Abstract
MicroRNAs (miRs) serve important roles in cancer genesis and progression. The expression of miR-1908 has been reported in a number of types of cancer; however, the clinical significance of miR-1908 in human ovarian cancer (OC) remains unclear. A total of 491 patients with OC from The Cancer Genome Atlas project cohort were selected and divided into two groups according to the median expression level of miR-1908. Univariate and multivariate analyses, using the Kaplan-Meier method and Cox regression, were performed to identify the characteristics that predict OC prognosis. Bioinformatics tools were used to identify potential targets of miR-1908. It was identified that the low expression of miR-1908 is associated with a poor prognosis for OC (P<0.05). The potential target genes of miR-1908 included podocan-like 1, JunB AP-1 transcription factor subunit, homeobox B8, SET binding factor 1 and sirtuin 2; high expression of these five genes additionally predicted a poor prognosis. These results suggest that miR-1908 may be a suitable target for the development of novel approaches in OC diagnosis and therapy in the future.
Keywords: microRNAs, prognosis, ovarian cancer
Introduction
Ovarian cancer (OC) is the most lethal of all gynecological tumors; there are >204,000 newly diagnosed patients and 125,000 mortalities annually, worldwide (1). The 5-year survival rate for OC is <50%; the main reason is the high frequency of OC recurrence subsequent to therapy (2). In order to improve estimates of the prognosis of OC and to develop novel treatments, novel OC biomarkers are required.
MicroRNAs (miRNAs/miRs) are non-protein coding genes that consist of <30 nucleotides; they serve important roles in a large number of cell behaviors, typically by downregulating target protein-coding genes. The dysregulation of miRNA expression is associated with a range of diseases, including the development of tumors (3). miR-1908 is regarded as an oncogene in numerous types of tumor (4). The miRNA functions as an oncogene through suppressing phosphatase and tensin homolog (PTEN), a tumor suppressor; this function promotes the proliferation and invasiveness of tumor cells in glioblastoma and osteosarcoma (5). miR-1908 expression is also regarded as a potential biomarker for breast cancer (6). Overall, a number of studies have identified that miR-1908 may be useful as a prognostic biomarker (7–9). However, it is necessary to clarify the functional and clinical importance of each miRNA in specific types of cancer, so that it is able to offer an improved proposal for therapeutics in the future. As the 5-year survival rate of OC is poor, biomarkers to predict the prognosis of patients with OC are required to improve the diagnosis of this disease. Therefore, the present study used data on patients with OC from The Cancer Genome Atlas (TCGA) cohort to identify whether miR-1908 was significantly associated with the prognosis of OC.
Patients and methods
Patients and samples
Patients with OC were selected from the TCGA cohort (https://portal.gdc.cancer.gov/). Patients included in the present study were those with fully characterized tumors, complete overall and disease-free survival time information, and complete miRNAseq and mRNAseq data. The clinical characteristics that were considered in the present study included age, tumor grade, clinical stage (American joint committee on cancer) (10), tumor size, lymphatic invasion, anatomical neoplasm subdivision and ethnicity. Python version 2.7.5 (https://www.python.org/download/releases/2.7.5/) and R version 3.2.0 (https://cran.r-project.org/bin/windows/base/old/3.2.0/) were used to screen the data of the TCGA OC cohort according to their patient IDs.
Identification of potential targets for miR-1908
The bioinformatics tools Targetscan (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/) were used to predict target genes for miR-1908. The top 100 highest-scoring genes were initially selected from each database to give 200 potential targets. Duplicates were then removed from the list to give a list of 131 potential target genes.
Statistical analysis
SPSS version 21 (IBM Corp., Armonk, NY, USA) was used for the statistical analysis and the construction of statistical plots. The values are presented as the mean ± the standard error of mean, and the values between the two groups were compared with independent t-test. Overall survival time was defined as the time from the date of diagnosis to the date of cancer-associated patient mortality or last follow-up. Disease-free survival time was defined as the time from the date of diagnosis to the date of first recurrence or cancer-associated patient mortality. Patients without further events or recorded mortality were regarded as censored at the time of the last follow-up. Patients were separated into two groups depending on the expression of miR-1908, with the cut-off being the median expression level. Survival curves were produced using the Kaplan-Meier (KM) method, and log-rank tests were used to assess the survival differences between the high expression group and the low expression group. The adjusted hazard ratio (HR) was calculated using the Cox proportional hazards model. Univariate and multivariate Cox proportional HR of miRNA expression and overall survival were analyzed. A two-sided P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics of the OC cohort
miR-1908 expression and clinical data of patients with OC from the TCGA cohort were sorted to identify 491 cases where clinical and miRNA data were available. The basic clinical features of the 491 patients with OC are summarized in Table I. The mean age at the time of the initial pathological diagnosis was 59.5 years (standard deviation, ±11.5) and patients ranged from 30 to 87 years old. The majority of the patients from the cohort (70.7%) were at clinical stage IIIC and 84% had a tumor grade of G4, indicating that OC may be difficult to identity and diagnose in the early stages. The median follow-up time for the cohort was 30.3 months; during this time, 270 patients (55.0%) succumbed to the disease.
Table I.
Clinical characteristics of the 491 patients with ovarian cancer included in the study.
| Characteristic | Value |
|---|---|
| Age at initial pathological diagnosis, years | |
| Mean ± standard deviation | 59.5±11.5 |
| Range | 30–87 |
| ≥60, n (%) | 234 (47.7) |
| <60, n (%) | 257 (52.3) |
| Tumor size, mm | |
| ≥20, n (%) | 406 (82.7) |
| <20, n (%) | 85 (17.3) |
| Tumor grade, n (%) | |
| G1+2 | 61 (12.4) |
| G3+4 | 417 (84.9) |
| Other | 13 (2.7) |
| Vascular invasion, n (%) | |
| Yes | 75 (15.3) |
| No | 56 (11.4) |
| Unknown | 360 (73.3) |
| Lymphatic invasion, n (%) | |
| Yes | 112 (22.8) |
| No | 67 (13.6) |
| Unknown | 312 (63.5) |
| Position, n (%) | |
| Bilateral | 341 (69.5) |
| Unilateral | 124 (25.2) |
| Unknown | 26 (5.3) |
| Clinical stage, n (%) | |
| Stage I+II | 33 (6.7) |
| Stage III+IV | 454 (92.5) |
| Unknown | 4 (0.8) |
| Ethnicity, n (%) | |
| American Indian or Alaska native | 2 (0.4) |
| Asian | 16 (3.3) |
| Black or African American | 31 (6.3) |
| Caucasian | 428 (87.2) |
| Hawaiian or other Pacific island native | 1 (0.2) |
| Not available | 13 (2.6) |
| miR1908 expression, n (%) | |
| High | 246 (50.1) |
| Low | 245 (49.9) |
miR, microRNA.
High expression level of miR-1908 is an independent positive prognostic factor for patients with OC
By analyzing the association between the expression level of miR-1908 and the overall survival time of patients with OC, it was identified that the high expression level of miR-1908 may be a protective factor in the cancer. To investigate whether the expression status of miR-1908 has potential as an OC prognostic marker, univariate and multivariate analyses using the KM method and Cox regression were performed. The OC cases were divided into two groups with the cut-off being the median level of miR-1908 expression.
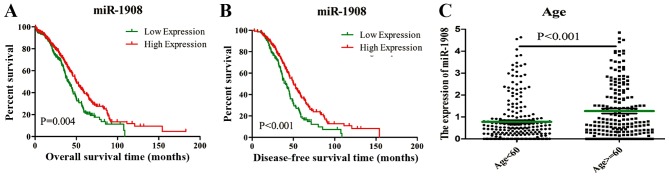
From the univariate Cox proportional hazards model analysis, it was determined that age, tumor size, clinical stage and miR-1908 expression level were significantly associated with the overall survival time of patients with OC in the TCGA cohort (P<0.05; Table II). A multivariate Cox proportional hazards model analysis was performed to identify independent factors for the prediction of prognosis in OC; the result indicated that miR-1908 expression level was an independent predictor of the overall survival time of patients with OC (HR, 0.673; 95% CI, 0.520–0.872; P<0.05; Table II). The KM plot for miR-1908 is presented in Fig. 1. The result indicates that the low expression of miR-1908 predicts a poor prognosis in terms of overall (Fig. 1A; P<0.05) and disease-free (Fig. 1B; P<0.05) survival time.
Table II.
Univariate and multivariate Cox proportional hazards analysis of the association between clinical characteristics, miR-1908 expression level and overall survival time.
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, ≥60/<60 years | 1.274 | 1.003–1.619 | 0.047 | 1.406 | 1.093–1.818 | 0.008 |
| Tumor size, ≥20/<20 mm | 0.690 | 0.513–0.929 | 0.014a | 0.831 | 0.489–1.774 | 0.101 |
| Tumor grade, G1+2/G3+4 | 1.346 | 0.948–1.910 | 0.096 | 1.406 | 0.913–2.971 | 0.053 |
| Position, unilateral/bilateral | 0.895 | 0.675–1.118 | 0.443 | 1.294 | 0.892–1.877 | 0.175 |
| Clinical stage, I+II/III+IV | 2.401 | 1.187–4.805 | 0.015a | 1.876 | 0.954–4.173 | 0.374 |
| miR-1908 expression, high/low | 0.698 | 0.548–0.891 | 0.004a | 0.673 | 0.520–0.872 | 0.003a |
P<0.05. miR, microRNA; HR, hazard ratio; CI, confidence interval.
Figure 1.
KM survival analysis based on miR-1908 expression level in ovarian cancer tissues, and distribution of miR-1908 expression by age. (A) KM plot of overall survival rate. (B) KM plot of disease-free survival rate. (C) Comparison of miR-1908 expression level between individuals aged ≥60 and <60 years (mean ± standard error of mean; green bar represents the mean). KM, Kaplan-Meier; miR, microRNA.
Multivariate logistic regression analyses of characteristics that may affect the expression of miR-1908 were performed. It was identified that miR-1908 expression was significantly associated with age (HR, 2.518; 95% CI, 1.305–4.860; P=0.006; Table III). In order to further analyze the age-associated expression difference of miR-1908, the cohort was divided into two groups according to the median age (≥60 vs. <60 years). The mean expression of miR-1908 was significantly higher in the ≥60 years group than that in the <60 years group (P<0.001; Fig. 1C).
Table III.
Multivariate logistic regression analysis of the association between patient characteristics and miR-1908 expression.
| miR-1908 | |||
|---|---|---|---|
| Characteristic | OR | 95% CI | P-value |
| Age, ≥60/<60 years | 2.518 | 1.305–4.860 | 0.006a |
| Tumor size, ≥20/<20 mm | 2.255 | 0.820–6.198 | 0.115 |
| Tumor grade, G1+2/G3+4 | 2.433 | 0.792–7.474 | 0.120 |
| Position, unilateral/bilateral | 1.548 | 0.781–3.067 | 0.211 |
| Clinical stage, I+II/III+IV | 0.466 | 0.160–1.356 | 0.161 |
P<0.05. miR, microRNA.
Prediction of miR-1908 target genes with bioinformatics analysis
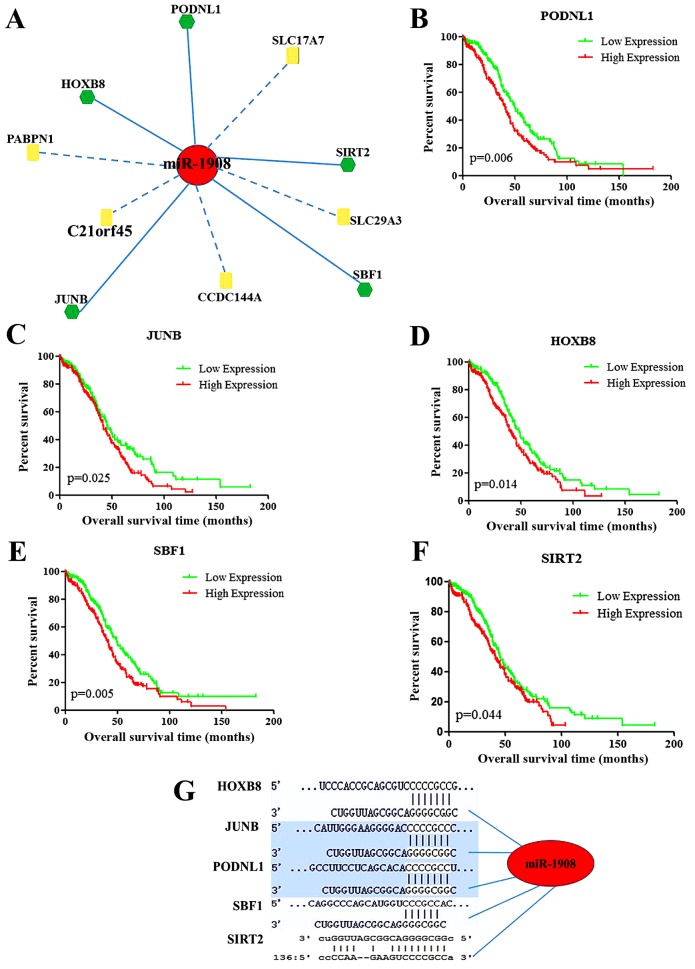
Following the survival analysis of 131 potential target genes, 10 genes were determined to be associated with prognosis in OC (Table IV; Fig. 2A). The low expression of the potential target genes podocan-like 1 (PODNL1), junB AP-1 transcription factor subunit (JUNB), homeobox B8 (HOXB8), SET binding factor 1 (SBF1) and sirtuin 2 (SIRT2) was associated with a significantly increased survival time (Fig. 2B-F). As the high expression of miR-1908 is associated with an improved prognosis and it has the ability to bind to the 3′-untranslated region (UTR) of these 5 genes, as illustrated in Fig. 2G, we hypothesize that these 5 genes are potential targets for miR-1908 that determine its effect on the prognosis of patients with OC.
Table IV.
Predicted target genes for miR-1908 that are associated with ovarian cancer survival time.
| Gene symbol | Full gene name | P-value |
|---|---|---|
| CCDC144A | Coiled-coil domain containing 144A | 0.032 |
| SLC29A3 | Solute carrier family 29, member 3 | 0.032 |
| C21orf45 | MIS18 kinetochore protein A | 0.046 |
| SIRT2 | Sirtuin 2 | 0.044 |
| PABPN1 | Poly(A) binding protein, nuclear 1 | 0.036 |
| SLC17A7 | Solute carrier family 17 (vesicular glutamate transporter), member 7 | 0.009 |
| SBF1 | SET binding factor 1 | 0.008 |
| JUNB | JunB AP-1 transcription factor subunit | 0.025 |
| HOXB8 | Homeobox B8 | 0.014 |
| PODNL1 | Podocan-like 1 | 0.005 |
Figure 2.
Predicted targets of miR-1908. (A) The predicted targets for miR-1908 that were associated with prognosis for patients with ovarian cancer; green hexagons represent genes for which high mRNA expression predicted a poor prognosis, whereas yellow rectangles represent the genes where low mRNA expression predicted a poor prognosis. Kaplan-Meier survival curves for (B) PODNL1, (C) JUNB, (D) HOXB8, (E) SBF1 and (F) SIRT2. (G) Binding sites for miR-1908 in the 3′-untranslated region of the target genes. miR, microRNA; PODNL1, podocan-like 1; JUNB, JunB AP-1 transcription factor subunit; HOXB8, homeobox B8; SBF1, SET binding factor 1; SIRT2, sirtuin 2.
Discussion
miRNA dysregulation is a common characteristic of numerous types of cancer, and miRNAs serve key functions in tumor genesis and progression, including cell proliferation, differentiation, migration and invasion (11). OC is associated with a poor prognosis and a high mortality rate due to its high likelihood of recurrence. Therefore, more prognostic markers are required to improved estimates of patient survival time and construct personalized plans for therapy. In the present study, miR-1908 was identified as a biomarker for predicting the survival time of patients with OC. Patients with OC from the TCGA cohort were selected, and their clinical, RNAseq and mRNAseq data were analyzed. It was identified that the high expression of miR-1908 may be a protective factor for OC, as high expression was associated with improved survival time for patients with the cancer.
In glioblastoma, miR-1908 functions as an oncogene by repressing the PTEN pathway (11). Overexpression of miR-1908 significantly promoted tumor growth, as demonstrated in vitro and in vivo. Additionally, the expression levels of miR-1908 were significantly increased in patient tumor samples and its high expression was associated with a reduction in survival time (11). Another study identified that the low expression of miR-1908 predicted a poor prognosis in glioma (12). In addition, miR-1908 has also been associated with the prognosis of chordoma, hepatoma and melanoma (13–16). miR-1908 has different functions in different cancer types, as it may target different genes.
According to the present study, SIRT2, PODNL1, SBF1, JUNB and HOXB8 are likely to be the targets of miR-1908 in OC. These genes are associated with the overall survival time of OC patients, as low expression of these genes predicts an improved prognosis, and miR-1908 is able to regulate them by binding their 3′UTRs. SIRT2 is a NAD-dependent deacetylase that is involved in a number of cellular processes, including cell proliferation, death, senescence and stress responses (17,18). PONDL1 is a transcription factor; however, its function remains unclear (19). SBF1 encodes a member of the protein-tyrosine phosphatase family; this protein contains a guanine nucleotide exchange factor domain, necessary for its role in cell growth and differentiation (16). JUNB promotes tumor metastasis and progress; silencing of JUNB resulted in reduced cell growth and colony formation associated with decreased activator protein-1 activity, and G1/S or G2/M cell cycle arrest (20). HOXB8 is a member of the ANTP homeobox family and encodes a nuclear protein with a homeobox DNA-binding domain. The high expression of cytoplasmic HOXB8 was associated with a significantly reduced progression-free survival time, whereas high nuclear HOXB8 expression was associated with significantly shorter overall survival time in an analysis limited to patients with post-chemotherapy effusions (21). miR-1908 may target these genes to suppress the progress of OC and increase the survival time of patients with OC.
In conclusion, the present study demonstrated that the expression of miR-1908 is significantly associated with the overall survival time of patients with OC. The high expression of miR-1908 predicts a better prognosis by conferring a longer survival time. The low expression of miR-1908 may be a valuable biomarker to identify patients with OC with a poor prognosis. Potential target genes of miR-1908 were identified; these targets are likely to have functions in tumor genesis and progression. These data may support novel approaches for the diagnosis and therapy of patients with OC in the future.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (grant no. 81470153) and the Projects of International Cooperation and Exchanges National Natural Science Foundation of China (grant no. 81320108022).
References
- 1.Zhang B, Cai FF, Zhong XY. An overview of biomarkers for the ovarian cancer diagnosis. Eur J Obstet Gynecol Reprod Biol. 2011;158:119–123. doi: 10.1016/j.ejogrb.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH, Park CW, Um D, Baek KH, Jo Y, An H, Kim Y, Kim TJ. Mass spectrometric screening of ovarian cancer with serum glycans. Dis Markers. 2014;2014:634289. doi: 10.1155/2014/634289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan H, Gao Y. MicroRNA-1908 is upregulated in human osteosarcoma and regulates cell proliferation and migration by repressing PTEN expression. Oncol Rep. 2015;34:2706–2714. doi: 10.3892/or.2015.4242. [DOI] [PubMed] [Google Scholar]
- 6.Rawlings-Goss RA, Campbell MC, Tishkoff SA. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics. 2014;7:53. doi: 10.1186/1755-8794-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lian D, Wang ZZ, Liu NS. MicroRNA-1908 is a biomarker for poor prognosis in human osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20:1258–1262. [PubMed] [Google Scholar]
- 9.Xie C, Shi K, Zhang X, Zhao J, Yu J. MiR-1908 promotes scar formation post-burn wound healing by suppressing Ski-mediated inflammation and fibroblast proliferation. Cell Tissue Res. 2016;366:371–380. doi: 10.1007/s00441-016-2434-6. [DOI] [PubMed] [Google Scholar]
- 10.Xia X, Li Y, Wang W, Tang F, Tan J, Sun L, Li Q, Sun L, Tang B, He S. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol Cancer. 2015;14:154. doi: 10.1186/s12943-015-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Xue F, Zhang W. miR-506: A regulator of chemo-sensitivity through suppression of the RAD51-homologous recombination axis. Chin J Cancer. 2015;34:485–487. doi: 10.1186/s40880-015-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shou J, Gu S, Gu W. Identification of dysregulated miRNAs and their regulatory signature in glioma patients using the partial least squares method. Exp Ther Med. 2015;9:167–171. doi: 10.3892/etm.2014.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghanbari M, Sedaghat S, de Looper HW, Hofman A, Erkeland SJ, Franco OH, Dehghan A. The association of common polymorphisms in miR-196a2 with waist to hip ratio and miR-1908 with serum lipid and glucose. Obesity (Silver Spring) 2015;23:495–503. doi: 10.1002/oby.20975. [DOI] [PubMed] [Google Scholar]
- 14.Long C, Jiang L, Wei F, Ma C, Zhou H, Yang S, Liu X, Liu Z. Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS One. 2013;8:e66676. doi: 10.1371/journal.pone.0066676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et al. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilic Eren M, Kilincli A, Eren Ö. Resveratrol induced premature senescence is associated with DNA damage mediated SIRT1 and SIRT2 down-regulation. PLoS One. 2015;10:e0124837. doi: 10.1371/journal.pone.0124837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grbesa I, Pajares MJ, Martinez-Terroba E, Agorreta J, Mikecin AM, Larráyoz M, Idoate MA, Gall-Troselj K, Pio R, Montuenga LM. Expression of sirtuin 1 and 2 is associated with poor prognosis in non-small cell lung cancer patients. PLoS One. 2015;10:e0124670. doi: 10.1371/journal.pone.0124670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurat CF, Lambert JP, Petschnigg J, Friesen H, Pawson T, Rosebrock A, Gingras AC, Fillingham J, Andrews B. Cell cycle-regulated oscillator coordinates core histone gene transcription through histone acetylation. Proc Natl Acad Sci USA. 2014;111:14124–14129. doi: 10.1073/pnas.1414024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atsaves V, Lekakis L, Drakos E, Leventaki V, Ghaderi M, Baltatzis GE, Chioureas D, Jones D, Feretzaki M, Liakou C, et al. The oncogenic JUNB/CD30 axis contributes to cell cycle deregulation in ALK+ anaplastic large cell lymphoma. Br J Haematol. 2014;167:514–523. doi: 10.1111/bjh.13079. [DOI] [PubMed] [Google Scholar]
- 21.Salmanidis M, Brumatti G, Narayan N, Green BD, van den Bergen JA, Sandow JJ, Bert AG, Silke N, Sladic R, Puthalakath H, et al. Hoxb8 regulates expression of microRNAs to control cell death and differentiation. Cell Death Differ. 2013;20:1370–1380. doi: 10.1038/cdd.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]