Significance
The human placenta is a dynamic and cellular heterogeneous organ, which is critical in fetomaternal homeostasis and the development of preeclampsia. Previous work has shown that placenta-derived cell-free RNA increases during pregnancy. We applied large-scale microfluidic single-cell transcriptomic technology to comprehensively characterize cellular heterogeneity of the human placentas and identified multiple placental cell-type–specific gene signatures. Analysis of the cellular signature expression in maternal plasma enabled noninvasive delineation of the cellular dynamics of the placenta during pregnancy and the elucidation of extravillous trophoblastic dysfunction in early preeclampsia.
Keywords: single-cell transcriptomics, cell-free RNA, noninvasive prenatal testing, placenta, preeclampsia
Abstract
The human placenta is a dynamic and heterogeneous organ critical in the establishment of the fetomaternal interface and the maintenance of gestational well-being. It is also the major source of cell-free fetal nucleic acids in the maternal circulation. Placental dysfunction contributes to significant complications, such as preeclampsia, a potentially lethal hypertensive disorder during pregnancy. Previous studies have identified significant changes in the expression profiles of preeclamptic placentas using whole-tissue analysis. Moreover, studies have shown increased levels of targeted RNA transcripts, overall and placental contributions in maternal cell-free nucleic acids during pregnancy progression and gestational complications, but it remains infeasible to noninvasively delineate placental cellular dynamics and dysfunction at the cellular level using maternal cell-free nucleic acid analysis. In this study, we addressed this issue by first dissecting the cellular heterogeneity of the human placenta and defined individual cell-type–specific gene signatures by analyzing more than 24,000 nonmarker selected cells from full-term and early preeclamptic placentas using large-scale microfluidic single-cell transcriptomic technology. Our dataset identified diverse cellular subtypes in the human placenta and enabled reconstruction of the trophoblast differentiation trajectory. Through integrative analysis with maternal plasma cell-free RNA, we resolved the longitudinal cellular dynamics of hematopoietic and placental cells in pregnancy progression. Furthermore, we were able to noninvasively uncover the cellular dysfunction of extravillous trophoblasts in early preeclamptic placentas. Our work showed the potential of integrating transcriptomic information derived from single cells into the interpretation of cell-free plasma RNA, enabling the noninvasive elucidation of cellular dynamics in complex pathological conditions.
The placenta plays an essential role in the establishment of the fetomaternal interface and the maintenance of fetal homeostasis during pregnancy (1). It is a heterogeneous organ composed of cells of maternal and fetal origins, organized in multilobulated villous units. The multinucleated syncytiotrophoblast (SCTB) layer on top of villous cytotrophoblasts (VCTBs) is in direct contact with maternal blood. The entire placental villous structure is supported by stromal cells containing fetal macrophages (Hofbauer cells) and is perfused by the fetal capillary vasculature. A distinct type of trophoblast cells, the extravillous trophoblasts (EVTBs), infiltrates the maternal decidua in a unique process of “controlled invasion” to remodel the maternal uterine spiral arteries and interact with maternal lymphocytes to prevent allorejection of the fetus. Placental cellular dysfunction, therefore, contributes to major gestational complications, such as preeclampsia (PE) (2).
Despite the clinical significance of the placenta, direct tissue monitoring of the placenta of women with placental pathologies has not been feasible because of safety concerns associated with invasive placental tissue sampling. Instead, ultrasonography and maternal serum protein markers have been pursued to noninvasively monitor placental function during pregnancy (3, 4). It has been shown that the placenta is the major source of circulating cell-free fetal nucleic acids in maternal plasma (5–7). Significantly elevated levels of total cell-free DNA and selected placenta-specific RNA transcripts have also been reported in the maternal plasma of women with PE (8–11), restricted fetal growth (12), and preterm birth (13–15), supporting a role for cell-free nucleic acids as a noninvasive tool for placental monitoring. Previous studies have attempted to provide a comprehensive assessment of maternal plasma nucleic acids by microarray analysis, massively parallel transcriptomic, or methylomic sequencing (16–22). Several groups have explored the use of fetal-specific DNA polymorphisms, organ-specific DNA methylation (21), nucleosome footprinting (23), DNA fragmentation patterns (24), and tissue-specific RNA transcripts (19, 20) to isolate the placental signal in the pool of circulating cell-free fetal nucleic acids and obtain changes of overall placental contribution. Nevertheless, these approaches have a low resolution in examining the dynamics of the different fetal and maternal components in the placenta and differentiating the specific pathological changes of the placenta in different gestational pathologies at the cellular level.
To address these challenges, we explored the use of microfluidic single-cell digital transcriptomic technology to comprehensively characterize the transcriptomic heterogeneity of the human placenta. We analyzed, in an unbiased manner, the single-cell transcriptomes of more than 24,000 nonmarker-selected placental cells from normal and early PE placentas. PE is a potentially lethal gestational condition characterized by new onset of hypertension and proteinuria at ≥20 wk of gestation. It is a leading cause of maternal and perinatal morbidity and mortality. Defective placental implantation has been proposed as the major pathological mechanism in early PE occurring before 34 wk of gestation (2, 25), and previous studies based on whole-tissue profiling have reported extensive changes in the gene-expression profiles of preeclamptic placentas (26, 27).
Using the comprehensive single-cell dataset, we first showed that individual placental cellular components can be monitored through the aggregated signals of sets of highly cell-type–specific signature genes, which revealed the longitudinal cellular dynamics in maternal plasma during pregnancy progression. This approach also allows the identification of the EVTB pathology in early PE placentas from maternal plasma cell-free RNA. Our study showed the potential of an integrative and synergistic analytical approach of single-cell and cell-free RNA transcriptomic studies.
Results
Dissection of the Cellular Heterogeneity of the Human Placenta.
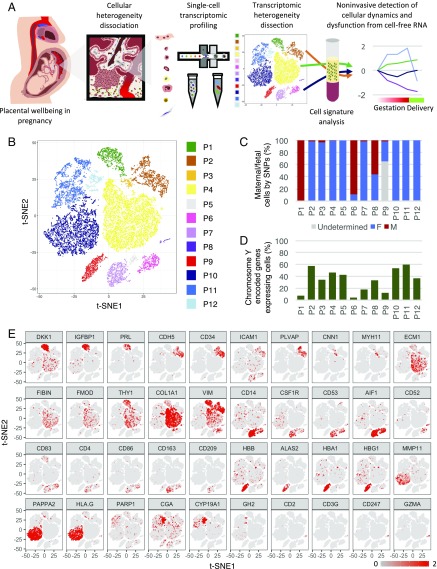
We set out to obtain a comprehensive understanding of the cellular heterogeneity of the human placenta using large-scale droplet-based single-cell digital transcriptomic profiling (28) (Fig. 1A). In this system, the individual cells are encapsulated in microfluidic droplets with a hydrogel bead containing reverse transcription and template-switching oligonucleotides. The oligonucleotides contain nucleotide barcodes unique to each hydrogel bead and identifiers [unique molecule identifiers (UMIs)] for tagging RNA molecules inside the encapsulated droplets. The UMIs allowed differentiation of amplified products from actual unique transcripts.
Fig. 1.
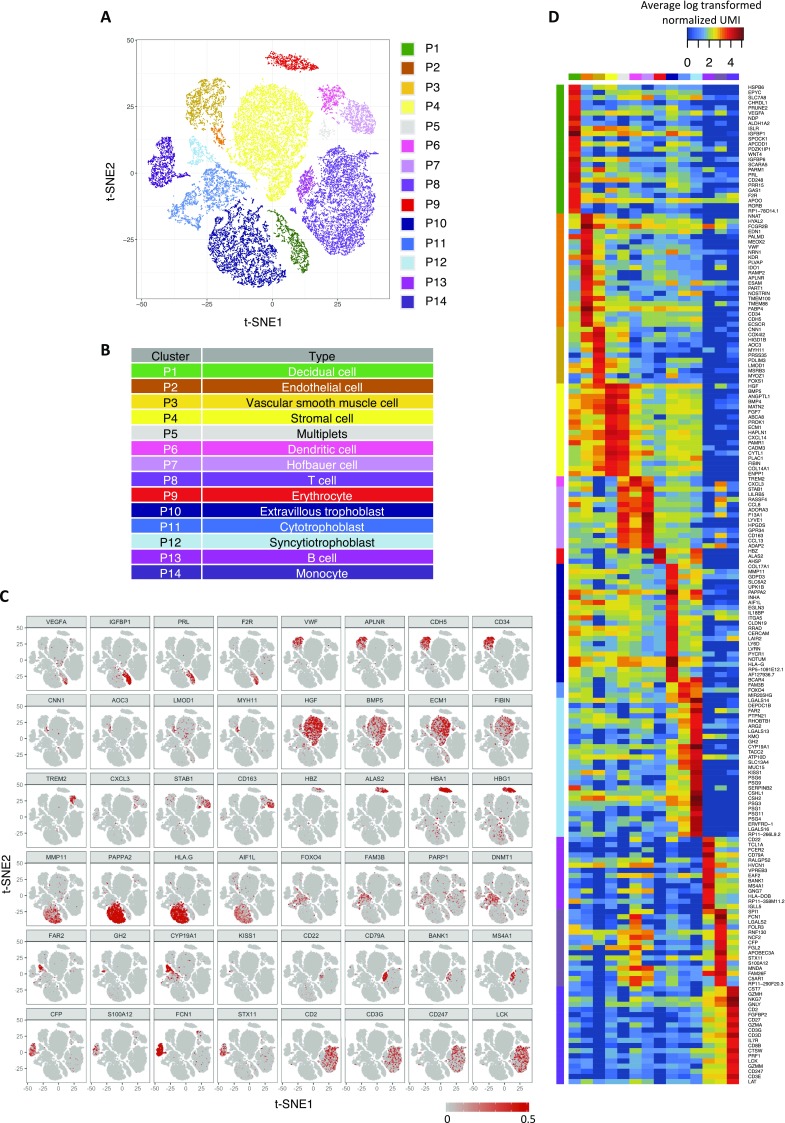
Single-cell transcriptomic profiling and the dissection of the cellular heterogeneity of human placenta. (A) Schematic diagram showing the experimental design in this study. Cellular heterogeneity of the human placenta is dissected by large-scale droplet-based single-cell transcriptomic profiling. Cell-type–specific signatures of different types of placental cells are identified and used to obtain information of cellular dynamics in the maternal plasma RNA profiles in pregnancy and PE. (B) Biaxial scatter plot showing single-cell transcriptomic clustering of placental cells from human term placentas by t-SNE analysis. Cells were further grouped into specific subgroups (P1–P12) and colored individually based on expression patterns of specific marker genes and spatial proximity in the biaxial plot. (C) Column chart comparing the fraction of maternal or fetal cells in each cellular subgroup. (D) Column chart indicating the percentage of cells expressing Y chromosome-encoded genes in each cellular subgroup. (E) Biaxial scatter plots showing the expression pattern of specific genes among different subgroups of placental cells.
We collected placental parenchymal biopsies at defined locations of multiple freshly Cesarean section-delivered placentas (from two male and two female babies) and dissociated the tissues into single-cell suspension without surface marker preselection. We obtained the single-cell transcriptome of 20,518 placental cells from six different full-term placenta parenchymal biopsies. The average number of genes detected per libraries was 1,006 (792–1,333), with a mean coverage of 21,471 (16,613–36,829) reads per cell (Table S1).
Table S1.
Descriptive statistics of the single-cell libraries
| Single-cell placental libraries | ||||||||||
| PN1 | PN2 | PN3C | PN3P | PN4C | PN4P | PE1 | PE2 | PE3 | PE4 | |
| Sample identification | M12475 | M12491 | M12548 | M12551 | M12604 | M12630 | M12632 | M12612 | ||
| Sampling distance relative to cord insertion | 5 cm | 5 cm | 5 cm | Placenta periphery | 5 cm | Placenta periphery | 5 cm | 5 cm | 5 cm | 5 cm |
| No. of valid cells in library | 3,151 | 7,621 | 2,754 | 1,391 | 2,653 | 2,948 | 5,849 | 2,294 | 4,669 | 5,167 |
| Mean raw reads per cell | 44,184.5 | 36,732.4 | 34,365 | 64,729.6 | 26,200.2 | 20,430.2 | 36,319 | 68,860 | 19,309 | 33,971 |
| Mean mapped reads per cell | 22,710 | 19,016.5 | 20,708.4 | 36,829.1 | 16,613.4 | 12,949.1 | 22,990 | 41,178.3 | 12,512.2 | 20,654.4 |
| Median genes per cell | 1,031 | 1,333 | 1,019 | 792 | 922 | 939 | 1,537 | 1,145 | 1,133 | 1,468 |
| Gestational age, wk | 38 | 38 | 38+2 | 38+2 | 38 | 38 | 31+2 | 32+4 | 29+6 | 28+1 |
Clustering analysis by t-stochastic neighborhood embedding (t-SNE) identified 12 major clusters of placental cells in our dataset (P1–P12) (Fig. 1B). Furthermore, we genotyped the genome-wide SNP patterns of the mother and the fetus to differentiate the fetomaternal origin of individual cells genetically by comparing the ratio of RNA molecules carrying fetal-specific SNP alleles with those carrying maternal-specific SNP alleles in each cluster. We also examined the presence of Y chromosome-encoded transcripts in the cells from the placentas of male fetus-carrying pregnancies (Fig. 1 C and D and Fig. S1A).
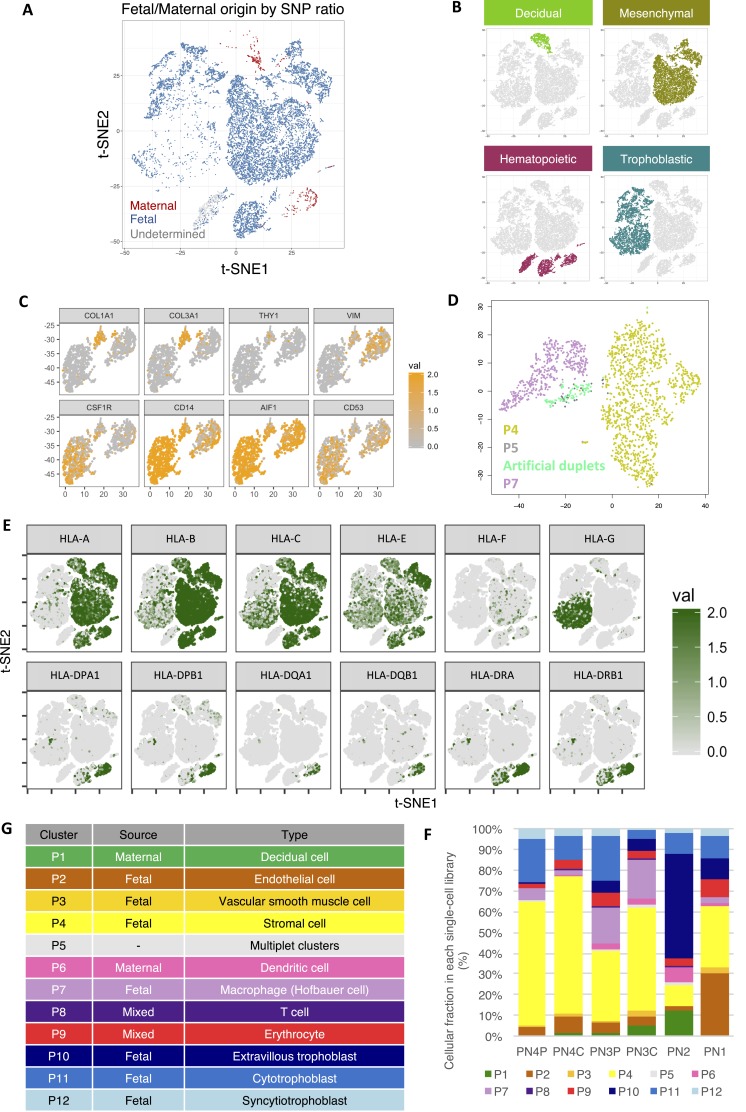
Fig. S1.
Dissection of the cellular heterogeneity and annotation of cellular identity in the human placenta. (A) Biaxial scatter plot showing the distribution of cells of predicted fetal/maternal origin in the original t-SNE clustering distribution as in Fig. 1B. Data from PN2 libraries have not been plotted, as no genotyping information was available for fetomaternal origin prediction. (B) The four major cellular groups in the human placenta: decidual (P1; green), stromal (P2–P4; olive), hematopoietic (P5–P9; red), and trophoblastic (P10–P12; turquoise). (C) Expression pattern of stromal (COL1A1, COL3A1, THY1, and VIM) and myeloid (CSF1R, CD14, AIF1, and CD53) markers in P5–P7 subgroups. (D) t-SNE analysis showed clustering of P5 cells (gray) with artificial P4/P7 duplets (light green) generated in silico, suggesting that P5 cells are likely multiplets. (E) Biaxial scatter plots showing the expression pattern of genes encoding for HLAs among different subgroups of placental cells. (F) Cellular subgroup composition heterogeneity in different single-cell transcriptomic datasets. PN3P/PN3C and PN4P/PN4C represent paired biopsies taken proximal to the umbilical cord insertion sites (PN3C/PN4C) and distal at the periphery of the placental disk (PN3P/PN4P). (G) Table summarizing the annotated nature of each cellular subgroup.
Our analysis showed that all clusters, except P1, P6, P8, and P9, were of predominant fetal origin (Fig. 1 C and D). P1 transcriptionally corresponded to maternal decidual cells, with strong expression of DKK1, IGFBP1, and PRL (Fig. 1E); P6 expressed dendritic markers CD14, CD52, CD83, and CD86, likely representing maternal uterine dendritic cells (Fig. 1E). Meanwhile, P8 expressed high levels of T-lymphocyte markers (e.g., CD3G and GZMA) (Fig. 1E). The fetomaternal SNP ratio analysis suggested that P8 was a mixture of both fetal and maternal lymphocytes (Fig. 1 C and D). Similarly, the homogeneous expression of adult and fetal hemoglobin genes, such as HBA1, HBB, and HBG1, and the gene encoding the heme biosynthetic enzyme ALAS2 in P9 suggested that they were composed of erythrocytic cells from the fetal cord and from a maternal source.
The rest of the fetal subgroups (P2–P5, P7, and P10–P12) could be broadly classified into four groups [i.e., vascular (P2 and P3), stromal (P4), macrophage-like (P5 and P7), and trophoblastic (P10–P12) cells] (Fig. S1B). P2 cells commonly expressed vascular endothelial genes (e.g., CD34, PLVAP, CDH5, and ICAM1) (Fig. 1E). A few endothelial cells of maternal origin could also be found in the P2 cluster (Fig. S1A). P3 cells showed features of vascular smooth muscle cells, with expression of MYH11 and CNN1 (Fig. 1E). The large cluster of P4 cells expressed mRNAs of the ECM1 protein and fibromodulin, both of which were markers of villous stromal cells. Similar to maternal P6 cells, fetal P5 and P7 clusters also highly expressed activated monocyte/macrophage-specific genes CD14, CSF1R (encoding CD115), CD53, and AIF1 (Fig. 1E). Nonetheless, fetal P5 and P7 subgroups showed additional expression of CD163 and CD209, both being markers of placental resident macrophages (Hofbauer cells) (Fig. 1E). Comparing with P7 cells, the P5 subgroups also showed prevalent expression of fibroblastic and mesenchymal genes shared with P4 villous stromal cells, such as THY1 (encoding CD90), collagen genes (COL3A1 and COL1A1), and VIM (Fig. S1C). These results raised the possibility that the P5 subgroup may be composed of duplets of P4 and P7 cells during single-cell encapsulation. To corroborate this hypothesis, we performed in silico duplet simulation analysis (Fig. S1D), and our result indicated that the P5 cells closely resembled the simulated data and hence, likely represented artificial duplets.
The trophoblastic clusters (P10–P12) could be divided into three subgroups [i.e., EVTBs (P10), VCTBs (P11), and SCTBs (P12)], with distinctive expression patterns of PAPPA2, PARP1, and CGA, respectively (Fig. 1E). Genes involved in the production of important gestational hormones, including CYP19A1 (encoding aromatase for estrogen synthesis), CGA (human CG), and GH2 (human placental growth hormone), were all specifically expressed in P12 (SCTBs) (Fig. 1E). It is known that placental EVTBs expressed nonclassical form of HLAs, such as HLA-G, to promote maternal immunotolerance of the fetus with uterine natural killer cells (29–31). Indeed, we detected strong expression of HLA-G in the EVTBs (P10) subgroup (Fig. S1E). Expression of HLA genes in VCTBs and SCTBs was generally scarce, whereas classical HLA-A is specifically expressed in nontrophoblast cells (P1–P9). Expression of genes encoding the HLA class II molecules, such as HLA-DP, HLA-DQ, and HLA-DR, was concentrated in P6 and P7, which was consistent with the antigen-presenting functions in the maternal dendritic cells and fetal macrophages.
Previous bulk tissue transcriptomic profiling has shown significant spatial heterogeneity between biopsies taken from different sites of the placenta (32). Comparison of the compositional heterogeneity of different libraries in our dataset also reflected such variations (Fig. S1F). We included two paired biopsies of the placental parenchyma at sites proximal (PN3C and PN4C) and distal (PN3P and PN4P) to the umbilical cord insertion from two different individuals (Table S1). We found that P1 decidual cells were significantly underrepresented in the PN1 library compared with others. Instead, the P2 fetal endothelial cells fraction was significantly higher in PN1 than other libraries, suggesting high contribution from the umbilical vasculature on the fetal surface of the placenta in the PN1 biopsy. In contrast, the PN2 library contained the highest fraction of P1 decidual cells, P6 maternal uterine dendritic cells, and P10 EVTBs. The PN2 library likely captured more cells at the deeper fetomaternal interface. Cellular compositions of biopsies obtained from paired proximal and distal middle sections were more comparable, with only significant reduction in decidual cells, and increased in EVTBs at the distal site, but the interindividual variation remained high (Fig. S1F). These findings highlighted the cellular heterogeneity in the placenta and the necessities of a single-cell analytical approach.
In short, our dataset delineated 11 transcriptomically and genetically distinct major cellular subtypes in the human placenta (Fig. S1G).
Reconstructing the Cellular Relationship of Trophoblasts.
Our dataset captured a significant number of trophoblastic cells (7,450; 36%). We observed that the P10–P12 subgroups clustered in continuum, despite the clear topological expression of certain trophoblast subtype markers (Fig. 1 B and C). In fact, it has been proposed that these trophoblast subtypes are developmentally connected: VCTBs (P11) are the progenitor compartment for the generation of EVTBs (P10) by decidual invasion and SCTBs (P12) by syncytium formation during placental development.
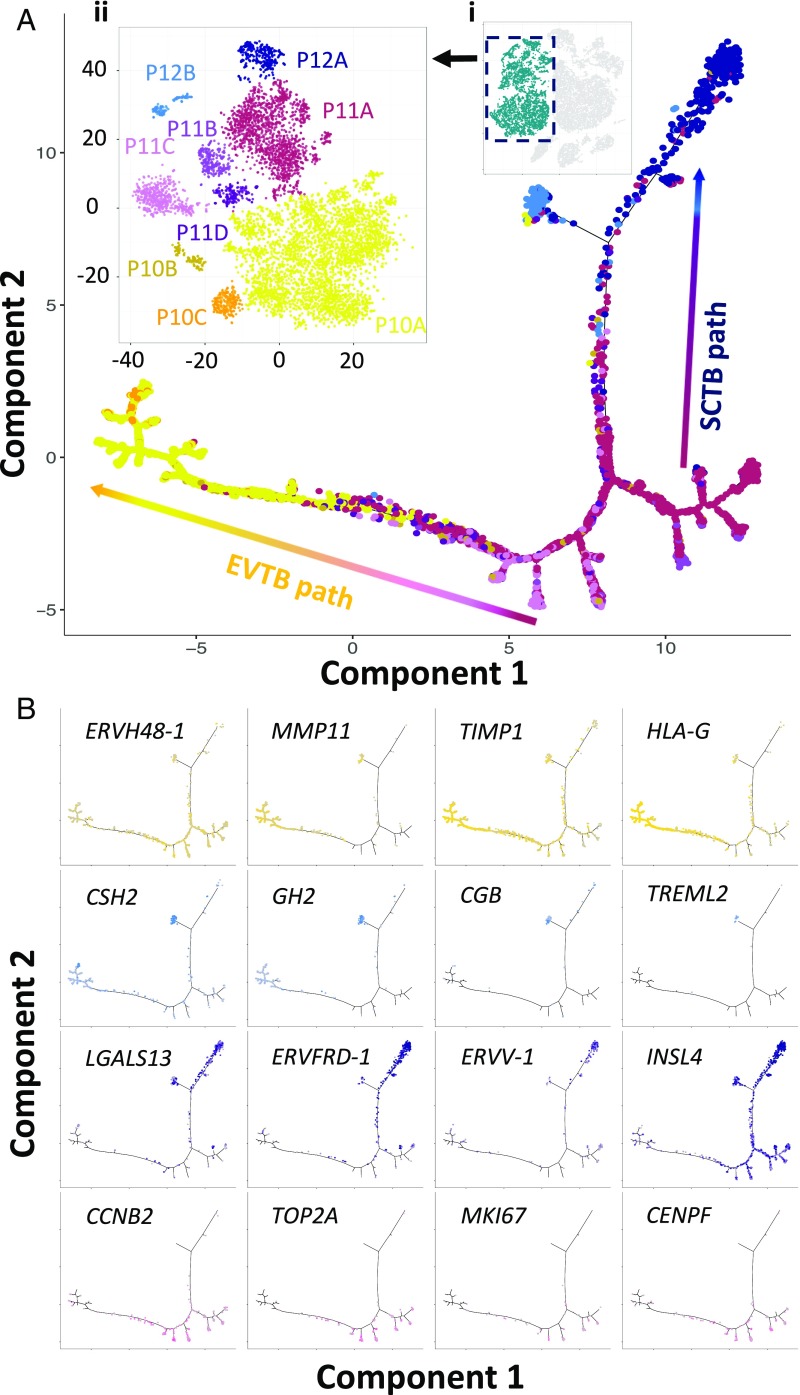
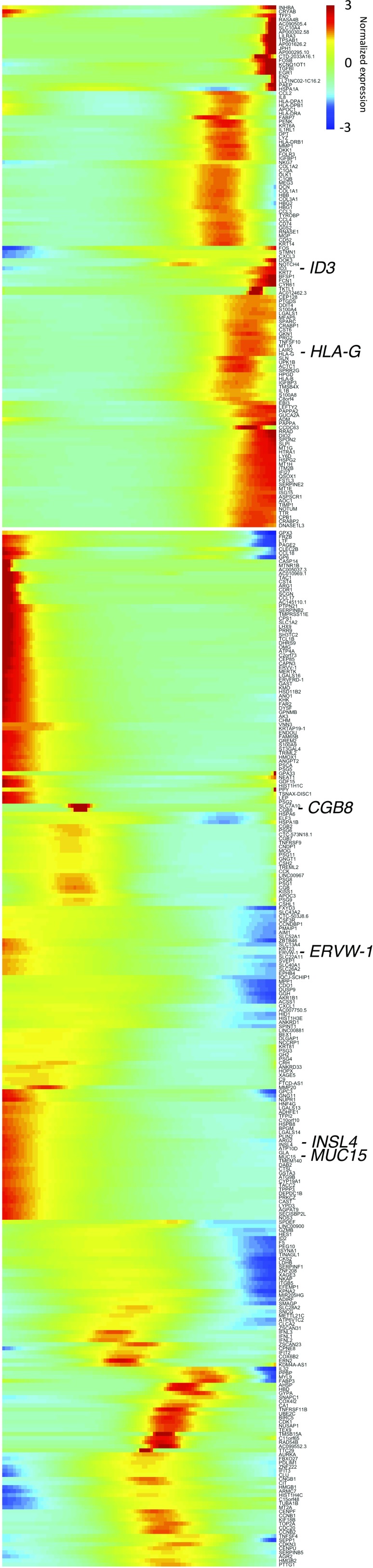
To delineate this relationship transcriptomically and to study the genes that regulate this process, we first identified the highly variable genes among these three clusters for t-SNE reclustering analysis (Fig. 2A, Inset i). In such analysis, multiple smaller new clusters emerged (Fig. 2A, Inset ii). We then ordered individual trophoblast cells computationally in a 2D “pseudotime” trajectory following the method described by Trapnell et al. (33) to reconstruct their differentiation relationship. We found that the trophoblasts formed a continuous “u-shaped” trajectory (Fig. 2A), with EVTB (P10) and SCTB (P12) cells occupying the two heads and VCTBs (P11) at the turn and tail. The villous SCTB branch further bifurcated into two subbranches (Fig. 2A). One subbranch was occupied by cells expressing high levels of gestational hormone genes, GH2 and CGB, whereas the other one showed strong expression of genes involved in cell fusion. Endogenous retroviral transcripts (ERVs) are involved in syncytial cell fusion [e.g., ERVW-1 (syncytin-1), ERVFRD-1 (syncytin-2), and ERVV-1] and increased along the SCTB differentiation pathway (34) (Fig. 2B and Fig. S2). In contrast, another ERV transcript, ERVH48-1, is depleted in SCTBs (Fig. 2B), concordant with a recent report that it functions as an inhibitor of syncytium formation in VCTBs (35). To further study the cell fusion process, we identified genes that showed variation similar to ERVFRD-1 along the SCTB pathway (Fig. S2). These included an SCTB invasion suppressor (MUC15) (36), a member of the placental galactin family (LGALS13), and a direct gene target of ERVs (INSL4) (37). We also identified multiple genes (e.g., OMG, SLC1A2, ADHFE1, and DEPDC1B) as potential regulators in SCTB development, laying ground for function characterization in future studies.
Fig. 2.
Reconstruction of the developmental relationship of trophoblast cells by pseudotime analysis. (A) Developmental trajectory visualization in biaxial scatter plot. The trophoblastic cell subgroups (P10–P12) were selected for trajectory reconstruction (Inset i). Trophoblastic cells were reclustered based on highly variable genes only by t-SNE analysis, and smaller trophoblastic subgroups (P10A–C, P11A–D, and P12A and -B colored individually) could be visualized in the biaxial plot (Inset ii). Pseudotime reconstruction revealed a bifurcating trajectory of trophoblast development. The coloring of individual cell along the optimized embedding path corresponded to that in Inset ii. (B) Biaxial scatter plots showing the distribution patterns of cells expressing specific marker genes in the pseudotemporal trajectories. For clear visualization, only cells with nonzero expression were plotted.
Fig. S2.
Pseudotemporal change of gene expression in trophoblasts. Genes were organized into two major groups by their similar pseudotemporal covariation of expression. Notable gene examples involved in trophoblast development were marked and enlarged.
Pseudotime analysis of the EVTB developmental pathway also revealed specific up-regulation of various extracellular enzymes involved in the invasive and migratory phenotype of EVTBs [e.g., proteinases and their regulators (MMP11 and TIMP1), nuclease (DNASE1L3), ECM (SPON2), and migration inhibitor (GKN1)] (38) (Fig. 2B and Fig. S2). Minor branches stemming from the area occupied by P11 VCTB cells were lined by cells with high expression of genes involved in cell division (e.g., CCNB1, TOP2A, MKI67, and CENPF). These minor branches likely represented entry to the cell cycle of proliferative VCTBs in the process of EVTB differentiation. In contrast, proliferative cells were scarce along the SCTB pathways, consistent with their nondividing nature (Fig. 2B). In short, our dataset enabled the delineation of the trophoblast lineage relationship and shed light on the regulatory mechanism of lineage development.
Noninvasive Elucidation of Placental Cellular Dynamics During Pregnancy.
Previous maternal plasma transcriptomic profiling studies have shown that certain trophoblast-specific transcripts and the overall fractional placental contribution increase with gestation (20, 39, 40). The fraction of fetal-derived RNA increases from only 3.7% in early pregnancy to 11.28% in late pregnancy (19, 20). We reasoned that establishment of the cell-type–specific gene signatures at the single-placental cell level would allow us to isolate and dissect the dynamic changes of both trophoblastic and nontrophoblastic cellular components in the maternal plasma. However, it is known that fetal-derived cell-free RNA in maternal plasma circulates in mixture with cell-free RNA derived from the maternal hematopoietic system. Liver-specific transcripts, such as ALB, are also readily detectable in the plasma (41). We, therefore, identified and filtered the cell signature genes of individual placental cell types in our dataset by reanalyzing peripheral blood mononucleated cells (PBMCs) single-cell transcriptomic profiles and the tissue transcriptome data of leukocytes and liver from public databases (28, 42) (Fig. S3 A–E).
Fig. S3.
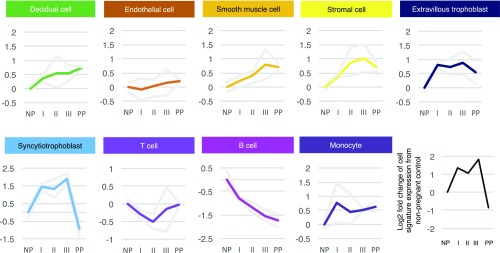
Identification of cell-type–specific signature genes sets and noninvasive elucidation of placental cellular dynamic in maternal cell-free RNA. (A) Biaxial t-SNE plot showing the clustering pattern of PBMCs and placental cells. The PBMC data (donor B) from Zheng et al. (28) were plotted with the placental dataset. (B) Table summarizing the annotated nature of each cellular subgroup in the placenta/PBMC merged dataset. (C) Biaxial scatter plots showing the expression pattern of specific marker genes among different subgroups of placental cells and PBMCs. (D) Heat map showing the average expression of cell-type–specific signature genes in different PBMCs and placental cells clusters. The column side color corresponds to the cell cluster coloring in A. The row side color indicates the cell-type specificity of the gene. (E) Box plots comparing the expression levels of different cell-type–specific signature genes in human leukocytes, the liver, and the placenta. (F) Cell signature expression analysis of the maternal plasma RNA profiles of Koh et al. (20). Line plots showing the change of the average expression of individual cell-type–specific signature in different stages of pregnancy with respective to first trimester maternal plasma. The gray lines demarcate the range of the data. FPKM, fragments per kilobase of transcript per million mapped reads.
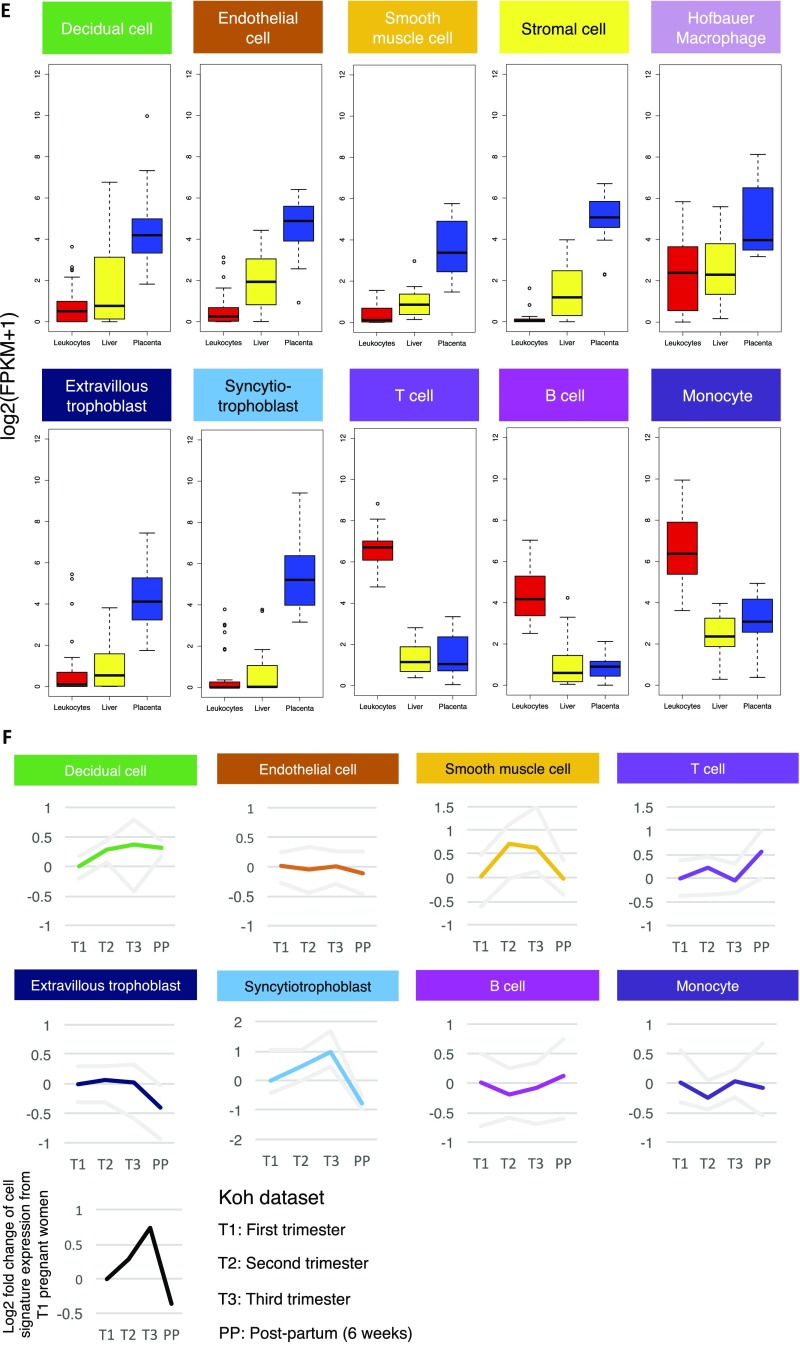
We then studied the expression dynamics of the corresponding cell-type–specific signature in the maternal plasma RNA profiles from different stages of pregnancy by Tsui et al. (19). In the dataset by Tsui et al. (19), we observed a dramatic up-regulation of SCTB signature in the maternal plasma RNA of early pregnancy compared with nonpregnant controls (Fig. 3). The trend peaked at predelivery maternal plasma before rapidly dropping to levels of nonpregnant controls 24 h after delivery. A similar pattern could also be found in the trophoblastic EVTB, placental stromal cell, and vascular smooth muscle cell signatures. These patterns corresponded to the rapid growth of the stromal, SCTB, and EVTB components of the placenta in early pregnancy and its clearance after placental delivery. Meanwhile, the change of the gene signature expression of endothelial cell is minimal. Intriguingly, the signature of decidual cells remained observable in maternal plasma up to 24 h after delivery. This can be explained by the fact that release of cell-free RNA from residual maternal decidual tissues degeneration may continue after placental delivery. In contrast, we found that the signature of B cell continued to drop throughout pregnancy, whereas signature of T cell first dropped and then recovered to nonpregnant levels before delivery. Consistently, previous studies on pregnancy-associated lymphopenia by flow cytometry have shown that T- and B-cell levels decline with the progression of pregnancy (43–45) and that recovery of peripheral B-cell levels may occur later than those for T cells (44). Meanwhile, the signature of monocytes showed a more variable pattern, up-regulating in early pregnancy, dipping, and rebounding before delivery, in line with the findings of myeloid immunity activation during pregnancy (43, 46–48). To further confirm these findings, we reanalyzed another independent maternal plasma RNA dataset by Koh et al. (20) collected at three trimesters of pregnancy and within 6-wk postpartum. We observed dynamic patterns of cell signature consistent with what we had found in the dataset by Tsui et al. (19) (Fig. S3F). These findings showed the ability of cell-type–specific signatures analysis to dissect the dynamics of individual cellular components in the maternal plasma RNA profiles.
Fig. 3.
Elucidation of placental cellular dynamic in maternal plasma RNA profiles during pregnancy. Line plots showing the change of the average cell signature expression of individual placental cell type in different gestational groups. The maternal plasma RNA profiles were retrieved from Tsui et al. (19). The gray lines demarcate the range of the data. I, early pregnancy (13–20 wk); II, mid/late pregnancy (24–30 wk); III, predelivery; NP, nonpregnant; PP, 24-h postpartum.
Deciphering Cellular Aberrations in Preeclamptic Placentas from Maternal Plasma Cell-Free RNA.
We next expanded our study to uncover the placental cellular dysfunction in the maternal cell-free RNA of early severe preeclamptic patients. We reasoned that cellular pathology in the preeclamptic placentas affects the cell turnover and hence, the release of the cell-type–specific RNAs into the maternal plasma. The cellular origin of the pathology can, therefore, be revealed by comparing the expression levels of different cell-type–specific signatures in the maternal plasma of early severe preeclamptic patients with those of healthy pregnant controls.
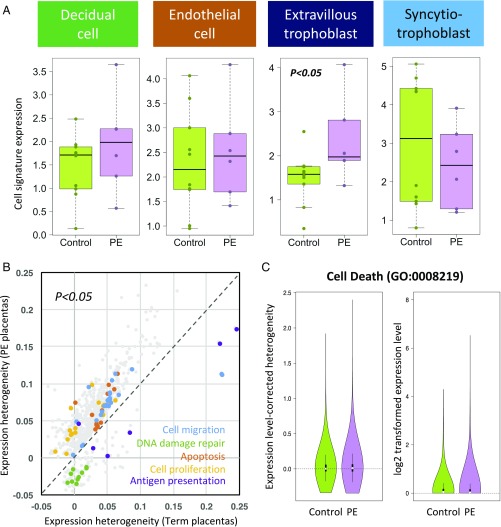
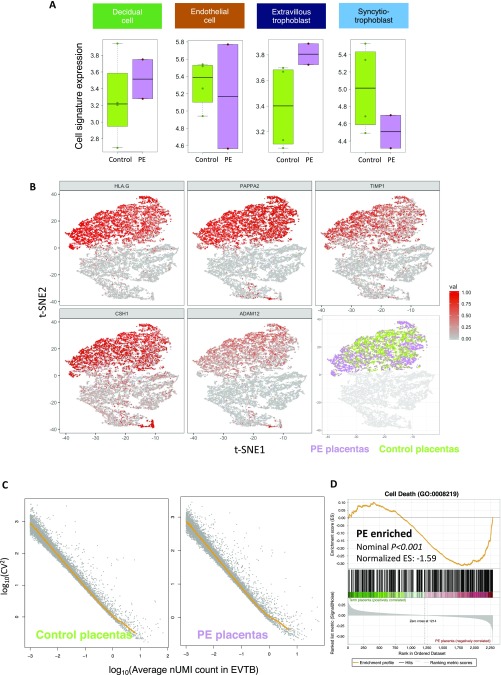
Strikingly, we found that the EVTB signature is specifically and consistently up-regulated in early severe preeclamptic patients in two separate cohorts assayed with different plasma RNA library preparation chemistries (P = 0.042, two-tailed two-sample Wilcoxon signed rank test) (Fig. 4A and Fig. S4A). These results pointed to an increased release of EVTB-derived cell-free RNA into the maternal circulation in early PE. We then validated this finding directly at the tissue level. We characterized the single-cell transcriptome of placental biopsies from four of the early preeclamptic patients and compared the intracluster transcriptomic heterogeneity in the EVTB clusters between normal-term and early preeclamptic placentas to reveal the abnormalities in different biological processes (Fig. S4B). As gene-expression heterogeneity decreased with increasing expression levels (49, 50) (Fig. S4C), we quantified the expression heterogeneity of each gene as the vertical distance of its squared coefficient of variations (CV2) from the median CV2 [distance to median (DM)] at its corresponding expression level in a double log-transformed space (50). We compared the DM values of gene sets annotated in different gene ontology (GO) terms between PE and normal-term placental EVTBs to identify differential biological processes (Fig. S4C); we observed that the transcriptional heterogeneity of genes involved in cell migration, cell death, and proliferation is significantly more variable in early preeclamptic placentas, whereas DNA damage repair and antigen presentation ontology terms are more variable in term placentas (Fig. 4 B and C). Indeed, genes annotated in “cell death” showed not only higher expression variability but also, higher levels of gene expression in early preeclamptic EVTBs (Fig. 4C). Alternative gene set enrichment analysis (51) also supported significant enrichment of expression of cell death-related genes in the preeclamptic EVTB cluster (Fig. S4D). These results suggested that EVTB in early preeclamptic placentas might have higher levels of cell death. This conclusion is in line with previous reports that trophoblastic apoptosis is increased in PE (52–59). Despite the invasion insufficiency of EVTB in PE, the increased cell death in EVTB may also contribute to the up-regulation of the EVTB signatures in the maternal plasma of early preeclamptic patients. In short, we showed the ability of plasma cell-free RNA cellular signature analysis, with early PE as an example, to serve as a noninvasive hypothesis-free exploratory tool in revealing hidden cellular pathology of a complex organ and to provide a potential noninvasive approach for molecular testing of PE.
Fig. 4.
Uncovering placental cellular aberrations in early preeclamptic maternal plasma RNA profiles. (A) Box plot comparing the cell signature expression of different cell types in the maternal plasma RNA profiles of third trimester pregnancy (control) and early PE patients. Statistical testing was performed by two-tailed two-sample Wilcoxon signed rank test. (B) Biaxial scatter plot showing the average single-cell expression heterogeneity of different GO annotated gene sets in third trimester term and early preeclamptic placentas. Only data points with statistically significant differences are shown (P < 0.05). GO terms associated with cell proliferation, cell migration, apoptosis, antigen presentation, and DNA damages are colored and highlighted. (C) Violin plots comparing the expression level-corrected heterogeneity (DM values) and average expression levels of genes annotated in the GO term “Cell Death” between PE patients and normal controls.
Fig. S4.
Noninvasive detection of cellular aberrations in early preeclamptic placentas. (A) Box plot comparing the cell signature expression of decidual cells, placental endothelial cells, EVTB cells, and SCTB cells in the maternal plasma RNA profiles of third trimester pregnancy (control) and early preeclamptic patients (PE) from a separate small cohort assayed using the RNA library preparation method as described by Tsui et al. (19). (B) Biaxial scatter plots showing the trophoblast clusters in the merged PE/control placenta single-cell transcriptomic dataset and the expression of EVTB marker genes HLA-G, PAPPA2, TIMP1, CSH1, and ADAM12. The EVTB cells from PE/control placentas are colored in light green (control) and purple pink (PE). (C) Scatter plot showing the relationship of log-transformed CV2 and log-transformed average expression levels of the cells in the EVTBs cluster: term placenta (Left) and PE (Right). The rolling medians across the expression range are plotted (orange line), and the expression level-corrected expression variability of a gene is calculated as the vertical distance of its CV2 value from the rolling median regression line (DM). (D) Gene set enrichment analysis result (51) confirming enrichment of cell death-related genes expression in early preeclamptic EVTB cells.
Discussion
The discovery of circulating cell-free fetal nucleic acids in maternal plasma has enabled the development of noninvasive prenatal diagnosis of fetal aneuploidy and monogenic diseases through detection of the pathogenic mutations, allelic imbalances, and chromosomal imbalances (60, 61). However, it remains difficult to study placental pathology using cell-free fetal nucleic acids. One difficulty is the ascertainment of the origin of RNA transcripts. It has been shown that fetal RNA in maternal plasma is placenta-derived (16, 39), and RNA transcripts derived from other fetal tissues have also been reported in maternal plasma (20). The tissue origins of these RNA transcripts are inferred from comparison of whole-tissue gene-expression profiles of multiple tissues types. However, biological tissues are composed of multiple types of cells originating from different developmental lineages and serving different specialized functions. The averaged expression profile from whole tissues may, therefore, distort the actual heterogeneous composition of the tissue and bias toward cells with the highest cell number in the tissue sample, such as the trophoblasts in the placenta. It is, therefore, imperative to connect the circulating pool of cell-free nucleic acids with their cellular origins to reveal the complex dynamics of both trophoblastic and the nontrophoblastic components of the placenta during pregnancy. The advance in single-cell transcriptomic technology provides an opportunity for us to bridge the study of the placenta with circulating cell-free nucleic acids during pregnancy.
The potential of single-cell transcriptomic analysis on placental biology can be seen in two recent small-scale studies in the mouse and human (62, 63). In this study, we harnessed the power of microfluidic single-cell transcriptomic technology to establish a large-scale cellular transcriptomic atlas of the human placenta, profiling more than 24,000 nonmarker-selected cells from normal-term and early preeclamptic placentas. We annotated the fetomaternal origin of individual cells using both genetic and transcriptional information to provide a comprehensive picture of placental cellular heterogeneity, including decidual cells, resident immune cells, and vascular and villous stromal cells. The high number of trophoblast cells recovered in our dataset allowed computational reconstruction of the trophoblastic differentiation trajectories and identification of highly specific cell signature genes. Our analysis supported a bifurcating model with VCTB differentiating bidirectionally into SCTB and EVTB (64). The trajectories recapitulated many known regulator interactions during trophoblast development and discovered potential players that laid a foundation for future investigation. Furthermore, our analysis highlighted the high degree of cellular heterogeneity among placental parenchyma biopsies, even at defined sampling locations, reinforcing the need for a single-cell approach in the transcriptomic study of the human placentas. Future bioinformatics tools can be developed to exploit the cellular transcriptomic signature established in this placental atlas to normalize cellular composition heterogeneity in whole-tissue profiles.
Finally, we showed the feasibility of integrating single-cell transcriptomic analysis with plasma-circulating RNA analysis in dissecting the complex fetal and maternal cellular dynamics during normal pregnancy progression and revealing the cellular pathology in early preeclamptic placentas noninvasively. We discovered cell-type–specific signature genes from large-scale single-cell transcriptomic profiling de novo and aggregated signals of all cell-type–specific genes to reconstruct the cellular information. With this approach, comparable cellular dynamic patterns can be observed in two independent maternal plasma RNA datasets (19, 20). The dynamic patterns of trophoblastic and hematopoietic cell types revealed are consistent with current knowledge on the hematopoietic system and the placenta during pregnancy. More importantly, this approach allowed the recovery of differential expression of the EVTB signatures in maternal plasma, which reflected the actual cellular abnormalities in early preeclamptic patients at the tissue level. As invasive placental biopsy in gestation age-matched healthy pregnant women is not feasible, we compared the post-Cesarean section placental biopsies from early preeclamptic patients with those from healthy full-term pregnancies. Since placental cell death increases with gestation progression and placental maturation (54), one would expect an even stronger difference in cell death between early preeclamptic and healthy early third trimester EVTB cells. This challenge also underlines the need of cell-free RNA cell signature analysis as a noninvasive molecular tool in exploratory in vivo studies to differentiate cellular pathology in different forms of placental dysfunction and offer clinical diagnostic information. With continuous improvement in the cost-effectiveness of large-scale single-cell transcriptomic technology and the effort of the Human Cellular Atlas Initiative in profiling the cellular transcriptomic heterogeneity of all cellular subtypes in different major human organs (28, 65–67), it can be envisioned that the same analytical approach can be extended to other situations, such as intratumoral dissection in cell-free tumoral RNAs and noninvasive exploration of the cellular pathology in other gestational diseases.
In short, our study has established a large-scale single-cell transcriptomic atlas of the normal and early preeclamptic placentas and showcased the power of integrative analysis of single-cell transcriptomics and plasma cell-free RNA to reconstruct cellular information from plasma. This opens an avenue for noninvasive elucidation of cellular dynamics and aberrations in complex biological systems and molecular diagnostics.
Materials and Methods
Subjects, Sample Collection, and Processing.
This study was approved by the institutional ethics committee, and informed consent was obtained after the nature and possible consequences of the studies were explained. Healthy and early preeclamptic pregnant women (Table S2) were recruited from the Department of Obstetrics and Gynecology, Prince of Wales Hospital, Hong Kong with informed consent. We recruited patients with early-onset PE requiring delivery at 24- to 33+6-wk gestation. PE was defined as blood pressure ≥140/90 mmHg on at least two occasions 4 h apart developing after 20-wk gestation with proteinuria of ≥300 mg in 24 h, ≥30 mg/mmol in protein/creatinine ratio, or two readings of ≥2+ on dipstick analysis of midstream or catheter urine specimens if no 24-h collection was available. Only patients not in active labor with delivery by Cesarean section were recruited to avoid cellular contamination from the birth canal and to ensure placental cellular viability.
Table S2.
Sample information of plasma RNA data: Plasma RNA samples
| Sample | Group | Gestation age, wk | Library type | Source |
| M12700 | PE | 26+3 | NuGEN | This study |
| M12612 | PE | 28+1 | NuGEN | This study |
| M12604 | PE | 31+2 | NuGEN | This study |
| M12630 | PE | 32+4 | NuGEN | This study |
| M12632 | PE | 29+6 | NuGEN | This study |
| M12649 | PE | 28+2 | NuGEN | This study |
| M12701 | Healthy | 38+3 | NuGEN | This study |
| M12475 | Healthy | 38 | NuGEN | This study |
| M12491 | Healthy | 38 | NuGEN | This study |
| M12548 | Healthy | 38+2 | NuGEN | This study |
| M12551 | Healthy | 38 | NuGEN | This study |
| M12675 | Healthy | 32+2 | NuGEN | This study |
| M12677 | Healthy | 32+4 | NuGEN | This study |
| M12678 | Healthy | 28+6 | NuGEN | This study |
| M12685 | Healthy | 28+4 | NuGEN | This study |
| M12691 | Healthy | 28 | NuGEN | This study |
| M9113P | PE | 24+3 | Illumina | This study |
| M9359P | PE | 29+1 | Illumina | This study |
| CTR081 | Nonpregnant woman | — | Illumina | Tsui et al. (19) |
| CTR082 | Nonpregnant woman | — | Illumina | Tsui et al. (19) |
| M9600 | I | 13+6 | Illumina | Tsui et al. (19) |
| M9549 | I | 18+1 | Illumina | Tsui et al. (19) |
| M6335P | II | 24 | Illumina | This study |
| M7879P | II | 24+3 | Illumina | This study |
| M7316P | II | 24+5 | Illumina | This study |
| M6268P | II | 28 | Illumina | This study |
| M9356 | III | 37+2 | Illumina | Tsui et al. (19) |
| M9415 | III | 38 | Illumina | Tsui et al. (19) |
| M9356BP | Postdelivery 24 h | — | Illumina | Tsui et al. (19) |
| M9415BP | Postdelivery 24 h | — | Illumina | Tsui et al. (19) |
For each case, 20 mL of maternal peripheral blood was collected into EDTA-containing tubes before Cesarean section. Plasma was isolated by a double-centrifugation protocol as previously described (19). For placental parenchymal biopsy, 1-cm3 placental tissue was dissected freshly after delivery from a region 2-cm deep and 5 cm away from the umbilical cord insertion after peeling of the fetal membranes. In some cases (PN3P and PN4P), biopsies were also taken from the placental periphery. The dissected tissues were then washed in PBS and subjected to enzymatic digestion using the Umbilical Cord Dissociation Kit (Miltenyi Biotech) according to the manufacturer’s protocol. Red blood cells were lysed and removed by ACK buffer (Invitrogen). Cell debris was removed by a 100-µm filter (Miltenyi Biotech), and the single-cell suspension was washed again three times in PBS (Invitrogen). Successful dissociation was confirmed under a microscope.
Plasma RNA Extraction and Library Preparation.
Plasma RNA was preserved by mixing TRIzol (Ambion) with plasma in a ratio of 3:1 immediately after plasma isolation. Plasma RNA was then extracted using the RNeasy Mini Kit (Qiagen). All extracted RNA was quantified by NanoDrop ND-2000 Spectrophotometer (Invitrogen) and real-time quantitative PCR on a LightCycler 96 System (Roche). cDNA reverse transcription, second-strand synthesis, and RNA-sequencing (RNA-seq) library construction were performed using the Ovation RNA-seq System V2 (NuGEN) kit according to the manufacturer’s protocol. Amplified and purified cDNA was sonicated into 250-bp fragments using a Covaris S2 Ultrasonicator (Covaris). All libraries were then quantified by Qubit (Invitrogen) and real-time quantitative PCR on a LightCycler 96 System (Roche) and sequenced on a NextSeq 500 system (Illumina).
Single-Cell Encapsulation, In-Droplet RT-PCR, and Sequencing Library Preparation.
Single-cell RNA-seq libraries were generated using the Chromium Single Cell 3′ Reagent Kit (10X Genomics) as described (28). Briefly, single-cell suspension (cell concentration between 200 and 1,000 cells per 1 µL PBS) was mixed with RT-PCR master mix and loaded together with Single Cell 3′ Gel Beads and Partitioning Oil into a Single Cell 3′ Chip (10X Genomics) according to the manufacturer’s instructions. RNA transcripts from single cells were uniquely barcoded and reverse-transcribed within droplets. cDNA molecules were preamplified and pooled followed by library construction according to the manufacturer’s instructions. All libraries were quantified by Qubit and real-time quantitative PCR on a LightCycler 96 System (Roche). The size profiles of the preamplified cDNA and sequencing libraries were examined by the Agilent High Sensitivity D5000 and High Sensitivity D1000 ScreenTape Systems (Agilent), respectively.
Sequencing, Alignment, and Gene-Expression Quantification.
All single-cell libraries were sequenced with a customized paired end with dual indexing (98/14/8/10-bp) format according to the recommendation by 10X Genomics. All single-cell libraries were sequenced on a MiSeq system (Illumina) or a NextSeq 500 system (Illumina) using the MiSeq Reagent v3 Kit (Illumina) or the NextSeq 500 High Output v2 Kit (Illumina), respectively. The data were aligned and quantified using the Cell Ranger Single-Cell Software Suite (version 1.0) as described by Zheng et al. (28). In short, samples were demultiplexed based on the 8-bp sample index, 10-bp UMI tags, and the 14-bp GemCode barcode. The 98-bp-long read 1 containing the cDNA sequence was aligned using STAR (68) against the hg19 human reference genome. UMI quantification, GemCode, and cell barcodes filtering based on error detection by Hamming distance were performed as described by Zheng et al. (28).
For alignment of the plasma RNA library, adaptor sequences and low-quality bases on the fragment ends (i.e., quality score < 5) were trimmed, and reads were aligned to the human reference genome (hg19) using the TopHat (v2.0.4) software with the following parameters: transcriptome mismatches = 3; mate-std-dev = 50; genome-read-mismatches = 3 with the pair end alignment option as well as the annotated gene model file downloaded from University of California, Santa Cruz, Genome Browser (genome.ucsc.edu/). Gene-expression quantification was performed by an in-house script quantifying the number of reads overlapping with exonic regions on genes annotated in the Ensembl GTFs (GRCh37.p13).
Fetal and Maternal Origin Determination.
To differentiate the genetic origin of a cell, maternal and fetal genotypes were first determined by the iScan system (Illumina) using buffy coat and placenta tissues, respectively. Genotype information of case M12491 (PN2) was not available because of the limitation of biopsy materials. Informative SNPs covered by sequencing reads were then identified, in which a SNP is classified as maternal-specific when it is heterozygous in the mother (A/B) and homozygous in the fetus (A/A). Fetal-specific SNPs were classified vice versa. Next, we calculated the allelic ratio (R) as follows; B (allelic count of the origin-specific SNP B) and A (allelic count of the common SNP A):
Fetal-specific allelic ratio () and maternal-specific allelic ratio were obtained for each cell. A cell would be annotated as (i) fetal origin if > , (ii) maternal origin if > , and (iii) undetermined if = or if there are no reads covering any informative SNPs.
Duplet Simulation.
Gene-expression matrix of 1,365 P4 cells and 526 P7 cells was first extracted from the PN3C dataset. To emulate 100 duplet data points, the transcriptome of the duplet was modeled as random mixture of one P4 cell and one P7 cell. The gene-expression levels of the artificial duplets were set as the average of the two cells. Principal component analysis (PCA) and t-SNE clustering were performed using the prcomp and Rtsne package in R, respectively.
Identification of Cell-Specific Gene Signature.
Single-cell transcriptomic data of PBMCs were retrieved from the public domain of 10X Genomics at the link https://support.10xgenomics.com/single-cell/datasets. The dataset was previously published (28). The PBMC dataset (donors A and B) was merged with the placenta dataset and normalized by random read subsampling using the cellrangerRkit version 0.99.0 package. t-SNE clustering was performed with built-in functions in the cellrangerRkit package using the first 10 principal components. Cells clusters were identified and cellular types were annotated in the biaxial t-SNE plots based on known marker gene expression and spatial proximity. To identify cell-type–specific gene signature, we used gene-level and gene set-level filtering.
We calculated the gene-expression z score as a measure of cell-type expression specificity with the formula
where zg is the z score for gene , is the mean expression level of gene in cell-type A (log-transformed normalized UMI count), is the mean expression level of gene in non-A cells, and is the SD of gene expression of gene in non-A cells.
Genes with z score greater than three and mean log-transformed normalized UMI expression in testing cell type greater than 0.1 and less than 0.01 in nontesting cell types were classified as the cell-type–specific signature genes. Expression levels of each cell-type–specific gene in the whole-tissue profile of the placenta, liver, and leukocytes were then compared, and only genes that showed the highest expression levels in their corresponding source organs, placenta, or leukocytes were selected. We then excluded signature gene sets that recover less than 10 genes and sets that did not show adequate placenta and leukocyte/liver separation (Fig. S3E). Among the 14 cell clusters in the PBMC–placenta datasets, no specific genes were identified for cluster P5, and only less than five genes passed the filter for cluster P6, P9, and P11. P7 signature set representing placental Hofbauer macrophage was excluded from additional analysis because of inadequate separation from leukocytes. The whole-tissue expression datasets were retrieved online from the Human lincRNAs Catalog project (42): portals.broadinstitute.org/genome_bio/human_lincrnas/.
Cellular Signature Expression Analysis.
We reanalyzed the maternal plasma RNA profiles from Tsui et al. (19). In addition, we generated plasma RNA data from four healthy pregnant women (24–30 wk of gestation) and two pregnant women suffering from early PE following the method described by Tsui et al. (19) (Table S1). The plasma RNA profiles were normalized by size factor normalization using DESeq2 (69). The cell signature expression of each plasma RNA profile was calculated as the average normalized count levels of the specific cell signature gene set. The maternal plasma samples were grouped into five groups [nonpregnant, early pregnancy (13–20 wk), mid/late pregnancy (24–30 wk), predelivery, and 24-h postpartum]. The average signature scores of each group were then compared as the change with respective to nonpregnant level to illustrate the cellular dynamics in pregnancy progression. Similarly, maternal plasma RNA-seq profiles of Koh et al. (20) were retrieved using Sequence Read Archive study accession number SRP042027. The data were aligned using Tophat2 as described above. Patients with mappable reads >1 million and samples across four different time points (first trimester, second trimester, third trimester, and 6 wk postpartum) were selected for additional analysis (patients 2, 15, 24, and 32). The cell signature expression in each group was calculated as described above. The change is then visualized as the change with respective to first trimester pregnant women level. Dynamics of P4 (stromal cells) was not analyzed because of the low number (<50%) of signature genes detected in the plasma profiles.
Cellular Signature Expression Comparison in PE and Normal Maternal Plasma.
The plasma RNA cell-type–specific signature expression levels were compared between the mid/late pregnancy plasma group (Fig. 3A) and two early preeclamptic patients as an exploratory cohort (Fig. S4A). A new cohort of 6 early preeclamptic patients and 10 healthy third trimester pregnant women was recruited to validate the finding of differential EVTB cell signature expression in the exploratory dataset. In this new cohort, the plasma RNA profiles were generated using the Ovation RNA-Seq System V2 (NuGEN) (20) and analyzed as described above. The statistical significance of the differences of EVTB signature expression between early PE and healthy controls was determined by two-tailed two-sample Wilcoxon signed rank test.
Identification of Highly Variable Genes.
UMIs were integrated into the single-cell transcriptome library preparation chemistry in the single-cell Chromium system (10X Genomics). Technical variability from library amplification was removed by collapsing mappable reads with the same UMI barcode. We followed the method described by Kolodziejczyk et al. (50) in quantifying the transcriptional heterogeneity of individual gene by the DM approach. In short, the rolling median CV2 values were calculated across the range of the mean gene-expression levels to provide a linear fitted line in a double log-transformed space. The DM value of each gene is then calculated as the vertical distance of the CV2 of the gene from the median CV2 values of all genes with the same average expression level. In trophoblast differentiation trajectory reconstruction, genes with DM values greater than 0.3 were labeled as highly variable genes and selected for t-SNE clustering analysis and pseudotime analysis using Monocle2 (33).
Quantitative Analysis of Transcriptomic Heterogeneity of EVTB in PE Placentas.
In the intracluster heterogeneity comparison between term and early preeclamptic EVTB, the four single-cell libraries of early preeclamptic placentas and six term placentas were merged and normalized using the cellrangerRkit package by downsampling as described above. EVTB cells were identified by the specific expression of HLA-G, PAPPA2, TIMP1, CSH1, and ADAM12 on the biaxial t-SNE plot. DM values of each genes were calculated separately in the term and early preeclamptic placenta EVTB cells. Genes set information of each GO term (Biological Process) was retrieved from the org.Hs.eg.db package. GO terms containing less than 10 annotated genes with available DM values were removed from paired t test comparison of DM values using R (version 3.3.2). GO terms with P values less than 0.05 are regarded as significantly different between term and early preeclamptic placentas (Dataset S1).
Microarray Genotyping and SNP Identification.
Genomic DNA extracted from maternal buffy coat and placental tissue biopsies was genotyped with the Infinium Omni2.5–8 V1.2 Kit and the iScan system (Illumina). SNP calling was performed using the Birdseed v2 algorithm. The fetal genotypes of the placentas were compared with the maternal buffy coat genotypes to identify the fetal-specific SNP alleles. A SNP was considered informative if it was homozygous in the mother and heterozygous in the fetus.
Statistical Analysis.
Details of statistical analyses are described above. We regard a P value less than 0.05 as statistically significant.
Supplementary Material
Acknowledgments
We thank Ms. Carol Szeto for her technical assistance in tissue genotyping; Ms. Wing-Shan Lee for bioinformatics support; Dr. Macy Heung, Dr. Nancy Tsui, Ms. Cherry S. T. Leung, and Mr. K. W. Chan for technical assistance; and Ms. C. Y. Lai for patient recruitment. This work was supported by the Research Grants Council of the Hong Kong SAR Government under Theme-Based Research Scheme T12-403/15-N and Chinese University of Hong Kong Faculty Innovation Award 2016 FIA2016/B/01. Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation.
Footnotes
Conflict of interest statement: J.C.H.T., J.S.L.V., L.J., P.J., and Y.M.D.L. have filed patent applications based on the data generated from this work. P.J. is a consultant to Xcelom and Cirina. R.W.K.C. and Y.M.D.L. were consultants to Sequenom, Inc. R.W.K.C. and Y.M.D.L. hold equities in Sequenom and Grail. R.W.K.C. and Y.M.D.L. are founders of Xcelom and Cirina. Y.M.D.L. is a scientific cofounder of Grail.
Data deposition: The sequence data for the subjects studied in this work who had consented to data archiving have been deposited in the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk/ega/) hosted by the European Bioinformatics Institute (EBI; accession no. EGAS00001002449).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710470114/-/DCSupplemental.
References
- 1.Burton GJ, Fowden AL. The placenta: A multifaceted, transient organ. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140066. doi: 10.1098/rstb.2014.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaiworapongsa T, Chaemsaithong P, Yeo L, Romero R. Pre-eclampsia part 1: Current understanding of its pathophysiology. Nat Rev Nephrol. 2014;10:466–480. doi: 10.1038/nrneph.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vintzileos AM, Ananth CV, Smulian JC. Using ultrasound in the clinical management of placental implantation abnormalities. Am J Obstet Gynecol. 2015;213(4) Suppl:S70–S77. doi: 10.1016/j.ajog.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 4.Zeisler H, et al. Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med. 2016;374:13–22. doi: 10.1056/NEJMoa1414838. [DOI] [PubMed] [Google Scholar]
- 5.Chim SS, et al. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci USA. 2005;102:14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberry M, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: Confirmation that the origin is the trophoblast. Prenat Diagn. 2007;27:415–418. doi: 10.1002/pd.1700. [DOI] [PubMed] [Google Scholar]
- 7.Faas BH, et al. Non-invasive prenatal diagnosis of fetal aneuploidies using massively parallel sequencing-by-ligation and evidence that cell-free fetal DNA in the maternal plasma originates from cytotrophoblastic cells. Expert Opin Biol Ther. 2012;12(12 Suppl 1):S19–S26. doi: 10.1517/14712598.2012.670632. [DOI] [PubMed] [Google Scholar]
- 8.Lo YMD, et al. Quantitative abnormalities of fetal DNA in maternal serum in preeclampsia. Clin Chem. 1999;45:184–188. [PubMed] [Google Scholar]
- 9.Ng EK, et al. The concentration of circulating corticotropin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin Chem. 2003;49:727–731. doi: 10.1373/49.5.727. [DOI] [PubMed] [Google Scholar]
- 10.Martin A, Krishna I, Badell M, Samuel A. Can the quantity of cell-free fetal DNA predict preeclampsia: A systematic review. Prenat Diagn. 2014;34:685–691. doi: 10.1002/pd.4416. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123:2113–2118. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 12.Pang WW, et al. A strategy for identifying circulating placental RNA markers for fetal growth assessment. Prenat Diagn. 2009;29:495–504. doi: 10.1002/pd.2230. [DOI] [PubMed] [Google Scholar]
- 13.Leung TN, Zhang J, Lau TK, Hjelm NM, Lo YMD. Maternal plasma fetal DNA as a marker for preterm labour. Lancet. 1998;352:1904–1905. doi: 10.1016/S0140-6736(05)60395-9. [DOI] [PubMed] [Google Scholar]
- 14.Farina A, et al. High levels of fetal cell-free DNA in maternal serum: A risk factor for spontaneous preterm delivery. Am J Obstet Gynecol. 2005;193:421–425. doi: 10.1016/j.ajog.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsen TR, Clausen FB, Rode L, Dziegiel MH, Tabor A. High levels of fetal DNA are associated with increased risk of spontaneous preterm delivery. Prenat Diagn. 2012;32:840–845. doi: 10.1002/pd.3917. [DOI] [PubMed] [Google Scholar]
- 16.Tsui NB, et al. Systematic micro-array based identification of placental mRNA in maternal plasma: Towards non-invasive prenatal gene expression profiling. J Med Genet. 2004;41:461–467. doi: 10.1136/jmg.2003.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lun FM, et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin Chem. 2013;59:1583–1594. doi: 10.1373/clinchem.2013.212274. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsui NB, et al. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem. 2014;60:954–962. doi: 10.1373/clinchem.2014.221648. [DOI] [PubMed] [Google Scholar]
- 20.Koh W, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci USA. 2014;111:7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun K, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. 2015;112:E5503–E5512. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin Y, et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016;22:111–128. doi: 10.1261/rna.054809.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KCA, et al. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci USA. 2016;113:E8159–E8168. doi: 10.1073/pnas.1615800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol. 2015;213(4 Suppl):S115–S122. doi: 10.1016/j.ajog.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sõber S, et al. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci Rep. 2015;5:13336. doi: 10.1038/srep13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinrouweler CE, et al. Differentially expressed genes in the pre-eclamptic placenta: A systematic review and meta-analysis. PLoS One. 2013;8:e68991. doi: 10.1371/journal.pone.0068991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovats S, et al. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 30.Djurisic S, Hviid TV. HLA class Ib molecules and immune cells in pregnancy and preeclampsia. Front Immunol. 2014;5:652. doi: 10.3389/fimmu.2014.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trowsdale J, Moffett A. NK receptor interactions with MHC class I molecules in pregnancy. Semin Immunol. 2008;20:317–320. doi: 10.1016/j.smim.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci USA. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 35.Sugimoto J, Sugimoto M, Bernstein H, Jinno Y, Schust D. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci Rep. 2013;3:1462. doi: 10.1038/srep01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shyu MK, et al. Mucin 15 is expressed in human placenta and suppresses invasion of trophoblast-like cells in vitro. Hum Reprod. 2007;22:2723–2732. doi: 10.1093/humrep/dem249. [DOI] [PubMed] [Google Scholar]
- 37.Bièche I, et al. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod. 2003;68:1422–1429. doi: 10.1095/biolreprod.102.010322. [DOI] [PubMed] [Google Scholar]
- 38.Fahlbusch FB, et al. The tumor suppressor gastrokine-1 is expressed in placenta and contributes to the regulation of trophoblast migration. Placenta. 2013;34:1027–1035. doi: 10.1016/j.placenta.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Ng EK, et al. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100:4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu RWK, et al. Time profile of appearance and disappearance of circulating placenta-derived mRNA in maternal plasma. Clin Chem. 2006;52:313–316. doi: 10.1373/clinchem.2005.059691. [DOI] [PubMed] [Google Scholar]
- 41.Chan RWK, et al. Aberrant concentrations of liver-derived plasma albumin mRNA in liver pathologies. Clin Chem. 2010;56:82–89. doi: 10.1373/clinchem.2009.133355. [DOI] [PubMed] [Google Scholar]
- 42.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valdimarsson H, Mulholland C, Fridriksdottir V, Coleman DV. A longitudinal study of leucocyte blood counts and lymphocyte responses in pregnancy: A marked early increase of monocyte-lymphocyte ratio. Clin Exp Immunol. 1983;53:437–443. [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe M, et al. Changes in T, B, and NK lymphocyte subsets during and after normal pregnancy. Am J Reprod Immunol. 1997;37:368–377. doi: 10.1111/j.1600-0897.1997.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 45.Lima J, et al. Characterization of B cells in healthy pregnant women from late pregnancy to post-partum: A prospective observational study. BMC Pregnancy Childbirth. 2016;16:139. doi: 10.1186/s12884-016-0927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrews WC, Bonsnes RW. The leucocytes during pregnancy. Am J Obstet Gynecol. 1951;61:1129–1135. doi: 10.1016/0002-9378(51)90315-8. [DOI] [PubMed] [Google Scholar]
- 47.Pitkin RM, Witte DL. Platelet and leukocyte counts in pregnancy. JAMA. 1979;242:2696–2698. [PubMed] [Google Scholar]
- 48.Balloch AJ, Cauchi MN. Reference ranges for haematology parameters in pregnancy derived from patient populations. Clin Lab Haematol. 1993;15:7–14. doi: 10.1111/j.1365-2257.1993.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 49.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 50.Kolodziejczyk AA, et al. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiFederico E, Genbacev O, Fisher SJ. Preeclampsia is associated with widespread apoptosis of placental cytotrophoblasts within the uterine wall. Am J Pathol. 1999;155:293–301. doi: 10.1016/S0002-9440(10)65123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reister F, et al. Macrophage-induced apoptosis limits endovascular trophoblast invasion in the uterine wall of preeclamptic women. Lab Invest. 2001;81:1143–1152. doi: 10.1038/labinvest.3780326. [DOI] [PubMed] [Google Scholar]
- 54.Leung DN, Smith SC, To KF, Sahota DS, Baker PN. Increased placental apoptosis in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2001;184:1249–1250. doi: 10.1067/mob.2001.112906. [DOI] [PubMed] [Google Scholar]
- 55.Ishihara N, et al. Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation. Am J Obstet Gynecol. 2002;186:158–166. doi: 10.1067/mob.2002.119176. [DOI] [PubMed] [Google Scholar]
- 56.Lala PK, Chakraborty C. Factors regulating trophoblast migration and invasiveness: Possible derangements contributing to pre-eclampsia and fetal injury. Placenta. 2003;24:575–587. doi: 10.1016/s0143-4004(03)00063-8. [DOI] [PubMed] [Google Scholar]
- 57.Kadyrov M, Kingdom JC, Huppertz B. Divergent trophoblast invasion and apoptosis in placental bed spiral arteries from pregnancies complicated by maternal anemia and early-onset preeclampsia/intrauterine growth restriction. Am J Obstet Gynecol. 2006;194:557–563. doi: 10.1016/j.ajog.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 58.Tomas SZ, Prusac IK, Roje D, Tadin I. Trophoblast apoptosis in placentas from pregnancies complicated by preeclampsia. Gynecol Obstet Invest. 2011;71:250–255. doi: 10.1159/000320289. [DOI] [PubMed] [Google Scholar]
- 59.Longtine MS, Chen B, Odibo AO, Zhong Y, Nelson DM. Villous trophoblast apoptosis is elevated and restricted to cytotrophoblasts in pregnancies complicated by preeclampsia, IUGR, or preeclampsia with IUGR. Placenta. 2012;33:352–359. doi: 10.1016/j.placenta.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 61.Hui WW, et al. Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clin Chem. 2017;63:513–524. doi: 10.1373/clinchem.2016.268375. [DOI] [PubMed] [Google Scholar]
- 62.Pavličev M, et al. Single-cell transcriptomics of the human placenta: Inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017;27:349–361. doi: 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson AC, Mould AW, Bikoff EK, Robertson EJ. Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal-foetal interface during pregnancy. Nat Commun. 2016;7:11414. doi: 10.1038/ncomms11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji L, et al. Placental trophoblast cell differentiation: Physiological regulation and pathological relevance to preeclampsia. Mol Aspects Med. 2013;34:981–1023. doi: 10.1016/j.mam.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gierahn TM, et al. Seq-Well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.