Abstract
Encoded microparticles have become a powerful tool for a wide array of applications, including high-throughput sample tracking and massively parallel biological multiplexing. Spectral encoding, where particles are encoded with distinct luminescence spectra, provides a particularly appealing encoding strategy because of the ease of reading codes and assay flexibility. To date, spectral encoding has been limited in the number of codes that can be accurately resolved. Here, we demonstrate an automated 5-dimensional spectral encoding scheme using lanthanide nanophosphors that is capable of producing isotropic spherical microparticles with up to 1,100 unique codes, which we term MRBLEs (Microspheres with Ratiometric Barcode Lanthanide Encoding). We further develop a quantitative framework for evaluating global ability to distinguish codes and demonstrate that for six different sets of MRBLEs ranging from 106 to 1,101 codes in size, > 98% of MRBLEs can be assigned to a code with 99.99% confidence. These > 1,000 code sets represent the largest spectral code libraries built to date. We expect that these MRBLEs will enable a wide variety of novel multiplexed assays.
Keywords: Spectral Encoding, Multiplexing, Microparticles, Lanthanides
Introduction and Body
With the rise of genomic and proteomic data, high content multiplexed bioassays, which allow multiple analytes of interest to be probed and tracked in a single experiment, have become widely used for both basic research and clinical diagnosis[1–16]. While this area has traditionally been dominated by spatial microarrays, bead-based assays offer several key advantages, including rapid mixing for near fluid-phase kinetics, easier handling and manipulation, and smaller required sample volumes[15–17]. Encoded particles are attractive for biological multiplexing because they can serve as a solid-phase support for a given biological probe, thus linking the particle code to the identity of the probe and its associated analyte. Furthermore, encoded particles provide the ability to selectively cleave off probes to assess their quality or identify probe-bound material after assay completion. However, while spatial arrays of hundreds of thousands of probes have been demonstrated[18,19], assays using encoded particles have been significantly more limited due to technical difficulties in producing more than a few hundred unique codes.
Current particle encoding strategies include spatial barcodes[7,20–26] and luminescence spectral encoding, where the spectrum of light emitted by each particle varies. Although some spatial barcoding schemes have achieved large numbers of codes, bead shape and orientation requirements during code readout have precluded widespread adoption of these schemes, especially for bioassays[5,21,27]. Spectral codes isotropically embedded in spherical beads can be read in any orientation, making them ideal for benchtop assays. However, the available number of codes in spectrally encoded libraries has typically been relatively small. Bead-based technologies that rely on fluorescent dyes for spectral encoding (e.g. Luminex xMAP) have been reported to achieve 100–500 codes[28–30], but these numbers approach the limit of what is theoretically possible due to spectral overlap between species. Technologies employing quantum dots (QDs) have also reached a published limit of ~100 codes due to energy transfer between QDs when incorporated into beads[24,31–37]. Additional technical difficulties associated with using fluorophores and QDs for spectral encoding include spectral interference between the luminescent species used for encoding and for biological assay detection, the need for multiple costly excitation sources, photobleaching of the encoding species, and incompatibility with chemical reagents required for on-bead solid-phase synthesis of probe libraries[38,39].
Lanthanide nanophosphors (LNs) offer several advantages over organic fluorophores or QDs for spectral encoding. LNs possess large Stokes shifts, resist photobleaching, and emit visible light in narrow spectral bands, making species easily distinguishable from one another[40–44]. LNs are chemically inert and relatively insensitive to environmental changes, making them compatible with common chemical conditions for bead functionalization[40,45]. Additionally, all LNs are excited at a single UV wavelength, reducing instrumentation costs and preserving the ability to use the full range of conventional fluorescent dyes for multiplexed analyte detection in downstream assays[46]. As a result, several groups have employed lanthanide-based encoding to create spatially invariant code sets up to tens of codes[12,25,47–53]. In our groups, we previously synthesized and discriminated beads containing 24 unique spectral codes created via the ratiometric incorporation of Eu-, Sm-, and Dy-doped YVO4 nanophosphors within each bead[54].
Here, we demonstrate the ability to produce and distinguish LN-doped microspheres for a code set of 1,023 distinct codes with 99.8% of beads assigned to a spectral code at 99.99% confidence with high reproducibility, representing, to our knowledge, the largest code set achieved by pure spectral encoding ever demonstrated and a 50-fold improvement from our previous work[54]. To achieve this code space, we synthesized and incorporated 5 brightly luminescent LN species and developed a next-generation microfluidic device for controlled, high throughput automated bead synthesis. We apply a novel computational framework to identify the embedded codes and quantitatively assess the microsphere code assignment accuracy. We anticipate that these 1000-plex encoded microspheres, which we term MRBLEs (Microspheres with Ratiometric Barcode Lanthanide Encoding), will have broad utility for high content biological assays, from genomic and proteomic biological library testing to clinical diagnosis and assessment.
Results and Discussion
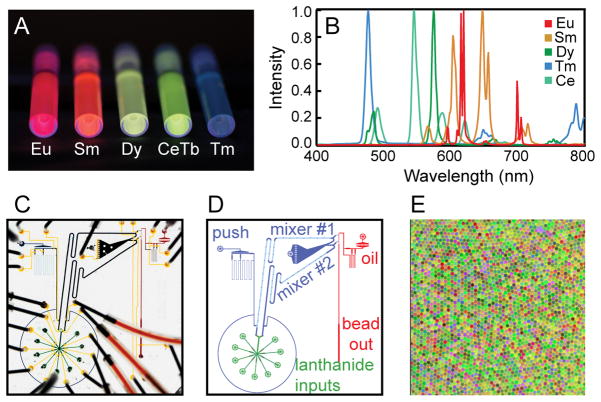
MRBLEs are generated by the microfluidic production and photopolymerization of polymer droplets containing precise ratios of embedded LN species into solid polymer beads. A ratiometric encoding scheme improves code quality by enabling correction for small variations in excitation intensity and light collection efficiency across the field of view and between images. This scheme can yield a large number of unique spectral codes with a theoretical maximum limit of IC, where I represents the number of distinct intensity levels that can be distinguished, and C represents the number of encoding species. To maximize I, we increased the brightness of individual YVO4:Eu, YVO4:Sm, and YVO4:Dy LNs ~3–14-fold from our previous work by removal of a bismuth co-dopant previously added to red-shift the emission spectra (Figure S1A)[54]. We then increased C by synthesizing additional LN species (YVO4:Ho, YVO4:Er, YVO4:Tm, and LaPO4:CeTb)[55,56]. Of these, YVO4:Tm and LaPO4:CeTb yielded homogeneous aqueous suspensions that are brightly luminescent when excited with deep UV light (Figures 1A, S1), and are spectrally well-resolved from other emitting species (Figures 1B, S1).
Figure 1.
Lanthanide nanophosphor species and microfluidic devices used to generate MRBLEs. (A) Photograph showing vials of YVO4:Eu, YVO4:Sm, YVO4:Dy, LaPO4:CeTb, and YVO4:Tm excited by 305 nm light from a handheld UV lamp. (B) Normalized emission spectra for YVO4:Eu, YVO4:Sm, YVO4:Dy, LaPO4:CeTb, and YVO4:Tm LNs. All LNs were excited at 285 nm except for LaPO4:CeTb, which was excited at 275 nm. (C) Photograph of microfluidic bead synthesizer device used for MRBLE production. (D) Cartoon schematic showing bead synthesizer device modules. (E) False color image showing monodisperse microspheres from a 551 MRBLE code set.
We then developed a next-generation microfluidic bead synthesizer capable of high-throughput production of the millions of MRBLEs required for a >1000 code set. To produce MRBLEs, individual mixtures containing polymer (polyethylene glycol diacrylate), photoinitiator[57], a single coding LN species, and a reference LN (Eu:YVO4) are loaded into one of 8 inputs (Figure 1C,D). Precisely controlling the pressures driving each input (and hence their relative flow rates) generates unique ratios of lanthanides, each of which comprises a distinct spectral code. Once loaded, LN/polymer solutions are mixed via passage through a grooved herringbone channel, formed into droplets at a T-junction with a perpendicular channel flowing a surfactant/mineral oil solution (2% v/v Abil EM 90 and 0.05% v/v Span 80), and irradiated with UV light to drive polymerization into monodisperse solid beads (Figure 1E). To compensate for small code-dependent differences in solution viscosity (which can affect droplet size and code resolution), beads are synthesized in a two-step process in which LN/polymer solutions are first mixed and then pushed toward droplet generation using a single water source (Figure S2). The next-generation synthesizer presented here includes two mixers and two droplet generators (Figures 1C, 1D, S2), increasing throughput >3-fold for production of 3,000 individual MRBLEs in 2.5 minutes.
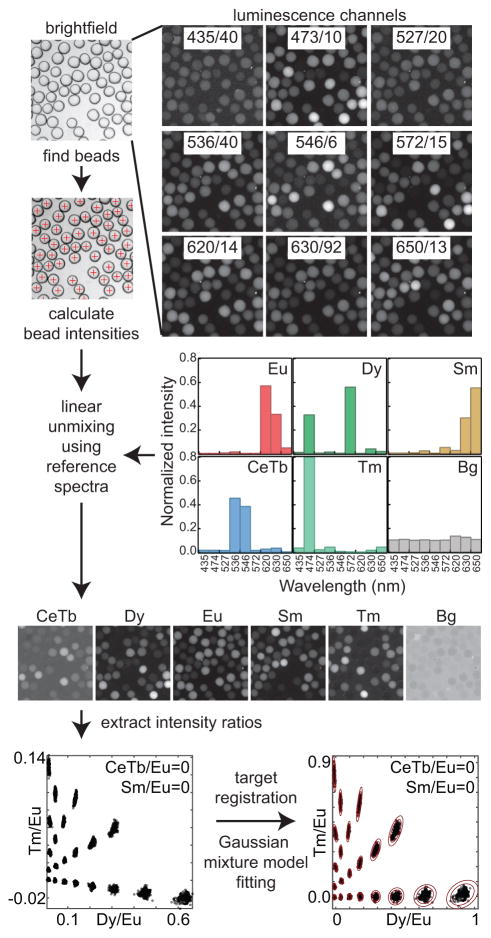
Embedded codes within MRBLEs are read via excitation with deep UV light (292 nm) and imaged at 9 wavelengths chosen to best discriminate between individual LN emission spectra (Figure 2A). The raw images are converted to intensity images for each lanthanide by linear unmixing to determine the most likely linear combination of LNs to have produced the observed spectra at each pixel (Figure 2B). Individual MRBLE codes are then reported as the ratio of intensities of each coding LN relative to the YVO4:Eu internal standard LN (Figure 2C). A transformation matrix is applied to register the measured ratios onto the known programmed ratios, and a Gaussian mixture model (GMM) is used to fit the mean ratios and covariance matrices that describe each code cluster, and then assign each bead to its most likely code cluster (Figure 2C). Our analysis pipeline and optical instrumentation is discussed extensively in the Supplementary Methods.
Figure 2.
Image analysis workflow. (A) Brightfield and luminescence images of MRBLEs are recorded with the indicated filters; beads are identified from the brightfield image using the circular Hough transformation. (B) Measured luminescence images are transformed into LN intensity images by linear unmixing using spectra acquired from reference MRBLEs containing a single LN species. For each bead, the median intensity and median intensity ratio are recorded. (C) A transformation matrix registers measured ratios to programmed ratios and a Gaussian mixture model is used to assign individual beads to a particular code. Red ellipses are the 3 and 4 standard deviation contours derived from the Gaussian mixture model covariance matrix.
Expanding a spectral encoding system to large numbers of codes requires careful determination of a theoretical code spacing that maximizes the number of distinct code clusters that can be resolved accurately. This optimal code spacing, in turn, depends on an accurate model of the standard deviation (SD) associated with each code cluster. In previous work using two LNs (Dy and Sm), we found that the SD of each cluster in each channel depended linearly upon only the LN level in that channel[54]. To explore whether this independence held for a larger set of lanthanide nanophosphors (Dy, Sm, Tm, and CeTb), we generated and imaged a sparse 106-code MRBLE set containing 2 levels of CeTb, 6 levels each of Dy and Sm, and 5 levels of Tm. Although embedded codes are identified by fitting a GMM to all dimensions simultaneously, we visualize all clusters within this four-dimensional data set by first classifying each MRBLE by CeTb/Eu and Sm/Eu ratios (shown as columns and ratios, respectively), and then plotting their Tm/Eu ratios versus Dy/Eu ratios (Figure S3). These data (from 3,185 MRBLEs) demonstrate that each code forms a tight, well-separated cluster. For all LN ratios except Tm/Eu, the SD for each cluster, extracted from the GMM covariance matrix, is well fit by a linear model depending only on the mean ratio of that LN (Figure S4, Table S1); for Tm/Eu, the SD for each cluster depends on both Tm/Eu and Dy/Eu ratios, likely due to overlapping emission peaks at ~470 nm (Figures 1B, S1D). These empirically derived SD models allow prediction of intensity level spacings that separate code clusters by at least n SD (where n is specified by the user) to attain the maximum number of code clusters within a 4-dimensional space.
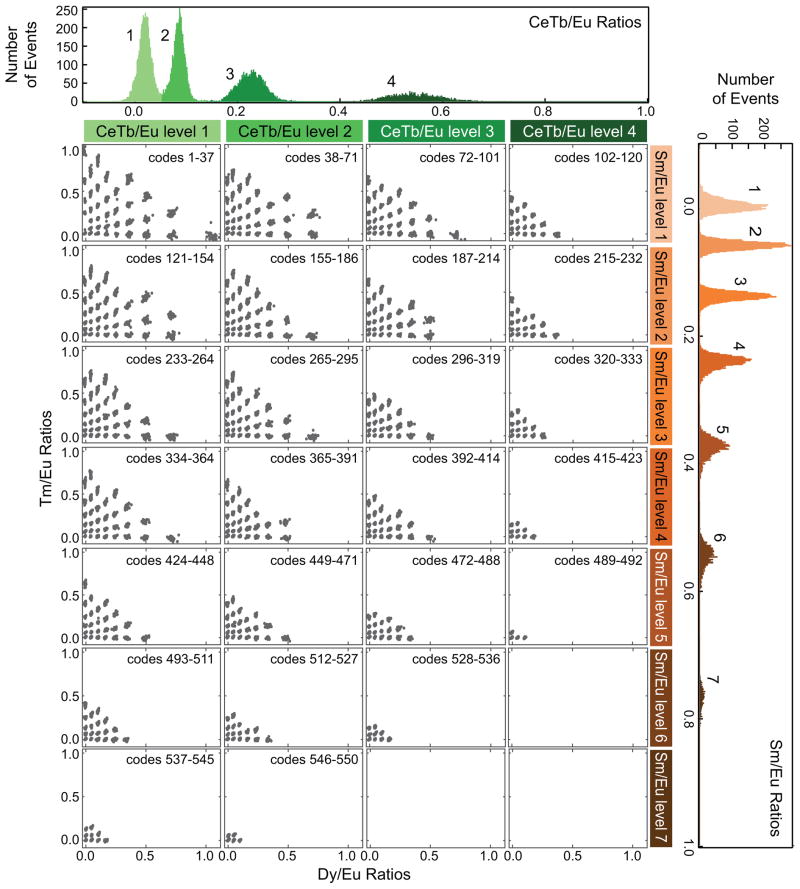
Using this framework, we iteratively synthesized increasingly larger code sets (Table S2). We used the initial 106-code data to generate a set of 285 MRBLE codes separable by 8 standard deviations (Figure S5), used this 285-code data to predict a set of 341 codes separable by 7 standard deviations (Figure S6), and used this 341 code data to predict a set of 551 codes separable by 5.7 standard deviations. Figure 3 shows the four-dimensional data from 20,801 MRBLEs from this 551-code MRBLE set comprised of 4 CeTb levels, 9 Dy levels, 7 Sm levels, and 7 Tm levels; the full set of intensity histograms for each channel is shown in Figure S7. As with the sparser code sets, MRBLEs fall into discrete clusters that are well separated from their neighbors, although one cluster is missing due to a production error. This demonstration of 550 codes surpasses all known purely spectral encoding libraries published to date.
Figure 3.
Measured intensity ratios in four dimensions for 20,801 MRBLEs from a 550 code set. MRBLEs are first separated by CeTb/Eu ratio (four columns, with each column corresponding to a single peak within the histogram of all CeTb/Eu intensities shown at top) and Sm/Eu ratio (seven rows, with each row corresponding to a single peak within the histogram of all Sm/Eu intensities shown at right). For each unique combination of CeTb/Eu and Sm/Eu ratios, a panel displays Tm/Eu ratios plotted vs Dy/Eu ratios to show individual clusters.
We hypothesized that our library size could be further increased by synthesizing several sets of MRBLEs, each containing a different Eu reference level as an additional coding parameter. To test this, we verified that different Eu levels could be easily resolved from one another (Figure S8) and synthesized additional code sets containing either 2 or 3 different levels of Eu. First, we synthesized an additional 551 code MRBLEs containing 40% of the standard Eu reference level and combined it with the 550 code MRBLE above to create a composite set of 1,101 codes (Figures 3, S9, and Movie S1). Second, we created three 341-code MRBLE sets containing either 100%, 50%, or 25% of the standard Eu reference and combined them to create a composite set of 1,023 codes (Figures S10–12).
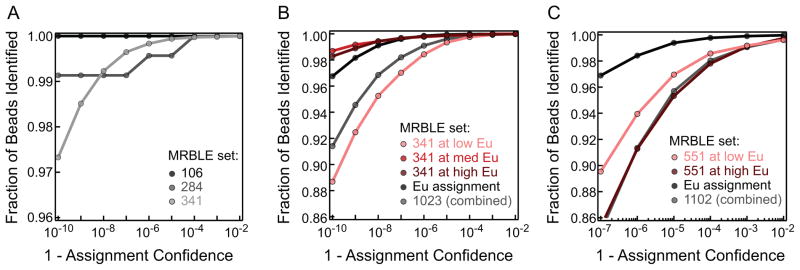
For small numbers of codes, code quality is often established by simply plotting histograms or clusters of intensities and visually assessing whether it seems possible to unambiguously assign a code to any given bead. To evaluate the quality of these large code sets spanning 5-dimensional space, we developed a new quantitative framework similar to a receiver operating characteristic (ROC) curve. First, we assess the likelihood of a MRBLE belonging to each code using the posterior probabilities returned by the Gaussian mixture model described above. For MRBLEs with varying reference Eu levels, we multiply this posterior code probability by the probability of the Eu level assignment. For a given probability, we then globally assess code quality by determining the fraction of MRBLEs assigned to their most likely code with that probability or higher. For the 1,023 code set, 99.8% of MRBLEs can be assigned to a code with 99.99% confidence and 99.9% of MRBLEs to a code with 99.9% confidence (Figure 4B, Table S2). For the 1,101 code set, 98.0% of MRBLEs can be assigned to a code with 99.99% confidence and 99.1% of MRBLEs to a code with 99.9% confidence (Figure 4C, Table S2). Smaller code sets can be assigned with even higher accuracies; e.g. for a 341-code set, we can assign 99.97% of MRBLEs to a code with 99.99% confidence (Figure 4A, Table S2). Taken together, these assignment rates far exceed all existing technologies to date known to us from the literature.
Figure 4.
Confidence plots for MRBLE code assignment. (B) Plot showing the fraction of MRBLEs that can be identified with a given confidence level for 50,564 MRBLEs from a 1,101 code set containing two distinct Eu levels (20,801 with 100% Eu and 29,853 with 50% Eu). (C) Plot showing the fraction of MRBLEs that can be identified with a given confidence level for MRBLEs from a 1,023 code set containing three distinct Eu levels (16,419 with 25% Eu, 7,008 with 50% Eu, and 16,932 with 100% Eu).
In conclusion, we have demonstrated the ability to synthesize Microspheres with Ratiometric Barcode Lanthanide Encoding (MRBLEs) with over 1,000 unique spectral barcodes, representing the largest code set created using purely spectral encoding to date. We further developed a quantitative metric for globally assessing code quality, which is broadly applicable to benchmarking many encoded particle technologies beyond that presented here. In total, we have shown data for six different sets of MRBLEs ranging from 106 to 1,101 codes in size, with assignment accuracies ranging from 100% assignment to 98% assignment at 99.99% confidence.
While commercial fluorophore-based technologies are approaching their theoretical encoding limit due to spectral overlap, this lanthanide-based encoding system has significant potential for future growth to even larger code sets. First, coding capacity can be increased by incorporating upconverting LNs, which are excited with infrared light (typically at 980 nm) and emit visible light in narrow, well-defined spectral bands[58–62]. Two popularly synthesized upconverting species (NaYF4:YbEr and NaYF4:YbTm) are spectrally orthogonal to the downconverting LNs employed here as well as fluorophores commonly used for bound analyte detection, rendering them ideal for achieving > 104 distinct codes. Second, accounting for the observed covariance between LN ratios within each code would allow tighter packing of code clusters off of the orthogonal grid we currently use. Finally, we present here a rigorous code separation method to demonstrate the potential of downconverting LNs for creating extremely large code spaces. In future practical applications, it is likely that we can tolerate a lower assignment confidence that what we observe here, thereby achieving even larger numbers of codes. In summation, MRBLEs represent a promising lanthanide-based encoding technology that enables spectral encoding to reach unprecedented numbers of codes, and anticipate this system will ultimately prove useful for a broad range of microsphere-based multiplexed bioassays.
Supplementary Material
Acknowledgments
H.Q. Nguyen and B.C. Baxter contributed equally to this work. The authors thank Marshall Burke for device photography and Zev Bryant for helpful comments on the manuscript. Funding for this work was provided in part by NIH/NIGMS grant R01GM107132, the Howard Hughes Medical Institute, and by a W.M. Keck Foundation grant. Portions of this work were performed as a user project at the Molecular Foundry and was supported by the Office of Basic Energy Science, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supporting Information is available online from the Wiley Online Library.
References
- 1.Birtwell S, Morgan H. Integr Biol. 2009;1:345. doi: 10.1039/b905502a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broder GR, Ranasinghe RT, She JK, Banu S, Birtwell SW, Cavalli G, Galitonov GS, Holmes D, Martins HFP, MacDonald KF, Neylon C, Zheludev N, Roach PL, Morgan H. Anal Chem. 2008;80:1902. doi: 10.1021/ac7018574. [DOI] [PubMed] [Google Scholar]
- 3.Cederquist KB, Dean SL, Keating CD. WIREs Nanomed Nanobiotechnol. 2010;2:578. doi: 10.1002/wnan.96. [DOI] [PubMed] [Google Scholar]
- 4.Lawrie GA, Battersby BJ, Trau M. Adv Funct Mater. 2003;13:887. [Google Scholar]
- 5.Lee H, Kim J, Kim H, Kim J, Kwon S. Nat Mater. 2010;9:745. doi: 10.1038/nmat2815. [DOI] [PubMed] [Google Scholar]
- 6.Le Goff GC, Srinivas RL, Hill WA, Doyle PS. European Polymer Journal. 2015;72:386. doi: 10.1016/j.eurpolymj.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim LN, Kim M, Jung K, Bae HJ, Jang J, Jung Y, Kim J, Kwon S. Chem Commun. 2015;51:12130. doi: 10.1039/c5cc02048d. [DOI] [PubMed] [Google Scholar]
- 8.Chapin SC, Appleyard DC, Pregibon DC, Doyle PS. Angew Chem Int Ed. 2011;50:2289. doi: 10.1002/anie.201006523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Appleyard DC, Chapin SC, Doyle PS. Anal Chem. 2011;83:193. doi: 10.1021/ac1022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolan JP, Mandy F. Cytometry. 2006;69A:318. doi: 10.1002/cyto.a.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim CT, Zhang Y. Biosensors and Bioelectronics. 2007;22:1197. doi: 10.1016/j.bios.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Haushalter RC, Haushalter RW, Shi Y, Zhang Y, Ding K, Zhao D, Stucky GD. Small. 2011;7:1972. doi: 10.1002/smll.201100629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, Shi Q, Zhang Y, Shi Y, Ding K, Zhao D, Stucky GD. Advanced Materials. 2011 doi: 10.1002/adma.201101868. n. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Cheng Y, Shang L, Wang J, Xie Z, Gu Z. Small. 2015;11:151. doi: 10.1002/smll.201401600. [DOI] [PubMed] [Google Scholar]
- 15.Braeckmans K, De Smedt SC, Leblans M, Pauwels R, Demeester J. Nat Rev Drug Discov. 2002;1:447. doi: 10.1038/nrd817. [DOI] [PubMed] [Google Scholar]
- 16.Finkel NH, Lou X, Wang C, He L. Anal Chem. 2004;76:352 A. doi: 10.1021/ac0416463. [DOI] [PubMed] [Google Scholar]
- 17.Wilson R, Cossins AR, Spiller DG. Angew Chem Int Ed Engl. 2006;45:6104. doi: 10.1002/anie.200600288. [DOI] [PubMed] [Google Scholar]
- 18.Heller MJ. Annu Rev Biomed Eng. 2002;4:129. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 19.Legutki JB, Zhao ZG, Greving M, Woodbury N, Johnston SA, Stafford P. Nature Communications. 2014;5:1. doi: 10.1038/ncomms5785. [DOI] [PubMed] [Google Scholar]
- 20.Nicewarner-Peña SR, Freeman RG, Reiss BD, He L, Peña DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Science. 2001;294:137. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 21.Dejneka MJ, Streltsov A, Pal S, Frutos AG, Powell CL, Yost K, Yuen PK, Müller U, Lahiri J. Proceedings of the …. 2003;100:389. doi: 10.1073/pnas.0236044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fournier Bidoz S, Jennings TL, Klostranec JM, Fung W, Rhee A, Li D, Chan WCW. Angew Chem Int Ed. 2008;47:5577. doi: 10.1002/anie.200800409. [DOI] [PubMed] [Google Scholar]
- 23.Ji XH, Zhang NG, Cheng W, Guo F, Liu W, Guo SS, He ZK, Zhao XZ. J Mater Chem. 2011;21:13380. [Google Scholar]
- 24.Zhao Y, Shum HC, Chen H, Adams LLA, Gu Z, Weitz DA. J Am Chem Soc. 2011;133:8790. doi: 10.1021/ja200729w. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Bisso PW, Srinivas RL, Kim JJ, Swiston AJ, Doyle PS. Nat Mater. 2014;13:524. doi: 10.1038/nmat3938. [DOI] [PubMed] [Google Scholar]
- 26.Falconnet D, She J, Tornay R, Leimgruber E, Bernasconi D, Lagopoulos L, Renaud P, Demierre N, van den Bogaard P. Anal Chem. 2015;87:1582. doi: 10.1021/ac502741c. [DOI] [PubMed] [Google Scholar]
- 27.Braeckmans K, De Smedt SC, Roelant C, Leblans M, Pauwels R, Demeester J. Nat Mater. 2003;2:169. doi: 10.1038/nmat828. [DOI] [PubMed] [Google Scholar]
- 28.Houser B. Archives of Physiology and Biochemistry. 2012;118:192. doi: 10.3109/13813455.2012.705301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Birru R, Di YP. Molecular Toxicology Protocols. Humana Press; Totowa, NJ: 2014. pp. 43–57. [Google Scholar]
- 30.Dunbar SA. Clinica Chimica Acta. 2006;363:71. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fournier Bidoz S, Jennings TL, Klostranec JM, Fung W, Rhee A, Li D, Chan WCW. Angew Chem Int Ed. 2008;47:5577. doi: 10.1002/anie.200800409. [DOI] [PubMed] [Google Scholar]
- 32.Gao X, Nie S. J Phys Chem B. 2003;107:11575. [Google Scholar]
- 33.Han SW, Jang E, Koh WG. Sensors & Actuators: B Chemical. 2015;209:242. [Google Scholar]
- 34.Ji XH, Cheng W, Guo F, Liu W, Guo SS, He ZK, Zhao XZ. Lab on a Chip. 2011;11:2561. doi: 10.1039/c1lc20150f. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Xie Z, Gu H, Jin L, Zhao X, Wang B, Gu Z. NPG Asia Mater. 2012;4:e25. [Google Scholar]
- 36.Shojaei-Zadeh S, Morris JF, Couzis A, Maldarelli C. Journal of Colloid and Interface Science. 2011;363:25. doi: 10.1016/j.jcis.2011.06.073. [DOI] [PubMed] [Google Scholar]
- 37.Han M, Gao X, Su JZ, Nie S. Nat Biotechnol. 2001;19:631. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 38.Wang F, Tan WB, Zhang Y, Fan X, Wang M. Nanotechnology. 2005 [Google Scholar]
- 39.Leng Y, Sun K, Chen X, Li W. Chemical Society Reviews. 2015;44:5552. doi: 10.1039/c4cs00382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronda CR, Jüstel T, Nikol H. Journal of Alloys and Compounds. 1998;275–277:669. [Google Scholar]
- 41.Xu H, Wang H, Meng Y, Yan H. Solid State Communications. 2004;130:465. [Google Scholar]
- 42.Xu W, Song H, Yan D, Zhu H, Wang Y, Xu S, Bai X, Dong B, Liu Y. J Mater Chem. 2011;21:12331. [Google Scholar]
- 43.Lim X. Nature News. 2016;531:26. doi: 10.1038/531026a. [DOI] [PubMed] [Google Scholar]
- 44.Petoud S, Cohen SM, Bünzli JCG, Raymond KN. J Am Chem Soc. 2003;125:13324. doi: 10.1021/ja0379363. [DOI] [PubMed] [Google Scholar]
- 45.Lin M, Zhao Y, Wang SQ, Liu M, Duan ZF, Chen YM, Li F, Xu F, Lu TJ. Biotechnology Advances. 2012;30:1551–1561. doi: 10.1016/j.biotechadv.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Bünzli JCG, Piguet C. Chem Soc Rev. 2005;34:1048. doi: 10.1039/b406082m. [DOI] [PubMed] [Google Scholar]
- 47.Gorris HH, Ali R, Saleh SM, Wolfbeis OS. Advanced Materials. 2011;23:1652. doi: 10.1002/adma.201004697. [DOI] [PubMed] [Google Scholar]
- 48.Schuetz P, Caruso F. Chem Mater. 2002;14:4509. [Google Scholar]
- 49.Wartenberg N, Raccurt O, Imbert D, Mazzanti M, Bourgeat-Lami E. J Mater Chem C. 2013;1:2061. doi: 10.1002/chem.201203657. [DOI] [PubMed] [Google Scholar]
- 50.Haushalter RC Parallel Synthesis Technologies, Inc. 8,673,107. US. 2014
- 51.Haushalter RC Parallel Synthesis Technologies, Inc. 8,796,030. US. 2014
- 52.Haushalter RW, Haushalter RC Parallel Synthesis Technologies, Inc. 8,927,892. US. 2015
- 53.Parallel Synthesis Technologies. 2016 www.parallel-synthesis.com.
- 54.Gerver RE, Gómez-Sjöberg R, Baxter BC, Thorn KS, Fordyce PM, Diaz-Botia CA, Helms BA, DeRisi JL. Lab Chip. 2012;12:4716. doi: 10.1039/c2lc40699c. [DOI] [PubMed] [Google Scholar]
- 55.Buissette V, Moreau M, Gacoin T, Boilot JP, Chane-Ching JY, Le Mercier T. Chem Mater. 2004;16:3767. [Google Scholar]
- 56.Buissette V, Giaume D, Gacoin T, Boilot JP. J Mater Chem. 2006;16:529. [Google Scholar]
- 57.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng W, Han C, Li F. Advanced Materials. 2013;25:5287. doi: 10.1002/adma.201301946. [DOI] [PubMed] [Google Scholar]
- 59.Jiajia Zhou SXJZJQ. Nanoscale. 2015;7:15026. doi: 10.1039/c5nr02979a. [DOI] [PubMed] [Google Scholar]
- 60.Zhang F. Photon Upconversion Nanomaterials. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 233–253. [Google Scholar]
- 61.Lin M, Zhao Y, Wang S, Liu M, Duan Z, Chen Y, Li F, Xu F, Lu T. Biotechnology Advances. 2012;30:1551. doi: 10.1016/j.biotechadv.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Huang K, Idris NM, Zhang Y. Small. 2015;12:836. doi: 10.1002/smll.201502722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.