Abstract
Background
The cytochrome P450 monooxygenase (P450) superfamily is involved in the biosynthesis of various primary and secondary metabolites. However, little is known about the effects of whole genome duplication (WGD) and tandem duplication (TD) events on the evolutionary history and functional divergence of P450s in Brassica after splitting from a common ancestor with Arabidopsis thaliana.
Results
Using Hidden Markov Model search and manual curation, we detected that Brassica species have nearly 1.4-fold as many P450 members as A. thaliana. Most P450s in A. thaliana and Brassica species were located on pseudo-chromosomes. The inferred phylogeny indicated that all P450s were clustered into two different subgroups. Analysis of WGD event revealed that different P450 gene families had appeared after evolutionary events of species. For the TD event analyses, the P450s from TD events in Brassica species can be divided into ancient and recent parts. Our comparison of influence of WGD and TD events on the P450 gene superfamily between A. thaliana and Brassica species indicated that the family-specific evolution in the Brassica lineage can be attributed to both WGD and TD, whereas WGD was recognized as the major mechanism for the recent evolution of the P450 super gene family. Expression analysis of P450s from A. thaliana and Brassica species indicated that WGD-type P450s showed the same expression pattern but completely different expression with TD-type P450s across different tissues in Brassica species. Selection force analysis suggested that P450 orthologous gene pairs between A. thaliana and Brassica species underwent negative selection, but no significant differences were found between P450 orthologous gene pairs in A. thaliana–B. rapa and A. thaliana–B. oleracea lineages, as well as in different subgenomes in B. rapa or B. oleracea compared with A. thaliana.
Conclusions
This study is the first to investigate the effects of WGD and TD on the evolutionary history and functional divergence of P450 gene families in A. thaliana and Brassica species. This study provides a biology model to study the mechanism of gene family formation, particularly in the context of the evolutionary history of angiosperms, and offers novel insights for the study of angiosperm genomes.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4094-7) contains supplementary material, which is available to authorized users.
Keywords: Cytochrome P450s, Brassica, Tandem duplication, Whole genome duplication, Evolution mechanism, Expression profiling
Background
The cytochrome P450 monooxygenase (P450) superfamily comprises heme-thiolate proteins that are mostly attached to the cytoplasmic surface of the endoplasmic reticulum and are involved in different NADPH- and O2-dependent hydroxylation reactions [1]. P450 proteins serve diverse biological functions in plants. Many P450s are involved in the syntheses of pigments, antioxidants, structural polymers, and defense-related compounds, such as flavonoids, phenylpropanoids, phenolic esters, coumarins, alkaloids, terpenoids, lipids, cyanogenic glycosides, glucosinolates, benzoxazinones, and isoprenoids, whereas others are crucial for the synthesis and catabolism of plant growth regulators and signaling molecules. Similar to their animal counterparts, some P450s are also involved in the detoxification of pollutants, herbicides, and insecticides [2].
The first cytochrome P450 gene was found in the rat liver 56 years ago [3]. Subsequently, it was identified in all domains of life, such as plants, mammals, fish, insects, fungi, bacteria, and viruses. To date, more than 18,500 sequences of P450s have been reported from hundreds of species [4]. The universal guidelines for the nomenclature of P450 genes suggest that a newly discovered P450 protein sharing a sequence similarity greater than 40% to a previously identified P450 family should be grouped into the same family but should be classified under a new family when the sequence similarity is 40% or less. Similarly, same-family P450s with a sharing identity greater than 55% are grouped in the same subfamily but are separated when the sharing identity is less than 55%. In addition, P450s with a sharing identity of 97% or more are considered allelic variants of the same gene, and genes encoding the same P450 protein from different species are assigned to the same subfamily [5, 6]. According to the evolutionary relationship of P450s in different organisms, plant P450s are generally categorized into two groups: A type and non-A type. The first category includes P450s that are specific to plants and have evolved since the divergence of plants from animals and fungi; the second category includes non-plant-specific P450s that share the sequence similarity to animals and fungal enzymes [7]. Nowadays, in the current plant P450 phylogeny, plant P450 families are grouped into nine different clans, including CYP51, CYP71, CYP710, CYP711, CYP72, CYP74, CYP85, CYP86, and CYP97 clans [8]. CYP 71 clan was classified into A type P450s, and the remaining eight clans were classified into non-A type. These analyses brought critical information for studying the phylogeny of cytochrome P450 gene family.
So far, the available genome sequences in plants have provided more than 3000 P450 genes, making them a vast gene superfamily in plant genomes. For instance, previous studies reported approximate arrays of 356 cytochrome P450 genes in Oryza sativa, 312 in Populus trichocarpa, 457 in Vitis vinifera, 39 in Chlamydomonas reinhardtii, 71 in Physcomitrella patens, 225 in Selaginella moellendorffii, and 313 in Lotus japonicas [6, 9, 10]. The number of cytochrome P450 gene datasets is expected to increase with the development of genome sequencing technologies, which will allow researchers to study these genes from the perspective of comparative genomics or evolutionary biology.
During the past decade, the model plant Arabidopsis from the Brassicaceae family has broadened the understanding of plant biology, especially genome evolution in angiosperms. Evolutionary patterns suggest the occurrence of three rounds (i.e., γ, β, and α) of whole genome duplication or triplication (WGD or WGT) in A. thaliana and all Brassicaceae taxa [11]. The first WGT (γ) has been linked to the diversification of eudicots and angiosperms [12, 13].The second duplication (β), which was estimated to have occurred between 235 and 112 Mya [11, 14, 15]. The most recent α duplication occurred between 100 and 20 Mya [11, 16]. After the α duplication, Arabidopsis lineage separated from its Brassica ancestor approximately 43.2 MYA, and a WGT event was experienced in Brassica ancestor approximately 22.5 MYA [17]. Furthermore, two model Brassica diploid species, Brassica rapa and Brassica oleracea, diverged from a common ancestor about 3.75 MYA [16, 18–20] (Fig. 1). The evolutionary history of angiosperms enriches the content of cytochrome P450 gene evolution group, which provides a clear evolutionary roadmap for the evolutionary history of cytochrome P450 genes. Aside from WGD or WGT, tandem duplication (TD) is another important mechanism that creates tandem duplicated genes. Tandem duplicated genes reside within segments of DNA that are repeated head-to-tail a number of times and are necessary for the maintenance of large gene families for expanding and contracting rapidly in response to demand of changing environment [21]. Tandem duplicated genes represent a large portion of the total genes in a genome, i.e., 14–17% of animal genomes [22]. A previous report revealed that tandem duplicated genes can rapidly change in response to the needs of the environment, causing an increase in genetic complexity [23]. These two types of evolutionary mechanisms play important roles in the evolution of cytochrome P450 supergene family.
Fig. 1.
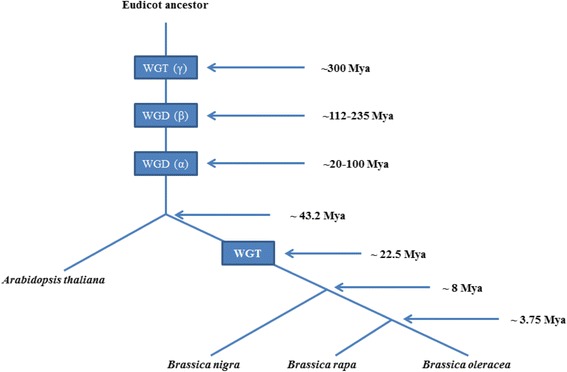
Ancestral polyploidy events and corresponding occurrence times in eudicots. Blue rectangles represent whole genome duplication or whole genome triplication events
To date, scholars have yet to elucidate the mechanism by which WGD and TD influence the expansion or reduction of cytochrome P450 gene families in the genus Brassica; they also have yet to identify which experienced stronger selection pressures of P450 orthologous gene pairs between or within A. thaliana–B. rapa and A. thaliana–B. oleracea lineages. The difference in expression patterns of cytochrome P450 gene families in Brassica species also remains undetermined. In this study, we identified the members of the cytochrome P450 gene superfamily in B. rapa and B. oleracea and compared them with those in A. thaliana on the basis of P450 gene superfamily size, genomic distribution, and phylogenetic analysis. The evolutionary history for the P450 gene super family and the contributions of WGD and TD to the evolution of different P450 gene families in A. thaliana and Brassica species were also investigated. We analyzed the expression patterns of different P450 gene families and the different types of P450s in different P450 gene families in Brassica species. According to the syntenic relationships between the A. thaliana and Brassica genomes, we detected the selection forces for inter- or intra-genome P450 gene pairs between or within A. thaliana–B. rapa and A. thaliana–B. oleracea lineages. Finally, we illustrated the evolution mechanisms of cytochrome P450 gene families in an attempt to uncover a clear evolutionary dynamic path that shaped this large and divergent gene superfamily accompanied with the evolutionary history of the genus Brassica.
Methods
Data resources
Genomic and annotation data for A. thaliana, B. rapa, and B. oleracea were retrieved from TAIR10 (http://www.arabidopsis.org) [24], BRAD (http://Brassicadb.org/brad/) [25], and Bolbase (http://ocri-genomics.org/bolbase) [26], respectively. The putative datasets of tandem duplicated genes in A. thaliana, B. rapa, and B. oleracea were downloaded from PTGBase (http://ocri-genomics.org/PTGBase/) [27]. The hidden Markov model (HMM) profile of the cytochrome P450 domain (PF00067) was retrieved from Pfam 26.0 (http://pfam.xfam.org/) [28]. The RNA-seq data of B. rapa and B. oleracea were obtained from the Gene Expression Omnibus database with accession numbers GSE43245 and GSE42891, respectively [29]. The microarray data of A. thaliana were obtained from AtGenExpress resource [30].
Sequence retrieval
Cytochrome P450 genes for the A. thaliana and Brassica genomes were identified through the HMM profile corresponding to the Pfam (p450) family PF00067 domain by using HMMER V3.0 [31], with the “trusted cutoff” as threshold. From the selected protein sequences filtered through the p450 domain, high-quality sequences between A. thaliana and Brassica species were aligned through ClustalW [32]. These sequences were then utilized to construct the species-specific HMM profile of cytochrome P450 by using the “hmmbuild” module of HMMER V3.0. With this species-specific HMM profile, all putative cytochrome P450 genes were identified in the A. thaliana and Brassica genomes. Finally, all cytochrome P450 genes in the A. thaliana and Brassica genomes were checked by using the conserved domain (Pfam: PF00067 and GENE3D: G3DSA:1.10.630.10) of cytochrome P450 genes.
Physical map location
On the basis of the general feature format (GFF) file of the predicted genes in the A. thaliana, B. rapa, and B. oleracea genomes, 12 in-house Perl and Python scripts were used to process the data and draw the graphic portrayal of cytochrome P450s on chromosomes or pseudo-molecules with the SVG module.
Alignments and phylogenetic analysis
For the phylogenetic analysis, the complete predicted protein sequences of cytochrome P450s that contain the full p450 domain in A. thaliana, B. rapa, and B. oleracea were aligned using MUSCLE with default options [33]. Using location of conserved P450 domains on P450 protein sequences, the alignment of the conserved domains of all cytochrome P450s among 3 species were processed through manual curation using Jalview [34]. The resulting alignment of conserved P450 domain of all P450s were used to construct a phylogenetic tree by using the maximum likelihood method with 1000 replications and the substitution model with the Jones–Taylor–Thornton model in MEGA 5.0 [35].
Statistical tests
Significant difference in the ratios of the rates of non-synonymous to synonymous substitutions (Ka/Ks) of P450 orthologous gene pairs between two data samples was evaluated using the Mann–Whitney U test in the R programming language.
RNA-seq data analysis of cytochrome P450s
For the expression values of cytochrome P450 genes, transcript abundance was calculated by fragments per kilobase of exon model per million mapped reads (FPKM) from CuffLinks v2.1.0, and the FPKM values were log2 transformed [36]. A hierarchical cluster was generated by using Cluster 3.0, and a heat map was displayed using TreeView 1.60 [37].
Substitution rate estimates for P450s
The ratio of the rates of non-synonymous to synonymous substitutions (Ka/Ks) of all P450 gene pairs was evaluated for each branch of the phylogenetic tree by using PAML to estimate the selection mode for the P450 orthologous gene pairs in A. thaliana compared with B. rapa and B. oleracea [38]. For each subtree of P450 orthologous gene pairs between A. thaliana–B. rapa and A. thaliana–B. oleracea lineages, model 1 with Ka/Ks ratios was evaluated separately for the branches. The Ka/Ks values with terminal branches between modern species and their most recent reconstructed ancestors were used for the subsequent analyses.
Results
Brassica species has a larger P450 gene superfamily than A. thaliana
We identified the members of the P450 super gene family from the released protein sequences of A. thaliana, B. rapa, and B. oleracea to process an in-depth comparison between the cytochrome P450 gene family in A. thaliana and Brassica species. According to the conserved domains or motifs (Pfam: PF00067 and G3DSA: 1.10.630.10) of cytochrome P450 genes, we extracted 251 P450 genes from 27,416 A. thaliana representative protein-encoding genes based on TAIR10 genome release, which represented 0.916% of total representative protein-encoding genes in A. thaliana; these genes were distributed into 47 P450 gene families [28, 39]. Using consistent procedures, we identified 354 and 343 P450 genes from 41,174 B. rapa and 45,758 B. oleracea protein-encoding genes which represented 0.8598 and 0.7496% of total protein-encoding genes in B. rapa and B. oleracea, respectively. From comparison, P450s in B. rapa and B. oleracea were 1.41- and 1.37-fold as many as those in A. thaliana (Additional file 1: Table S1). After manually curation and combing the classification criteria of cytochrome P450 by previous studies, we classified the 948 P450 genes into 47 P450 gene families among the three species [4]. The Evolutionary analysis of P450s in different organisms revealed that P450 gene families are grouped into two classes: non-A type and A-type [7]. P450 gene families of non-A type share a sequence similarity to analogs in animal and fungi, but those of A-type are plant specific; the second type was generated after the divergence of plants from animals and fungi. The identified 948 P450s among the three species were classified into two classes. In A. thaliana, 94 and 157 P450s were clustered into non-A type and A-type, respectively. For Brassica species, 143 B. rapa genes and 136 B. oleracea genes were classified into non-A type and further divided into 28 families; the remaining 211 B. rapa and 207 B. oleracea genes were grouped into A-type P450s, which were divided into 19 families (Table 1). Comparison of A-type and non-A type P450s in Brassica species revealed the greater number of P450s in A-type than in non-A type, which suggested that plant-specific P450s underwent family member evolution unlike the ancestral P450s shared with animal and fungi in Brassica species. Due to classification of different clans of plant P450 families, 47 P450 gene families were classified into nine different clans of P450 gene families among A. thaliana, B. rapa and B. oleracea.
Table 1.
Statistics of cytochrome P450 gene families in A. thaliana and Brassica species
| Classifications | Family | Arabidopsis thaliana | Brassica rapa | Brassica oleracea | |
|---|---|---|---|---|---|
| Different types | Different clans | ||||
| Non-A type | CYP51 clan | CYP51 | 1 | 3 | 5 |
| CYP72 clan | CYP714 | 2 | 1 | 2 | |
| CYP715 | 1 | 4 | 3 | ||
| CYP709 | 3 | 6 | 7 | ||
| CYP72 | 9 | 18 | 6 | ||
| CYP721 | 1 | 1 | 1 | ||
| CYP734 | 1 | 1 | 1 | ||
| CYP735 | 2 | 3 | 2 | ||
| CYP710 clan | CYP710 | 4 | 3 | 2 | |
| CYP711 clan | CYP711 | 1 | 1 | 1 | |
| CYP74 clan | CYP74 | 2 | 2 | 2 | |
| CYP85 clan | CYP702 | 6 | 4 | 3 | |
| CYP707 | 4 | 8 | 9 | ||
| CYP708 | 4 | 6 | 3 | ||
| CYP716 | 3 | 0 | 0 | ||
| CYP718 | 1 | 1 | 1 | ||
| CYP720 | 1 | 1 | 1 | ||
| CYP722 | 1 | 1 | 2 | ||
| CYP724 | 1 | 2 | 2 | ||
| CYP85 | 2 | 4 | 4 | ||
| CYP87 | 1 | 2 | 2 | ||
| CYP88 | 2 | 2 | 2 | ||
| CYP90 | 4 | 9 | 9 | ||
| CYP86 clan | CYP704 | 3 | 5 | 4 | |
| CYP86 | 11 | 13 | 14 | ||
| CYP94 | 6 | 10 | 10 | ||
| CYP96 | 14 | 30 | 32 | ||
| CYP97 clan | CYP97 | 3 | 2 | 6 | |
| Total non-A type | 94 | 143 | 136 | ||
| A-type | CYP71 clan | CYP701 | 1 | 1 | 1 |
| CYP703 | 1 | 2 | 2 | ||
| CYP705 | 25 | 22 | 3 | ||
| CYP706 | 8 | 10 | 9 | ||
| CYP71 | 50 | 73 | 77 | ||
| CYP712 | 2 | 2 | 2 | ||
| CYP73 | 1 | 6 | 5 | ||
| CYP75 | 1 | 1 | 1 | ||
| CYP76 | 9 | 12 | 12 | ||
| CYP77 | 5 | 5 | 5 | ||
| CYP78 | 6 | 11 | 12 | ||
| CYP79 | 10 | 11 | 11 | ||
| CYP81 | 18 | 32 | 37 | ||
| CYP82 | 5 | 4 | 9 | ||
| CYP83 | 2 | 6 | 6 | ||
| CYP84 | 2 | 4 | 6 | ||
| CYP89 | 7 | 4 | 3 | ||
| CYP93 | 1 | 1 | 2 | ||
| CYP98 | 3 | 4 | 4 | ||
| Total A-type | 157 | 211 | 207 | ||
| Total | 251 | 354 | 343 | ||
Genomic distribution on chromosomes/pseudo-molecular chromosomes
According to the GFF file of cytochrome P450 genes between the A. thaliana and Brassica genomes, 251 (100%) P450s in A. thaliana, 337 (95.2%) in B. rapa, and 281 (81.92%) in B. oleracea were distributed on chromosomes or pseudo-molecule chromosomes, and the remaining 17 (4.8%) P450s in B. rapa and 62 (18%) P450s in B. oleracea were distributed on unanchored contigs or scaffolds [25, 26, 39]. For the distribution of P450s in the A. thaliana genome, Bak et al. finished this analysis based on previous analysis results of P450s [40]. In present study, Chr3 contained the greatest number of P450s (e.g., 68; 27.1% of the total P450s) among all the studied chromosomes, a consistent with Dr. Bak’s reports [40]. In the B. rapa genome, A03 (53; 15.63%) and A10 (16; 4.72%) contained the greatest and the least numbers of P450s, respectively. For the B. oleracea genome, C07 (55; 19.43%) and C05 (12; 4.24%) carried the greatest and the least numbers of P450s, respectively. The distribution of the members of P450 gene families on chromosomes or pseudo-molecules did not exhibit any of the same or similar regularity. For the members of the CYP72 gene family in the A. thaliana genome, 8 of 9 P450s were located on Chr2; the remaining 1 P450 was located on Chr1. For the members of the CYP72 gene family in the B. rapa genome, 18 P450s were located on six chromosomes (6 P450s on A05, 5 P450s on A03, 2 P450s in A01 and A06, 1 on each P450 on A08 and A09, and only 1 P450 on unanchored contigs). For the 5 members of the CYP72 gene family in the B. oleracea genome, 3 P450s were located on C08, 1 unanchored on chromosomes, and another was on C05 (Additional file 2: Figure S1).
Phylogenetic analysis of cytochrome P450 genes in B. rapa, B. oleracea, and A. thaliana
On the basis of the phylogenetic analysis of cytochrome P450 genes among A. thaliana, B. rapa, and B. oleracea, 948 P450s were clustered into groups I and II (Additional file 3: Figure S2). Group I contained all non-A type P450 gene families, which were clustered into CYP51 clan, CYP710 clan, CYP711 clan, CYP72 clan, CYP74 clan, CYP85 clan, CYP86 clan, and CYP97 clan. Group II contained all A type P450 gene families, which were clustered into CYP71 clan. These results were consistent with previous study [8, 40]. Group I included 94, 143, and 134 P450s in A. thaliana, B. rapa, and B. oleracea, respectively, which covered 29 P450 gene families. Among the 29 P450 gene families, the CYP96 gene family (belonged to CYP86 clan) contained the most number of P450s in the three species, i.e., 14, 30, and 31 P450s in A. thaliana, B. rapa, and B. oleracea, respectively. The CYP716 gene family (belonged to CYP85 clan) had 3 P450s in A. thaliana based on TAIR10 but none in B. rapa and B. oleracea. This finding suggests that this family is lost in B. rapa and B. oleracea. There were two single specific family subgroups detected in group I; each of them contained one P450 gene in B. oleracea, which were Bol038708 and Bol016308, respectively. Analysis of protein sequences of these two P450s revealed these two P450s contained partial sequence of p450 conserved domain. Based on the basis of the classification criteria of P450 gene families, Bol038708 was classified into CYP82 gene family (belonged to CYP71 clan), and Bol016308 was classified into CYP96 gene family (belonged to CYP86 clan) in B. oleracea.
The A-type of P450 gene families (recognized as CYP71 clan) was classified into group II, which included 577 P450 genes among the three species. These P450 genes in II group were clustered into 23 P450 gene families, which represented 62.55% (157), 59.6% (211), and 60.93% (209) of the total P450s in A. thaliana, B. rapa, and B. oleracea, respectively. Of the 23 P450 gene families in group II, the CYP71 gene family, the largest A-type P450 gene family, contained 50, 73, and 77 P450 genes in A. thaliana, B. rapa, and B. oleracea, respectively. For the phylogenetic relationship of the members in the CYP71 gene family, members of the CYP83 and CYP71 gene families were clustered together. Members of the CYP83 gene family exhibited a closer evolutionary relationship than some members of the CYP71 gene family, indicating that members of the former showed closer relative relationship than some members of the latter. Except for clusters from 23 P450 gene families, one single specific family subgroup compared to A. thaliana was detected in the A-type P450 gene families, which contained 1 P450 gene (Bra014459) in B. rapa. Concerning Bra014459, sequence analysis revealed that it was formed by fragments of two different conserved domains: the first 125 amino acids correspond to a L-arabinokinase-like protein (“Glyco_trans_1–3 superfamily” domain), and the rest of the sequence correspond to a partial P450 protein. There was another single specific family subgroups existed in group II; this group contained 3 P450s (Bol042895, Bol009797, and Bol016015) in B. oleracea. These three P450s also contained partial sequences of p450 conserved domain of standard P450 protein through analysis of protein sequences. According to the basis of the classification criteria of P450 gene families, Bra014459 was classified into CYP76 gene family (belonged to CYP71 clan), and the three P450s (Bol042895, Bol009797, and Bol016015) from single specific family subgroup were classified into CYP709 (belonged to CYP72 clan), CYP72 (belonged to CYP72 clan), CYP96 (belonged to CYP86 clan) gene families in B. oleracea respectively.
Synteny analysis of P450s between A. thaliana and Brassica species
A. thaliana and all the Brassicaceae taxa have experienced three rounds (i.e., γ, β, and α) of WGD or WGT in the genome evolutionary history of angiosperms. To trace the evolutionary history of the cytochrome P450 gene superfamily, we used the duplicated genes from three WGD or WGT events in A. thaliana [11]. After curation, we obtained 3530, 1421, and 505 syntenic gene pairs in the A. thaliana genome from the α, β, and γ WGD or WGT events, respectively. According to the new curated duplicated gene datasets based on TAIR10 genome release, only one P450 gene pair from the CYP94 gene family was retained after the γ WGT event, indicating that these two members of the CYP94 gene family existed before the γ WGD of eudicot ancestor. For the β WGD, we obtained three cytochrome P450 gene pairs distributed in the CYP86, CYP72, and CYP81 gene families, implying that these cytochrome P450 genes existed before the β WGD and retained the duplicated gene pairs after the β WGD. For duplicated gene pairs of the α WGD, we retrieved 22 P450 genes from the syntenic gene pairs caused by the α WGD, which were distributed in 11 P450 gene families. This result suggests that these 22 cytochrome P450 genes were retained after the most recent α duplication of Brassicaceae.
After the split of A. thaliana and Brassica from a common ancestor, the Brassica ancestor experienced WGT and then diverged to yield the modern B. rapa and B. oleracea. The WGT brought triplicated genomic regions in the B. rapa and B. oleracea genomes. After the completion of the B. rapa and B. oleracea genome sequencing project, the triplicated regions were constructed in the B. rapa and B. oleracea genomes relating to the 24 ancestral crucifer blocks (A–X) in the A. thaliana genome [41, 42]. On the basis of the difference in gene density, the triplicated blocks in the two Brassica species were classified into three subgenomes: MF1 (medium fractionated), MF2 (most fractionated), and LF (least fractionated) [41]. According to the above-mentioned classification criteria of syntenic blocks, we reconstructed the triplicated regions in the B. rapa and B. oleracea genomes compared with that in the A. thaliana genome. On the basis of the syntenic relationships between A. thaliana and Brassica species, 131 A. thaliana P450s were found orthologous in Brassica species, which were distributed in 42 cytochrome P450 gene families. Then, 124 and 118 A. thaliana P450s were orthologous in 187 and 178 P450s in B. rapa and B. oleracea, respectively. From the syntenic regions in B. rapa and B. oleracea compared with A. thaliana, 162 orthologous gene pairs were identified between the B. rapa and B. oleracea genomes (Fig. 2). Of the 120 P450s in A. thaliana, 11 cytochrome P450s were detected in 3 orthologous genes and 41 P450s were detected in 2 orthologous genes in the B. rapa genome. The remaining 72 genes were detected in only one orthologous gene in the B. rapa genome. For 118 P450s in A. thaliana compared with the B. oleracea genome, 10, 39, and 69 A. thaliana P450s showed 3, 2, and 1 orthologous gene (s) in the B. oleracea genome, respectively (Additional file 4: Table S2).
Fig. 2.
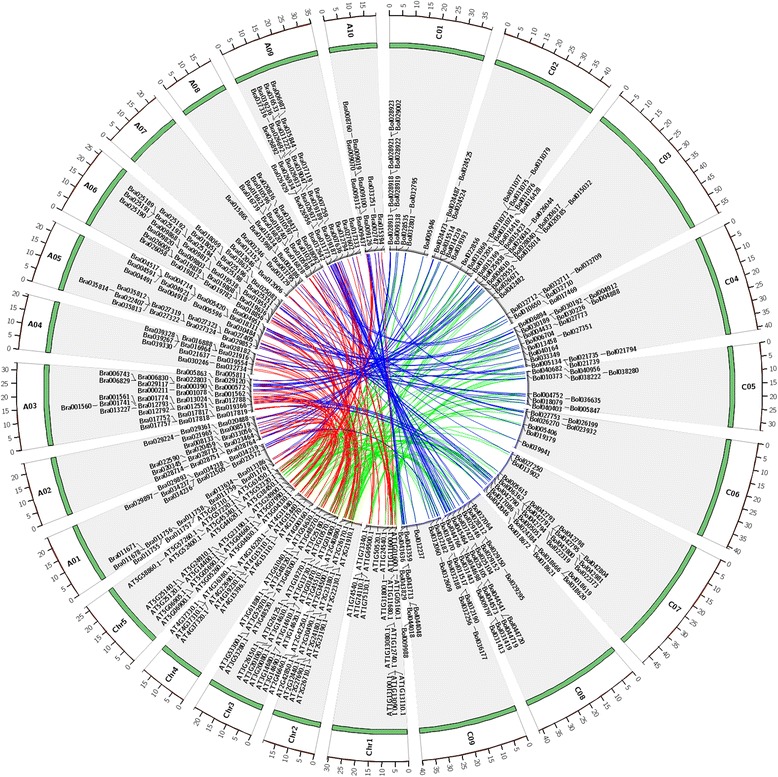
Syntenic relationships of cytochrome P450 genes between A. thaliana and Brassica species. Green bars represent pseudochromosomes of three species. A01–A10 represent pseudochromosomes in B. rapa, C01–C09 represent pseudochromosomes in B. oleracea, and Chr1–Chr5 represent pseudochromosomes in A. thaliana. Red lines represent the collinear relationships of P450 gene pairs between A. thaliana and B. rapa. Blue lines represent the collinear relationships of P450 gene pairs between B. rapa and B. oleracea. Green lines represent the collinear relationships of P450 gene pairs between A. thaliana and B. oleracea
According to the syntenic relationship between A. thaliana and Brassica species, the duplicated gene pairs from different polyploidy events in A. thaliana were orthologous in different subgenomes in Brassica species. One duplicated gene pair of the CYP94 gene family (AT2G27690.1 and AT3G56630.1) from the γ WGT was detected in 6 corresponding orthologous genes in Brassica species. AT2G27690.1 was detected in 1 orthologous gene and AT3G56630.1 was detected in 2 orthologous genes in different subgenomes in B. rapa or B. oleracea. For the β WGD, three duplicated gene pairs were detected in 21 orthologous P450 genes in Brassica species. For example, one member (AT1G17060.1) of the duplicated gene pairs (AT1G17060.1 and AT3G14690.1) from the CYP72 family was detected in 3 orthologous P450 genes in B. rapa and B. oleracea, but another P450 gene (AT3G14690.1) was detected in 3 orthologous P450 genes in B. rapa and 1 orthologous P450 gene in B. oleracea. This result indicates that these two A. thaliana P450s showed significant expansion in the genus Brassica. The 11 duplicated gene pairs from the α WGD were also detected in 63 orthologous P450 genes in Brassica species, and each duplicated gene has its corresponding P450s in B. rapa or B. oleracea. Of the 22 duplicated genes in A. thaliana, only AT4G37370.1 and AT5G45340.1 from the CYP81 and CYP707 gene families showed 3 copies that were retained in B. rapa and B. oleracea, respectively; other A. thaliana P450s showed 1 or 2 copies that were retained in Brassica species (Table 2).
Table 2.
Retention or loss of P450 orthologous gene pairs between A. thaliana and Brassica species from different polyploidy events
| Polyploidy events | Brassica rapa | Brassica oleracea | Gene | Gene name | Family | Orthologous genes | Gene name | Brassica rapa | Brassica oleracea | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BraLF | BraMF1 | BraMF2 | BolLF | BolMF1 | BolMF2 | BraLF | BraMF1 | BraMF2 | BolLF | BolMF1 | BolMF2 | ||||||
| α | Bra033251 | Bra032642 | NA | Bol040682 | Bol018470 | NA | AT1G01600.1 | CYP86A4 | CYP86 | AT4G00360.1 | CYP86A2 | Bra037336 | NA | Bra008519 | Bol011517 | NA | Bol040623 |
| Bra015394 | NA | NA | Bol040956 | NA | NA | AT1G05160.1 | CYP88A3 | CYP88 | AT2G32440.1 | CYP88A4 | Bra005596 | NA | NA | Bol013458 | NA | NA | |
| Bra018856 | NA | Bra030484 | NA | NA | NA | AT1G50520.1 | CYP705A27 | CYP705 | AT3G20130.1 | CYP705A22 | Bra035812 | NA | NA | Bol004752 | NA | NA | |
| NA | Bra034021 | NA | NA | NA | NA | AT1G67110.1 | CYP735A2 | CYP735 | AT5G38450.1 | CYP735A1 | NA | NA | NA | Bol044541 | NA | NA | |
| NA | Bra031222 | NA | NA | Bol044766 | NA | AT2G21910.1 | CYP96A5 | CYP96 | AT4G39490.1 | CYP96A10 | Bra007636 | NA | NA | Bol044573 | Bol037477 | NA | |
| Bra030246 | NA | NA | Bol031784 | NA | NA | AT2G22330.1 | CYP79B3 | CYP79 | AT4G39950.1 | CYP79B2 | Bra037336 | NA | Bra008519 | Bol011517 | NA | Bol040623 | |
| NA | Bra039189 | Bra000572 | NA | Bol044719 | Bol015032 | AT2G23190.1 | CYP81D7 | CYP81 | AT4G37370.1 | CYP81D8 | Bra013634 | Bra019366 | Bra020836 | Bol028535 | Bol042046 | Bol010650 | |
| NA | Bra039330 | NA | NA | Bol021735 | NA | AT2G45550.1 | CYP76C4 | CYP76 | AT3G61040.1 | CYP76C7 | NA | NA | Bra010214 | NA | NA | Bol017469 | |
| Bra004517 | Bra039267 | NA | Bol004912 | Bol021794 | NA | AT2G46660.1 | CYP78A6 | CYP78 | AT3G61880.1 | CYP78A9 | NA | NA | Bra005811 | NA | NA | Bol015369 | |
| Bra029852 | NA | NA | Bol005847 | NA | NA | AT3G10570.1 | CYP77A6 | CYP77 | AT5G04660.1 | CYP77A4 | Bra028182 | NA | NA | Bol040164 | NA | NA | |
| Bra013386 | Bra012551 | NA | Bol009338 | Bol024384 | NA | AT4G19230.1 | CYP707A1 | CYP707 | AT5G45340.1 | CYP707A3 | Bra025083 | Bra027602 | Bra021965 | Bol005615 | Bol032060 | Bol022856 | |
| β | Bra026005 | Bra026692 | Bra016621 | Bol038280 | Bol009797 | Bol029295 | AT1G17060.1 | CYP72C1 | CYP72 | AT3G14690.1 | CYP72A15 | Bra027319 | Bra021572 | Bra001562 | Bol002804 | NA | NA |
| Bra011755 | NA | NA | Bol028923 | NA | NA | AT4G37310.1 | CYP81H1 | CYP81 | AT5G67310.1 | CYP81G1 | Bra012152 | Bra037119 | NA | Bol027250 | NA | NA | |
| Bra033251 | Bra032642 | NA | Bol040682 | Bol018470 | NA | AT1G01600.1 | CYP86A4 | CYP86 | AT2G45970.1 | CYP86A8 | Bra004951 | NA | NA | Bol030226 | NA | NA | |
| γ | NA | Bra012006 | NA | NA | Bol013448 | NA | AT2G27690.1 | CYP94C1 | CYP94 | AT3G56630.1 | CYP94D2 | Bra007259 | NA | Bra003246 | Bol045464 | NA | Bol004209 |
Contributions of tandem duplication to family size
Gene families organized in tandem arrays of two or more units have been described in A. thaliana, B. rapa, and B. oleracea [41–43]. The putative tandem duplicated genes of cytochrome P450 in A. thaliana, B. rapa, and B. oleracea were extracted from PTGBase (http://ocri-genomics.org/PTGBase/) [27]. According to the classification criteria of cytochrome P450 gene families, we curated the putative datasets of tandem duplicated genes among the three species. We obtained 128, 137, and 112 tandem duplicated P450 genes in A. thaliana, B. rapa, and B. oleracea, respectively (Additional file 5: Table S3). Analysis of P450s in the A. thaliana genome revealed that 128 P450s, representing 51% of the total P450s in A. thaliana, were generated by TD and were distributed in 40 tandem arrays with 2–9 P450s. Among the 128 cytochrome P450s in the A. thaliana genome, 39 P450s were members of the CYP71 gene family, representing 30.47% of the total tandem duplicated genes of cytochrome P450s in the A. thaliana genome. This result indicates that the CYP71 gene family was influenced more strongly by the TD event compared with the other P450 gene families in A. thaliana. Among the 354 B. rapa P450 genes, 137 (38.70%) were generated by TD and were distributed in 47 tandem arrays with 2–10 P450s. All tandem duplicated P450s in the B. rapa genome were distributed in 18 gene families. The CYP71 gene family in B. rapa contained 44 tandem duplicated P450s, representing 12.36% of the total tandem duplicated P450s, which contained most of the tandem duplicated P450s compared with the other P450 gene families in B. rapa. In the B. oleracea genome, 112 of the total 343 identified P450 genes, representing 32.65% of the total B. oleracea cytochrome P450s, were detected in 40 tandem arrays of 2–6 genes. Similarly, the CYP71 gene family also contained more tandem duplicated P450 genes compared with other P450 families in B. oleracea. These results suggest that the P450 genes in A. thaliana were influenced more strongly by the TD event compared with the other genes in Brassica species and that the CYP71 gene family was influenced more strongly by the TD event compared with the other gene families among the three species (Table 3).
Table 3.
Tandem duplicated genes of cytochrome P450 in A. thaliana, B. rapa, and B. oleracea
| Categories | Arabidopsis thaliana | Brassica rapa | Brassica oleracea |
|---|---|---|---|
| CYP702 | 4 | 2 | 0 |
| CYP704 | 0 | 2 | 0 |
| CYP705 | 19 | 7 | 0 |
| CYP706 | 6 | 2 | 2 |
| CYP708 | 0 | 2 | 0 |
| CYP709 | 2 | 4 | 5 |
| CYP71 | 39 | 44 | 40 |
| CYP710 | 4 | 2 | 0 |
| CYP714 | 2 | 0 | 0 |
| CYP715 | 0 | 2 | 2 |
| CYP716 | 2 | 0 | 0 |
| CYP72 | 8 | 13 | 4 |
| CYP73 | 0 | 4 | 4 |
| CYP76 | 8 | 7 | 7 |
| CYP77 | 4 | 0 | 0 |
| CYP78 | 0 | 2 | 0 |
| CYP79 | 6 | 0 | 0 |
| CYP81 | 10 | 20 | 23 |
| CYP82 | 3 | 0 | 2 |
| CYP83 | 0 | 2 | 2 |
| CYP84 | 0 | 0 | 2 |
| CYP86 | 2 | 0 | 0 |
| CYP89 | 3 | 2 | 0 |
| CYP94 | 0 | 2 | 2 |
| CYP96 | 4 | 18 | 17 |
| CYP98 | 2 | 0 | 0 |
| Total tandem duplicated genes | 128 | 137 | 112 |
| Total P450 genes | 251 | 354 | 343 |
| Percentage | 51% | 38.7% | 32.65% |
We traced the ancient tandem duplicated genes of cytochrome P450 before the split of A. thaliana and its Brassica ancestor to investigate the effects of WGD on the tandem duplicated P450s. Basing from the syntenic relationships among the B. rapa, B. oleracea, and A. thaliana genomes, we used syntenic gene pairs among the three species to classify the ancient units of tandem duplicated genes. After the curation of orthologous gene pairs among the three species, we detected that 24 A. thaliana P450s, distributed in tandem arrays of 2–6 P450 genes, obtained 28 and 17 corresponding orthologous genes in the B. rapa and B. oleracea genomes, respectively, which were located on different subgenomes in the Brassica genomes. Among the 24 A. thaliana P450s, 12 race specific P450s co-retained the corresponding orthologous P450 genes in the Brassica genomes after the TD event, and 12 lineage-specific P450s co-retained 1 corresponding orthologous P450 gene in the B. rapa or B. oleracea genome. These ancient units of tandem duplicated genes have existed before the split of A. thaliana and its Brassica ancestor and have been retained after the divergence of B. rapa and B. oleracea. For 128 P450s in A. thaliana, 24 P450s existed before the divergence of A. thaliana and its Brassica ancestor, and 104 P450s were species-specific tandem duplicated genes in the A. thaliana genome. This result indicates that cytochrome P450 gene families experienced species-specific expansion by TD after species divergence. A total of 109 and 95 P450s in the B. rapa and B. oleracea genomes, respectively, were found to be species specific after excluding the ancient tandem duplicated P450s from the two species. The tandem duplicated P450 genes in the B. rapa and B. oleracea genomes were composed of two parts. One part contained the ancient P450s generated before the split of A. thaliana from its Brassica ancestor; the other part contained the recent P450 members generated after the divergence of B. rapa and B. oleracea from a common ancestor (Table 4).
Table 4.
Ancient tandem arrays of cytochrome P450 genes in A. thaliana compared with B. rapa and B. oleracea
| Block | AGI | Gene name | Family | Location | Brassica rapa | Brassica oleracea | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| BraLF | BraMF1 | BraMF2 | BolLF | BolMF1 | BolMF2 | |||||
| A | AT1G13080.1 | CYP71B2 | CYP71 | Chr1 | / | / | / | / | Bol031411 | / |
| A | AT1G13090.1 | CYP71B28 | CYP71 | Chr1 | / | Bra026934 | / | / | Bol031412 | / |
| A | AT1G13100.1 | CYP71B29 | CYP71 | Chr1 | / | Bra026933 | / | / | Bol031413 | / |
| F | AT3G14610.1 | CYP72A7 | CYP72 | Chr3 | Bra027324 | / | / | / | / | / |
| F | AT3G14620.1 | CYP72A8 | CYP72 | Chr3 | Bra027323 | / | / | / | / | / |
| F | AT3G14630.1 | CYP72A9 | CYP72 | Chr3 | Bra027322 | / | / | / | / | / |
| F | AT3G14680.1 | CYP72A14 | CYP72 | Chr3 | / | / | Bra001561 | / | / | / |
| F | AT3G14690.1 | CYP72A15 | CYP72 | Chr3 | / | / | Bra001562 | / | / | / |
| F | AT3G20130.1 | CYP705A22 | CYP705 | Chr3 | Bra035812 | / | / | / | / | / |
| F | AT3G20140.1 | CYP705A23 | CYP705 | Chr3 | Bra035813 | / | / | / | / | / |
| L | AT3G26170.1 | CYP71B19 | CYP71 | Chr3 | Bra025189 | / | / | Bol042791 | / | / |
| L | AT3G26180.1 | CYP71B20 | CYP71 | Chr3 | Bra025190 | / | / | Bol042790 | / | / |
| L | AT3G26190.1 | CYP71B21 | CYP71 | Chr3 | Bra025191 | / | / | Bol042793 | / | / |
| L | AT3G26200.1 | CYP71B22 | CYP71 | Chr3 | Bra025192 | Bra036345 | / | Bol042794 | Bol031073 | / |
| L | AT3G26210.1 | CYP71B23 | CYP71 | Chr3 | Bra025193 | Bra036344 | / | Bol042795 | Bol031074 | / |
| L | AT3G26220.1 | CYP71B3 | CYP71 | Chr3 | Bra025194 | Bra036343 | / | / | Bol031075 | / |
| L | AT3G26280.1 | CYP71B4 | CYP71 | Chr3 | / | / | Bra034236 | / | / | / |
| L | AT3G26290.1 | CYP71B26 | CYP71 | Chr3 | / | / | Bra034237 | / | / | / |
| L | AT3G26320.1 | CYP71B36 | CYP71 | Chr3 | / | / | Bra034238 | / | / | / |
| L | AT3G26330.1 | CYP71B37 | CYP71 | Chr3 | / | / | Bra034239 | / | / | / |
| U | AT4G37320.1 | CYP81D5 | CYP81 | Chr4 | / | Bra017817 | / | / | Bol018621 | / |
| U | AT4G37330.1 | CYP81D4 | CYP81 | Chr4 | / | Bra017818 | / | / | Bol018620 | / |
| U | AT4G37400.1 | CYP81F3 | CYP81 | Chr4 | Bra011758 | / | Bra010597 | Bol028919 | / | Bol032711 |
| U | AT4G37410.1 | CYP81F4 | CYP81 | Chr4 | Bra011759 | / | Bra010598 | Bol028918 | / | Bol032712 |
Influence of WGD and TD events on the P450 gene superfamily
After the divergence of A. thaliana and Brassica lineage from a common ancestor, cytochrome P450 gene families experienced different evolutionary patterns under environmental selection pressure. The WGD and TD events played important roles in the evolution of cytochrome P450 gene families. A previous study revealed that all A. thaliana orthologous genes show a 1.2-fold retention rate in syntenic regions in Brassica species through syntenic analysis between the A. thaliana and Brassica genomes [42]. Comparison of the retention rates of P450 orthologous genes in A. thaliana with Brassica genomes revealed that 1.5-fold A. thaliana P450s were retained in different subgenomes in Brassica species, which was higher than the retention rate of all A. thaliana orthologous genes in syntenic regions in Brassica species. This result suggests that WGD or WGT events caused significant increase of the members of P450 gene families in Brassica species. In the present study, we compared the influences of WGD and TD events on P450 gene families. Comparison of the influences of WGD events on P450s in Brassica species demonstrated that 124 and 117 A. thaliana P450s existed in 187 and 177 orthologous P450 genes in B. rapa and B. oleracea, respectively. This result reflects an identical retention rate of 1.5-fold of A. thaliana orthologous genes in B. rapa and B. oleracea. These P450s influenced by WGD represented 52.82 and 51.6% of the total P450s in B. rapa and B. oleracea, respectively. Analysis of influences of TD on P450s among the three species revealed that 128, 137, and 112 P450s in A. thaliana, B. rapa, and B. oleracea were generated by TD, representing 51% (251), 38.7% (354), and 32.65% (343) of the total P450s in the three species, respectively. These results indicated that WGD caused more influence on the evolution of P450 gene families than TD in the Brassica lineage and that TD exerted more influence on the evolution of P450 gene families in A. thaliana than in Brassica species.
For each P450 gene family, we summarized the members of P450 gene families that were generated by WGT or TD events based on the two classes: non-A type and A-type. Of the 28 non A-type P450 gene families, 22 (8 families in A. thaliana, 9 families in B. rapa, and 5 families in B. oleracea) were influenced by TD, but 23 and 24 P450 gene families were influenced by WGD in B. rapa and B. oleracea, respectively. Of the 19 A-type P450 gene families, 10, 9, and 8 P450 gene families were influenced by TD in A. thaliana, B. rapa, and B. oleracea, respectively, but 18 P450 gene families were influenced by WGD in each B. rapa and B. oleracea. Of the 137 and 112 tandem duplicated P450 genes in B. rapa and B. oleracea, 90 and 82 were distributed in A-type P450 supergene families, which represented 65.69 and 73.21% of the total tandem duplicated P450 genes in Brassica species, respectively. Among the 187 and 177 P450s from WGD in B. rapa and B. oleracea, 57.75 and 59.89% were distributed in A-type P450 supergene families. These results indicate that A-type P450 gene families were influenced more strongly by TD and WGD compared with non-A type P450 gene families in the Brassica lineage (Table 5).
Table 5.
Comparison of cytochrome P450 genes in different types among A. thaliana, B. rapa, and B. oleracea
| Classification | Family | Arabidopsis thaliana | Brassica rapa | Brassica oleracea | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Different types | Different clans | Total P450s | P450s from WGD | P450s from TD | Total P450s | P450s from WGD | P450s from TD | Total P450s | P450s from WGD | P450s from TD | |
| Non-A Type | CYP51 clan | CYP51 | 1 | 1 | 0 | 3 | 2 | 0 | 5 | 2 | 0 |
| CYP72 clan | CYP714 | 2 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 0 | |
| CYP715 | 1 | 1 | 0 | 4 | 2 | 2 | 3 | 2 | 2 | ||
| CYP709 | 3 | 1 | 2 | 6 | 1 | 4 | 7 | 1 | 5 | ||
| CYP72 | 9 | 6 | 8 | 18 | 11 | 13 | 6 | 5 | 4 | ||
| CYP721 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| CYP734 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| CYP735 | 2 | 2 | 0 | 3 | 2 | 0 | 2 | 1 | 0 | ||
| CYP710 clan | CYP710 | 4 | 1 | 4 | 3 | 2 | 2 | 2 | 1 | 0 | |
| CYP711 clan | CYP711 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | |
| CYP74 clan | CYP74 | 2 | 1 | 0 | 2 | 1 | 0 | 2 | 1 | 0 | |
| CYP85 clan | CYP702 | 6 | 2 | 4 | 4 | 3 | 2 | 3 | 1 | 0 | |
| CYP707 | 4 | 3 | 0 | 8 | 7 | 0 | 9 | 7 | 0 | ||
| CYP708 | 4 | 0 | 0 | 6 | 0 | 2 | 3 | 0 | 0 | ||
| CYP716 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| CYP718 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| CYP720 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| CYP722 | 1 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | ||
| CYP724 | 1 | 1 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | ||
| CYP85 | 2 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | ||
| CYP87 | 1 | 1 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | ||
| CYP88 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | ||
| CYP90 | 4 | 3 | 0 | 9 | 7 | 0 | 9 | 7 | 0 | ||
| CYP86 clan | CYP704 | 3 | 2 | 0 | 5 | 3 | 2 | 4 | 3 | 0 | |
| CYP86 | 11 | 9 | 2 | 13 | 13 | 0 | 14 | 13 | 0 | ||
| CYP94 | 6 | 5 | 0 | 10 | 9 | 2 | 10 | 9 | 2 | ||
| CYP96 | 14 | 3 | 4 | 30 | 3 | 18 | 32 | 2 | 17 | ||
| CYP97 clan | CYP97 | 3 | 2 | 0 | 2 | 2 | 0 | 6 | 3 | 0 | |
| Total non-A type | 94 | 53 | 28 | 143 | 79 | 47 | 136 | 71 | 30 | ||
| A Type | CYP71 clan | CYP701 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| CYP703 | 1 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | ||
| CYP705 | 25 | 5 | 19 | 22 | 6 | 7 | 3 | 2 | 0 | ||
| CYP706 | 8 | 2 | 6 | 10 | 3 | 2 | 9 | 4 | 2 | ||
| CYP71 | 50 | 28 | 39 | 73 | 37 | 44 | 77 | 35 | 40 | ||
| CYP712 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | ||
| CYP73 | 1 | 1 | 0 | 6 | 3 | 4 | 5 | 3 | 4 | ||
| CYP75 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ||
| CYP76 | 9 | 5 | 8 | 12 | 4 | 7 | 12 | 5 | 7 | ||
| CYP77 | 5 | 4 | 4 | 5 | 5 | 0 | 5 | 5 | 0 | ||
| CYP78 | 6 | 5 | 0 | 11 | 8 | 2 | 12 | 9 | 0 | ||
| CYP79 | 10 | 3 | 6 | 11 | 4 | 0 | 11 | 4 | 0 | ||
| CYP81 | 18 | 12 | 10 | 32 | 23 | 20 | 37 | 23 | 23 | ||
| CYP82 | 5 | 2 | 3 | 4 | 2 | 0 | 9 | 2 | 2 | ||
| CYP83 | 2 | 1 | 0 | 6 | 1 | 2 | 6 | 1 | 2 | ||
| CYP84 | 2 | 2 | 0 | 4 | 2 | 0 | 6 | 3 | 2 | ||
| CYP89 | 7 | 1 | 3 | 4 | 1 | 2 | 3 | 1 | 0 | ||
| CYP93 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 0 | ||
| CYP98 | 3 | 2 | 2 | 4 | 4 | 0 | 4 | 4 | 0 | ||
| Total A-type | 157 | 78 | 100 | 211 | 108 | 90 | 207 | 106 | 82 | ||
| Total | 251 | 131 (52.19%) | 128 (51%) | 354 | 187 (52.82%) | 137 (38.7%) | 343 | 177 (51.6%) | 112 (32.65%) | ||
Family-specific evolution for P450 gene families
WGD and TD triggered the evolution of P450 gene families in B. rapa and B. oleracea compared with A. thaliana. We defined the expansion/reduction rate to measure the expansion or reduction of the members of P450 gene families in B. rapa and B. oleracea compared with A. thaliana. The expansion/reduction rate is the ratio between the number of members of P450 gene families in B. rapa or B. oleracea and the number of members of corresponding P450 gene families in A. thaliana. An expansion/reduction rate greater than 1, less than 1, or equal to 1 represents expansion, reduction, or no difference in P450 gene families in B. rapa and B. oleracea compared with A. thaliana, respectively. According to the definition of the expansion/reduction rate of P450 gene families, 26, 13, and 8 P450 gene families showed expansion, no difference, and reduction of P450 gene families in B. rapa compared with A. thaliana, respectively. CYP73 gene families had higher expansion/reduction rate and showed more significant expansion than other families in B. rapa compared with A. thaliana. A total of 27, 13, and 7 P450 gene families indicated expansion, no difference, and reduction in B. oleracea compared with A. thaliana, respectively. CYP51 and CYP73 gene families had higher expansion/reduction rate and showed more significant expansion than other families in B. oleracea compared with A. thaliana (Fig. 3a).
Fig. 3.
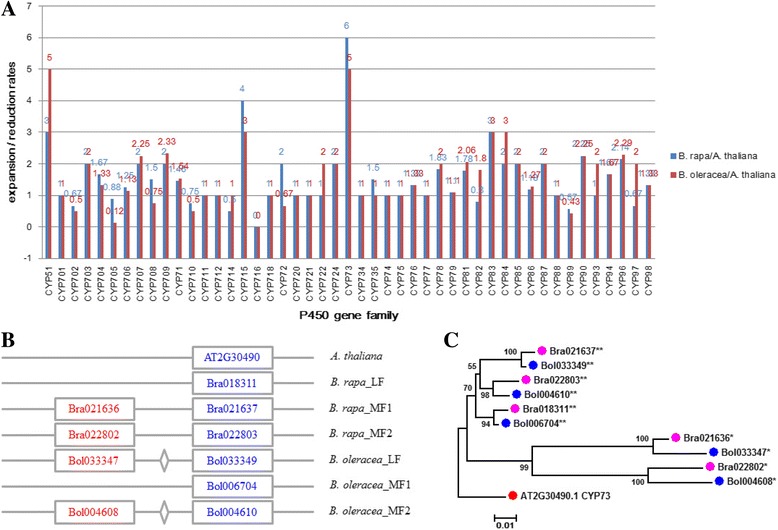
Comparison of expansion/reduction rates of P450 gene families and characteristics of CYP73 gene family members between A. thaliana and Brassica species. a Comparison of expansion/reduction rates of P450 gene families between A. thaliana and Brassica species. b Distribution of the member (s) of the CYP73 gene family in the A. thaliana and Brassica genomes. Gray lines represent the genomic or subgenomic regions among the A. thaliana, B. rapa, and B. oleracea genomes. Blue rectangles represent P450 genes, and diamonds represent non-P450 genes. c Phylogenetic relationship of CYP73 gene family members among A. thaliana, B. rapa, and B. oleracea. Red, pink, and blue solid circles represent P450 genes in A. thaliana, B. rapa, and B. oleracea, respectively. P450 genes with two stars represent P450s from whole genome duplication, and P450 genes with one star represent P450s from tandem duplication
We used the CYP73 gene family, which showed the highest expansion/reduction rate among the P450 gene families in Brassica species, as an example to analyze further the expansion of P450 gene families. The CYP73 gene family only have 1 P450s (AT2G30490.1, named CYP73A5) in A. thaliana. This P450 gene can encode a cinnamate-4-hydroxylase, and mutations in this gene affect phenylpropanoid metabolism, growth, and development [44]. In the Brassica lineage, we obtained five corresponding orthologous P450 genes in each B. rapa and B. oleracea compared with P450s in A. thaliana. Analysis of WGD revealed that the P450 gene (AT2G30490.1) in A. thaliana obtained three orthologous genes retained in triplicated blocks in B. rapa and B. oleracea. Of the three orthologous genes (Bra018311, Bra021637, and Bra022803 located on BraLF, BraMF1, and BraMF2, respectively) of the A. thaliana P450 gene (AT2G30490.1) in B. rapa, two (Bra021637 and Bra022803 located on BraMF1 and BraMF2) underwent TD and generated two paralogous genes (Bra021636 and Bra022802) in B. rapa. In B. oleracea, two P450s (Bol033349 and Bol004610 located on BolLF and BolMF2) experienced TD and generated two paralogous genes (Bol033347 and Bol004608) among triplicated genes (Bol033349, Bol006704, and Bol004610 located on BolLF, BolMF1, and BolMF2, respectively) (Fig. 3b). Phylogenetic analysis revealed that the triplicated P450 genes from Brassica species were clustered into one subgroup, and tandem duplicated P450 genes from triplicated P450 genes were clustered into the other subgroup, indicating closer relative relationships among triplicated P450 genes in the Brassica lineage (Fig. 3c). These results suggest that the A. thaliana orthologous P450 gene (AT2G30490.1) underwent the same evolutionary paths and that the WGD and TD evolutionary events were both responsible for the expansion of A. thaliana P450 gene (AT2G30490.1) in the Brassica lineage.
Expression analysis of the members of cytochrome P450 gene families in A. thaliana and the Brassica lineage
Using microarray data from the developmental set of A. thaliana, we selected four tissues (root with 21 days; stem, 2nd internode with 21+ days; cauline leaves with 21+ days and flowers stage 9 with 21+ days) for expression analysis of 251 P450s in A. thaliana [30]. After curation, 221 P450s in A. thaliana were expressed in the four tissues. According to the expression profiling analysis from the FPKM, 354 and 343 P450 genes in B. rapa and B. oleracea were used to investigate the expression patterns of P450s in four tissues (root, stem, leaf and flower) in the genus Brassica. In total, 296 and 278 P450s were expressed in the four tissues in B. rapa and B. oleracea, respectively (Additional file 6: Table S4). The expression distribution of these genes among A. thaliana, B. rapa and B. oleracea in the four tissues is detailed by the histogram (Fig. 4). From microarray data analysis of all A. thaliana P450s, 221 P450s were all expressed in the four tissues in A. thaliana although some P450s showed very low expression values in certain tissues. But for the RNA-seq analysis of P450s in Brassica lineage, some P450s indicated tissue-specific expression in certain tissues in B. rapa or B. oleracea. In leaf and stem tissues of Brassica species, relatively few expressed P450s were identified compared with other tissues in B. rapa and B. oleracea, indicating less P450s participating in the biosynthesis of various primary and secondary metabolites in these two tissues. The number of P450s expressed in the leaf tissue in B. rapa was higher than that in B. oleracea, suggesting relatively more P450s were expressed in the tissue in B. rapa. For the comparative analysis of expressed P450 gene family, all members of 20 P450s gene family were expressed in one or more tissues, and most of the members of 26 P450s gene family were expressed in one or more tissues among A. thaliana and Brassica species. For the CYP703 gene family, there was only one P450s identified in A. thaliana and this P450s (AT1G01280.1, named CYP703A2) was expressed in the four tissues in A. thaliana. There were 2 P450s identified in B. rapa and B. oleracea respectively, but no P450s was detected to express in B. rapa; only one P450s was detected to express in B. oleracea. For the CYP716 gene family, 2 of 3 P450s were detected to express in four tissues in A. thaliana, although the member of CYP716 gene family was not identified in Brassica species (Additional file 7: Table S5).
Fig. 4.
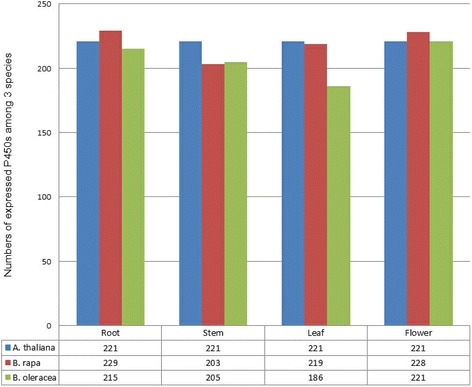
Comparison of the numbers of expressed P450s in different tissues in A. thaliana and Brassica species
Expression difference of P450s of different types in P450 gene families in A. thaliana and Brassica lineage
The discrete degree of expression level of P450s can reflect the functional divergence of different P450 genes, which can be used to investigate the difference in expression pattern among the different P450 gene families in A. thaliana and Brassica species. For the expression analysis of P450s in the four tissues in A. thaliana and Brassica species, we employed quartile map to display the expression difference of various P450 gene families (Fig. 5a). Results showed that the expression values of the members of CYP73 gene family in A. thaliana indicated more discrete than those in other P450 gene families. In Brassica lineage, the expression values of CYP73 genes in B. rapa or B. oleracea were more discrete than those of genes from most of P450 families in A. thaliana. For the members of CYP73 gene family, the expression values of P450s in B. oleracea were more discrete than those in B. rapa, but the difference was not significant (P = 0.745 < 0.05, Mann-Whitney U test). Analysis of expansion of the CYP73 gene family uncovered that WGD and TD were both responsible for the expansion of the A. thaliana P450 gene (AT2G30490.1) in the Brassica lineage. After curation of the members of P450 gene families that underwent WGD or TD, we classified P450 genes in Brassica species into two types: P450s generated by WGD (WGD-type) and P450s generated by TD (TD-type). Therefore, five corresponding orthologous P450 genes in B. rapa or B. oleracea were divided into two TD-type P450s and three WGD-type P450s in Brassica species.
Fig. 5.
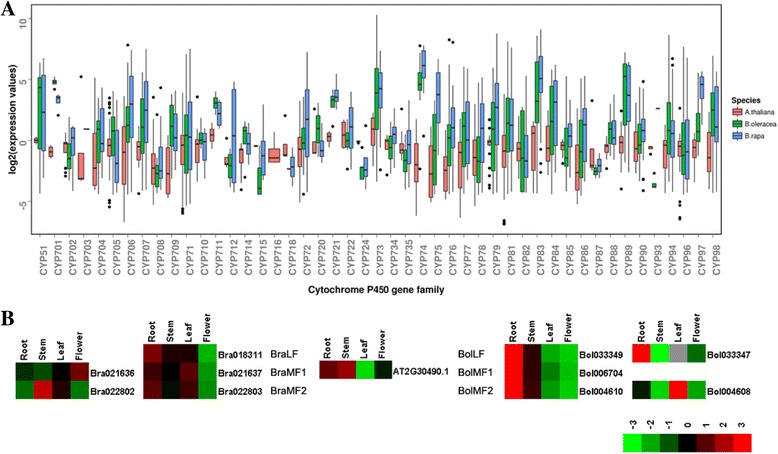
Boxplot comparisons of expression of P450s in different gene families and expression analysis of CYP73 gene family members in A. thaliana and Brassica species. a Boxplot comparisons of expression of P450s in different gene families in A. thaliana, B. rapa and B. oleracea. b Expression analysis of CYP73 gene family members across different tissues in A. thaliana, B. rapa and B. oleracea
To investigate functional divergence, we compared the expression difference of the members of CYP73 gene family between A. thaliana and Brassica species, as well as intra-genome different types of the P450s from CYP73 gene family in Brassica species. In A. thaliana, the A. thaliana P450 gene (AT2G30490.1) was highly expressed in the stem tissue but lowly expressed in the leaf tissue. For the P450s in Brassica lineage, the two different types of P450s from CYP73 gene family indicated different expression patterns in B. rapa and B. oleracea. In B. rapa, the expression levels of WGD-type P450s (Bra018311, Bra021637, and Bra022803) were nearly the same across different tissues. These P450s were highly expressed in the root and leaf but lowly expressed in the flower. The TD-type P450s (Bra021636 and Bra022802) displayed different expression patterns compared with their other member of two-copy tandem array in the WGD-type P450s. For example, Bra021636 and Bra021637 were two-copy tandem duplicated genes located on BraMF1 regions in B. rapa genome. Bra021636 showed high expression in the flower and low expression in the stem, but Bra021637 showed high expression in the leaf and low expression in the flower. Bra022802 and Bra022803 were also two-copy tandem duplicated genes located on BraMF2 regions in B. rapa genome. Bra022802 were highly expressed in the stem, but Bra022803 were highly expressed in the root. For the five members of the CYP73 gene family in B. rapa, the WGD- and TD-type P450s displayed different expression patterns across different tissues, indicating functional divergence among five P450s of the CYP73 gene family in B. rapa. In B. oleracea, the WGD-type P450s (Bol033349, Bol006704, and Bol004610) displayed high expression in the root and stem but reduced expression in the leaf and flower. The TD-type P450s (Bol033347 and Bol004608) showed completely different expression levels with WGD-type P450s in B. oleracea. For example, Bol033347 was highly expressed in the root and lowly expressed in the stem, but Bol004608 was highly expressed in the leaf and lowly expressed in the stem, which were different with expression patterns of their other member of two-copy tandem array in the WGD-type P450s from CYP73 gene family (Fig. 5b). From comparison, A. thaliana P450 gene (AT2G30490.1) indicated different expression patterns with its corresponding homologous P450 genes in Brassica species. The expression patterns of the members of the CYP73 gene family in B. rapa were the same as those in B. oleracea. The WGD-type P450s displayed the same expression trends and showed completely different expression with the TD-type P450s in Brassica species. Among the TD-type P450s, anyone P450 gene showed a different expression with the other P450 genes in the four tissues in Brassica species. These results suggest that the TD-type P450s show more functional divergence compared with the WGD-type P450s in Brassica.
Selection forces on P450 orthologs in A. thaliana and Brassica species
The Ka/Ks ratio was measured to investigate the strength of selection pressure for each branch of the phylogenetic tree using PAML [38]. According to the syntenic relationships between the A. thaliana and Brassica genomes, 124 and 118 A. thaliana P450s have 187 and 177 orthologous genes in B. rapa and B. oleracea, respectively. In total, 187 and 177 orthologous gene pairs in A. thaliana–B. rapa and A. thaliana–B. oleracea lineages were used to investigate the differential selection patterns in A. thaliana compared with the B. rapa and B. oleracea genomes. The mean Ka/Ks ratio of all orthologous gene pairs was 0.194 in B. rapa compared with the A. thaliana genome, which is greater than that of B. oleracea compared with the A. thaliana genome (0.191), although the difference was not significant (Mann–Whitney U-test, P = 0.7988 > 0.05). These results suggest that the P450 orthologous gene pairs in A. thaliana–B. rapa and A. thaliana–B. oleracea lineages underwent negative selection, but the two displayed no significant difference after Brassica species diverged from a common ancestor with A. thaliana.
To investigate the selection pressure of P450 orthologous gene pairs for intragenome in Brassica, the P450 gene pairs located on triplicated genomic regions in Brassica genomes compared with A. thaliana were employed to detect the difference in selection pressure between different subgenomes compared with the A. thaliana genome. A total of 83, 56, and 48 gene pairs in A. thaliana compared with the BraLF, BraMF1, and BraMF2 subgenomes in B. rapa were used to calculate the Ka/Ks values and evaluate the difference in P450 gene pairs among the A. thaliana–BraLF, A. thaliana–BraMF1, and A. thaliana–BraMF2 subgroups in the A. thaliana–B. rapa lineage, respectively. The mean Ka/Ks ratio of the P450 gene pairs was 0.189 in the A. thaliana–LF subgroup, which was larger than that of the A. thaliana–MF1 subgroup (0.188) and smaller than that of the A. thaliana–MF2 subgroup (0.21). These results indicate that the P450 gene pairs in the different subgenomes in B. rapa underwent negative selection compared with those in A. thaliana, but the difference was not significant (Mann–Whitney U test, PBraLF:BraMF1 = 0.6322 > 0.05; PBraLF:BraMF2 = 0.355 > 0.05; PBraMF1:BraMF2 = 0.2724 > 0.05). For the P450 orthologous gene pairs in A. thaliana compared with the different subgenomes in B. oleracea, 79, 56, and 43 orthologous gene pairs were detected in the A. thaliana–BolLF, A. thaliana–BolMF1, and A. thaliana–BolMF2 subgroups in the A. thaliana–B. oleracea lineage. Slight differences were found in their mean Ka/Ks ratios (BolLF: 0.1899, BolMF1: 0.1908, BolMF2: 0.1920), which also showed no significant differences between them (Mann–Whitney U test, PBolLF:BolMF1 = 0.6843 > 0.05; PBolLF:BolMF2 = 0.7372 > 0.05; PBolMF1:BolMF2 = 0.913 > 0.05). A similar inference of negative selection with the consensus evolutionary pattern for P450 gene pairs was detected in A. thaliana compared with the different subgenomes in B. rapa and B. oleracea, although the P450 gene pairs were distributed in different Brassica genomes compared with the A. thaliana genome.
Discussion
Comparison of P450 gene families in A. thaliana and Brassica lineage
On the basis of the syntenic relationships between subgenomes of A. thaliana, duplicated gene pairs from the α, β, and γ WGD events were identified in the A. thaliana genome. Retained ancient genes in different evolutionary periods can be identified by using the ancient relationship evidence from the three rounds of WGD events in the evolutionary history of angiosperms. On the basis of the syntenic relationships in A. thaliana, 1, 3, and 11 duplicated gene pairs were identified from the γ, β, and α WGT or WGD events to trace the ancient cytochrome P450 gene families. As mentioned above, one duplicated gene pair that was retained from the γ WGD event was the more ancient P450 duplicated gene pair in A. thaliana. These two ancient P450 genes were members of the CYP94 gene family, which meant that the CYP94 gene family was relatively more ancient than the P450 gene family. CYP51 is an ancient gene family in lower monocellular eukaryotes [9]. According to the classification criteria of cytochrome P450 nomenclature by Dr. Nelson [45], the CYP51 gene family was distributed in Part C (covers CYP10–CYP69 and 501–699 and 5001 and higher), which included the P450 gene families identified in animals, fungi, and other lower eukaryotes [4]. Except for CYP51, all P450 gene families in the present study were distributed in Part D (covers CYP71–CYP99 and 701–772), which only contained cytochrome P450 gene families in plants. After detecting duplicated gene pairs from the three rounds of WGD in A. thaliana, members of the CYP51 gene family were not detected, indicating that the duplicated genes of the CYP51 gene family were quickly lost after each WGD event in angiosperms. This phenomenon may be the reason why only one P450 gene was detected in the CYP51 gene family on the basis of the TAIR10 version, although this P450 gene family was older than other gene families. A total of 3 and 11 duplicated gene pairs were detected from the β and α WGD events in A. thaliana, which covered 3 and 11 P450 gene families for the two WGD events, respectively. However, members of the CYP94 gene family in the 14 gene families were not found, suggesting the loss of the gene pairs of CYP94 in the latter two WGD events. For the latter two WGD events, only two (CYP81 and CYP86) of the four P450 gene families retained in the β WGD event were examined in the α WGD event. Thus, the rest of the gene family was lost after the α WGD event. Finally, 13 P450 gene families were influenced by the three rounds of WGD events in angiosperms. The gene pairs of the remaining P450 gene families possibly experienced quick loss or the members of P450 gene families later emerged in A. thaliana after three rounds of WGD in angiosperms.
Taking the P450 gene families in A. thaliana as reference, 47 P450 gene families in A. thaliana retrieved 46 corresponding P450 gene families in each B. rapa and B. oleracea, and the CYP716 gene family was lost in two Brassica species. Members of the CYP716 gene family are involved in brassinosteroid biosynthesis, brassinosteroid homeostasis, multicellular organismal development, and sterol metabolism [46]. We speculated that the loss of the CYP716 gene family B. rapa and B. oleracea might indicate the incomplete genome sequencing and assembling for the two Brassica species. Moreover, there were four specific subgroups compared to A. thaliana detecting through phylogenetic analysis, which referred to one B. rapa and five B. oleracea P450s. These six cytochrome P450s were grouped into five different clans of cytochrome P450 gene families. Analysis of protein sequences of these six P450s revealed that these P450s contained partial sequences of p450 conserved domain of standard P450 protein. This result might indicate the incomplete gene prediction or gene function loss of P450s due to selection pressure.
Family-specific evolution of P450 gene families in Brassica
The Brassica lineage underwent WGT and generated triplicated genomic regions in B. rapa and B. oleracea after the Brassica lineage split from a common ancestor with A. thaliana. According to the syntenic relationships between the A. thaliana and B. rapa genomes, the orthologous P450 genes of 124 and 118 A. thaliana P450s were detected 187 B. rapa P450s and 177 B. oleracea P450s, which were distributed in 41 and 42 P450 gene families in B. rapa and B. oleracea, respectively. Compared with the 46 P450 gene families in Brassica species, five and four P450 gene families in B. rapa and B. oleracea have not detected syntenic orthologous P450s in A. thaliana, indicating the loss of duplicated genes of the remaining P450 gene families after the WGT in the Brassica lineage. Through the analysis of TD for the P450 supergene family, tandem duplicated genes for cytochrome P450s were identified (132/251, 52.59% in A. thaliana; 137/354, 38.7% in B. rapa; and 112/343, 32.65% in B. oleracea) in A. thaliana and Brassica species. Comparison of influence of the WGD and TD events for the P450 supergene family between A. thaliana and Brassica species indicated that TD exerted more effect on the evolution of the P450 supergene family in A. thaliana than in Brassica species, and WGD showed more influence than TD on the evolution of the P450 supergene family in Brassica lineage. Although previous study revealed that genes with essential functions are less duplicated across evolution [47], these two evolution mechanisms still caused the increase of the members of P450 gene families in A. thaliana and Brassica lineage, leading to the 1.4 times members of P450 gene families in Brassica species.
According to the expansion or reduction rates of P450 gene families, 43 P450 gene families were influenced by WGD or TD in A. thaliana, B. rapa, and B. oleracea. The remaining P450 gene families were not detected in any of the WGD-type and TD-type P450s in A. thaliana and Brassica species. These results suggested that not all P450 gene families were influenced by WGD and TD, although the two events played an important role in the evolution of the P450 supergene family in A. thaliana and Brassica lineage. After comparing the different evolution mechanisms of P450 gene families between A. thaliana and Brassica species, we can conclude that the evolution of each P450 gene family in Brassica species has its own family-specific mechanisms.
Dynamics of TD in the evolutionary history of the P450 gene family
According to syntenic relationships between A. thaliana and Brassica species, tandem duplicated genes were classified to confirm ancient tandem duplicated genes through analysis of location on syntenic genomic regions in A. thaliana compared with B. rapa and B. oleracea genomes. This result indicates that these ancient tandem duplicated genes were generated before the split of A. thaliana and Brassica ancestor from a common ancestor. Except for the ancient tandem duplicated genes, the remaining tandem duplicated genes in A. thaliana and Brassica lineage have not showed ancient syntenic relationships. This result indicates that these genes were generated after divergence of the Brassica ancestor. The cytochrome P450 genes that formed after the divergence of Brassica ancestor were species-specific tandem duplicated genes in B. rapa and B. oleracea. These results suggest that the tandem duplicated P450 genes formed in a continuous historical process, ending with the evolutionary history of cytochrome P450s. This phenomenon is consistent with a previous report that tandem duplication occurred continuously throughout the evolutionary history of corresponding species [42].
Conclusions
The cytochrome P450 gene superfamily is involved in the biosynthesis of various primary and secondary metabolites. This study identified 251, 354, and 343 P450 genes in A. thaliana, B. rapa, and B. oleracea, respectively. These P450 genes were classified into 47 P450 gene families. Evolutionary analysis of P450s in different organisms revealed that 948 P450s of P450 gene families were grouped into non-A-type and A-type classes among the three species. Chromosomal organization and composite phylogenetic analysis provided a clear distribution on the chromosomes and evolutionary relationship of cytochrome P450 genes among A. thaliana, B. rapa, and B. oleracea. The inferred phylogeny and syntenic relationships of the P450 genes between A. thaliana and Brassica species indicated that the family-specific evolution in the Brassica lineage can be attributed to both WGD and TD, whereas WGD was recognized as the major mechanism for the recent evolution of the P450 supergene family in the Brassica lineage. Expression analysis of the members of P450 gene family in A. thaliana and Brassica species revealed that the WGD-type P450s showed the same expression pattern, but the TD-type P450s showed completely different expression across different tissues in A. thaliana and Brassica species. The WGD- and TD-type P450s showed different expression patterns in the four tissues in A. thaliana and Brassica species. Selection force analysis suggested that the P450 orthologous gene pairs in A. thaliana compared with Brassica species underwent negative selection, but no significant differences were found between orthologous P450 gene pairs in the A. thaliana–B. rapa and A. thaliana–B. oleracea lineages. The P450 orthologous gene pairs in the different subgenomes in B. rapa or B. oleracea compared with A. thaliana also underwent negative selection, but Mann-Whitney U test results indicated no significant differences among them. Through comparative analysis of P450 genes among A. thaliana, B. rapa, and B. oleracea, we investigated the influence of WGD and TD on the evolutionary history and functional divergence of P450 gene families in the genus Brassica. This study provides a biology model to study the mechanism of gene family formation, particularly in the context of the evolutionary history of angiosperms, and offers novel insights for the study of angiosperm genomes.
Additional files
Members of cytochrome P450 gene families in A. thaliana, B. rapa, and B. oleracea. The basic information of cytochrome P450s in A. thaliana, B. rapa, and B. oleracea contained the name of gene family, the location of conserved domains and protein sequences. (XLSX 298 kb)
Distribution of cytochrome P450 genes in A. thaliana, B. rapa and B. oleracea. A. Chr1-Chr5 represent pseudochromosomes in A. thaliana. B. A01–A10 represent pseudochromosomes in B. rapa. C. C01–C09 represent pseudochromosomes in B. oleracea. Green bars represent pseudochromosomes of three species. Balck lines on pseudochromosomes represent the location of P450s on pseudochromosomes in A. thaliana, B. rapa and B. oleracea. (JPEG 6260 kb)
Phylogenetic analysis of cytochrome P450 gene families in A. thaliana and Brassica species. I and II represent different groups among three species. Red solid circles represent cytochrome P450 genes in A. thaliana, pink solid circles represent cytochrome P450 genes in B. rapa, and blue solid circles represent cytochrome P450 genes in B. oleracea. CYP represents cytochrome P450 gene family. B. rapa-specific represents specific cytochrome P450 genes in B. rapa. B. oleracea-specific represents specific cytochrome P450 genes in B. oleracea. Brassica-specific represents specific cytochrome P450 genes in Brassica species. (JPEG 886 kb)
Orthologous genes pairs from different whole genome duplication events in A. thaliana compared with different subgenomes in B. rapa and B. oleracea. This file contained the orthologous genes pairs of cytochrome P450s in A. thaliana compared to BraLF, BraMF1, and BraMF2 subgenomes in B. rapa, and BolLF, BolMF1, and BolMF2 subgenomes in B. oleracea, respectively. (XLSX 24 kb)
Tandem arrays of cytochrome P450 genes among A. thaliana, B. rapa, and B. oleracea. The basic information of tandem arrays of cytochrome P450s among three species included the gene family, location, name, gene number, and gene list of tandem arrays. (XLSX 31 kb)
Expression values of cytochrome P450 genes across different tissues among A. thaliana, B. rapa, and B. oleracea. This file contained the expressed cytochrome P450s of A. thaliana in root, stem, leaf, and flower tissues according to the analysis of microarray data, and the expressed cytochrome P450s of B. rapa, and B. oleracea in root, stem, leaf, and flower tissues according to the analysis of RNA-seq data. (XLSX 72 kb)
Expression statistics of the members of cytochrome P450 gene families among A. thaliana, B. rapa, and B. oleracea. This file contained the proportion of expressed P450s in different cytochrome P450 gene family among three species. (XLSX 12 kb)
Acknowledgments
Funding
This work was supported by the Agricultural Science and Technology Innovation Program, CAAS and China Agriculture Research System (no. CARS-15), and project on the Integration of Industry, Education and Research of Henan Province, China (no. 162107000007).
Availability of data and materials
All the supporting data are included as additional files.
Abbreviations
- HMM
Hidden Markov model
- ML
Maximum likelihood
- Mya
Million years ago
- TD
Tandem duplication
- WGD
Whole genome duplication
- WGT
Whole genome triplication
Authors’ contributions
JY analyzed the data and prepared the manuscript. ST, LW, KD, XZ, TK and BL participated in data analysis and the manuscript preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4094-7) contains supplementary material, which is available to authorized users.
Contributor Information
Jingyin Yu, Email: yujingyin@caas.cn.
Sadia Tehrim, Email: tehrim.sadia@gmail.com.
Linhai Wang, Email: linhai827@163.com.
Komivi Dossa, Email: dossakomivi@gmail.com.
Xiurong Zhang, Email: zhangxr@oilcrops.cn.
Tao Ke, Email: ketao1@163.com.
Boshou Liao, Email: lboshou@hotmail.com.
References
- 1.Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1(6):REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn RA, Durst F. Function and evolution of plant cytochrome P450. Recent Adv Phytochem. 2000;34:151–190. doi: 10.1016/S0079-9920(00)80007-6. [DOI] [Google Scholar]
- 3.Klingenberg M. Pigments of rat liver microsomes. Arch Biochem Biophys. 1958;75(2):376–386. doi: 10.1016/0003-9861(58)90436-3. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4(1):59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapple C. Molecular-genetic analysis of plant Cytochrome P450-dependent Monooxygenases. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- 6.Mizutani M, Ohta D. Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- 7.Durst F, Nelson DR. Diversity and evolution of plant P450 and P450-reductases. Drug Metabol Drug Interact. 1995;12(3–4):189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- 8.Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. Plant J. 2011;66(1):194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- 9.Nelson DR. Plant cytochrome P450s from moss to poplar. Phytochem Rev. 2006;5(2–3):193–204. doi: 10.1007/s11101-006-9015-3. [DOI] [Google Scholar]
- 10.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus Japonicus. DNA Res. 2008;15(4):227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422(6930):433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 12.De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20(11):591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, Depamphilis CW, Wall PK, Soltis PS. Polyploidy and angiosperm diversification. Am J Bot. 2009;96(1):336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 14.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis Thaliana. Proc Natl Acad Sci U S A. 2002;99(21):13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 16.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13(2):137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis Thaliana. Proc Natl Acad Sci U S A. 2010;107(43):18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. Comparative genomics of Brassica Oleracea and Arabidopsis Thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18(6):1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica Rapa. Plant Cell. 2006;18(6):1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15(4):516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham GJ. Tandem genes and clustered genes. J Theor Biol. 1995;175(1):71–87. doi: 10.1006/jtbi.1995.0122. [DOI] [PubMed] [Google Scholar]
- 22.Lajoie M, Bertrand D, El-Mabrouk N. Evolution of tandemly arrayed genes in multiple species. In: Comparative Genomics: RECOMB 2007 International Workshop, RECOMB-CG 2007, San Diego, CA, USA, September 16-18, 2007 Proceedings. Edited by Tesler G, Durand D. Berlin: Springer Berlin Heidelberg; 2007:96–109.
- 23.Lodish HABCKMKABHPAAMS . Beth McHenry molecular cell biology. New York: W.H. Freeman Company; 2013. Genes, genomics, and chromosomes; pp. 227–230. [Google Scholar]
- 24.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29(1):102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Zhao M, Wang X, Tong C, Huang S, Tehrim S, Liu Y, Hua W, Liu S. Bolbase: a comprehensive genomics database for Brassica Oleracea. BMC Genomics. 2013;14:664. doi: 10.1186/1471-2164-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Ke T, Tehrim S, Sun F, Liao B, Hua W. PTGBase: an integrated database to study tandem duplicated genes in plants. Database (Oxford). 2015;2015:bav017. doi:10.1093/database/bav017. [DOI] [PMC free article] [PubMed]
- 28.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis Thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 31.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clamp M, Cuff J, Searle SM, Barton GJ. The jalview java alignment editor. Bioinformatics. 2004;20(3):426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy JL, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 2016;7:735. doi:10.3389/fpls.2016.00735. [DOI] [PMC free article] [PubMed]
- 37.Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, et al. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7(4):255–269. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 39.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W. The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29(1):102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bak S, Beisson F, Bishop G, Hamberger B, Hofer R, Paquette S, Werck-Reichhart D. Cytochromes p450. Arabidopsis Book. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. The genome of the mesopolyploid crop species Brassica Rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S, et al. The Brassica Oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis Thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 44.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis Thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272(31):19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 45.Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12(1):1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 46.Fukushima EO, Seki H, Ohyama K, Ono E, Umemoto N, Mizutani M, Saito K, Muranaka T. CYP716A Subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 2011;52(12):2050–2061. doi: 10.1093/pcp/pcr146. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Defoort J, Tasdighian S, Maere S, Van de Peer Y, De Smet R. Gene duplicability of Core genes is highly consistent across all angiosperms. Plant Cell. 2016;28(2):326–344. doi: 10.1105/tpc.15.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Members of cytochrome P450 gene families in A. thaliana, B. rapa, and B. oleracea. The basic information of cytochrome P450s in A. thaliana, B. rapa, and B. oleracea contained the name of gene family, the location of conserved domains and protein sequences. (XLSX 298 kb)
Distribution of cytochrome P450 genes in A. thaliana, B. rapa and B. oleracea. A. Chr1-Chr5 represent pseudochromosomes in A. thaliana. B. A01–A10 represent pseudochromosomes in B. rapa. C. C01–C09 represent pseudochromosomes in B. oleracea. Green bars represent pseudochromosomes of three species. Balck lines on pseudochromosomes represent the location of P450s on pseudochromosomes in A. thaliana, B. rapa and B. oleracea. (JPEG 6260 kb)
Phylogenetic analysis of cytochrome P450 gene families in A. thaliana and Brassica species. I and II represent different groups among three species. Red solid circles represent cytochrome P450 genes in A. thaliana, pink solid circles represent cytochrome P450 genes in B. rapa, and blue solid circles represent cytochrome P450 genes in B. oleracea. CYP represents cytochrome P450 gene family. B. rapa-specific represents specific cytochrome P450 genes in B. rapa. B. oleracea-specific represents specific cytochrome P450 genes in B. oleracea. Brassica-specific represents specific cytochrome P450 genes in Brassica species. (JPEG 886 kb)
Orthologous genes pairs from different whole genome duplication events in A. thaliana compared with different subgenomes in B. rapa and B. oleracea. This file contained the orthologous genes pairs of cytochrome P450s in A. thaliana compared to BraLF, BraMF1, and BraMF2 subgenomes in B. rapa, and BolLF, BolMF1, and BolMF2 subgenomes in B. oleracea, respectively. (XLSX 24 kb)
Tandem arrays of cytochrome P450 genes among A. thaliana, B. rapa, and B. oleracea. The basic information of tandem arrays of cytochrome P450s among three species included the gene family, location, name, gene number, and gene list of tandem arrays. (XLSX 31 kb)
Expression values of cytochrome P450 genes across different tissues among A. thaliana, B. rapa, and B. oleracea. This file contained the expressed cytochrome P450s of A. thaliana in root, stem, leaf, and flower tissues according to the analysis of microarray data, and the expressed cytochrome P450s of B. rapa, and B. oleracea in root, stem, leaf, and flower tissues according to the analysis of RNA-seq data. (XLSX 72 kb)
Expression statistics of the members of cytochrome P450 gene families among A. thaliana, B. rapa, and B. oleracea. This file contained the proportion of expressed P450s in different cytochrome P450 gene family among three species. (XLSX 12 kb)
Data Availability Statement
All the supporting data are included as additional files.


