core-score SNPs. To control for the gene density around a SNP, which can impact the number of cis-associations, we added a covariate corresponding to the number of genes within 1 Mb of a SNP.
Using the same method, we studied the enrichment in each chromatin state among trans-eQTL for each tissue,
where is an indicator function equal to 1 if the SNP is a trans-eQTL and equal to 0 otherwise. We used all SNPs with a MAF greater than or equal to 5% as background.
Supplementary Material
Acknowledgments
This work was supported by grants from the US National Institutes of Health, including grants from the National Heart, Lung, and Blood Institute (5P01HL105339, 5R01HL111759, 5P01HL114501, and K25HL133599), the National Cancer Institute (5P50CA127003, 1R35CA197449, 1U01CA190234, and 5P30CA006516), and the National Institute of Allergy and Infectious Disease (5R01AI099204). Additional funding was provided through a grant from the NVIDIA foundation. This work was conducted under dbGaP approved protocol no. 9112.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707375114/-/DCSupplemental.
References
- 1.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tak YG, Farnham PJ. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:1–18. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from gwas. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres JM, et al. Cross-tissue and tissue-specific eqtls: Partitioning the heritability of a complex trait. Am J Hum Genet. 2014;95:521–534. doi: 10.1016/j.ajhg.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirsten H, et al. Dissecting the genetics of the human transcriptome identifies novel trait-related trans-eqtls and corroborates the regulatory relevance of non-protein coding loci. Hum Mol Genet. 2015;24:4746–4763. doi: 10.1093/hmg/ddv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzén O, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brynedal B, et al. Large-scale trans-eqtls affect hundreds of transcripts and mediate patterns of transcriptional co-regulation. Am J Hum Genet. 2017;100:581–591. doi: 10.1016/j.ajhg.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 14.The GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulson JN, et al. 2016. YARN: Robust Multi-Condition RNA-Seq Preprocessing and Normalization. R package version 1.0.1.
- 16.Paulson J, et al. 2016. Tissue-aware RNA-Seq processing and normalization for heterogeneous and sparse data. bioRxiv: dx.doi.org/10.1101/081802. Accessed October 20, 2016.
- 17.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stranger BE, De Jager PL. Coordinating GWAS results with gene expression in a systems immunologic paradigm in autoimmunity. Curr Opin Immunol. 2012;24:544–551. doi: 10.1016/j.coi.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platig J, Ott E, Girvan M. Robustness of network measures to link errors. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;88:062812. doi: 10.1103/PhysRevE.88.062812. [DOI] [PubMed] [Google Scholar]
- 22.Wang DJ, Shi X, McFarland DA, Leskovec J. Measurement error in network data: A re-classification. Social Networks. 2012;34:396–409. [Google Scholar]
- 23.Platig J, Castaldi PJ, DeMeo D, Quackenbush J. Bipartite community structure of eQTLs. PLoS Comput Biol. 2016;12:e1005033. doi: 10.1371/journal.pcbi.1005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz K, et al. The breathing heart—Mitochondrial respiratory chain dysfunction in cardiac disease. Int J Cardiol. 2014;171:134–143. doi: 10.1016/j.ijcard.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Welter D, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battle A, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abnet CC, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gronnier C, et al. The MUC1 mucin regulates the tumorigenic properties of human esophageal adenocarcinomatous cells. Biochim Biophys Acta. 2014;1843:2432–2437. doi: 10.1016/j.bbamcr.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Barber MJ. Modularity and community detection in bipartite networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:066102. doi: 10.1103/PhysRevE.76.066102. [DOI] [PubMed] [Google Scholar]
- 31.Hicks SC, et al. 2016. Smooth quantile normalization. bioRxiv: dx.doi.org/10.1101/085175.
- 32.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695 [Google Scholar]
- 36.Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theor Exp. 2008;2008:P10008. [Google Scholar]
- 37.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 38.Kudaravalli S, Veyrieras JB, Stranger BE, Dermitzakis ET, Pritchard JK. Gene expression levels are a target of recent natural selection in the human genome. Mol Biol Evol. 2009;26:649–658. doi: 10.1093/molbev/msn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.