Francesco Saverio Tedesco works with a big gene. In fact, the human dystrophin gene, with its whopping 2.4 million nucleotides, is one of the largest found so far in nature. A clinician-scientist at University College London, Tedesco hopes to use gene therapy to replace a faulty version of dystrophin in people with Duchenne muscular dystrophy. But the large deletions in some patients are too big to fix with gene editing, and the entire dystrophin gene is too large to fit inside the viruses normally used to deliver replacement DNA.
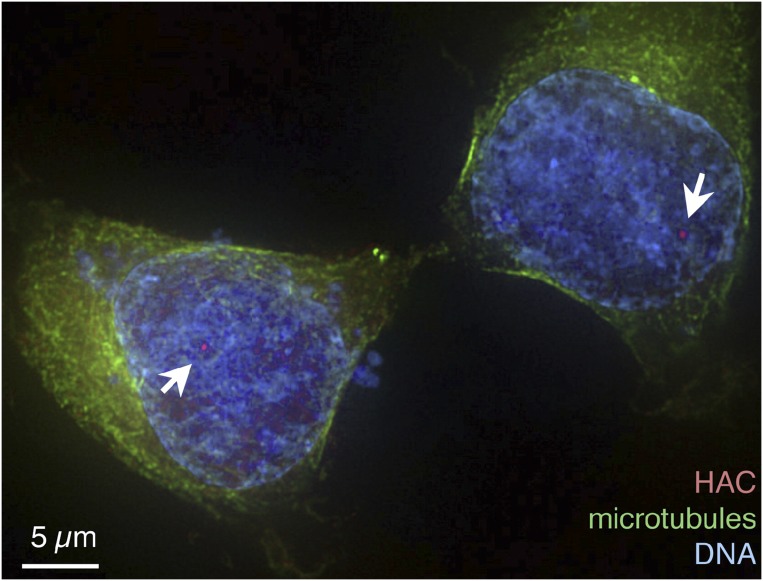
An artificial human chromosome (red) segregates along with natural chromosomes (blue) in a dividing cell. Reproduced from ref. 5.
Instead, Tedesco and his colleagues turned to human artificial chromosomes, or HACs, which can hold an essentially unlimited amount of DNA. They have already used HAC delivery to improve movement and other muscular dystrophy features in a mouse model (1), and are now trying it out in human cells.
HACs, whether designed from scratch or built using natural human chromosomes as a framework, are slowly gaining in popularity. These artificial DNA molecules, which can exist in human cells as an extra, 47th chromosome, are useful in a variety of applications, from basic studies of gene and chromosome function to potential stem cell and, perhaps, gene therapy treatments. Eventually, some synthetic biologists would like to use HACs to hold entire man-made biological pathways.
HACs can also complement the much-discussed CRISPR/Cas9 gene-editing tools, experts say. “CRISPR is good for creating specific and relatively small changes—deletions, insertions, mutations—in an existing genome,” says Alina Chan of Harvard Medical School in Boston. “HACs are for large, megabase-scale genome recoding. They have the power to house and test multiple pathways that would otherwise take an incredible amount of labor and time to engineer with CRISPR/Cas.” For example, Tedesco says CRISPR isn’t capable of fixing big dystrophin deletions in human patients.
But researchers face big hurdles with HACs. It’s difficult to get large genes to be expressed and regulated normally while being embedded in a HAC that itself must replicate and segregate with every cell division. Sometimes it’s hard to predict—from the DNA scientists start with—exactly what HAC will result. The human centromere, the chromosomal structure that links the chromatids and plays a crucial role during cell division, is a particular difficulty, notes Chan. “Centromeres remain poorly defined,” she says. “We do not know the size requirements, genetic sequence requirements remain ambiguous, and we know only a few proteins that can help to establish a de novo centromere.”
But perhaps the key caveat holding HACs back from widespread use is the difficulty moving them between cells. Efforts, already underway, to improve both the transfer protocols and the HACs themselves could make it possible for more researchers to incorporate HACs into their work.
Chromosome Flavors
Scientists began generating HACs in the 1990s, from synthetic DNA and artificial yeast chromosomes, to study centromeres and construct possible therapeutic gene carriers (2, 3). Like the bacterial and yeast versions of artificial chromosomes (BACs and YACs), HACs allow researchers to insert their desired DNA codes and replicate them in the namesake organism, or its cells in the case of HACs.
Scientists have long used YACs to modify or replicate large, isolated segments of the human genome within yeast. But while it’s possible to use YACs and BACs to deliver genes to human cell types, they’re “blunt tools,” says Chan, who is developing HACs. They usually insert their genes willy-nilly into the host genome, where those new sequences could potentially interrupt or interfere with native genes or their expression. “You need HACs if you want to be in human cells,” says cell biologist William Earnshaw of the University of Edinburgh in Scotland.
Even so, HAC development lags behind that of YACs and BACs because of a variety of challenges. Progress since the 1990s has been slow, as scientists still don’t have a very good understanding of the structure of the HACs they create. And until recently, the artificial chromosomes lacked an easy spot in which to add new genes. Another issue is that human chromosome structure complicates the DNA engineering—those repeat-heavy centromeres, as well as telomeres, which protect chromosome ends. The simple YAC centromere is just 125-base pairs long, compared with the multimegabase size of human centromeres. Telomeres also contain repeated sequences that can be difficult to reproduce or replicate; many HAC scientists get around the difficulty by making the HACs circular so no telomeres are needed.
HACs Two Ways
There are two main ways to build a HAC. The “top-down” approach starts with chromosomes and cuts out all of the genes, leaving nothing but the basic framework. The “bottom-up” approach assembles the
“The HAC offers this nice way to isolate the synthetic circuit.”
—Michael Elowitz
HAC from individual pieces, artificially synthesized or copied from natural chromosomes. Either way, scientists can add specific sites where they can then paste in their genes of choice.
Cell biologist Mitsuo Oshimura at Tottori University in Japan takes the top-down tactic. He starts with a full chromosome, such as number 21. He then cuts out everything except for the centromere. In the process, he adds telomeres. The result is a linear chromosome called 21HAC. To demonstrate its stability, the researchers transferred their HAC into mouse embryonic stem cells and generated mice, which were able to transmit 21HAC to their offspring (4).
Earnshaw and his collaborators take the bottom-up approach with their HAC, which they made by copying sequences from natural human centromeres. Geneticists Natalay Kouprina and Vladimir Larionov at the National Cancer Institute in Bethesda, Maryland, who specialize in working with long stretches of DNA, built a 50,000-base pair, artificial centromere by repeating these sequences. They kept their DNA circular to simplify construction and avoid the need for protective telomeres (5). Overall, Earnshaw says, the HAC is about as stable as a native chromosome. “Maybe at the lower end of the scale,” he says.
When the researchers first placed the tetracycline operator (tetO)-HAC into a human cell line, it merged, temporarily with chromosome 13, bringing about 400,000 of the chromosome’s base pairs with it when it detached again. The completed tetO-HAC thus contains this stretch of extra DNA, mostly not protein-coding. It also contains extra, scrambled copies of the centromeric sequences, says Earnshaw (6). “It is not fully sequenced, and as a result I would not be comfortable putting it into a person,” he says. Kouprina notes she’s made newer HACs lacking these extra inserts, but hasn’t yet fully characterized them.
Focus on Function
Although it’s not suitable for gene therapy in a person, the group’s original bottom-up tetO-HAC is ideal for studies of what different genes do in cultured cells, says Kouprina. That’s because in addition to the centromere, the designers added a snippet of synthesized DNA based on the Escherichia coli tetO, which can be bound by a protein called the tet repressor.
In the bacteria’s genome, the repressor turns off genes controlled by the tetO DNA. But scientists also use this DNA–protein pair as a convenient way to recruit other proteins to genes. By fusing various centromere-silencing proteins to the DNA-binding part of the tet repressor protein, they can attract those proteins to the tetO sequence, deactivating the HAC. The chromosome will fail to replicate, and its numbers will dwindle as cells divide.
To apply the HAC in gene function studies, Kouprina explains, scientists could first use the HAC to insert their gene of interest into a given cell type and examine how it changes that cell’s behavior. Then, as an additional control, they could inactivate the HAC to get rid of the chromosome and confirm that the behavior ceases.
Kouprina and her collaborators recently used this approach, in Chinese hamster ovary and human ovarian cancer cells, to show that the breast cancer-linked protein BRCA1 is involved in the function of the kinetochore, the group of proteins that attach to the centromere when cells divide and split up their DNA (7). This may help to explain why losing BRCA1 function causes chromosome instability, leading to cancer.
Still, it’s difficult to determine exactly how stable HACs would be in human cells, says Leslie Mitchell, a geneticist at New York University Medical Center in New York. Many experiments have taken place in cancer cell lines, which are known to maintain chromosome counts poorly to begin with. Others have been done in chimeric, mixed-genome mice, where only a subset of the cells should contain the HAC. That makes it difficult to know how many have lost the HAC. To more directly assess the loss rate, Mitchell is now engineering mice that contain the HAC in every cell, although she notes if cells lose the HAC, it could simply be because it’s unstable in mouse cells.
Even so, HACs have already allowed for a variety of investigations into the inner workings of chromosomes themselves. At the California Institute of Technology in Pasadena, Michael Elowitz and his team used Oshimura’s HACs to understand gene silencing. The researchers targeted different enzymatic regulators to a gene encoding a fluorescent protein on the HAC, and found that each silencer worked with different timing. Removing acetyl groups from the histone proteins in the chromosome, for example, led to fairly short-term silencing of the fluorescence gene. In contrast, methylating the DNA itself shut the gene down for up to a month (8).
By using the HAC, Elowitz didn’t have to insert the fluorescence gene within any other DNA, avoiding the possibility the transgene would influence nearby genes, and vice versa. “The HAC offers this nice way to isolate the synthetic circuit,” says Elowitz. The artificial chromosome also offers portability. Synthetic biologists could, he imagines, construct a complex, multigene system just once on a single HAC, and transfer it into whatever cell type they wish.
Tricky Transfer
However, Elowitz’s vision faces big hurdles. It’s not easy to move HACs—large pieces of DNA and associated proteins—from cell to cell. The standard method to transfer such a giant piece of nucleic acid is a 40-year-old, inefficient protocol known as microcell-mediated chromosome transfer (MMCT). “I really call it the bottleneck of the whole technology,” says Larionov.
Until scientists started playing with HACs, there was simply little reason to improve the procedure, explains Kouprina. Researchers must first break apart the nucleus of the chromosome-donor cells, so each chromosome ends up surrounded by its own individual nuclear membrane. Then, they fragment the cells themselves, leading to little vesicles containing individual chromosomes. These are the microcells that the scientists then fuse with the recipient cells. Recipients, then, should receive a single chromosome from the donor pool: either the HAC or one of the donor’s natural chromosomes.
One catch: only a couple of cell types, such as the Chinese hamster ovary line, are amenable to micronucleation. That’s because the MMCT protocol requires a long exposure to microtubule inhibitors, which kills most cells. However, Chinese hamster ovary and A9 mouse cells respond by making extra chromosomes, so they produce more micronuclei.
Both the National Cancer Institute researchers and Oshimura’s group are making progress in improving MMCT (9–11), which should allow HACs to become a more widespread tool. “We’re still at the beginning of this field,” says Elowitz. “It’s something that I think has a lot of potential.”
References
- 1.Tedesco FS, et al. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci Transl Med. 2011;3:96ra78. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- 2.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 3.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 4.Kazuki Y, et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 2011;18:384–393. doi: 10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouprina N, et al. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth Biol. 2012;1:590–601. doi: 10.1021/sb3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kononenko AV, et al. A portable BRCA1-HAC (human artificial chromosome) module for analysis of BRCA1 tumor suppressor function. Nucleic Acids Res. 2014;42:e164. doi: 10.1093/nar/gku870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bintu L, et al. Dynamics of epigenetic regulation at the single-cell level. Science. 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiratsuka M, et al. Retargeting of microcell fusion towards recipient cell-oriented transfer of human artificial chromosome. BMC Biotechnol. 2015;15:58. doi: 10.1186/s12896-015-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T, Kazuki Y, Oshimura M, Hara T. Highly efficient transfer of chromosomes to a broad range of target cells using Chinese hamster ovary cells expressing murine leukemia virus-derived envelope proteins. PLoS One. 2016;11:e0157187. doi: 10.1371/journal.pone.0157187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liskovykh M, Lee NC, Larionov V, Kouprina N. Moving toward a higher efficiency of microcell-mediated chromosome transfer. Mol Ther Methods Clin Dev. 2016;3:16043. doi: 10.1038/mtm.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]