Significance
Sister chromatids are tethered together by the cohesin complex from the time they are made until cell division. Acetylation of the Smc3 subunit of cohesin stabilizes its association with chromatin, and is critical for sister chromatid cohesion. In vertebrates, cohesin is acetylated by two related enzymes: Esco1 and Esco2. We show here that Esco1 is responsible for most Smc3 acetylation but has very little effect on sister cohesion. Esco1 is active throughout the cell cycle, while Esco2 modifies cohesin only during S phase, when sister chromatid cohesion is established. We propose that two distinct pathways regulate cohesin in vertebrates: one is dedicated to cohesion between sister chromatids, and one promotes other functions of cohesin, such as maintenance of chromosome structure.
Keywords: chromosome biology, sister chromatid cohesion, Esco enzymes, cell cycle
Abstract
Sister chromatids are tethered together by the cohesin complex from the time they are made until their separation at anaphase. The ability of cohesin to tether sister chromatids together depends on acetylation of its Smc3 subunit by members of the Eco1 family of cohesin acetyltransferases. Vertebrates express two orthologs of Eco1, called Esco1 and Esco2, both of which are capable of modifying Smc3, but their relative contributions to sister chromatid cohesion are unknown. We therefore set out to determine the precise contributions of Esco1 and Esco2 to cohesion in vertebrate cells. Here we show that cohesion establishment is critically dependent upon Esco2. Although most Smc3 acetylation is Esco1 dependent, inactivation of the ESCO1 gene has little effect on mitotic cohesion. The unique ability of Esco2 to promote cohesion is mediated by sequences in the N terminus of the protein. We propose that Esco1-dependent modification of Smc3 regulates almost exclusively the noncohesive activities of cohesin, such as DNA repair, transcriptional control, chromosome loop formation, and/or stabilization. Collectively, our data indicate that Esco1 and Esco2 contribute to distinct and separable activities of cohesin in vertebrate cells.
Cohesin is a multisubunit protein complex first identified based on its role in tethering together sister chromatids in M phase cells. Since that time, cohesin has also been shown to play critical roles in certain kinds of DNA repair and, in higher eukaryotes, in chromosome structure. All of cohesin’s activities depend on its ability to entrap or tether chromatin: in the case of sister chromatid cohesion, cohesin tethers together the two identical products of DNA replication as they emerge from the replication fork; in its structural role, cohesin is proposed to stabilize chromosome loops (1–5).
The stability of the interaction between cohesin and chromatin is controlled in part by acetylation of the head domain of the Smc3 subunit of the complex. This acetylation inhibits opening of the cohesin ring by the protein Wapl, thereby stabilizing cohesion (6, 7). In budding yeast, cohesin is acetylated by the Eco1 acetyltransferase (8–10). Vertebrates express two related acetyltransferase enzymes, called Esco1 and Esco2, but their relative contributions to cohesin regulation are not clear. In embryonic extracts, the two Esco enzymes are not functionally redundant. Depletion of Esco2 from Xenopus egg extracts results in loss of cohesion. Supplementation of extracts with recombinant Esco1, which is not normally expressed in the early frog embryo, rescues Smc3 acetylation, but does not restore sister chromatid tethering (11). In contrast, some reports using cultured somatic cells have suggested that both Esco1 and Esco2 contribute to sister chromatid cohesion, as simultaneous depletion of both enzymes resulted in cohesion defects that were more severe than either single depletion (12).
Esco1 and Esco2 have distinct patterns of expression relative to cell cycle progression. While Esco1 is present at nearly constant levels throughout the cell cycle, Esco2 is a substrate of the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that is activated at mitotic exit (11–13). Thus, Esco2 levels are low in G1, and only increase as APC activity drops during S phase.
Finally, chromatin immunoprecipitation experiments in somatic cells indicate that Esco1 and Esco2 have distinct chromosomal addresses. Colocalization of Esco1 with the insulator protein CTCF and cohesin at the base of chromosome loops suggests that Esco1 promotes normal chromosome structure (14, 15). Consistent with this, depletion of Esco1 in somatic cells results in dysregulated transcriptional profiles (15). In contrast, Esco2 is localized to distinctly different sites, perhaps due to association with the CoREST repressive complex (15, 16).
Here, using a combination of siRNA-mediated depletion, rescue, and CRISPR/Cas9-mediated genome editing, we define the contributions of Esco1 and Esco2 to sister chromatid cohesion and Smc3 acetylation during cell cycle progression. We show that the majority of Smc3 acetylation is due to the activity of Esco1, while cohesion establishment during S phase requires Esco2. Inactivation of the ESCO1 gene has insignificant impact on mitotic cohesion. We propose that cohesin acetylation by Esco1 promotes normal chromosome structure throughout interphase and provides epigenetic memory during cell division by ensuring cohesin stabilization at appropriate loci upon mitotic exit. In contrast Esco2-dependent cohesin modification is essential during DNA replication for the establishment of cohesion between sister chromatids.
Results
The Contributions of Esco1 and Esco2 to Sister Chromatid Cohesion.
Like the founding member of the family, budding yeast Eco1, the vertebrate Esco enzymes both contain a PCNA interacting protein (PIP) box, a C2H2 zinc finger, and a catalytic region at the C terminus (12, 17). In contrast to the yeast protein, both Esco1 and Esco2 contain long N-terminal extensions, whose functions are poorly characterized. These regions show no obvious sequence or structural similarities between them (Fig. 1A).
Fig. 1.
Contributions of Esco1 and Esco2 to mitotic cohesion. (A) Schematic of Saccharomyces cerevisiae Eco1p and the vertebrate Esco1 and Esco2 enzymes with conserved domains indicated. The catalytic acetyltransferase domain is shown in blue, and the conserved zinc fingers and PCNA interacting protein (PIP) box are shown in black and gold, respectively. Nonhomologous N-terminal extensions in Esco1 and Esco2 are shown in green and light blue, respectively. (B) HeLa cells were transfected with siRNA against the indicated proteins, and whole cell lysates were analyzed by immunoblot for the indicated proteins. The cytosolic scaffold protein Nck was used as a loading control. * indicates background band detected by Esco2 antibody (Fig. S1). (C) Chromosome spreads were prepared from the same samples shown in B and cohesion phenotypes were classified as one of four states of cohesion, I–IV, as shown. (I) Unresolved sister chromatid arms; (II) resolved arms, with tight centromeres, (III) separated sisters that remain near to each other; and (IV) scattering of individual sister chromatids. (Scale bar, 20 µm.) (D) The percentage of cells with defective cohesion in categories III (separated) and IV (scattered) was determined for cells depleted of the indicated proteins. Data are presented as the percentage of mitotic spreads with the indicated phenotype, n ≥ 100 for all samples (E). Total loss of cohesion (categories III and IV): cumulative data from six separate experiments that were scored as in D. Boxes: 25–75% data range; whiskers: total data range. ****P < 0.0001; **P = 0.0021; n.s., not significant; ANOVA, n ≥ 100 all samples.
To define the contributions of Esco1 and Esco2 to sister chromatid cohesion, we scored mitotic cohesion in HeLa cells that had been depleted of Esco1 and Esco2, either singly or together, by siRNA-mediated depletion (Fig. 1B). Chromosome spreads were scored as representing one of four states of cohesion: I–IV (Fig. 1C). Spreads that fell into categories III and IV were considered defective in cohesion. Depletion of the cohesion regulator Sororin was used as positive control for loss of cohesion (18). Depletion of Esco2 caused loss of cohesion in mitotic cells, with ∼30% of mitotic spreads showing defective cohesion (Fig. 1 D and E). In contrast, acute depletion of Esco1 did not cause significant loss of cohesion, consistent with the analysis of a genetic knockout (see below). Codepletion of Esco1 with Esco2 caused an increase in defective cohesion compared with cells depleted of Esco2 only (Fig. 1E). In some experiments, >65% of mitotic spreads showed defective cohesion when depleted of both enzymes. We conclude that mitotic cohesion requires Esco2-dependent cohesin modification.
Mitotic Progression Is Delayed in Cells Depleted of Esco2 but Not Esco1.
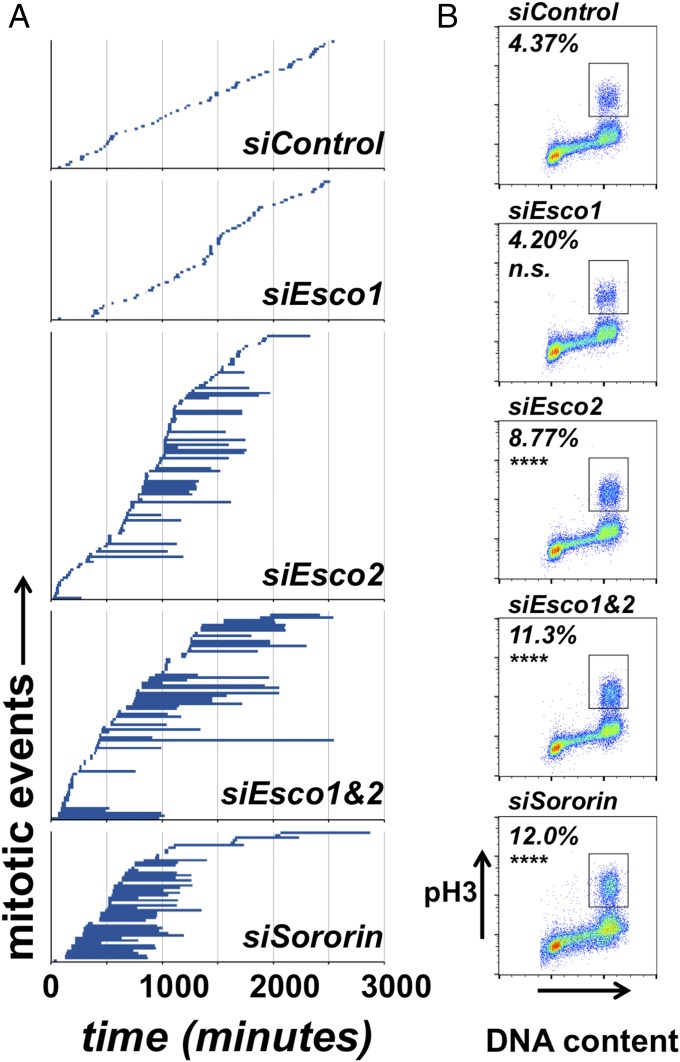
We observed little effect of Esco1 depletion on mitotic cohesion (Fig. 1), although loss of cohesion had previously been reported in Esco1 RNAi experiments (8, 12, 14). We therefore sought more sensitive methods to detect subtle cohesion defects. Because reduced cohesion can lead to activation of the spindle checkpoint, a signal transduction pathway that prevents mitotic exit in the presence of unattached chromosomes, we tested the effects of Esco1 and/or Esco2 depletion on mitotic progression. HeLa cells stably expressing red fluorescent protein (RFP)-histone H2B were transfected with siRNAs and analyzed by time-lapse microscopy. The duration of mitosis in each of the cell populations from nuclear envelope breakdown (NEBD) until segregation of chromosomes into two masses was measured. Control cells progressed through mitosis in ≤40 min (mean = 34.2 min; n = 50), as did the cells depleted of Esco1 (mean = 33.7 min; n = 54) (Fig. 2A). Cells depleted of Esco2 frequently showed sustained mitotic arrest (mean = 185 min; n = 102), as did cells that were depleted of both Esco1 and Esco2 (mean = 367 min; n = 79). The mitotic arrest in these cells was similar to that seen in the samples depleted of Sororin (18). These results are broadly consistent with the analysis of chromosome spreads (Fig. 1); loss of cohesion was evident in the absence of Esco2, and this effect was exacerbated in the absence of Esco1. Depletion of Esco1 alone had no significant effect on mitotic cohesion or progression through mitosis.
Fig. 2.
Mitotic progression is delayed in cells depleted of Esco2. HeLa cells stably expressing RFP-H2B were transfected with siRNA against the indicated proteins and time-lapse images were collected starting 24 h after transfection. (A) The duration of mitosis was measured starting 24 h after siRNA transfection. Each blue line represents a single mitotic event, with the length of the line indicating the duration of mitosis from nuclear envelope breakdown until the separation of two RFP-positive DNA masses, shown on x axis. Data from one representative experiment are shown. (B) Parallel samples were harvested 48 h after siRNA transfection, fixed, and stained for phosphorylated histone H3 (pH3) as a marker of mitosis, and analyzed by flow cytometry for both DNA content (x axis) and pH3 signal (y axis). The percentages of cells in mitosis based on pH3 staining (boxed) are indicated for each sample. Significant differences in pH3-positive cells compared with control cells are indicated: ****P < 0.0001; n.s., not significant; χ2 with Yates correction.
As an independent means of determining whether depletion of the Esco enzymes results in an increased mitotic index, we analyzed the Esco1- and/or Esco2-depleted cells by flow cytometry using phosphorylated histone H3 (pH3) as a marker for M phase combined with DNA content analysis. Control and Esco1-depleted cells had similar percents of cells in mitosis, 4.37% and 4.24% (Fig. 2B). In contrast, depletion of Esco2, codepletion of Esco1 and Esco2, or depletion of Sororin all caused an increase of the percentage of cells in M phase compared with controls (Fig. 2B). We conclude that Esco2 activity is sufficient to promote full cohesion, and that Esco1’s ability to promote cohesion is only detectable when Esco2 levels are reduced.
Esco1 Knockout Cell Lines Have Essentially Normal Mitotic Cohesion.
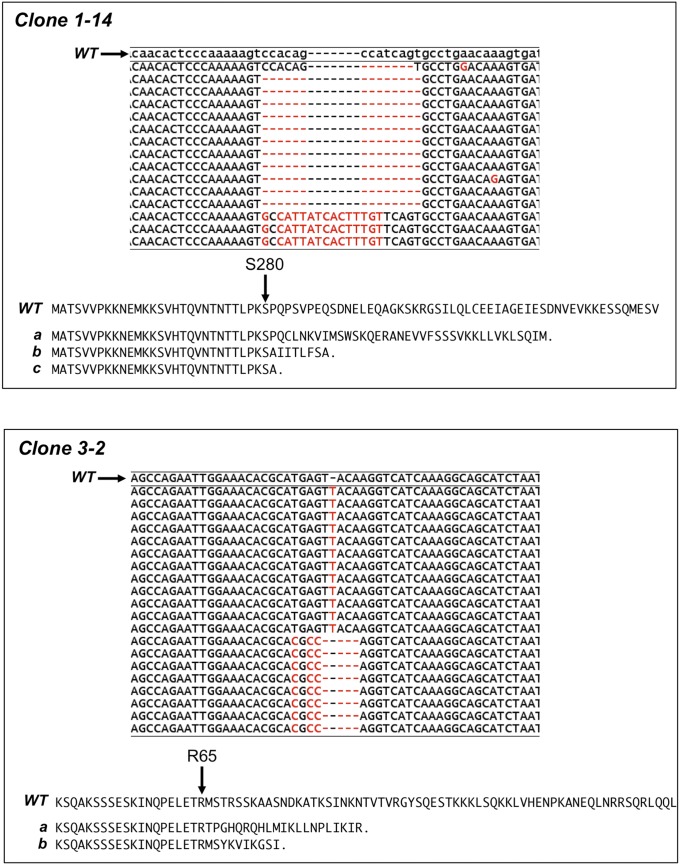
We did not detect an impact of Esco1 depletion on sister chromatid cohesion, in contrast to previous reports (11, 12). Because incomplete depletion following RNAi is particularly problematic when manipulating expression of enzymes as small amounts of catalytic activity can suffice, we used CRISPR/Cas9 technology to inactivate the ESCO1 gene in HeLa cells. The Cas9 nuclease was targeted to two different regions of the Esco1 gene using gRNA sequences unique in the human genome. Individual clones were screened by immunoblot for loss of Esco1 expression, and inactivation of the gene was confirmed by sequence analysis of genomic DNA. Two independent lines were isolated containing inactivating mutations in the ESCO1 gene (Fig. S2). Perhaps surprisingly, the ESCO1KO cells grew indistinguishably from their parental counterparts under standard culture conditions, and bulk DNA replication appeared unaffected (Fig. S3). Consistent with its crucial function for cohesion, repeated attempts to knock out both ESCO2 alleles using a similar strategy were unsuccessful (Discussion).
Fig. S2.
Sequence analysis of ESCO1KO cell lines. DNA fragments were amplified from ESCO1KO candidates with primers flanking the CRISPR/Cas9 target site. PCR products were cloned into plasmids and 15–25 individual colonies were picked and sequenced. Shown are alignments between the sequenced fragments and the wild-type Esco1 genomic sequence. Clone 1-14 contains three alleles of Esco1. All are predicted to result in a frameshift and premature stop codon. Predicted protein sequences and truncations due to frameshifts are illustrated, showing that all three mutations result in truncation shortly after S280. Clone 3-2 contained only two alleles. Both are predicted to result in truncations shortly after R65, as shown. Karyotype analysis suggests that chromosome 18, on which the ESCO1 gene is found, is present in two to three copies in HeLa cells (42).
Fig. S3.
Analysis of DNA replication by EdU incorporation. ESCO1KO cells and parental controls were pulse labeled with EdU and fixed. Following fluorescent labeling of the EdU with click chemistry, the cells were analyzed by flow cytometry to measure both total DNA content (x axis) and EdU incorporation (y axis). Both cell lines showed very similar profiles, as was evident when the data are overlaid.
The ESCO1KO cells were used to determine the contributions of Esco1 and Esco2 to Smc3 acetylation. Cells were synchronized in M phase by sequential thymidine-nocodazole arrest, released to resume cell cycle progression, and assayed for Smc3 acetylation on K105/106 and DNA content (Fig. 3 A and B). Progression through S phase was similar in the parental and ESCO1KO cells (Fig. 3B). In mitotic cells (t = 0), Smc3 acetylation was relatively low, as seen previously (14). Smc3 acetylation in the parental cells rose early in G1, well before DNA replication was evident (Fig. 3A). In contrast, in the ESCO1KO cell line, Smc3 acetylation remained low until bulk DNA replication occurred, ∼8 h after nocodazole release. Thus, Esco1-dependent acetylation occurs throughout interphase, both before and during DNA replication, and at all time points the majority of Smc3 acetylation on K105/106 is dependent upon Esco1 (Fig. 3C), while Esco2-dependent acetylation occurs only coincident with DNA replication.
Fig. 3.
Esco1-dependent Smc3 acetylation occurs throughout interphase. Parental and ESCO1KO cells were synchronized in M phase by sequential thymidine and nocodazole treatment. Following washout, samples were collected at the indicated times for immunoblot analysis (A) and to measure DNA content by flow cytometry (B). All samples were run on the same gel, and blots were cut horizontally (Fig. S4). The relative amount of Smc3Ac (normalized to total Smc3) in parental and ESCO1KO cells was quantified by comparing chemiluminescent signal intensities in A and is presented in C. As, asynchronously growing cells. T = 0 samples were collected before nocodazole washout. * indicates background band detected by Esco2 antibody (Fig. S1).
Fig. S4.
Blots used to generate Fig. 3A. Shown are the chemiluminescence and bright field images of immunoblots used to build Fig. 3A. Samples were run on one gel, and images were collected in two sessions. (A) Blots probed for Esco1, Esco2, and Nck. (B) Blot used to probe for Smc3Ac and subsequently probed for Smc3.
Consistent with our siRNA experiments (Fig. 1), we found that cohesion was largely unaffected by inactivation of the ESCO1 gene alone (Fig. 4A). Depletion of Esco2 from the parental HeLa cell line resulted in significant loss of cohesion (∼45%). Strikingly, depletion of Esco2 from the ESCO1KO cell line resulted in catastrophic loss of cohesion, similar to depletion of the essential cohesion regulator Sororin. The mobility of Sororin was reduced in these samples, consistent with mitotic arrest (Fig. 4B). These data indicate that Esco1 enhances cohesion when Esco2 is present, but does not normally contribute to sister chromatid cohesion.
Fig. 4.
ESCO1KO cells have largely normal sister chromatid cohesion. (A) Cohesion assay. Chromosome spreads from parental and ESCO1KO cells were scored for sister chromatid cohesion following transfection with the indicated siRNAs. Shown are the mean and SD of three replicate experiments. (B) Immunoblot showing levels of the indicated proteins from a representative experiment included in A. Both histone H3 and Nck were used as loading controls. * indicates background band. (C) Intercentromere distance. The distance between sister centromeres was measured in parental and ESCO1KO cells stained with CREST serum. Error bars represent minimum/maximum, box includes 25th–75th percentiles. Unpaired t test P = 0.06 n = 116 (parental) and 112 (ESCO1KO). n.s., not significant. (D) Cohesion fatigue. Analysis of cohesion fatigue in parental and ESCO1KO cell lines. The spindle-dependent separation of sister chromatids during M phase arrest was assessed by comparing samples treated with MG132 (blue bars) to cells treated with both MG132 and nocodazole (green bars). The graph indicates percent of spreads with scattered chromatids at 0, 4, and 6 h of treatment. n ≥ 100 for all samples. **P = 0.0137 Fisher’s exact.
Although ESCO1 gene inactivation alone caused no obvious loss of cohesion, it remained possible that cohesion was reduced, but not sufficiently to activate the spindle checkpoint. To explore this possibility, we measured the distance between sister centromeres, comparing parental and ESCO1KO cells that were stained with calcinosis, Reynaud’s phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia (CREST) serum to label centromeric proteins. The intercentromere distance in parental and ESCO1KO cells was not significantly different (Fig. 4C), indicating that cohesion at the centromere region is not greatly affected by loss of Esco1 function.
During sustained mitotic arrest, cohesion between sister chromatids eventually fails, and the rate at which this failure occurs is inversely correlated with the overall level of mitotic cohesion (19, 20). We measured “cohesion fatigue” in the ESCO1KO cells compared with parental controls. The ESCO1KO cells showed slightly elevated rate of fatigue, suggesting that Esco1 makes a measurable contribution to the strength of mitotic cohesion established by Esco2 activity (Fig. 4D).
Esco Activity Depends on N-Terminal Sequences.
To determine whether the ability of Esco2 to promote cohesion is dependent upon the unique sequences in its N terminus, we performed rescue experiments using chimeric fusion proteins. The N terminus of Esco1 was fused at the PIP box to the C terminus of Esco2 to generate a fusion protein called Esco1-2 (Fig. 5A). The converse cDNA fusion, Esco2-1, was also constructed (Fig. S5A). The genes were integrated into the ESCO1KO cell line in a manner that allowed transgene expression in a tetracycline-inducible manner (Fig. 5B). Expression of the transgenes was induced with doxycycline, and cells were transfected with siRNA directed against the endogenous Esco2. To avoid depletion of the transgenes being tested, siRNAs were chosen to target the regions of the endogenous Esco2 transcript not present in the transgene. Analyzing chromosome spreads as in Fig. 1, we found that Esco2-1 was able to promote cohesion, while Esco1-2 was not (Fig. 5C). Thus, the unique capacity of Esco2 to promote sister chromatid cohesion lies in its N terminus, which cannot be substituted with the N terminus of Esco1. The level of Smc3 acetylation was lower in the cells expressing Esco1-2 compared with Esco2-1. This may reflect nearly complete cohesion failure and mitotic arrest in the Esco1-2 cells, as in M phase, the overall acetylation is relatively low (Fig. 3). Alternatively, this result may suggest that Esco1-2 is intrinsically unable to promote Smc3 acetylation. To distinguish between these two possibilities, we synchronized cells in S phase and found that Esco1-2 chimera still did not promote Smc3 acetylation, while Esco2-1, which was able to rescue cohesion (Fig. 5D), did. Additional Esco1-2 clones had the same phenotype (Fig. S5B). Thus, the C-terminal, catalytic domain of Esco1 is functional for both Smc3 acetylation and cohesion establishment when fused to the N terminus of Esco2. We do not yet fully understand why the Esco1-2 fusion is inactive; perhaps dimerization, possibly important for Esco function (21, 22), is critically impaired in the Esco1-2 chimera.
Fig. 5.
The N terminus of Esco2 promotes cohesion establishment. Chimeric cDNAs were generated to express fusions between the Esco1 (green) and Esco2 (blue) proteins. (A) Cartoon illustrating the wild-type proteins, as well as chimeric derivatives, exchanged precisely at the PIP boxes (shown in black), as shown (dotted lines). Bars below each cartoon indicate the region recognized by antibodies against Esco2 (dark green) and Esco1 (dark blue). (B) Immunoblot analysis of expression of chimeric proteins. ESCO1KO cells were engineered to express Esco1, Esco1-2, or Esco2-1 in a doxycycline-inducible manner. The expression of each transgene following treatment with doxycycline was assessed using appropriate antibodies as indicated in A. Samples were all blotted together from the same gel; dotted line was added for clarity. To assess the ability of the chimeras to promote sister chromatid cohesion, cells were also treated with one of two siRNAs, selected to react only against the endogenous Esco2 gene. These siRNAs, “a” (N terminal) and “b” (C terminal) are illustrated in A; “c” = control siRNA. (C) The samples probed in B were assayed for sister chromatid cohesion and scored as described in Fig. 1. (D) ESCO1KO cells expressing the indicated transgenes were synchronized in S phase by treatment with thymidine for 20 h and analyzed by immunoblot for protein expression. Shown also is subcellular fractionation analysis of endogenous Esco1 and Esco2 in cells growing asynchronously (E) or arrested in M phase with nocodazole (F). N, chromatin associated; S, cytosolic supernatant; T, total cell lysate. The Esco2 band in M phase cells is difficult to detect due to the presence of a background band (*) (see also Fig. S1).
Fig. S5.
Chimeric Esco proteins. (A) Chimeric protein sequences. Esco1-2 (Top) contains N-terminal Esco1 sequence (yellow) fused at the PIP box (Q604) to Esco2 C-terminal sequences (gray). Similarly Esco2-1 contains N-terminal Esco2 sequence (gray) fused at its PIP box to the C terminus of Esco1 (yellow). (B) Expression of Esco1-2 in ESCO1KO cells. Multiple Esco1-2 expressing clones promote the same level of Smc3 acetylation. Shown are three clones (nos. 2, 3, and 5) transfected with the indicated siRNA (c: control siRNA, and a: N-terminal Esco2 siRNA, as in Fig. 5) and grown in doxycycline and 2 mM thymidine for 20 h. Whole cell lysate was probed for the indicated proteins. All clones were able to promote only weak Smc3 acetylation in the absence of Esco2.
To further characterize the differences between Esco1 and Esco2 we analyzed their localization in fractionated cells. In asynchronously growing cells, both enzymes were associated with the chromatin fraction (Fig. 5E). In contrast, although Esco1 recruitment to chromatin is dependent upon the cohesin complex (6, 7, 15), Esco1 remained associated with chromatin in M phase, when cohesin is largely dispersed (Fig. 5F), as noted previously (8–10, 12). Esco2 was difficult to detect in M phase samples (see also Fig. 3 and Fig. S1), suggesting that it may be at least partially degraded before metaphase (Fig. 5E). Importantly, Esco1 is retained on chromosomes in M phase where it can promote Smc3 acetylation directly upon mitotic exit. These data are consistent with a model in which Esco1 regulates cohesin beginning in early telophase.
Fig. S1.
Characterization of Esco2 antibody. HeLa cells were transfected with a plasmid encoding untagged Esco2 (OE, overexpress), with siRNA against Esco2 (siE2), or not transfected (unt). Each sample was divided in three, and left either untreated (−), treated with 100 ng/ml nocodazole to cause accumulation of mitotic cells (N), or treated with thymidine (2 mM) to cause accumulation of cells in S phase, all for 13 h beginning 24 h following transfection. The samples were then probed for Esco2 with three different antibodies: nos. 708, 216 (both custom made), or Bethyl A301-689A. The strong lower band is only detected with the Bethyl antibody, is enriched in all mitotic samples, and is independent of the Esco2 levels. We conclude that the Upper band is Esco2, while the Lower band is a cross-reacting antigen detected only in mitotic samples. Smc3 served as a loading control. NB, the relative abundance of the background band varied in different lots of the antibody. The Esco2 band is indicated with black arrows.
Discussion
Acetylation of the Smc3 subunit of cohesin by members of the Eco1 family of acetyltransferases stabilizes the interaction of cohesin with chromatin, and is essential for cohesion establishment. The advent of CRISPR-based genome editing has allowed us to assess unambiguously the relative contributions of the two vertebrate Eco enzymes, Esco1 and Esco2, to mitotic tethering of sister chromatids. Our results suggest that Esco1 is only able to promote cohesion when cohesion has already been established by the action of Esco2 during S phase. Loss of Esco1, either acutely, by siRNA-mediated depletion, or genetically, by gene inactivation, did not cause overt loss of mitotic cohesion. This observation is reminiscent of G2 cohesion establishment in response to DNA damage in budding yeast, which is only effective in tethering sister chromatids if replicative establishment has already occurred (23). Vertebrate Esco1 then, may contribute to or strengthen sister chromatid cohesion, but is not dedicated to replicative cohesion establishment. In contrast, depletion of Esco2 alone caused significant loss of cohesion. Importantly, in the absence of Esco1, depletion of Esco2 led to complete loss of cohesion. These data are consistent with earlier work, in which we showed that Esco1 does not promote cohesion in Esco2-depleted Xenopus egg extracts (13). In fact Esco1 is not expressed in the early frog embryo until the onset of zygotic transcription. Thus, Esco2 is sufficient to promote proper tethering of sister chromatids during early embryonic development. Our work here contradicts previous work suggesting that Esco1 makes critical contributions to mitotic sister chromatid cohesion in somatic cells (12). This prior result may reflect off-target effects of siRNAs, which were used at higher concentration than used here, or perhaps differences among strains of HeLa cells. The consistency of our data from both ESCO1 knockout cells and siRNA-mediated depletion experiments makes us confident in our conclusions.
If the contribution of Esco1 to mitotic sister chromatid tethering is minimal in both embryonic and somatic cells, what then is its function? Esco1 acetylates cohesin throughout interphase, even well before DNA replication is evident. As cohesin plays critical roles in formation or maintenance of interphase chromosome structure (2, 3, 14, 15, 24–29), one attractive model is that Esco1 contributes to these events, perhaps through impacts on chromosome loop formation or stabilization. In chromatin immunoprecipitation experiments, cohesin, Esco1, and the insulator protein CTCF all show significant colocalization at the bases of chromosome loops (3, 5, 14, 15, 29, 30). This localization is independent of DNA replication and evident in G1 and G2 cells. The principle role of Esco1 then may be in regulation of this “structural cohesion” (Fig. 6). Esco1 may, for example, enhance loop extrusion or formation during exit from mitosis, ensuring that interphase chromosome structure is properly reestablished during chromosome decondensation. Depletion of Esco1 results in increased expression of a number of genes, perhaps affecting the ability of insulator proteins such as CTCF to prevent enhancer–promoter interactions (15, 16). In the absence of Esco1, inappropriate loop structures may be formed, leading to dysregulation of gene expression. Chromosome conformation capture experiments will provide important information about how Esco1 might enhance or regulate loop formation.
Fig. 6.
Model depicting the contributions of Esco1 and Esco2 to cohesin regulation. The blue lines represent chromatids, and cohesin is depicted as green rings. The activity of Esco1 throughout interphase, including in G1 before DNA replication has occurred, promotes cohesin’s role in defining chromosome structure. In contrast, Esco2 activity during S phase ensures cohesion between sister chromatids, and this stable cohesion (marked with black triangles) can be reinforced by the activity of Esco1. In the absence of Esco2, Esco1 is unable to stabilize cohesion between sister chromatids, and sister chromatids separate.
The impacts of Esco1 on chromosome structure may be imposed throughout the cell cycle, including M phase, as Esco1 is retained on chromatin as cells transit through mitosis (12, 15–17, and this work). The retention of Esco1 on mitotic chromosomes may provide epigenetic memory and contribute to the reestablishment of chromosome structure and the resumption of gene expression profiles as cells reenter interphase, thus ensuring the retention of cellular identity even in cells that are actively dividing. Esco1 might also contribute to the establishment of the replication timing profile, which is established in G1 (31) and may be dependent upon cohesin (14, 24).
Although it was relatively straightforward to generate Esco1 knockout cells, we were unable to generate an Esco2 knockout cell line using the same CRISPR/Cas9 technology, although we had clear evidence of mutagenesis. In our hands ESCO2 mutant cells grew poorly or were not genetically stable. This was surprising, given that the developmental disorder Roberts syndrome results from loss of Esco2 function (32). The robust spindle checkpoint of HeLa cells may prevent their proliferation in the absence of Esco2, while other cell lines might not arrest so strongly. Alternatively, Roberts syndrome mutations may be hypomorphic, or there may be mosaicism in affected individuals.
We have shown that it is the N terminus of Esco2 that specifies its distinct ability to promote sister chromatid cohesion. The C-terminal catalytic region, which contains the PIP box and zinc finger motif, may not contribute to the mechanistic specificity of the enzymes, as the C terminus of Esco1 is a functional surrogate when fused to the Esco2 N terminus. Prior work has shown that the C-terminal PIP box is essential for Esco2 function (13), but the purpose of the conserved PIP box in Esco1 remains mysterious, especially considering that Esco1 is active outside of S phase. In both Esco1 and Esco2, short conserved stretches within their N termini have previously been shown to promote normal function but the mechanistic basis for this is not known (12, 13, 15, 33). Interestingly, the N termini of both Esco1 and Esco2 are sufficient to promote their association with chromatin, independently of the C-terminal domain (12, 14). In budding yeast, the Eco1 PIP box was shown to be sufficient for binding to chromatin (17, 19, 20). The identity of the chromatin-associated binding partners of Esco1 and Esco2 will provide important clues about how these enzymes are differentially regulated to contribute to cohesin’s distinct functions. The ability of Esco1 to partially compensate when Esco2 is depleted may suggest that Esco1 competes for interaction with the same chromatin-associated factor, although perhaps inefficiently.
Methods
Please see SI Methods for additional experimental details.
Cell Culture, RNAi, and Flow Cytometry.
HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (Corning) supplemented with 10% FBS, and transfected with Lipofectamine 2000 (for DNA) or Lipofectamine RNAiMAX (for RNA) (both from Invitrogen) following the manufacturer’s guidelines. HeLa cells were synchronized in M phase by sequential treatment with 2 mM thymidine for 24 h, 4 h release, and 100 ng/mL nocodazole for 12 h. Flow cytometry data were acquired using FACSCalibur (BD Biosciences) and analyzed with FloJo software (Tree Star). Statistical analyses were done with Prism (GraphPad).
Cohesion Assays and Microscopy.
Cohesion assays were prepared as previously described (34). Loss-of-cohesion phenotypes, scored blind, were assigned to spreads in which ≥10 chromosomes showed the indicated morphology. These data were removed during the revision process. Cohesion fatigue was assayed by incubation of asynchronously growing cells with 10 μM MG132, both with and without 100 ng/mL nocodazole.
SI Methods
Antibodies and Immunoblots.
Antibodies to Esco2 (Bethyl Labs), Nck (Neomarkers), histone H2B (Upstate Biotechnology), histone H3 (Cell Signaling), and pH3 (Upstate Biotechnology) were commercially obtained. Antibodies to Sororin, Smc3 (34), and Smc3Ac (35) were reported previously. Anti-hEsco1 antibody was made by immunizing rabbits with a bacterially expressed purified fragment of human Esco1 (amino acids 1–139) and was affinity purified from whole serum. CREST serum was obtained from Antibodies, Inc., and cells were labeled as described previously (36). For immunoblots, protein samples were resolved on 7–15% gradient SDS/PAGE gels, transferred to nitrocellulose membranes, incubated with 5% milk in Tris-buffered saline, and probed with empirically determined concentrations of primary antibodies overnight at 4 °C. Horseradish peroxidase-labeled secondary antibodies were detected with chemiluminescent substrate (Licor Biosciences) and signals were collected using an Azure C600 CCD imager (Azure Biosystems), with the exception of Figure 1D, which was collected on radiographic film and subsequently scanned. Chemiluminescent image quantification was performed using Image Studio Lite (Licor Biosciences). The extraction method of Méndez and Stillman (37) followed by immunoblot was used to assess chromatin binding.
Flow Cytometry Details.
To assay for pH3, cells were fixed using ice-cold 70% ethanol, stored at −20 °C overnight, permeabilized with Perm Buffer (1× PBS, 0.5% BSA and 0.25% Triton) on ice for 15 min, pelleted, washed several times with PBS + 1% BSA, incubated with primary antibody for 1 h at RT, washed in PBS + 1% BSA, and then incubated with FITC-labeled secondary antibody (Jackson ImmunoResearch) for 30 min. Cells were washed in PBS + 1% BSA and resuspended in prodidium iodide (PI) staining solution (50 μg/mL propidium iodide, 100 μg/mL RNase A) for 30 min at room temperature. Analysis of DNA replication by pulse labeling asynchronously growing cells with EdU was done as previously described (38).
RNAi Details.
For RNAi, cells were transfected with pools of siRNA duplexes (Dharmacon Smartpools) at 20 nM, unless otherwise noted. For single siRNA transfections in rescue experiments, we used single RNA duplexes targeting the N terminus (504–523 bp of the coding sequence: CGAGUGAUCUAUAAGCCAA = “a” in Fig. 5 B–D) or the C terminus (1,457–1,476 bp: GAGAGUAGUUGGGUGUUUA = “b” in Fig. 5 B–D) of the Esco2 coding sequence at 10 nM. Negative control siRNA duplexes based on sequences not present in the human genome (Bioneer) were used at the same concentration as gene-specific siRNAs.
Chromosome Spread Details.
Cells were harvested with trypsin-EDTA, washed in media and then PBS, and swelled in 0.075 M KCl for 20 min at room temperature. Cells were fixed by suspension in ice-cold 3:1 methanol:acetic acid, and then placed at −20 °C overnight. After several exchanges with fresh methanol:acetic acid, the cells were dropped on glass slides and incubated at 65 °C for 1–2 h, and then air dried overnight. The spreads were visualized using Giemsa stain (VWR) according to the manufacturer’s protocol, and images were collected on a Zeiss Axioimager Z1 upright microscope equipped with an Axiocam color camera.
Construction of Knockout Cell Lines.
Unique or near unique Esco1 and Esco2 CRISPR/Cas9 target sites were identified using ChopChop (https://chopchop.rc.fas.harvard.edu; ref. 39). Plasmids encoding sgRNAs were engineered by cloning double-stranded adapters into plasmid pX330-U6-chimeric_BB-CBh-hSpCas9, a gift from Feng Zhang, The Broad Institute, Massachusetts Institute of Technology, Cambridge, MA (Addgene plasmid no. 42230) (40). The following primers were used to generate adapters: To make clone 3-2: SR694 (5′-CACCGTGGAAACACGCATGAGTACA) and SR695 (5′-AAACTGTACTCATGCGTGTTTCCAC); to make clone 1-14: SR690 (5′-CACCGTGTTCAGGCACTGATGGCTG) and SR691 (5′-AAACCAGCCATCAGTGCCTGAACAC). To generate knockout cell lines, 2.5e5 HeLa FlpIn T-Rex cells were plated in each well of a six-well dish, and the next day transfected with 1.5 μg per well of pX330 derivative plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The next day, the cells were plated in 10-cm dishes at ∼10, 20, and 50 cells per plate. Individual colonies that arose were collected on trypsin-EDTA–soaked filter paper, amplified, and screened by immunoblot for protein expression. Genomic DNA was isolated from clones with reduced protein expression, and fragments including the relevant Cas9 target site were amplified by PCR, cloned into plasmid vectors, and sequenced. Sequence analysis was performed using the Lasergene Molecular Biology Suite (DNASTAR).
Rescue Experiments.
All plasmids were made by isothermal assembly (41) and confirmed by sequence analysis. Plasmids derived from pcDNA5/FRT/TO (Invitrogen) were transfected into HeLa T-Rex cells (Invitrogen) containing an integrated Flp-In targeting site. Individual clones containing integrated plasmids were selected with 200 μg/mL hygromycin B (Gold Biotechnology), isolated on trypsin-soaked filters, and transgene induction following treatment with 1–5 μg/mL doxycycline was confirmed by immunoblot.
Acknowledgments
We thank K. Shirahige for the anti-Smc3Ac antibody and members of Cell Cycle & Cancer Biology at the Oklahoma Medical Research Foundation for many helpful discussions. E.M.L.d.S. was supported by a fellowship from the National Council for Scientific and Technological Development of Brazil. This work was supported by NIH Grant R01GM101250 and the Oklahoma Center for Adult Stem Cell Research (both to S.R.) and NIH Grant R01GM121703 (to C.L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708291114/-/DCSupplemental.
References
- 1.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 4.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckouët F, et al. Releasing activity disengages cohesin’s Smc3/Scc1 interface in a process blocked by acetylation. Mol Cell. 2016;61:563–574. doi: 10.1016/j.molcel.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Çamdere G, Guacci V, Stricklin J, Koshland D. The ATPases of cohesin interface with regulators to modulate cohesin-mediated DNA tethering. Elife. 2015;4:e11315. doi: 10.7554/eLife.11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Unal E, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 10.Rolef Ben-Shahar T, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 11.Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA. 2010;107:20364–20369. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou F, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell. 2005;16:3908–3918. doi: 10.1091/mbc.E04-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J, et al. Cohesin acetylation promotes sister chromatid cohesion only in association with the replication machinery. J Biol Chem. 2012;287:34325–34336. doi: 10.1074/jbc.M112.400192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamino M, et al. Esco1 acetylates cohesin via a mechanism different from that of Esco2. Curr Biol. 2015;25:1694–1706. doi: 10.1016/j.cub.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Rahman S, Jones MJK, Jallepalli PV. Cohesin recruits the Esco1 acetyltransferase genome wide to repress transcription and promote cohesion in somatic cells. Proc Natl Acad Sci USA. 2015;112:11270–11275. doi: 10.1073/pnas.1505323112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim BJ, et al. Esco2 is a novel corepressor that associates with various chromatin modifying enzymes. Biochem Biophys Res Commun. 2008;372:298–304. doi: 10.1016/j.bbrc.2008.05.056. [DOI] [PubMed] [Google Scholar]
- 17.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–1024. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens D, Gassmann R, Oegema K, Desai A. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One. 2011;6:e22969. doi: 10.1371/journal.pone.0022969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouznetsova E, et al. Sister chromatid cohesion establishment factor ESCO1 operates by substrate-assisted catalysis. Structure. 2016;24:789–796. doi: 10.1016/j.str.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 22.Onn I, Guacci V, Koshland DE. The zinc finger of Eco1 enhances its acetyltransferase activity during sister chromatid cohesion. Nucleic Acids Res. 2009;37:6126–6134. doi: 10.1093/nar/gkp656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ström L, et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 24.Guillou E, et al. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010;24:2812–2822. doi: 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Günal-Sadık G, et al. Stage-specific binding profiles of cohesin in resting and activated B lymphocytes suggest a role for cohesin in immunoglobulin class switching and maturation. PLoS One. 2014;9:e111748. doi: 10.1371/journal.pone.0111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt AD, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 32.Vega H, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 33.Higashi TL, et al. The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in Xenopus egg extracts. Curr Biol. 2012;22:977–988. doi: 10.1016/j.cub.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Wu FM, Nguyen JV, Rankin S. A conserved motif at the C terminus of sororin is required for sister chromatid cohesion. J Biol Chem. 2011;286:3579–3586. doi: 10.1074/jbc.M110.196758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiyama T, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 36.Sivakumar S, Daum JR, Tipton AR, Rankin S, Gorbsky GJ. The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol Biol Cell. 2014;25:594–605. doi: 10.1091/mbc.E13-07-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sansam CG, Goins D, Siefert JC, Clowdus EA, Sansam CL. Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes Dev. 2015;29:555–566. doi: 10.1101/gad.246827.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 42.Macville M, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]