Abstract
Autophagy is a critical cellular process that generally protects cells and organisms from stressors such as nutrient deprivation. In addition to its role in normal physiology, autophagy plays a role in pathological processes such as cancer. Indeed, there has been substantial work exploring the complex and context-dependent role of autophagy in cancer. One of the emerging themes is that in certain cancer types, autophagy is important to support tumor growth and therefore inhibiting autophagy as a therapeutic approach is actively being tested in clinical trials. A key mechanism of how autophagy promotes the growth and survival of various cancers is its ability to support cellular metabolism. The diverse metabolic fuel sources that can be produced by autophagy provides tumors with metabolic plasticity and can allow them to thrive in what can be an austere microenvironment. Therefore, understanding how autophagy can fuel cellular metabolism will enable more effective combinatorial therapeutic strategies.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a catabolic process whereby intracellular components (cargo) are enveloped in double-membraned vesicles, known as an autophagososomes, which ultimately fuse with lysosomes where the contents are degraded and recycled into the cytosol (Levine and Klionsky, 2004). In addition to macroautophagy, there are two additional types of autophagy, microautophagy and chaperone-mediated autophagy, which differ in how the particular cargo is delivered to lysosomes and are the subject of several excellent reviews (Bejarano and Cuervo, 2010; Kon and Cuervo, 2010; Mijaljica et al., 2011).
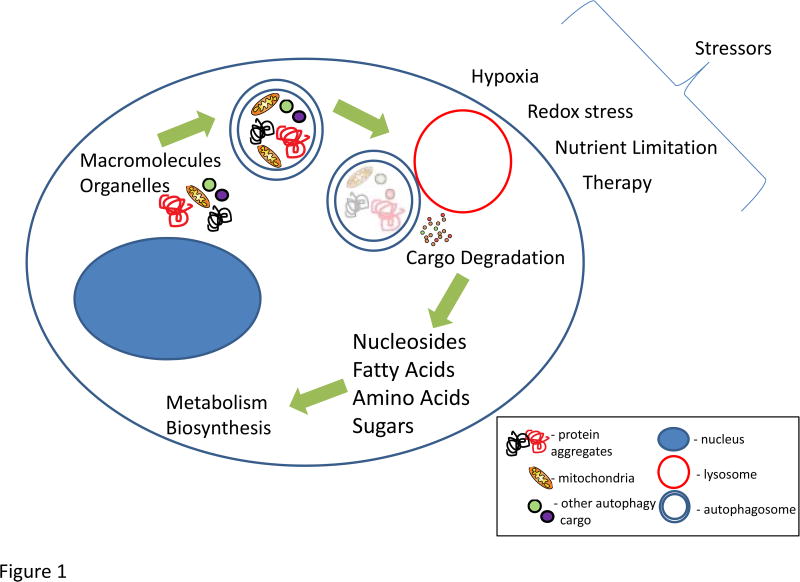
While initially thought to be only a mechanism of cell death, it is now recognized through multiple in vivo experiments that autophagy is a major reactive survival mechanism, although there are situations where it can contribute to cell death (Debnath et al., 2005; Levy and Thorburn, 2011; White, 2016). Autophagy promotes cellular and mammalian survival during periods of stress, particularly metabolic stress induced by nutrient deprivation (Figure 1). While most tissues have low levels of basal autophagy, it is significantly stimulated by the stressed state – the most well studied is starvation and this is tightly regulated by the mTOR and AMP kinase pathways (Kim and Guan, 2015; Mihaylova and Shaw, 2011; Neufeld, 2010).
Figure 1.
The process of autophagy leads to the degradation of cargo which amongst other functions serves to maintain organelle quality control (mitochondria as an example via mitophagy) as well as produce a variety of substrates to fuel cellular metabolism. This can help promote proliferation and survival of tumors during stressors such as hypoxia, redox stress, nutrient limitation, and a variety of anti-cancer therapies. The blue double-membraned vesicle -autophagosome; red vesicle – lysosome; blue shaded oval structure – nucleus; cargo in the autophagosome (mitochondria in yellow, protein aggregates in black and red, other cargo is generally represented in green and purple).
Through the seminal work of multiple groups, we have a significant understanding of the autophagic machinery and how it functions in the formation of autophagosomes through the fusion to lysosomes and culminating in the degradation of the autophagosome cargo and recycling of the breakdown products. Indeed, this has been the subject of multiple excellent reviews (Feng et al., 2014; Klionsky and Codogno, 2013; Ktistakis and Tooze, 2016). The process is directed and executed by a series of proteins encoded by autophagy-related genes (ATG) and there are currently more than 30 ATG genes that are involved in all aspects of the process (Klionsky et al., 2011; Mizushima, 2007). The process begins with the formation of the autophgosome from various sources of cellular membranes. During this process, cytosol, organelles and proteins become trapped in the forming autophagosome. Initially, autophagy was thought to be a non-selective degradative process whereby bulk cytoplasm and the proteins and organelles contained within it were sequestered by proximity into forming autophagosomes. More recently, it has been shown that there is significant selectivity for cargo and there are multiple selective autophagy pathways that are named for the particular cargo that is degraded (e.g. mitochondria – mitophagy; ferritin – ferritinophagy; ER – reticulophagy; bacteria – xenophagy) (Khaminets et al., 2016; Mancias and Kimmelman, 2016). Cargo is recognized by specific receptor proteins that bind to cargo and then often interact with ATG8 proteins that are inserted into the autophagsomal membrane. Once formed, autophagosomes fuse with lysosomes where the cargo is degraded via lysosomal enzymes and the resultant degraded products are recycled into the cytosol for use as substrates in metabolic and biosynthetic pathways.
Autophagy and Cancer
Given its important role in tissue homeostasis, it is not surprising that the dysregulation of autophagy has been linked to multiple disease states such as cancer and neurodegenerative disease. The role of autophagy in cancer has been of particular interest and the work in this area has greatly expanded over the past several years (reviewed in (Amaravadi et al., 2016; Galluzzi et al., 2015; Guo and White, 2017; Liu and Debnath, 2016; White, 2015)). Autophagy has a complex role in cancer and its function can be dependent on biological factors such as the tumor type, driving oncogene and tumor suppressor gene constellation of a tumor, as well as technical aspects such as the model system used to investigate its function (Amaravadi et al., 2016; Nyfeler and Eng, 2016). Initially, autophagy was thought to have a tumor suppressive role. This was based on two major lines of evidence. First, many of the activating mutations in oncogenes (e.g. PIK3CA) or inactivation of tumor suppressor genes (e.g. PTEN) would be predicted to inhibit autophagy. Second, deletion of autophagy genes in the setting of certain mouse models can result in the initiation of neoplasia. The initial identification of this phenotype was in Beclin1 (ATG6 ortholog) heterozygous mice, whereby these mice developed various neoplasms (Qu et al., 2003; Yue et al., 2003). An important aspect to note is that in this case autophagy was only partly attenuated as a functional copy of Beclin1 was intact in the mice. In contrast, when ATG5 was completely deleted in a mosaic fashion in the whole mouse, thereby completely inhibiting autophagy in those cells with the deletion, the results were different (Takamura et al., 2011). Interestingly, the only tissue that developed any neoplastic change was the liver, indicating that there are significant susceptibilities based on tissue type. Additionally, the lesions that developed were benign liver tumors, which indicates that autophagy is required for progression to malignancy and explains why Beclin1 heterozygous mice with diminished but intact autophagy can develop malignant tumors. Similar results were obtained when Atg7 deletion was targeted to the liver (Inami et al., 2011). The need for intact autophagy to progress to the malignant state may explain the apparent lack of mutations in canonical autophagy genes in human cancers. Beclin1 was initially identified as a haploinsufficient tumor suppressor gene given that multiple breast and ovarian tumors demonstrated loss of one allele (Liang et al., 1999), although indication that these are passenger deletions given its proximity to the BRCA1 tumor suppressor has recently been suggested (Laddha et al., 2014).
In contrast to its role in constraining tumor initiation, autophagy has been shown to have a critical pro-tumorigenic role in multiple cancer types. Initial studies demonstrated that autophagy was elevated in hypoxic regions of tumors and that the process could promote tumor cell survival upon a variety of stressors such as nutrient and oxygen deprivation (Degenhardt et al., 2006). These findings have been extended to show that, in many cases, autophagy can promote survival during the stress of therapies (chemotherapy, radiotherapy, and targeted agents) and thus promotes therapeutic resistance (Amaravadi et al., 2011; Rebecca and Amaravadi, 2016; Thorburn et al., 2014). In addition to therapeutic resistance, autophagy has been shown to play a critical role in tumor maintenance. Many tumor types demonstrate elevated basal autophagy and this can be seen in a cell autonomous manner, even under nutrient replete conditions (Kimmelman, 2011; White, 2015). Indeed, inhibition of autophagy genetically or pharmacologically can slow tumor growth in various model systems. Genetically engineered mouse models (GEMMs) of cancers have been particularly informative and helped define the critical role of autophagy in multiple cancers, including pancreatic cancer (Rosenfeldt et al., 2013; Yang and Kimmelman, 2014), lung cancer (Guo et al., 2013; Karsli-Uzunbas et al., 2014; Rao et al., 2014; Strohecker et al., 2013; Strohecker and White, 2014), prostate cancer (Santanam et al., 2016), melanoma (Xie et al., 2015), glioblastoma (Gammoh et al., 2016), and breast cancer (Huo et al., 2013; Wei et al., 2011). Additionally, the contribution of non-cell autonomous autophagy in tumor growth was also demonstrated in model organisms such as Drosophila (Katheder et al., 2017). This body of work has defined important cell autonomous and non-autonomous roles of autophagy in tumor maintenance. While the role of autophagy in promoting tumor growth is likely multifaceted, one of its key functions, and the subject of this review, is its contribution to the metabolism of tumors.
Metabolism and Cancer
It has long been known that cancers have altered metabolism to help meet the needs of cells that have the potential for unconstrained proliferation (Pavlova and Thompson, 2016; Vander Heiden and DeBerardinis, 2017). This includes a significant shift towards anabolic metabolism to biosynthesize the building blocks needed to support proliferation. Tumor cells have been shown to use a variety of fuel sources to accomplish these goals. Indeed, work has shown that some tumors have increased aerobic glycolysis (Warburg Effect) (Vander Heiden et al., 2009). Additionally, mitochondrial metabolism has been demonstrated to be critical for ATP production, redox balance, as well as the biosynthesis of other key metabolites in various tumor types (Weinberg and Chandel, 2015). Because of these changes in metabolism, there is potential for therapeutic targeting and preclinical studies have supported this approach with a variety of targets. Some of these efforts are making their way into early-phase clinical trials in human patients (Kishton and Rathmell, 2015; Ross and Critchlow, 2014; Vander Heiden, 2011). Two major concerns with this approach are the metabolic plasticity of cancer cells, allowing rapid metabolic rewiring as a compensatory response, as well as potential toxicity of targeting fundamental metabolic pathways in rapidly proliferating “normal” cells which also may rely on them for growth and survival. However, there is a long track record of targeting metabolic pathways in cancer therapy with anti-metabolites continuing to be a major component of the successful treatment of multiple cancer types.
Autophagy and cancer metabolism
Given the diverse substrates that can be degraded via autophagy, it is not surprising that autophagy has the potential to fuel nearly all aspects of central carbon metabolism (Guo and White, 2017; Rabinowitz and White, 2010) (Figure 1). For example, degradation of carbohydrates into sugars can fuel glycolysis; proteins into amino acids can fuel the TCA cycle; DNA into nucleosides can fuel glycolysis; and lipids into fatty acids can fuel the TCA cycle. Given the multitude of metabolic pathways that autophagy can feed into, it can provide normal and tumor cells with tremendous metabolic plasticity. This is particularly relevant to the multiple metabolic stresses a growing tumor faces ranging from hypoxia, to nutrient limitation, and even that from the therapies themselves.
Early studies that examined the importance of autophagy in cancer implicated its potential role in supporting tumor metabolism. Debnath and colleagues demonstrated that autophagy is important for Ras transformation and this is in part, due to its maintenance of glycolytic capacity (Lock et al., 2011). A similar role for autophagy in glycolysis was shown in polyoma middle T antigen (PMyT) driven breast cancer (Wei et al., 2011). The importance of autophagy in sustaining glycolysis was also demonstrated in hematological malignancies including chronic myeloid leukemia (Karvela et al., 2016). In a related fashion, our labs demonstrated that autophagy was important in growth of Ras-driven tumors (Guo et al., 2011; Yang et al., 2011). However, in the systems we used, maintenance of mitochondrial metabolism was significantly impaired in the setting of autophagy loss. Indeed, in both pancreatic cancer cells where autophagy was inhibited either pharmacologically with the lysosomal inhibitor chloroquine (CQ) or genetically using RNAi to essential autophagy genes, oxidative phosphorylation was significantly decreased. Consistent with this, growth of these cells could be partially rescued by adding mitochondrial fuel sources, such as pyruvate. In a similar fashion, oxidative phosphorylation and a variety of TCA cycle metabolites were decreased in autophagy incompetent, Hras transformed immortalized mouse kidney cells. Here, autophagy was also critical for maintaining mitochondrial quality control through mitophagy and the autophagy deficient transformed cells accumulated abundant defective mitochondria. The role of autophagy in sustaining amino acid pools in pancreatic cancers via control by the MiT/TFE transcription factors has also been demonstrated (Perera et al., 2015). Similarly, autophagy was important for mitochondrial function in Hodgkin lymphoma (Birkenmeier et al., 2016).
A series of studies using GEMMs for cancer have defined the role of autophagy in tumor progression and maintenance, including its role in the metabolism of various tumor types (reviewed in (Amaravadi et al., 2016)). In a Kras-driven lung cancer (GEMM), loss of Atg7 shifted the fate of cancers into benign tumors called oncocytomas and significantly decreased the tumor burden compared to autophagy-intact tumors (Guo et al., 2013). These oncocytomas show an accumulation of defective mitochondria as was previously demonstrated in the Hras transformed autophagy deficient cells. Atg7 loss also decreased tumor burden and shifted the histology in tumors where the tumor suppressor gene p53 was deleted. These tumors however had an interesting metabolic phenotype. In addition to the accumulation of defective mitochondria, these tumors accumulated significant amounts of neutral lipids, particularly cholesterol esters. Cells derived from these tumors were more sensitive to starvation and lipid levels were maintained during starvation, suggesting that they could not be efficiently used as a fuel source. A series of experiments demonstrated that the major defect was a problem with fatty acid oxidation (FAO). In fact, FAO inhibitors had a profound effect on autophagy-null tumor cell growth. Together, this data strongly implicates a role for autophagy in lipid homeostasis.
These findings in lung cancer were extended to mutant Braf-driven models, where again, loss of autophagy significantly impaired tumor development (Strohecker et al., 2013). Like the Kras-driven model, autophagy deficiency also increased the buildup of defective mitochondria and a shift to benign oncocytoma histology. Tumor growth and development were suppressed regardless of p53 status. Cells lines derived from both the Kras and Braf models were sensitive to starvation and oxygen consumption (OCR) under basal and starvation conditions was impaired in autophagy incompetent tumor cells. Both the OCR and growth defects could be profoundly rescued by the addition of exogenous glutamine, suggesting that autophagy may play a critical role for producing metabolic fuel sources. Interestingly, the genotype of a tumor may have a role in determining the metabolic contributions of autophagy, as the Braf-driven lung cancers with autophagy loss did not exhibit the same lipid accumulation defect that was seen in the Kras-driven model.
In Kras-driven pancreatic cancer GEMMs where autophagy was inhibited by Atg5 or Atg7 deletion, tumor development was significantly attenuated by autophagy loss (Rosenfeldt et al., 2013; Yang et al., 2014). When p53 loss was engineered into the model, autophagy inhibition delayed tumor progression in the setting of p53 LOH, analogous to the human situation (Amaravadi and Debnath, 2014; Yang et al., 2014). When p53 homozygous loss occurred during embryogenesis with both copies deleted simultaneously (a limitation of this particular mouse model), autophagy loss had a paradoxical effect that hastened tumorigenesis (Rosenfeldt et al., 2013). Cell lines derived from these models were sensitive to acute autophagy inhibition by genetic or pharmacologic approaches as were patient derived xenograft models treated with hydroxychloroquine irrespective of p53 status. Consistent with previous studies, autophagy inhibition in cells derived from the GEMMs significantly decreased OCR, indicating a role in mitochondrial metabolism.
A recent study using cells lines derived the Kras-driven lung cancer GEMM with and without ATG7 and sophisticated metabolic tracing studies sought to define the critical metabolic contributions of autophagy to tumor cells (Guo et al., 2016). Cells were labeled with heavy isotopes to saturation using a mixture of labeled nutrients for several days to maximize labeling of intracellular of macromolecules. They then developed a methodology using a chase with unlabeled media in combination with a short starvation period to identify which metabolites were produced through the autophagic degradation of macromolecules. This work made several critical observations. First, with the exception of small increases in glucose uptake and lactate production in full nutrient conditions, as has been seen previously in some studies, autophagy loss mainly impacted metabolic fluxes in nutrient-depleted conditions. One of the critical pathways that was supported by autophagy in both fed and starved states was redox balance, but restoration of this alone was not critical for the survival of cells during starvation. Consistent with work showing the importance of autophagy in maintaining oxidative phosphorylation in these models, autophagy was critical to maintain energy charge during starvation. In fact, there was a significant decrease in total nucleotide pools during starvation in autophagy-null cells and combinations of nucleosides could actually rescue survival in these cells during starvation. The authors proposed that the energy crisis in the starved autophagy-deficient cells causes nucleotide degradation to limit AMP accumulation as a futile escape mechanism. Thus, a critical function of autophagy in tumor cells is to prevent this fatal nucleotide depletion during periods of starvation.
While the aforementioned studies have focused predominantly on the direct contribution of autophagy to the metabolism in the tumor cell itself, several studies have also shown a critical impact of autophagy on tumor growth in a cell non-autonomous manner and in many cases this is related to effects on both tumor cell and host metabolism (Endo et al., 2017; Karsli-Uzunbas et al., 2014; Katheder et al., 2017; Sousa et al., 2016). Using a tamoxifen inducible cre recombinase, Atg7 was systemically deleted in the whole body of adult mice (Karsli-Uzunbas et al., 2014). Interestingly, and in contrast to embryonic deletion where mice die right after weaning, these mice were able to survive several months without autophagy. Autophagy was critical for mice to survive fasting and maintenance of fat and glycogen stores, as well as muscle mass during fasting. Indeed, fasted Atg7-null adult mice die of hypoglycemia. Even during the fed state, autophagy loss caused a significant depletion of white adipose tissue, indicating that there may be a systemic alteration in lipid metabolism when autophagy is inhibited.
The authors went on to use this model with a duel recombinase system (Kras and p53 FRT alleles) to test the effect of acute, systemic autophagy inhibition (analogous to a potent autophagy inhibitor administered systemically) on lung tumor growth. Tumors were induced using adenoviral Flpase and when they were fully formed, and Atg7 was deleted in the whole body using tamoxifen. Lung tumors showed profound treatment effects in terms of size, tumor burden, and tumor cell death where autophagy was acutely deleted throughout the mice. These effects were noted to be much more robust than the previous studies where Atg7 was deleted only in tumors. There are multiple explanations for this – the acute nature of the deletion in the fully formed tumor (rather than during the development of the tumor) may allow less time for adaptation; the systemic ablation of autophagy may induce anti-tumor effects. Defective host autophagy may possibly modulate the immune system to compromise tumor growth, or as was shown by the authors, alterations in whole body metabolism may contribute. In support of the systemic metabolism concept, calorie restriction was shown in a recent study to cooperate with autophagy loss in tumors cells grown as xenografts in mice (Lashinger et al., 2016). Thus, it is tempting to speculate that the systemic metabolic changes caused by defective autophagy may cooperate with the need for autophagy in tumor cells themselves to maintain energy balance and nucleotide pools.
Other recent work investigating a possible metabolic cross-talk in pancreatic cancer also highlights the importance of tumor cell non-autonomous autophagy in the metabolic support of these tumors (Sousa et al., 2016). Pancreatic cancers are highly desmoplastic and this leads to an hypoxic and nutrient poor microenvironment. One of the key cell types in the stroma is a specialized type of fibroblast known as a pancreatic stellate cell. This work showed that stellate cells secreted metabolites that supported pancreatic cancer mitochondrial metabolism. Through a series of metabolomics studies, it was determined that stellate cells rapidly secrete the non-essential amino acid alanine which is then taken up by pancreatic cancer cells. Alanine carbon fuels the TCA cycle of PDAC and allows glucose to be utilized for other anabolic processes such as serine/glycine biosynthesis. In the process of investigating how the stellate cells produce the secreted alanine, it was discovered that the secretion of the alanine was autophagy-dependent. Inhibition of autophagy in stellate cells inhibited alanine secretion and the conditioned media no longer impacted pancreatic cancer metabolism. The secreted alanine was able to support cell proliferation in nutrient limited media, which recapitulated the tumor microenvironment. This activity was lost in conditioned media collected from autophagy impaired stellate cells, despite the fact that autophagy loss did not impact growth or survival of the stellate cells themselves. Using a series of co-transplantation studies, it was shown that coinjection of pancreatic cancer cells with stellate cells improved tumor take and growth. Importantly, inhibition of autophagy specifically in the stellate cell compartment via RNAi abrogated this effect. Thus, this work demonstrates an autophagy-dependent metabolic crosstalk between pancreatic cancer and stellate cells that supports pancreatic cancer metabolism and promotes tumor growth. In line with this work, Endo et al. have recently shown that autophagy inhibition in pancreatic stellate cells decreases tumor growth in transplantation models (Endo et al., 2017). While this work did not focus on the metabolic cross-talk and focused on the role of autophagy in stellate cell activation, it confirms the importance of both stromal and tumor cell autophagy in pancreatic cancer growth. Further work is needed as to whether stellate cell activation is linked to alanine secretion. Additionally, while it is not yet clear whether this same type of autophagy-dependent metabolic crosstalk will extend to other tumor types, previous work has shown that lung cancer associated fibroblasts have elevated basal autophagy as compared to those derived from non-cancerous tissue thereby raising the possibility that other tumor types may share similar metabolic phenotypes (Chaudhri et al., 2013). Lastly, work in prostate cancer has shown that the autophagy adaptor, p62, plays an important role in the metabolic cross-talk in the tumor stroma (Valencia et al., 2014).
Conclusions/Perspectives
A great deal of evidence implicates the importance of autophagy in the metabolism of various cancers. Recent studies have helped more precisely define the metabolic contributions of autophagy and how these may factor in the pro-tumorigenic activities of this complex process. Interestingly, the involvement of autophagy in metabolism extends beyond the cancer cells themselves as it appears to play a role in the metabolic cross-talk between tumor and stromal cells in tumor types such as pancreatic cancer. The diverse nature of autophagic cargo allows for the rapid production of needed metabolic fuel sources that can feed into nearly every pathway in central carbon metabolism and beyond. Given the tremendous stresses that tumor cells undergo, this provides a reservoir to help cope with the ever changing metabolic needs of a tumor. This includes cell intrinsic stresses such as the need to maintain energy homeostasis and nucleotide pools during unconstrained proliferation and increased ROS production. On top of these intrinsic stresses, tumor cells encounter metabolic stresses created by harsh conditions in the tumor microenvironment. These include hypoxia, low pH, and decreased nutrient supply due to dysfunctional vasculature. Autophagy provides cells the ability to adapt to a series of stresses, allowing the tumor to continue to proliferate and survive.
Indeed, efforts to target autophagy given its aforementioned role in tumor cell growth are already underway (Amaravadi et al., 2016; Rebecca and Amaravadi, 2016). While most of these were not designed specifically in terms of inhibiting metabolism, they nonetheless will likely have this consequence on the tumors (Figure 2). The first series of trials have used antimalarial and rheumatological drugs such as hydroxychloroquine (HCQ) as part of the treatment regimen for a variety of cancer histologies and several have been reported with mixed results (Rebecca and Amaravadi, 2016). HCQ is not a specific autophagy inhibitor and acts at the level of the lysosome by preventing its acidification and therefore blocking the final step of autophagy, degradation of cargo in the lysosome (Piao and Amaravadi, 2016). Therefore, in addition to blocking autophagy, it also impacts other lysosomal processes. In the area of tumor metabolism, HCQ likely inhibits macropinocytosis, which along with autophagy converges at the lysosome. Interestingly, in Ras-driven cancers, macropinocytosis has also been shown to be critical for tumor cell growth at least in part through providing alternative metabolic fuel sources through the degradation of external proteins (Commisso et al., 2013). Therefore, HCQ likely has impacts on tumor cell metabolism through both of these processes (autophagy and micropinocytosis) as well as potentially other pathways (Maes et al., 2016). One important limitation of HCQ is its potency, as high micromolar levels are required to inhibit autophagy completely and to impact tumor cell growth. While micromolar levels can be achieved in patients (Munster et al., 2002; Tett et al., 1993), and it appears that autophagy levels can be modulated in patients, it is not likely a complete block and is not seen in all patients (Boone et al., 2015; Wolpin et al., 2014). However, when used in certain therapeutic combinations in some cancer types, there appears to be activity (Boone et al., 2015). More potent and selective autophagy inhibitors are in various phases of development (Lebovitz et al., 2015; Wang et al., 2016).
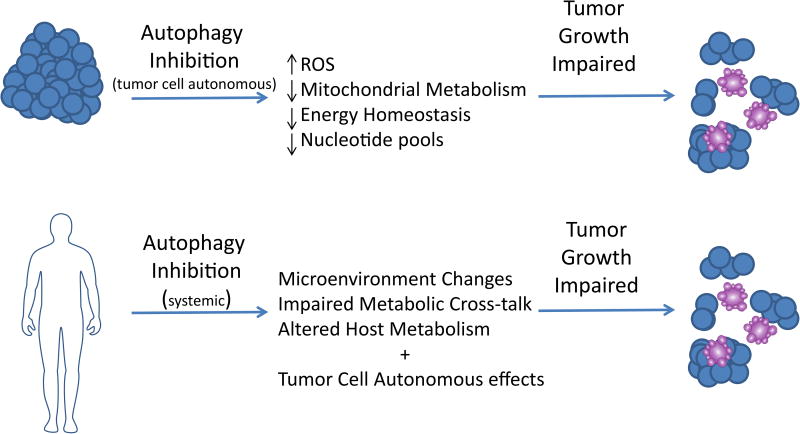
Figure 2.
Autophagy inhibition in the tumor cells can disrupt tumor metabolism leading to a variety of metabolic consequences including impaired mitochondrial metabolism, redox imbalance, depletion of nucleotide pools, and a decrease in energy charge. In certain settings, this can lead to an impairment in tumor growth as well as tumor cell death. Similarly, systemic autophagy inhibition may have anti-tumor effects through both the aforementioned tumor cell autonomous effects as well as its impact on the tumor microenvironment, host metabolism, and disruption of metabolic cross-talk circuits between tumor and stroma cells. Impairment of these can also lead to a disruption of tumor growth.
Future trials may be designed to utilize therapeutic combinations that take advantage of the role of autophagy in tumor metabolism. Therapies such as chemotherapy and radiation therapy can induce redox stress and the metabolic contributions from autophagy can help ameliorate the detrimental cellular effects of ROS as well as provide substrates to assist with redox balance. Therefore, inhibiting autophagy in conjunction with other therapeutic perturbations may disrupt these metabolic compensations and provide therapeutic utility in certain settings (Figure 2). These would also include using autophagy inhibition with inhibitors of cell metabolism to prevent a compensatory response. While HCQ may provide an additional benefit by targeting other lysosomal processes, more potent autophagy inhibitors will allow these clinical concepts to be tested in a more rigorous fashion.
The role of autophagy in cancer has gained increasing attention in recent years and there is growing interest in targeting the process as a cancer therapeutic. In this review, Kimmelman and White discuss the emerging importance of how autophagy supports tumor growth through its involvement in cellular metabolism.
Acknowledgments
We apologize for the omission of any primary references. ACK was supported by National Cancer Institute Grants R01CA157490, R01CA188048 and P01CA117969; ACS Research Scholar Grant RSG-13-298-01-TBG, and NIH grant R01GM095567. EW is supported by the NIH under award numbers R01CA163591, R01CA130893, R01CA188096, and R01CA193970, the Robert Wood Johnson Foundation and P30CA72720 to the Rutgers Cancer Institute of New Jersey.
Footnotes
Disclosure of Potential Conflicts of Interest: ACK and EW have financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach, and the autophagic control of iron metabolism. A.C.K is on the SAB of Cornerstone Pharmaceuticals. EW is on the SAB of Forma Therapeutics
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaravadi R, Debnath J. Mouse models address key concerns regarding autophagy inhibition in cancer therapy. Cancer Discov. 2014;4:873–875. doi: 10.1158/2159-8290.CD-14-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier K, Moll K, Newrzela S, Hartmann S, Drose S, Hansmann ML. Basal autophagy is pivotal for Hodgkin and Reed-Sternberg cells' survival and growth revealing a new strategy for Hodgkin lymphoma treatment. Oncotarget. 2016;7:46579–46588. doi: 10.18632/oncotarget.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, Singhi AD, Bao P, Bartlett DL, Liotta LA, et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri VK, Salzler GG, Dick SA, Buckman MS, Sordella R, Karoly ED, Mohney R, Stiles BM, Elemento O, Altorki NK, et al. Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor. Mol Cancer Res. 2013;11:579–592. doi: 10.1158/1541-7786.MCR-12-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, Koikawa K, Okumura T, Sada M, Horioka K, et al. Autophagy is Required for Activation of Pancreatic Stellate Cells, Associated With Pancreatic Cancer Progression and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammoh N, Fraser J, Puente C, Syred HM, Kang H, Ozawa T, Lam D, Acosta JC, Finch AJ, Holland E, et al. Suppression of autophagy impedes glioblastoma development and induces senescence. Autophagy. 2016;12:1431–1439. doi: 10.1080/15548627.2016.1190053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, White E. Autophagy, Metabolism, and Cancer. Cold Spring Harb Symp Quant Biol. 2017 doi: 10.1101/sqb.2016.81.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, Barnard N, Ganesan S, Karantza V, White E, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov. 2013;3:894–907. doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvela M, Baquero P, Kuntz EM, Mukhopadhyay A, Mitchell R, Allan EK, Chan E, Kranc KR, Calabretta B, Salomoni P, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy. 2016;12:936–948. doi: 10.1080/15548627.2016.1162359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhasz G, et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishton RJ, Rathmell JC. Novel therapeutic targets of tumor metabolism. Cancer J. 2015;21:62–69. doi: 10.1097/PPO.0000000000000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. (2nd) 2011;7:1273–1294. doi: 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5:427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis NT, Tooze SA. Digesting the Expanding Mechanisms of Autophagy. Trends Cell Biol. 2016;26:624–635. doi: 10.1016/j.tcb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12:485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashinger LM, O'Flanagan CH, Dunlap SM, Rasmussen AJ, Sweeney S, Guo JY, Lodi A, Tiziani S, White E, Hursting SD. Starving cancer from the outside and inside: separate and combined effects of calorie restriction and autophagy inhibition on Ras-driven tumors. Cancer Metab. 2016;4:18. doi: 10.1186/s40170-016-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz CB, DeVorkin L, Bosc D, Rothe K, Singh J, Bally M, Jiang X, Young RN, Lum JJ, Gorski SM. Precision autophagy: Will the next wave of selective autophagy markers and specific autophagy inhibitors feed clinical pipelines? Autophagy. 2015;11:1949–1952. doi: 10.1080/15548627.2015.1078962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131:130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liu J, Debnath J. The Evolving, Multifaceted Roles of Autophagy in Cancer. Adv Cancer Res. 2016;130:1–53. doi: 10.1016/bs.acr.2016.01.005. [DOI] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes H, Kuchnio A, Carmeliet P, Agostinis P. Chloroquine anticancer activity is mediated by autophagy-independent effects on the tumor vasculature. Mol Cell Oncol. 2016;3:e970097. doi: 10.4161/23723548.2014.970097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Kimmelman AC. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J Mol Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Munster T, Gibbs JP, Shen D, Baethge BA, Botstein GR, Caldwell J, Dietz F, Ettlinger R, Golden HE, Lindsley H, et al. Hydroxychloroquine concentration-response relationships in patients with rheumatoid arthritis. Arthritis Rheum. 2002;46:1460–1469. doi: 10.1002/art.10307. [DOI] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B, Eng CH. Revisiting autophagy addiction of tumor cells. Autophagy. 2016;12:1206–1207. doi: 10.1080/15548627.2016.1170265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Amaravadi RK. Targeting the lysosome in cancer. Ann N Y Acad Sci. 2016;1371:45–54. doi: 10.1111/nyas.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2016;35:1–11. doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- Ross SJ, Critchlow SE. Emerging approaches to target tumor metabolism. Curr Opin Pharmacol. 2014;17:22–29. doi: 10.1016/j.coph.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Santanam U, Banach-Petrosky W, Abate-Shen C, Shen MM, White E, DiPaola RS. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 2016;30:399–407. doi: 10.1101/gad.274134.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, White E. Targeting mitochondrial metabolism by inhibiting autophagy in BRAF-driven cancers. Cancer Discov. 2014;4:766–772. doi: 10.1158/2159-8290.CD-14-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tett SE, Day RO, Cutler DJ. Concentration-effect relationship of hydroxychloroquine in rheumatoid arthritis--a cross sectional study. J Rheumatol. 1993;20:1874–1879. [PubMed] [Google Scholar]
- Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell. 2014;26:121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Hu Q, Shen HM. Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol Res. 2016;105:164–175. doi: 10.1016/j.phrs.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6:a026120. doi: 10.1101/cshperspect.a026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, Fuchs CS, McCleary NJ, Meyerhardt JA, Ng K, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kimmelman AC. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy. 2014;10:1683–1684. doi: 10.4161/auto.29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]