Abstract
Conventional functional connectivity (FC) and corresponding networks focus on characterizing the pairwise correlation between two brain regions, while the high-order FC (HOFC) and networks can model more complex relationship between two brain region “pairs” (i.e., four regions). It is eye-catching and promising for clinical applications by its irreplaceable function of providing unique and novel information for brain disease classification. Since the number of brain region pairs is very large, clustering is often used to reduce the scale of HOFC network. However, a single HOFC network, generated by a specific clustering parameter setting, may lose multifaceted, highly complementary information contained in other HOFC networks. To accurately and comprehensively characterize such complex HOFC towards better discriminability of brain diseases, in this paper, we propose a novel HOFC based disease diagnosis framework, which can hierarchically generate multiple HOFC networks and further ensemble them with a selective feature fusion method. Specifically, we create a multi-layer HOFC network construction strategy, where the networks in upper layers are formed by hierarchically clustering the nodes of the networks in lower layers. In such a way, information is passed from lower layers to upper layers by effectively removing the most redundant part of information and, at the same time, retaining the most unique part. Then, the retained information/features from all HOFC networks are fed into a selective feature fusion method, which combines sequential forward selection and sparse regression, to further select the most discriminative feature subset for classification. Experimental results confirm that our novel method outperforms all the single HOFC networks corresponding to any single parameter setting in diagnosis of mild cognitive impairment (MCI) subjects.
Keywords: Functional connectivity, High-order network, Hierarchical clustering, Brain network, Resting state, Functional magnetic resonance imaging
1 Introduction
Alzheimer’s disease (AD) is the most prevalent dementia, accounting for about 60–80 % of dementia cases among the worldwide elderly population. Mild cognitive impairment (MCI), as a prodromal stage of AD, tends to convert to clinical AD with an average annual conversion rate of 10–15 % [1]. Early diagnosis of MCI is of great importance for possibly delaying the AD progression, and is always a hot spot in the translational clinical research.
Resting-state functional magnetic resonance imaging (RS-fMRI) can be used to infer functional connectivity (FC) among brain regions, and to characterize brain network abnormalities in MCI. Here, the FC is traditionally defined as the temporal correlation of the blood-oxygenation-level-dependent (BOLD) time series between two brain regions [2]. The whole brain network can further be constructed by accounting for FC of every brain region pairs. Recently, static FC calculated based on the entire BOLD time series has been challenged by dynamic FC and dynamic network studies. A well-adopted dynamic FC analysis strategy is to calculate FC in sliding windows [4–6]. Furthermore, multiple time-varying FC networks can be constructed and used for MCI classification [5]. Nevertheless, these time-varying FC networks are still low-order, since they are estimated based on the raw BOLD time series, reflecting just the relationship among brain regions in a pairwise manner.
The series of time-varying low-order FC (LOFC) networks can be equivalently represented as a set of dynamic FC time series, each of which is associated with a pair of brain regions and characterizes their time-evolving FC. Then, we can further compute the correlation between two FC time series (each of them involves two brain regions) and obtain a high-order FC (HOFC) which involves four brain regions (i.e., two pairs of brain regions). With this strategy, an HOFC network can be constructed in the whole brain. The graph theory based high-order features characterizing the properties of HOFC network can be extracted. HOFC has following merits: (1) it is computed based on FC time series, instead of raw BOLD time series, thus representing high-level features; (2) it reflects a more complex relationship among brain regions by characterizing how different brain region pairs, instead of two brain regions functionally interact with each other; (3) it is time invariant, solving the problem of phase mismatch among subjects.
However, the side-effect of HOFC (i.e., the increase of network scale from N2 to (N2)2, where N is the total number of brain regions) requires clustering-based dimension reduction and thus results in inevitable information loss when a single HOFC network (corresponding to a specific number of clusters) is used for classification. It is obvious that the clustering pattern in a high dimensional space is NOT a discrete structure; instead, rich information underlying in a continuous relationship between the network nodes in the space when viewing from different spatial scales could be used to boost classification accuracy. Accordingly, we first generate multiple HOFC networks for each subject; each network has different discriminative ability for disease identification. More importantly, these HOFC networks are organized in a hierarchical fashion, which means the network in each layer is generated by merging some nodes while retaining other nodes of the HOFC network in a previous layer. By doing so, the HOFC networks in two consecutive layers are highly overlapped. As a result, the features extracted from the HOFC network of each layer can be decomposed into two parts (blocks): one is redundant and the other is informative or complementary with respect to the features extracted from a previous layer. To further refine those informative feature blocks from all HOFC networks, a feature fusion strategy based on sequential forward selection and sparse regression [10] is developed and the resulting feature subset is used for classification with linear support vector machine (SVM).
2 Approach
The proposed method consists of the following 8 steps. (1) For each subject, a sliding window with length N and step size s is applied to partition the entire BOLD time series into multiple overlapping segments [4, 5]. (2) Multiple LOFC networks are constructed, each of which is based on a respective segment of BOLD time series. By doing so, we actually obtain a set of FC time series, each describing the temporal variation of correlation between two brain regions. (3) All subjects’ FC time series associated with the same brain region pair are concatenated together to form a long FC time series. (4) The long FC time series from all brain region pairs are grouped into U clusters by a clustering algorithm, thus yielding consistent clustering results across different subjects. (5) For each subject, the mean of the FC time series within the same cluster is computed and then a HOFC network is constructed based on the correlation between the mean FC time series of different clusters. (6) Repeating steps (4) and (5) multiple times with different Us generates multiple HOFC networks. (7) The features extracted from all HOFC networks are analyzed based on correlation, and then a feature subset is selected by a feature selection method that combines sequential forward selection and sparse regression. (8) Support vector machine (SVM) [8] with linear kernel is finally trained with the selected features to classify MCI and NC subjects. The main flowchart of our hierarchical HOFC networks construction and feature extraction is shown in Fig. 1, where four brain regions denoted as A, B, C, and D are illustrated.
Fig. 1.
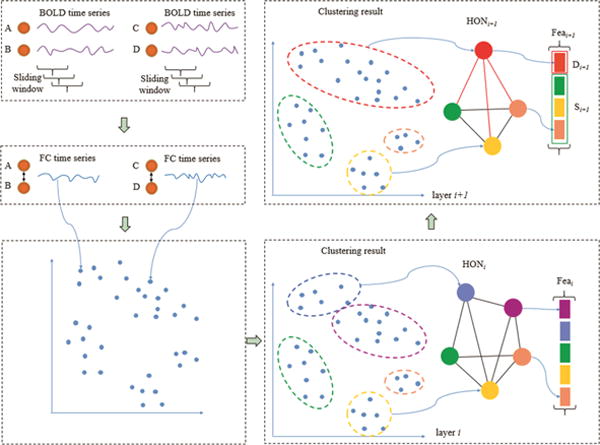
Hierarchical HOFC networks construction and feature decomposition.
2.1 Hierarchical Clustering and Feature Decomposition
As shown in the top left panel of Fig. 1, we can obtain FC time series corresponding to each pair of brain regions by following the above steps (1) and (2). When repeating step (4) with different numbers of clusters, we use an agglomerative hierarchical clustering [9] to group these FC time series into ui and ui+1 clusters (ui>ui+1), respectively, in layer i and layer i+1. In Fig. 1, we have ui=5 and ui+1=4. When the difference between ui and ui+1 is small, which is true using the agglomerative hierarchical clustering, a few clusters in the layer i + 1 are newly formed by merging some closer clusters in the layer i, while other clusters in the layer i + 1 are the same as those in the layer i. This is illustrated in the right panels of Fig. 1, where the blue and pink clusters in the layer i are merged into a new red cluster in the layer i + 1 while other clusters are kept the same. Based on the clustering results in the layers i and i + 1, the HOFC networks HONi and HONi+1 are constructed, respectively, where each node corresponds to each cluster and the weight for each edge is the Pearson’s correlation between the mean FC time series of two different clusters. As we can see from the upper right panel of Fig. 1, only the newly formed nodes and the associated edges (shown in red) in HONi+1 may contain extra information with respect to HONi.
Afterwards, the feature vectors and can be extracted from HONi and HONi+1, respectively. In this paper, weighted local clustering coefficients (WLCC) [7] is used as features. Each entry in Feai and Feai+1 corresponds to a node in HONi and HONi+1, respectively, thus also corresponding to a cluster in layers i and i + 1. Since only a small number of nodes and edges in HONi+1 are different from those in HONi, Feai+1 can be decomposed as Feai+1 = [Di+1, Si+1], where D and S respectively refer to the nodes newly formed from and the nodes already existed in the previous layer. As a result, Si+1 in layer i + 1 should be highly correlated with some features in Feai, while only Di+1 may contain some novel information with respect to Feai.
Generalizing the above observation to L levels, for each subject, the features extracted from all hierarchical HOFC networks can be condensed and expressed as Fea = [D0,D1,D2,⋯,DL], where D0 = S1. Each Di is called a feature block.
2.2 Selective Feature Fusion
Although the above agglomerative hierarchical clustering and correlation analysis can reduce the dimensionality of features to a large extent, the redundancy between different layers may still exist, especially when taking into account multiple layers. In addition, not all of the features in Fea are discriminative in terms of MCI classification. To benefit from the information contained in Fea and reduce redundancy, we propose a feature fusion method, by combining sequential forward selection and sparse regression [10], under the framework of wrapper-based feature selection [11]. Sequential forward selection can find discriminant feature block progressively, while sparse regression can select individual features that are predictive for classification.
Specifically, given a current set A of feature blocks, a new feature block Di from Fea – A can be selected and combined with A, thus producing an enlarged feature subset A∪Di. Then, l1-norm based sparse regression, i.e., least absolute shrinkage and selection operator (LASSO) is performed on all training samples with features within A∪Di to find a small subset C that is beneficial for classification. Next, the selected features of all training subjects are used to train a linear SVM model, and the classification accuracy on the validation subjects is used to guide the selection of Di. That is, the one yielding optimal accuracy is finally selected. In such a way, the feature block selected in the previous iteration will be kept and guild the selection of new feature block in the next iteration. The procedure above is repeated until either the optimal performance or the pre-defined number of feature blocks is reached.
3 Experiments
3.1 Data
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset is used in this study. 50 MCI subjects and 49 normal controls (NCs) are selected from ADNI-GO and ADNI-2 datasets. Subjects from both datasets were age- and gender-matched and were scanned using 3.0T Philips scanners. The voxel size is 3.13 × 3.13 × 3.13 mm3. SPM8 software package (http://www.fil.ion.ucl.uk/spm/software/spm8) was used to preprocess the RS-fMRI data.
The first 3 volumes of each subject were discarded before preprocessing for magnetization equilibrium. A rigid-body transformation was used to correct head motion in subjects (and the subjects with head motion larger than 2 mm or 2° were discarded). The fMRI images were normalized to the Montreal Neurological Institute (MNI) space and spatially smoothed with a Gaussian kernel with full width at half maximum (FWHM) of 6 × 6 × 6 mm3. We did not perform scrubbing to the data with frame-wise displacement larger than 0.5 mm, as it would introduce additional artifacts. We excluded the subjects who had more than 2.5 min (the maximum is 7 min) RS-fMRI data with large frame-wise displacement from further analysis. The RS-fMRI images were parcellated into 116 regions according to the Automated Anatomical Labeling (AAL) template. Mean RS-fMRI time series of each brain region was band-pass filtered (0.015 ≤ f ≤ 0.15 Hz). Head motion parameters (Friston24), mean BOLD time series of the white matter and the cerebrospinal fluid were regressed out.
3.2 HOFC Network and Feature Correlation Analysis
In this study, we use a sliding window with s = 1 and N = 50. Denote the number of clusters as U. To generate multiple layers, we start from one layer with a relatively large number of clusters (U = 220) because it can retain sufficient information. Then, the subsequent layers are added by gradually reducing U by 30 until the optimal performance is achieved. In such a way, we can eventually generate 4 HOFC networks from layer 1 to layer 4: HON1, HON2, HON3, and HON4, where the number of clusters equals 220, 190, 160, and 130, respectively. The averaged HOFC networks in layer 1 for MCI and NC subjects are shown in the left and middle of Fig. 2. The corresponding high-order feature vectors (WLCC) Fea1 ∈ R220, Fea2 ∈ R190, Fea3 ∈ R160, and Fea4 ∈ R130 are extracted. Since our method is a feature fusion method, the correlation between features from different HOFC networks provides important information about redundancy. To empirically verify the rationality of feature decomposition (Sect. 2.1), we compute the correlation between Fea1 and Fea2 and show the correlation matrices in the right of Fig. 2. The numbers of rows and columns of this matrix equal to 220 and 190, respectively. As shown by the red straight lines in this matrix, most features in Fea2 are highly correlated with features in Fea1. This observation is consistent for all HOFC networks, thus implying most features in the current layer are redundant with respect to those in the previous layer and thus should be eliminated before feature fusion. Based on this correlation, each feature vector Feai (i = 1, 2, 3, 4) can be decomposed into two feature blocks such as Feai = [Di, Si], where D1 ∈ R29, S1 ∈ R191, D2 ∈ R30, S2 ∈ R160, D3 ∈ R29, S3 ∈ R131, and D4 ∈ R30, S4 ∈ R100. Note that an extra level with 250 clusters is used to decompose Fea1 and then discarded. Consequently, only about 30 features, i.e., Di, of each layer (i > 1) are less correlated with the previous layer while others are redundant. To include sufficient information and meanwhile reduce the redundancy, five feature blocks S1 ∈ R191, D1 ∈ R29, D2 ∈ R30, D3 ∈ R29, and D4 ∈ R30 are engaged in the subsequent feature fusion, while others are discarded. Finally, the total number of features decreases from 700 to 309 by this unsupervised correlation analysis.
Fig. 2.
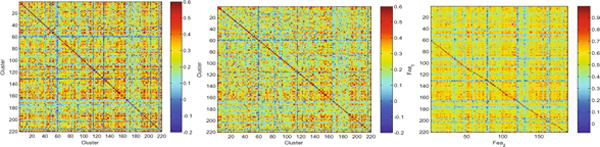
Averaged HOFC network in layer 1 for MCI (left) and NC (middle) subjects and feature correlation matrices between HOFC networks from layer 1 and layer 2 (right).
3.3 Classification Accuracy
The proposed sequential forward selection and sparse regression based hierarchical HOFC networks feature fusion (HHON-SFS) is compared with some closely related methods, including (1) a feature fusion method (HHON-CON), which concatenates all features extracted from four HOFC networks, (2) four individual HOFC networks (HON1, HON2, HON3, and HON4), and (3) two LOFC networks based on partial correlation (LON-PAC) and Pearson’s correlation (LON-PEC), respectively. All methods were implemented in MATLAB 2012b environment. The SLEP [10] and Libsvm tool‐boxes were utilized, respectively, to implement sparse regression and SVM classification. The leave-one-out cross validation (LOOCV) is adopted to evaluate performance of different methods. For the hyper-parameter in each method, we tune its value on the training subjects by using the nested LOOCV. To measure performances of different methods, we use the following indices: accuracy (ACC), area under ROC curve (AUC), sensitivity (SEN), specificity (SPE), Youden’s Index (YI), F-score, and balanced accuracy (BAC).
The experimental results are shown in Table 1. The HOFC networks achieve better accuracy than the two LOFC networks, indicating that the HOFC networks provide more discriminative biomarkers for MCI identification. Comparing across four individual HOFC networks, we can observe their performance is rather sensitive to the number of clusters. For example, too large or too small U will adversely affect the performance. Although the HOFC network HON2 (U = 190) achieves better performance than other individual HOFC networks, this does not mean that the information contained in other networks is completely useless in distinguishing MCI from NC subjects. To make full use of information in the four HOFC networks, HHON-CON directly concatenates Fea1, Fea2, Fea3, and Fea4 to form a combined feature vector of length 700. Although this method uses all features, the accuracy falls just between the best and the worst ones. This may be due to too many redundant features being used which makes the relationship between features complex, thus causing difficulty in individual feature selection and the potential over-fitting. In contrast, the proposed method, HHON-SFS, achieves the best performance among all competing methods. On one hand, this improvement can be attributed to the feature correlation analysis and also the resulting feature decomposition, which eliminates many redundant features. On the other hand, the combination of sequential forward selection and sparse regression makes it possible to evaluate the importance of feature blocks and individual features progressively. As a result, those crucial and complementary features have more probability to be selected and fused for classification.
Table 1.
Performance comparison of different methods in MCI classification.
| Method | ACC | AUC | SEN | SPE | YI | F-score | BAC |
|---|---|---|---|---|---|---|---|
| LON-PEC | 57.58 | 0.6008 | 58.00 | 57.14 | 15.14 | 58.00 | 57.57 |
| LON-PAC | 60.61 | 0.6249 | 58.00 | 63.27 | 21.27 | 59.79 | 60.63 |
| HON1 | 80.81 | 0.8567 | 86.00 | 75.51 | 61.51 | 81.90 | 80.76 |
| HON2 | 81.82 | 0.8743 | 82.00 | 81.63 | 63.63 | 82.00 | 81.82 |
| HON3 | 68.69 | 0.8094 | 74.00 | 63.27 | 37.27 | 70.48 | 68.63 |
| HON4 | 72.73 | 0.7857 | 74.00 | 71.43 | 45.43 | 73.27 | 72.71 |
| HHON-CON | 78.79 | 0.8702 | 80.00 | 77.55 | 57.55 | 79.21 | 78.78 |
| HHON-SFS | 84.85 | 0.9057 | 88.00 | 81.63 | 69.63 | 85.44 | 84.82 |
4 Conclusion
In this paper, we propose to fuse information contained in multiple HOFC networks for a better MCI classification. To this end, hierarchical clustering is utilized to generate multiple HOFC networks, each being located at one layer. With such a framework, features extracted from the network at each layer can be refined, and only the informative feature block is taken into account. Specifically, by combining the sequential forward selection and sparse regression, a novel feature fusion method is developed. This method is able to selectively integrate informative feature blocks from different HOFC networks and further detect a small set of individual features that are discriminative for early diagnosis. Finally, SVM with linear kernel is used for MCI classification. The experimental results demonstrate the capability of the proposed approach in making full use of information contained in multiple scales of HOFC networks. Also, combing multiple HOFC networks is demonstrated to yield better classification performance than simple use of a single HOFC network.
Acknowledgments
This work was supported by National Institutes of Health (EB006733, EB008374, and AG041721) and National Natural Science Foundation of China (Grand No: 61203244).
References
- 1.Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friston K, Frith C, Liddle P, Frackowiak R. Functional connectivity: the principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993;13:5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- 3.Brier MR, Thomas JB, Fagan AM, Hassenstab J, Holtzman DM, Benzinger TL, Morris JC, Ances BM. Functional connectivity and graph theory in preclinical Alzheimer’s disease. Neurobiol Aging. 2014;35:757–768. [Google Scholar]
- 4.Leonardi N, Richiardi J, Gschwind M, Simioni S, Annoni JM, Schluep M, Vuilleumier P, Van De Ville D. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. NeuroImage. 2013;83:937–950. doi: 10.1016/j.neuroimage.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Wee CY, Yang S, Yap P-T, Shen D, Alzheimer’s Disease Neuroimaging Initiative Sparse temporally dynamic resting-state functional connectivity networks for early MCI identification. Brain Imaging Behav. 2015;10:1–15. doi: 10.1007/s11682-015-9408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, de Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 9.Ward JH., Jr Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 10.Liu J, Ji S, Ye J. SLEP: sparse learning with efficient projections. Vol. 6. Arizona State University; 2009. p. 491. [Google Scholar]
- 11.Kohavi R, John GH. Wrappers for feature subset selection. Artif Intell. 1997;97:273–324. [Google Scholar]


