Abstract
Recent technological advances allow for high throughput profiling of biological systems in a cost-efficient manner. The low cost of data generation is leading us to the “big data” era. The availability of big data provides unprecedented opportunities but also raises new challenges for data mining and analysis. In this review, we introduce key concepts in the analysis of big data, including both “machine learning” algorithms as well as “unsupervised” and “supervised” examples of each. We note packages for the R programming language that are available to perform machine learning analyses. In addition to programming based solutions, we review webservers that allow users with limited or no programming background to perform these analyses on large data compendia.
New high throughput technologies allow biologists to generate enormous amounts of data ranging from measurements of genomic sequence to images of physiological structures. In between sequence and structure, measurements are performed through, and even beyond, the components of the central dogma. Such measurements include mRNA expression, transcription factor binding, protein expression, and metabolite concentrations. A given sample may include one or more of these levels of information. Now that the technology exists to probe each of these levels in high-throughput, two key questions must be addressed: “what should we measure?” and “how should we analyze collected data to discover underlying biology?” This review deals with the latter question.
We will introduce key concepts in the analysis of big data, including both “machine learning” algorithms as well as “unsupervised” and “supervised” examples of each. We will define important machine learning terms such as “gold standards.” By the conclusion of this review, the reader will have been introduced to key concepts in machine learning, as well as to motivating examples of analytical strategies that successfully discovered underlying biology. We refer readers elsewhere for a more general introduction to bioinformatics (Moore, 2007). We note packages for the R programming language that are available to perform machine learning analyses. In addition to programming based solutions, we will review available webservers that allow users with limited or no programming background to perform these analyses on large data compendia. We anticipate that servers which place advanced machine learning algorithms and large data compendia into the hands of biologists will be the conduit for the first wave of computational analyses of big data directed by wet bench biologists.
What Is Machine Learning and What Role Does it Have in Biology?
Effectively analyzing all currently available data through directed statistical analysis (e.g., constructing statistical tests of control and experimental conditions) is impossible. The amount of available data swamps the ability of an individual to perform, much less interpret, the results of all possible tests. Machine learning strategies are now beginning to fill the gap. These are computational algorithms capable of identifying important patterns in large data compendia. Machine learning algorithms fall into two broad classes: unsupervised and supervised algorithms. Both classes are best suited to addressing distinct questions, and both will be required to effectively focus “big data” on pressing biomedical problems.
In this review, we will introduce both supervised and unsupervised machine learning methods through case-study analyses of single datasets. For both classes of methods, our overall focus will be on integrative approaches that combine many measurements of different levels (e.g., mRNA expression, miRNA expression, and transcription factor binding), which we term “deep” data integration (Kristensen et al., 2014), or on integrative analyses over many distinct datasets, which we term “broad” data integration. Deep analyses let us understand how regulatory systems work together, while broad approaches lead to the discovery of general principles that are maintained across distinct conditions. Machine learning methods can be applied to perform effective integrative analyses, which are expected to fulfill the promise of big data in biology.
Big Data(sets)
An enormous amount of biological and clinical data has been generated in the past decade, particularly, after the advent of the next-generation sequencing technologies. The largest “deep” datasets to date are The Cancer Genome Atlas (TCGA; 2014) and The Encyclopedia of DNA Elements (ENCODE; 2012). TCGA contains measurements of somatic mutations (sequencing), copy number variation (array based and sequencing), mRNA expression (array based and sequencing), miRNA expression (array based and sequencing), protein expression (array based), and histology slides for approximately 7000 human tumors (http://cancergenome.nih.gov/). The ENCODE project has generated more than 2600 genomic datasets from ChIP-seq, RNA-seq, ChIA-PET, CAGE, high-C, and other experiments (The ENCODE Project Consortium, 2012). A total of 1479 ChIP-seq datasets have been produced to capture transcription factor binding and histone modification patterns in different human cell lines. Among them, 1242 datasets are ChIP-seq datasets for 199 transcription factors. The continuously increasing accumulation of big data has put big burden on storing and analyzing them. The European Bioinformatics Institute (EBI) in Hinxton, UK, part of the European Molecular Biology Laboratory and one of the world's largest biology-data repositories, currently stores 20 petabytes (1 petabyte is 1015 bytes) of data and back-ups about genes, proteins and small molecules. Genomic data account for 2 petabytes of that, a number that more than doubles every year (Marx, 2013).
Despite their considerable size, these deep datasets produced by large consortia are dwarfed by broad data compendia produced in the labs of individual investigators and made available upon publication. For example, Array Express, a compendium of publicly available gene expression data, contains more than 1.3 million genome-wide assays from more than 45,000 experiments (Rustici et al., 2013).
Supervised Versus Unsupervised Machine Learning
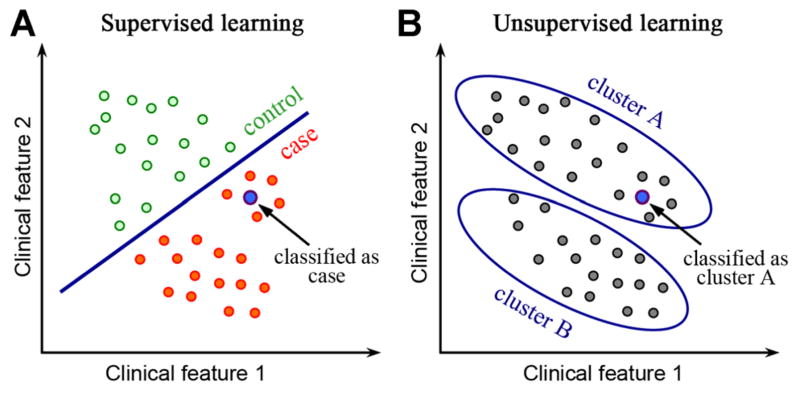
Machine learning algorithms can be divided into supervised and unsupervised based on the desired output and the type of input available for training the model. In a supervised learning model, the input consists of a set of training example with known labels (e.g., case and control). A supervised learning algorithm analyzes the training data and produces an inferred function, which can be used for classifying new samples (Fig. 1A). In an unsupervised learning model, the input is a set of unlabeled examples without predefined classes. The objective of unsupervised learning is to discover hidden signals within the data. After unsupervised learning is used to detect clusters, supervised algorithms can be run to classify new samples. For example, when a new sample is given, it can be assigned to the closest cluster (Fig. 1B). There are additional classes of machine learning algorithms beyond simply supervised and unsupervised. For example, semi-supervised algorithms combine both labeled and unlabeled data. These specialized algorithms exhibit promise but they are not yet widely applied in biology, so our review focuses only on the supervised and unsupervised algorithms. We will provide example case studies for both classes.
Fig. 1.
The difference between supervised and unsupervised machine learning. A: In supervised machine learning, a training dataset with labeled classes, for example, case or control, is provided. A model is trained to maximally differentiate between cases and controls, and then the classes of new samples are determined. B: In an unsupervised machine learning model, all samples are unlabeled. Clustering algorithms, an example of unsupervised methods, discover groups of samples that are highly similar to each other and distinct from other samples.
Unsupervised Methods Discover New Patterns
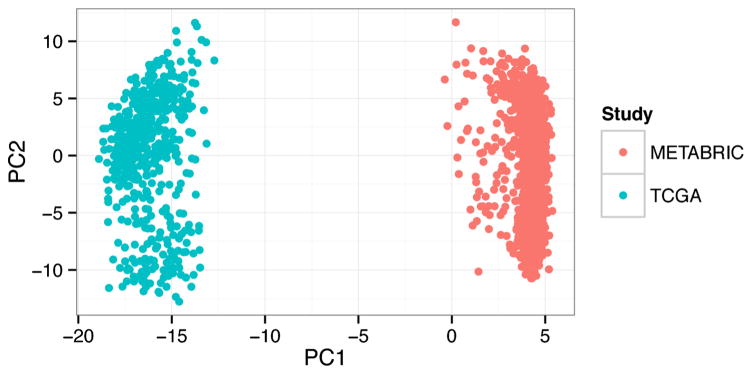
Unsupervised methods are used with the overarching goal to uncover the underlying organizational structure of the data, for example, “What patterns exist in gene expression of cancers?” These algorithms tend to discover dominant recurrent features in the data. They can be particularly susceptible to confounding factors. For example, principle components analysis (PCA) is an unsupervised approach to identify hidden features in the data that provide the most signal. The first principle component is the feature that explains most of the variability in the data. When we perform PCA analysis on a dataset combined from two large studies of breast cancer (Cancer Genome Atlas, 2012; Curtis et al., 2012), the algorithm identifies the study as the first principle component (Fig. 2). Such study, platform, or batch effects can confound unsupervised analyses, so these algorithms are often best applied within a single homogenous dataset. We discuss two such cases below.
Fig. 2.
Unsupervised analyses discover the predominant signals in the data. For example, principle component analysis applied to a dataset combined from two large studies of breast cancer identifies the study (METABRIC or TCGA) as the most important principle component (PC1). Such confounding factors have thus far made applying unsupervised analysis methods to broad compendia challenging, so this class of methods is most frequently used within large homogenous datasets.
Discovery of molecular subtypes of cancers
The challenge of discovering molecular subtypes is a problem that is best addressed with unsupervised methods. Given gene expression measurements for a set of cancers, we want to know if there exist shared gene expression patterns. Many clustering algorithms exist but those that divide the data into a predefined number of groups/clusters are most commonly used to discover cancer subtypes. An example of one of these algorithms is “k-means” clustering. K-means clustering attempts to divide measurements into groups that show similar patterns, but the number of groups, “k,” must be specified a priori. There are statistical ways to estimate the appropriate number of groups such as Tibshirani’s GAP statistic (Tibshirani et al., 2001), and these methods are implemented in the Cluster package (http://cran.r-project.org/web/packages/cluster/) for the R programming language. As an example, Tothill et al. (2008) demonstrate how to use k-means clustering to identify molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Unsupervised analyses are best judged by the reproducibility of their subtypes in independent data. For example, the TCGA ovarian cancer group reproduced the subtypes discovered by Tothill et al. in an independent cohort (Cancer Genome Atlas Research, 2011).
Genome segmentation based on histone modification patterns
Unsupervised machine learning techniques have been used to separate human genome into different functional segmentations based on genome wide histone modification patterns from the ENCODE (The Encyclopedia of DNA Elements) project. Specific histone modifications correlate with transcription factor binding, transcriptional initiation and elongation, enhancer activity and repression. Combinations of different modification types can provide even more precise insight into chromatin state (Ernst et al., 2011). Ernst et al. proposed an unsupervised machine learning method—a multivariate hidden Markov model called ChromHMM that uses combinatorial patterns of chromatin marks to distinguish chromatin states of the human genome (Ernst and Kellis, 2012).
The ChromHMM algorithm takes as input the ChIP-seq data for a collection of histone modifications. It receives a list of aligned reads for each chromatin mark and automatically converted into presence or absence calls for each mark across the genome. Chromatin states are analyzed at 200 bp intervals that roughly approximate nucleosome sizes. The output of ChromHMM is the learned chromatin-state model parameters and the chromatin-state assignments for each genomic position. The number of chromatin states can either be pre-specified or determined by a parameter-initialization procedure that allows inference of comparable models across different numbers of states. Chromatin states with similar emission parameters or proximal genomic locations are grouped together by ChromHMM. Given the chromatin states of genomic locations, one can further associate them with the likely biological roles of each chromatin state based on enrichment in diverse external annotations and experimental data. For example, certain chromatin states are enriched in histone marks for active enhancers and are thus annotated as active enhancer related states.
In one analysis, Ernst et al. applied the ChromHMM algorithm to map and analyze the chromatin state dynamics in nine human cell lines (Ernst and Kellis, 2012). They defined 15 different chromatin states, which correspond to repressed, poised and active promoters, strong and weak enhancers, putative insulators, transcribed regions, and large-scale repressed and inactive domains. These results provide a useful cell line specific functional annotation of the human genome using different cell lines. Biochemical and functional distinctions between these states have been experimentally validated.
Supervised Methods Build From Known Examples
Supervised algorithms answer questions that could be worded starting with the phrase “given that I know...” Because these algorithms start from a base of knowledge, they are frequently the most powerful choice to address to predict new pathway members, cell lineage specificity, or new interactions. Because these algorithms build from known biology, they can be substantially better at ignoring dataset-specific noise and are thus particularly suitable for integrative analysis of broad data collections (i.e. collections that span multiple datasets). Prior knowledge is provided to the algorithms through something called a “gold standard.” An ideal gold standard perfectly captures the essence of the question, but in biology our knowledge is often incomplete and is occasionally inaccurate. It is important that machine learning methods used in biology be robust to these inaccuracies.
Supervised methods can be targeted towards constructing decision rules that separate examples (e.g., genes) of two or more classes (e.g., in pathway vs. not in pathway). This task is called a “classification” task, and commonly used methods are support vector machine classification (SVM) (Cortes and Vapnik, 1995) or penalized logistic regression (Tibshirani, 1996). For gold standards are divided into “positive standards” which represent items that we want the algorithm to discover, and often “negative standards” which represent items we would like the algorithms to avoid. Intuitively both penalized logistic regression and SVM are straightforward. They attempt to separate the positive standards from negative standards while preserving simple models. The simplicity of the models is controlled by a user-defined value, which trades off classification ability for simplicity. Simplicity is often desired because simpler models are thought to be more likely to generalize.
Supervised methods can also be towards matching some continuous measurements. This is called a “regression” task, and commonly used methods are regularized linear regression or support vector regression. The least squares best fit line applied to many measurements is an example of a model built through supervised regression.
Using supervised methods, Ju et al. (2013) identified genes expressed in a single cell lineage from data measuring a complex mixture of cells in a solid tissue. Specifically the question addressed was “Given that I know these genes are expressed only in podocytes, what other genes are also only expressed in podocytes?” Ju et al. used a procedure termed “in silico nano dissection” to distinguish podocyte genes in a microarray gene expression dataset of renal biopsies. This approach combined the machine learning algorithm called support vector machines (SVM) with a decision rule capable of selecting the most informative examples from a set of gold standards. Nano-dissection built a rule that effectively divided gold standard podocyte genes (the positive standard) from genes expressed in other lineages (the negative standard). The same rule was applied to genes with unknown lineage specificity to predict which of those genes were also podocyte-specific. Results observed with the machine learning approach were significantly better than both an experiment using a murine podocyte model as well as a control method of selecting genes randomly.
While supervised methods are powerful methods to analyze simple datasets, they are particularly well suited to performing integrative analyses. We will illustrate how supervised methods can be used for both deep and broad integrative analyses. As an example of a deep integrative approach for a regression task comes from Cheng and Gerstein (2012). The goal of this analysis is to precisely predict the expression levels of genes in mouse embryonic stem cells (mESCs) from knowledge of 12 transcription factors and 7 histone modifications.
Integrating gene expression with TF binding and histone modification
Gene expression is precisely regulated by transcription factors (TFs) and histone modifications. Recently, genome wide TF binding and histone modification patterns have been profiled by ChIP-seq experiments. Given these datasets, Cheng et al. have developed supervised machine learning models to investigate the quantitative relationships between gene expression and the TF binding and/or histone modification signals (Cheng and Gerstein, 2012; Cheng et al., 2012). The models take the TF binding and histone modification signals as the “input” and relate them to the “output,” the expression levels of genes.
Given the genomic binding data for a total of N chromatin factors (e.g., TFs or histone marks), the supervised learning model predicts gene expression using the following steps. First, the DNA regions around transcriptional start site (TSS) of genes were divided into 80 bins, each of 100 bp in size. For each bin, the average signal for each chromatin factor was calculated, resulting in a matrix of G × N (G is the total number of genes and M is the number of factors). Second, in each bin the signals for all chromatin factors were utilized as predictors to predict the expression levels of genes. Finally, the predicted expression values by all of the 80 bins were combined by a second layer model to make the final prediction. A number of different machine learning methods have been employed, including Random Forest (RF), Support Vector Regression (SVR) and multivariable linear regression model (MLR) (Ouyang et al., 2009; Karlic et al., 2010; Cheng et al., 2012).
In one of the analysis, Cheng et al. have applied the above-described models to predict the expression level of genes in mouse embryostem cells(mESCs). As predictors, thechromatin factors include 12 TFs and 7 histone modifications (Cheng and Gerstein, 2012). Cheng et al. constructed and compared three models that respectively used TF binding signals only (TF model), histone modification signals only (HM model) and a combination of them (TF + HM model). Based on this analysis, the following findings have been achieved: (1) TF binding and histone modification achieve comparable predictive accuracy to gene expression, each accounting for >50% of variation in gene expression levels; (2) TF binding and histone modification signals are highly redundant in gene expression prediction—a model using both as predictor cannot further improve the prediction accuracy; (3) TF binding signals achieve the highest prediction accuracyatthe TSS of genes and the accuracy decays rapidly away from the TSS, whereas the histone modifications result in high accuracy in a relatively wide DNA regions from upstream to transcribed regions of genes; (4) models trained solely on protein-coding genes can predict microRNA expression with fairly high accuracy, suggesting a similar mechanism of transcriptional regulation between them.
Computational prediction of tissue specific human enhancers
Supervised machine learning models have also been applied to predict tissue specific enhancer regions in the human genome based on ENCODE data (The ENCODE Project Consortium, 2012). In Yip et al, the human genome was divided into 100 bp bins and the average signal of each chromatin feature was computed feature across the 100 bp of each bin. The features included DNase I hypersensitivity, FAIRE, and histone modifications (Yip et al., 2012). To predict tissue specific human enhancers, the following procedure was performed.
First, a supervised Random Forest model was built to predict the binding active regions (BAR) across the whole human genome. A BAR is defined as the genomic region where transcription-related factors (TRFs) tend to bind, which is associated with open chromatin structure that is highly accessible to transcription factors. To train the model, 100 bp bins overlapping with the binding peak of a TRF were collected as positive examples BAR regions, and non-positive bins were randomly sampled from the whole genome as negative examples. Chromatin data for GM12878 and K562 cell lines were used as predictors. The model predicted a list of candidate BAR bins in the human genome, presumably from DNA regions with open chromatin structure. Second, the predicted BAR bins that are proximal to known genes or promoters were excluded from the subsequent analysis. The resultant BAR bins were subject to further filtering to include only highly conserved ones. Finally, adjacent bins were merged into longer regions, and for each merged region the number of binding motifs for a set of embryo specific transcription factors (e.g., SOX and OCT family members) were counted. Conserved DNA regions that were predicted to be binding active and enriched in embryo TF binding motifs were reported as embryo specific enhancers.
Yip et al. selected 6 predicted enhancers out of the top 50 positive predictions for experimental validation. The enhancers were tested in embryos of transgenic mice on day E11.5 with a lacZ reporter gene fused with an hsp68 promoter as described in Pennacchio et al. (2006). Among the 6 tested predictions, 5 (83%) were found to have enhancer activities in various tissues with high reproducibility. The validation results suggested a high accuracy of the enhancer prediction protocol.
Supervised Mining of “Big Data” Without Programming
Large public data compendia contain substantial information about gene function, but these compendia have remained difficult for investigators to mine. Consequently such data are often used once to answer questions about response of genes to a treatment, or to identify subtypes. These data are then considered to have answered the question of interest, and are uploaded to public data repositories and promptly forgotten.
New webservers allow researchers to mine these compendia to construct hypotheses around specific processes, diseases or biological functions. The Platform for Interactive Learning by Genomics Results Mining (PILGRM) is one such server (Greene and Troyanskaya, 2011). PILGRM allows investigators to perform a supervised analysis of large gene expression compendia. The researcher defines a “positive standard” and a “negative standard.” The positive standard represents genes involved in the process in which the researcher would like to discover additional players. The negative standard indicate genes that are not desired, which could be either highly specific (e.g., genes that are known to be negative by an assay of the process) or general (e.g., genes selected randomly).
In summary, big data provide researchers with big opportunities for biological discovery and meanwhile confront them with big challenges in data storage, processing and analysis. Coordinated efforts from both experimental biologists and bioinformaticists are required to overcome these challenges.
Acknowledgments
This work was supported by NIH grants EY022300, LM009012, LM010098, AI59694, and GM103534.
Literature Cited
- The Cancer Genome Atlas. http://cancergenome.nih.gov/
- The R package “Cluster”. http://cran.r-project.org/web/packages/cluster/citation.html.
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Alexander R, Min R, Leng J, Yip KY, Rozowsky J, Yan KK, Dong X, Djebali S, Ruan Y, Davis CA, Carninci P, Lassman T, Gingeras TR, Guigo R, Birney E, Weng Z, Snyder M, Gerstein M. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22:1658–1667. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Gerstein M. Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:553–568. doi: 10.1093/nar/gkr752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Machine Learn. 1995;20:273–297. [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Troyanskaya OG. PILGRM: An interactive data-driven discovery platform for expert biologists. Nucleic Acids Res. 2011;39:W368–W374. doi: 10.1093/nar/gkr440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Greene CS, Eichinger F, Nair V, Hodgin JB, Bitzer M, Lee YS, Zhu Q, Kehata M, Li M, Jiang S, Rastaldi MP, Cohen CD, Troyanskaya OG, Kretzler M. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23:1862–1873. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen VN, Lingjarde C, Russnes HG, Vollan HM, Frigessi A, Borresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014;14:299–313. doi: 10.1038/nrc3721. [DOI] [PubMed] [Google Scholar]
- Marx V. Biology: The big challenges of big data. Nature. 2013;498:255–260. doi: 10.1038/498255a. [DOI] [PubMed] [Google Scholar]
- Moore JH. Bioinformatics. J Cell Physiol. 2007;213:365–369. doi: 10.1002/jcp.21218. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, Kurbatova N, Malone J, Mani R, Mupo A, Pedro Pereira R, Pilicheva E, Rung J, Sharma A, Tang YA, Ternent T, Tikhonov A, Welter D, Williams E, Brazma A, Parkinson H, Sarkans U. ArrayExpress update-trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B. 1996;58:267–288. [Google Scholar]
- Tibshirani R, Walther G, TH Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Ser B. 2001;63:411–423. [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I, Gertig D, DeFazio A, Bowtell DD Australian Ovarian Cancer Study G. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, Gerstein M. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]