Abstract
Background and Aims Plant roots growing underground are critical for soil resource acquisition, anchorage and plant–environment interactions. In wheat (Triticum aestivum), however, the target root traits to improve yield potential still remain largely unknown. This study aimed to identify traits of seedling root system architecture (RSA) associated with yield and yield components in 226 recombinant inbred lines (RILs) derived from a cross between the bread wheat Triticum aestivum ‘Forno’ (small, wide root system) and spelt Triticum spelta ‘Oberkulmer’ (large, narrow root system).
Methods A ‘pouch and wick’ high-throughput phenotyping pipeline was used to determine the RSA traits of 13-day-old RIL seedlings. Two field experiments and one glasshouse experiment were carried out to investigate the yield, yield components and phenology, followed by identification of quantitative trait loci (QTLs).
Key Results There was substantial variation in RSA traits between genotypes. Seminal root number and total root length were both positively associated with grains m–2, grains per spike, above-ground biomass m–2 and grain yield. More seminal roots and longer total root length were also associated with delayed maturity and extended grain filling, likely to be a consequence of more grains being defined before anthesis. Additionally, the maximum width of the root system displayed positive relationships with spikes m–2, grains m–2 and grain yield. Ten RILs selected for the longest total roots exhibited the same effects on yield and phenology as described above, compared with the ten lines with the shortest total roots. Genetic analysis revealed 38 QTLs for the RSA, and QTL coincidence between the root and yield traits was frequently observed, indicating tightly linked genes or pleiotropy, which concurs with the results of phenotypic correlation analysis.
Conclusions Based on the results from the Forno × Oberkulmer population, it is proposed that vigorous early root growth, particularly more seminal roots and longer total root length, is important to improve yield potential, and should be incorporated into wheat ideotypes in breeding.
Keywords: Phenology, QTL, root, spelt, Triticum aestivum, Triticum spelta, wheat, yield
INTRODUCTION
The roots, known as ‘the hidden half’ growing underground, are of fundamental importance to plant growth. For example, the roots acquire water and nutrients from soil, and can function as a storage site for nutrients (Kutman et al., 2012; Smith and De Smet, 2012; Gaju et al., 2014). They provide the anchorage for plants against lodging, and serve as a major interface sensing, and interacting with, biotic and abiotic factors in the rhizosphere (Smith and De Smet, 2012). An efficient root system is even more crucial for crops grown in poor soils, where water and nutrients are in short supply (Lynch, 2007). In addition, improving rooting efficiency could decrease the input of chemical fertilizers into the agroecosystem, leading to a reduction of both greenhouse gas emissions incurred during their production and the eutrophication of the natural environments from their application (White et al., 2013).
In wheat (Triticum aestivum), there are two types of roots: the seminal roots and the nodal roots (Kirby, 2002). While germinating, the radicle bursts through the coleorhiza of the seed, and then 3–6 other seminal roots emerge. Each seminal root has a root cap, followed sequentially by the zones of division, elongation and maturation. Numerous root hairs develop in the zone of maturation, and further back from the root hair zone the lateral roots grow from the stele. The seminal root system can reach 1·5–2·0 m in depth and support the plant throughout the crop’s life (Sylvester-Bradley et al., 2008). Unlike the seminal roots growing directly from the seed, the nodal (crown or adventitious) roots arise from the stem basal nodes and begin to extend when tillering starts. These roots are usually thicker than the seminal roots, and appear relatively horizontally. At anthesis, the whole root system consisting of seminal and nodal roots in the field reaches a maximum: approx. 17 km m–2 in root density (sample depth: 0–100 cm) and 0·8 t ha–1 in root dry weight, and this varies with genotype, sowing date, temperature, soil structure and environmental stress (Sylvester-Bradley et al., 2008; White et al., 2015). Roots then senesce during grain filling, though some in the deeper soil layers may continue growing.
The root system of wheat is complex, and a series of quantitative traits have been used to describe the root spatial configuration in soil, namely root system architecture (RSA). The architectural traits include root number and length, tip and emergence angles, rooting width and depth, convex hull area and root mass centre (Pound et al., 2013; Atkinson et al., 2015). However, quantifying these traits in the field is difficult since extraction of all intact roots from the soil is extremely laborious, time-consuming and expensive. Alternatively, a number of indoor approaches have been applied in practice, for instance using hydroponic culture (Ayalew et al., 2015; Kabir et al., 2015), gel-filled chambers (Manschadi et al., 2008; Christopher et al., 2013), germination bags (Robertson et al., 1979), sand-filled pots (Waines and Ehdaie, 2007; Hamada et al., 2012), soil-filled pots (Cao et al., 2014), clear pots (Richard et al., 2015), a germination paper-based ‘pouch and wick’ system (Atkinson et al., 2015), a paper-based ‘cigar roll’ system (Li et al., 2011; Bai et al., 2013) and soil-filled columns scanned by X-rays (Gregory et al., 2009). Coupled with digital image analysis software, some of these systems permit the fast measurement of RSA over a large number of genotypes, which have been used to investigate the diversity and genetics of RSA in wheat.
Root system architecture shows large genetic variation among different ploidy levels, between domesticated and wild forms, and within each of the diploids, tetraploids and hexaploids (Robertson et al., 1979; Manschadi et al., 2008; Ayalew et al., 2015). Hexaploid wheat has longer roots than the other ploidy levels (Ayalew et al., 2015); however, compared with the landraces, modern bread wheat cultivars have smaller root mass (Waines and Ehdaie, 2007), suggesting that the RSA needs to be optimized for further yield improvement, particularly under infertile soil conditions (Lynch, 2007). In addition, high-throughput phenotyping techniques also allow identification of the quantitative trait loci (QTLs) associated with the RSA. To date, about 339 QTLs have been detected for different root architectural traits, and distributed on all wheat chromosomes except 1D (Ren et al., 2012; Bai et al., 2013; Christopher et al., 2013; Liu et al., 2013; Atkinson et al., 2015; Kabir et al., 2015; Maccaferri et al., 2016).
Although many studies have initiated the first step to understand wheat RSA per se, the root architectural traits that are associated with increased yield remain largely unknown. The aim of the current study is to identify desirable root architectural traits at the seedling stage which could be associated with yield and yield components in the field. A mapping population, including 226 recombinant inbred lines (RILs) derived from a cross between the bread wheat Triticum aestivum ‘Forno’ and spelt Triticum spelta ‘Oberkulmer’, was quantified for the seedling RSA traits by using a ‘pouch and wick’ high-throughput phenotyping pipeline. The yield and yield components as well as phasic development of the population were determined in two field experiments and one glasshouse experiment. The physiological and genetic (i.e. QTL co-location) relationships between the RSA and yield traits were then analysed.
MATERIALS AND METHODS
Plant materials
Spelt is a relative of bread wheat. A preliminary experiment showed that spelt has a larger, deeper root system and resultant higher nitrogen uptake efficiency compared with bread wheat. To introduce the genetic variation in root traits from spelt, a cross between the bread wheat cultivar ‘Forno’ and spelt landrace ‘Oberkulmer’ was made, and the subsequent F5 RIL mapping population consisting of 226 lines was used in the present study (Messmer et al., 1999). Forno and Oberkulmer are both winter types and were widely grown in Switzerland.
Root phenotyping of the RIL seedlings in growth chamber
A germination paper-based ‘pouch and wick’ phenotyping system, coupled with digital image analysis, was adopted to measure the root architectural traits in 2014 (Bonser et al., 1996; Atkinson et al., 2015). All 226 RILs, together with Forno and Oberkulmer, were arranged in a randomized complete block (RCB) design with 20 plants per line. Due to limited space within the growth chamber, the 20 plants of each RIL were grown in eight runs: three plants per RIL in each of Run 1, 2, 3, 5, 6 and 7, as well as one plant per RIL in each of Run 4 and 8. In each run, intact, uniform and representative seeds of each RIL were selected from the combined grain samples in 2013, and grown at the top of the growth pouches (see below), with the embryos pointing downwards.
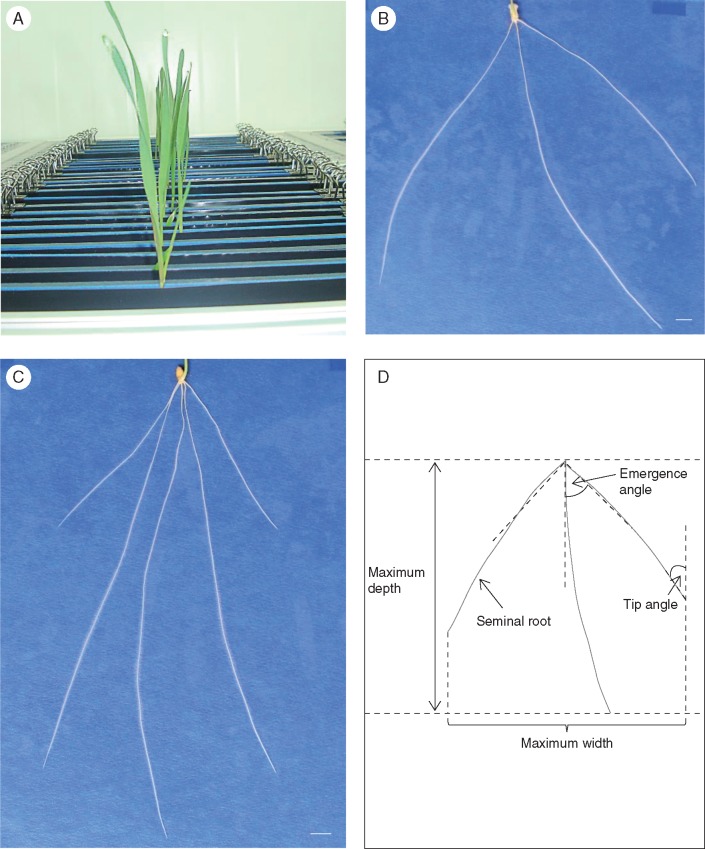
The growth system consisted of the pouches and tanks filled with deionized water. Each growth pouch included two black polythene sheets (24 × 28 cm; Cransford Polythene Ltd, Suffolk, UK), two blue germination papers (24 × 30 cm; Anchor Paper Company, St Paul, MN, USA), a plastic rod (Acrylic Online, Hull, UK) and two 19 mm fold-back clips (Fig. 1A). One polythene sheet and one germination paper were attached to one side of the rod, and the other polythene sheet and germination paper to the other side, fixed with two fold-back clips. Two seeds were grown on a growth pouch (one each side), and there were four tanks with a total of 360 pouches, allowing a maximum of 720 plants to be screened per run. Each tank was filled with 3 L of deionized water at the beginning of the experiment, and the water level was topped up at 2 d intervals to ensure that the germination papers remained saturated throughout the experiment. The tanks were placed in an environmentally controlled growth chamber, with 12 h light [photosynthetically active radiation (PAR) = 240 μmol m–2 s–1 at the tank level, temperature = 17 °C] and 12 h dark (temperature = 13 °C).
Fig. 1.
Root phenotyping pipeline for the Forno × Oberkulmer mapping population grown in a growth chamber during 2014. (A) Germination paper-based ‘pouch and wick’ system. (B and C) Example root images of Forno and Oberkulmer, respectively (scale bar = 1 cm). (D) Measurement of root architectural traits on a recombinant inbred line.
The images of the vertically grown root systems (Fig. 1B, C) were taken 13 d after transferring them onto the germination papers, using a Nikon D5100 DSLR camera and modified copy stand. The images were then analysed using the semi-automated software RootNav V1.7.5 (Pound et al., 2013). The parameters used in this study included: (1) seminal root number, excluding the lateral roots; (2) average seminal root length; (3) total root length, the total length of seminal and lateral roots for a given plant; (4) maximum width, the horizontal distance between the two widest points achieved by the root system for all depths; (5) maximum depth, the vertical distance from the base to the tip of the deepest seminal root; (6) width to depth ratio, the ratio of maximum width to maximum depth; (7) tip angle of seminal roots, the average angle of all seminal root tips relative to the vertical axis; and (8) emergence angle of seminal roots, the average angle of two outermost seminal root bases (by 50 % of root length) relative to the vertical axis (Fig. 1D). Among these parameters, total root length was considered to represent the size of the root system, with seminal root number and average seminal root length as components, while width to depth ratio was used to represent the shape of the root system, with maximum width and depth as components.
Field experiments
Yield and yield components.
The yield and yield components of the Forno × Oberkulmer mapping population were investigated in two seasons: 2011–2012 (referred to as 2012) and 2012–2013 (referred to as 2013). Field experiments were performed at the University of Nottingham Farm, Leicestershire, UK (52°50′N, 1°15′W, 50 m a.s.l.). Data on the air temperature, rainfall and solar radiation at the growing sites during the two seasons were collected from the nearby meteorological station and are summarized in Supplementary Data Fig. S1, together with the historic weather data from the Met Office (http://www.metoffice.gov.uk/). Both field sites were planted with winter oats in the previous cropping seasons. The soil was a sandy loam, with the nutrient indices similar over two sites: N = 0, P = 5, K = 4 and Mg = 4 (pH 7·5). The 226 RILs and two parents were grown in an RCB design with three replicates. The seeds were sown in 6 × 1·6 m plots on 19 October 2011 and in 12 × 1·6 m plots on 31 October 2012, with a sowing density of 250 seeds m–2 and row width 0·125 m in both seasons. Nitrogen fertilizer was applied as ammonium nitrate (NH4NO3, 34·5 % N) in three splits: early tillering, onset of stem elongation and flag leaf emergence at 40, 40 and 60 kg N ha–1 in 2012, and 40, 60 and 60 kg N ha–1 in 2013, respectively. Plant growth regulators were applied to reduce the risk of lodging, and prophylactic crop protection chemicals were applied to minimize diseases, weeds and pests.
Two sub-sets comprising 72 RILs in 2012 and 110 RILs in 2013 (including the former 72 lines) were selected in the light of their considerable differences in yield traits but similar flowering time (± 4 d in 2010 and ± 1 d in 2011). The sub-sets were used to calculate spikes m–2, grains per spike and biomass m–2 (in g), while machine-harvested grain yield (in t ha–1), grains m–2 and thousand grain weight (TGW, in g) were determined for the whole population (226 RILs).
At maturity, the plants in an area of 0·5 × 0·5 m (four central rows) were collected from each plot in the sub-sets. After cutting at ground level, the infertile shoots (bearing no spikes) were discarded as they were negligible in terms of dry mass in all RILs. The spike number of fertile shoots was then counted, which was multiplied by four to generate the spikes m–2. Subsequently, a sub-sample of approx. 70 g, with the fertile shoots counted, in 2012, and 20 fertile shoots in 2013, were obtained. For each sub-sample, the spikes were cut from these shoots, and the resultant spikes and stems were dried separately in an oven at 85 °C for 48 h. After weighing, above-ground biomass per shoot was calculated by dividing the sum of spike and stem dry weight by the fertile shoot number. The above-ground biomass m–2 was the biomass per shoot multiplied by the spikes m–2. The dried spikes were threshed completely, and the total grain dry weight was recorded. The average grain weight of an individual spike was calculated by dividing the total grain weight by shoot number in the sub-sample. Next, individual grain weight was calculated by counting 250 grains and weighing, and grains per spike were the result of grain weight per spike divided by individual grain weight.
After sampling, the actual machine-harvested area of each of the 226 RILs was measured (namely Ac, in m2). All the plots were then harvested with a combine harvester (2010, Sampo Rosenlew, Finland), and the combined grain samples were recorded for fresh weight (Wc, in g). Because of the non-free-threshing habit of spelt, the grain samples of many RILs could not be threshed completely. The machine-harvested grain yield, therefore, was derived from the threshability analysis. A sub-sample of 10 g was isolated randomly from each of the combined grain samples. Both threshed and unthreshed grains were counted by hand, recorded as N10. Total grain number in that combined grain sample (Nc) was estimated as: Nc = N10 × (Wc/10), and grain number m–2 (Npsm) as: Npsm = Nc/Ac. In addition, another grain sub-sample of approx. 1 kg from each plot was partially threshed, and 200 grains were counted, oven-dried and weighed for TGW (Wtg). Then, machine-harvested yield (Ym) was calculated as: Ym = Nc × (Wtg × 10–3) × 10–6/(Ac × 10–4).
Phasic development.
In the sub-sets, the key growth stages (GSs) of each RIL were determined in both seasons according to the decimal code compiled by Tottman and Broad (1987). For GS31 (onset of stem elongation), five main shoots selected randomly in the central rows of a plot were opened to measure the length of the first internodes every 4 d. A RIL was considered to enter GS31 when ≥3 shoots had their first internodes ≥1 cm. GS39 (flag leaf emergence), GS61 (beginning of anthesis) and GS92 (physiological maturity) were the dates when 50 % of the main shoots in a plot showed fully expanded flag leaves, visible anthers and yellow peduncles, respectively, as observed on a daily basis. The durations between different growth stages were also derived, including GS31−GS39, GS39−GS61, GS31−GS61 and GS61−GS92. The calendar dates for phenology were then converted into accumulated thermal time (degree-days, °Cd), with a base temperature 0 °C.
Glasshouse experiment
The 110 RILs in the sub-set, together with two parents, were grown in the glasshouse in 2013 − 2014 (referred as 2014) to investigate two key yield components: grains per spike and TGW. The seeds were sown in modular trays on 17 December 2013 for germination, followed by vernalization at 6 °C for 9 weeks. The plants were then individually transferred into 1 L pots, filled with a loam compost (No. 3, John Innes, Norwich, UK). The experiment was conducted in an RCB design with three replicates, and the blocks were arranged to account for the variation in PAR along the glasshouse. The plants were grown under natural daylight, watered every other day, and fed with 40 kg N ha–1 at early stem elongation. At maturity, the spikes of each plant were harvested, counted and threshed by hand. The grains were then dried at 85 °C for 48 h, weighed and counted to calculate grains per spike and TGW.
Statistical analysis of phenotypic data
Statistical analysis was performed using Genstat v17 and GraphPad Prism v6. Analysis of variance, multiple comparisons [Fisher’s unprotected least significant difference (LSD)] and Student’s t-test were applied to determine the significant differences between the parents and between the RILs. The average values over replicates were used for Pearson correlation analysis between different traits. All the phenotypic data were subjected to normality test before statistical analysis, and transformed if necessary.
Identification of quantitative trait loci
The genetic linkage map of Forno × Oberkulmer was downloaded from the GrainGenes database (http://wheat.pw.usda.gov/GG2/index.shtml), and consisted of 230 segregating loci (restriction fragment length polymorphisms and microsatellites) and 23 linkage groups covering 2469 cM. Analysis of QTLs was performed using MapQTL v6 (Van Ooijen, 2009), with the average values of phenotypic data over replicates. An initial interval mapping was carried out to identify the putative QTLs. A locus was considered significant when its peak of logarithm of the odds (LOD) was higher than the genome-wide threshold for the trait, which was computed from a permutation test with 1000 iterations. Subsequently, multiple-QTL model (MQM) mapping was performed by using the markers closest to the QTL peaks as the co-factors (P < 0·02). The nomenclature of QTLs followed the Catalogue of Gene Symbols for Wheat (http://wheat.pw.usda.gov/GG2/Triticum/wgc/2008/). The chart of linkage groups and 1 – LOD (the LOD peak values minus one) support intervals representing the QTL positions were drawn using MapChart v2.2 (Voorrips, 2002).
RESULTS
Phenotypic variation in seedling root traits between genotypes
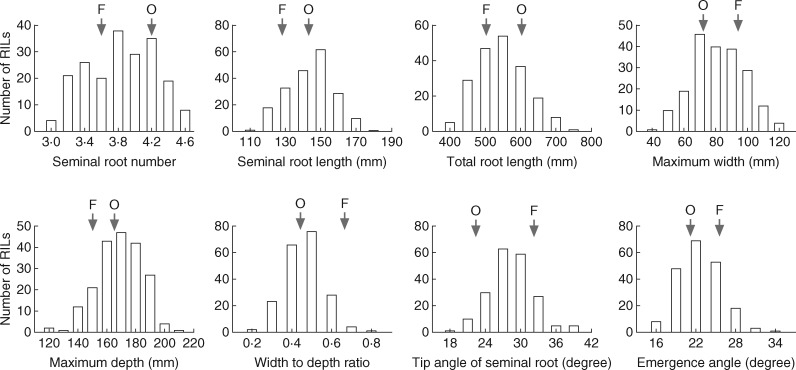
The bread wheat Forno and spelt Oberkulmer differed significantly in RSA examined in ‘the pouch and wick’ system in the growth chamber in 2014 (Figs 1 and 2; Table 1). Compared with Forno, Oberkulmer had more (17 %) and longer (12 %) seminal roots, and its total root length was also greater (20 %). No significant difference in root maximum depth was found, but the maximum width, width to depth ratio, and tip and emergence angles of seminal roots were smaller in Oberkulmer, indicating a narrow root system. As expected, there was a large variation in RSA among different RILs derived from Forno × Oberkulmer (Fig. 2; Table 1). Transgressive segregation (values beyond the parental phenotypic range) was seen for each trait. For example, the seminal root number of RIL-230 (4·7), seminal root length of RIL-164 (186 mm) and total root length of RIL-248 (766 mm) were 12, 30 and 27 % greater than those of the higher parent Oberkulmer.
Fig. 2.
Frequency distributions of the root architectural traits measured in the recombinant inbred line (RIL) mapping population of Forno (F) × Oberkulmer (O) in the ‘pouch and wick’ system in a growth chamber during 2014.
Table 1.
Descriptive statistics on seedling root traits in the ‘pouch and wick’ system in a growth chamber in 2014 as well as yield, yield components and phenology in field trials during 2012 and 2013 for the recombinant inbred line (RIL) mapping population of Forno (F) × Oberkulmer (O)
| Trait | Year | Parental lines |
RILs |
||||
|---|---|---|---|---|---|---|---|
| F | O | P-value | Mean (min; max) | P-value | s.e.d.* | ||
| Seedling root traits (n = 20) | |||||||
| Seminal root number | 2014 | 3·6 | 4·2 | <0·01 | 3·9 (3·0; 4·7) | <0·001 | 0·3 |
| Seminal root length (mm) | 2014 | 128 | 143 | <0·01 | 148 (118; 186) | <0·001 | 12 |
| Total root length (mm) | 2014 | 502 | 604 | <0·01 | 577 (404; 766) | <0·001 | 55 |
| Maximum width (mm) | 2014 | 94 | 72 | <0·001 | 86 (49; 127) | <0·001 | 16 |
| Maximum depth (mm) | 2014 | 150 | 165 | >0·05 | 174 (125; 215) | <0·001 | 13 |
| Width to depth ratio | 2014 | 0·66 | 0·44 | <0·001 | 0·51 (0·28; 0·80) | <0·001 | 0·10 |
| Tip angle of seminal root (°) | 2014 | 32·3 | 22·3 | <0·001 | 29·9 (18·1; 41·1) | <0·001 | 3·6 |
| Emergence angle of seminal root (°) | 2014 | 25·6 | 21·2 | <0·01 | 23·9 (16·7; 35·4) | <0·001 | 3·1 |
| Yield traits (n = 3 in each year) | |||||||
| Machine-harvested yield (t ha–1) | 2012 | 7·37 | 5·28 | <0·01 | 5·01 (2·62; 7·75) | <0·001 | 0·58 |
| 2013 | 9·56 | 5·60 | <0·01 | 6·42 (3·74; 10·39) | <0·001 | 0·97 | |
| Grains m–2 | 2012 | 20 296 | 12 393 | <0·01 | 12 478 (7244; 18 937) | <0·001 | 1309 |
| 2013 | 19 367 | 12 519 | <0·01 | 14 420 (8720; 24 864) | <0·001 | 2086 | |
| Spikes m–2 | 2012 | 726 | 472 | <0·01 | 545 (392; 848) | <0·001 | 77 |
| 2013 | 480 | 468 | >0·05 | 449 (300; 628) | <0·001 | 54 | |
| Grains per spike | 2012 | 40 | 33 | >0·05 | 37 (24; 51) | <0·001 | 4·6 |
| 2013 | 38 | 36 | >0·05 | 40 (26; 54) | <0·001 | 2·8 | |
| 2014† | 32 | 30 | >0·05 | 31 (17; 48) | <0·001 | 6 | |
| Thousand grain weight (g) | 2012 | 34·9 | 43·1 | <0·01 | 39·9 (31·9; 48·8) | <0·001 | 1·7 |
| 2013 | 48·5 | 43·9 | <0·01 | 45·0 (34·9; 53·6) | <0·001 | 1·3 | |
| 2014† | 52·5 | 47·5 | >0·05 | 47·3 (33·5; 58·8) | <0·001 | 4·4 | |
| Biomass m–2 (above-ground, g) | 2012 | 1765 | 1443 | >0·05 | 1570 (1018; 1914) | <0·05 | 182 |
| 2013 | 1719 | 1914 | >0·05 | 1829 (1391; 2428) | <0·001 | 249 | |
| Phenology (n = 3 in each year) | |||||||
| GS31 (onset of stem elongation, °Cd) | 2012 | 1184 | 1239 | <0·01 | 1214 (1175; 1267) | <0·001 | 20 |
| 2013 | 872 | 904 | <0·05 | 906 (843; 953) | <0·001 | 15 | |
| GS39 (full flag leaf emergence, °Cd) | 2012 | 1606 | 1672 | <0·01 | 1648 (1606; 1672) | <0·001 | 10 |
| 2013 | 1174 | 1228 | <0·01 | 1204 (1163; 1253) | <0·001 | 9 | |
| GS61 (anthesis, °Cd) | 2012 | 1876 | 1916 | <0·01 | 1900 (1840; 1944) | <0·001 | 9 |
| 2013 | 1442 | 1509 | <0·01 | 1485 (1434; 1547) | <0·001 | 9 | |
| GS92 (maturity, °Cd) | 2012 | 2658 | 2661 | >0·05 | 2680 (2553; 2811) | <0·001 | 27 |
| 2013 | 2066 | 2212 | <0·01 | 2106 (1975; 2268) | <0·001 | 30 | |
| GS31−GS39 (°Cd) | 2012 | 422 | 433 | >0·05 | 434 (389; 497) | <0·001 | 21 |
| 2013 | 302 | 324 | >0·05 | 298 (246; 358) | <0·001 | 18 | |
| GS39−GS61 (°Cd) | 2012 | 270 | 244 | <0·05 | 252 (209; 293) | <0·001 | 12 |
| 2013 | 268 | 281 | >0·05 | 281 (248; 320) | <0·001 | 12 | |
| GS31−GS61 (°Cd) | 2012 | 692 | 677 | >0·05 | 686 (628; 753) | <0·001 | 23 |
| 2013 | 570 | 605 | <0·05 | 579 (530; 652) | <0·001 | 17 | |
| GS61−GS92 (°Cd) | 2012 | 782 | 745 | >0·05 | 780 (658; 902) | <0·001 | 24 |
| 2013 | 624 | 703 | <0·01 | 622 (515; 753) | <0·001 | 28 | |
s.e.d., standard error of the difference;
Glasshouse experiment in 2014 (n = 3).
Phenotypic correlations between the seedling root traits
Significant correlations between different root traits were found (Table 2). Root system size represented by the total root length was positively correlated with both the number and length of seminal roots, and a closer relationship was found with the former. Root system shape represented by the ratio of width to depth was closely associated with the maximum width (r = 0·85, P < 0·01), rather than the depth. Total root length was positively associated with both the maximum width and depth, and had only a slight effect on root system shape. In addition, larger tip and emergence angles of seminal roots were associated with a wider root system, and a larger tip angle was also associated with a shallower root system.
Table 2.
Phenotypic correlations between the seedling root traits measured in the ‘pouch and wick’ system in a growth chamber during 2014 for the Forno × Oberkulmer mapping population
| SRN | SRL | TRL | MW | MD | W/D | TA | |
|---|---|---|---|---|---|---|---|
| SRL | −0 ·19** | ||||||
| TRL | 0 ·71** | 0 ·54** | |||||
| MW | 0 ·44** | 0 ·18* | 0 ·52** | ||||
| MD | 0 ·10 | 0 ·85** | 0 ·69** | 0 ·29** | |||
| W/D | 0 ·38** | −0 ·23** | 0 ·19** | 0 ·85** | −0 ·20** | ||
| TA | 0 ·14* | −0 ·24** | −0 ·05 | 0 ·35** | −0 ·34** | 0 ·52** | |
| EA | 0 ·21** | −0 ·11 | 0 ·14* | 0 ·49** | −0 ·08 | 0 ·54** | 0 ·36** |
Trait abbreviations: SRN, seminal root number; SRL, seminal root length; TRL, total root length; MW, maximum width; MD, maximum depth; W/D, width to depth ratio; TA, tip angle of seminal root; EA, emergence angle of seminal root.
Significant at P < 0 ·05, **significant at P < 0 ·01.
Phenotypic correlations of the root traits with yield and yield components
Field experiments showed that Oberkulmer had lower machine-harvested yield than Forno (Table 1). Also, grain number components, including grains m–2, spikes m–2 and grains per spike, tended to be lower in Oberkulmer. Thousand grain weight was higher in Oberkulmer in 2012, but not in 2013. In terms of the above-ground biomass m–2, there was no significant difference between the two parental lines. In the RIL population, a large variation was found for each yield trait investigated (Table 1).
Correlation analysis exhibited significant relationships between the seedling root traits, yield and yield components (Table 3). For the size parameters of the root system, total root length and its key component, seminal root number, were positively correlated with machine-harvested yield as well as grains m–2 in 2013. The two root parameters also showed positive relationships with grains per spike and biomass m–2, and negative relationships with TGW, in both years. For the shape parameters of the root system, both root maximum width and depth were positively correlated with machine-harvested yield in 2013. Consistent positive correlations of the maximum width with grains m–2 and spikes m–2 were found over both years. In general, there was no significant correlation between the tip and emergence angles of seminal roots, and yield traits, except for tip angle in 2013.
Table 3.
Phenotypic correlations of the seedling root traits in the ‘pouch and wick’ system with yield, yield components and phenology in (1) field trials during 2012 and 2013, and (2) in pot trials in a glasshouse in 2014 for the Forno × Oberkulmer mapping population
| SRN |
SRL |
TRL |
MW |
MD |
W/D |
TA |
EA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1)Field | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 | 2012 | 2013 |
| MHY | −0 ·01 | 0 ·22** | 0 ·09 | 0 ·17* | 0 ·05 | 0 ·31** | 0 ·09 | 0 ·20** | 0 ·10 | 0 ·31** | 0 ·07 | 0 ·05 | −0 ·06 | −0 ·14* | 0 ·05 | −0 ·03 |
| GPSM | 0 ·22 | 0 ·29** | 0 ·01 | 0 ·18 | 0 ·22 | 0 ·40** | 0 ·25* | 0 ·27** | 0 ·09 | 0 ·33** | 0 ·24* | 0 ·09 | 0 ·02 | −0 ·12 | 0 ·07 | −0 ·14 |
| SPSM | 0 ·21 | 0 ·18 | −0 ·03 | 0 ·02 | 0 ·19 | 0 ·17 | 0 ·25* | 0 ·21* | −0 ·02 | 0 ·02 | 0 ·25* | 0 ·16 | 0 ·01 | 0 ·02 | −0 ·05 | −0 ·01 |
| GPS | 0 ·32** | 0 ·31** | 0 ·06 | −0 ·05 | 0 ·35** | 0 ·22* | 0 ·22 | 0 ·04 | 0 ·18 | 0 ·06 | 0 ·18 | 0 ·03 | 0 ·09 | 0 ·05 | −0 ·03 | 0 ·01 |
| TGW | −0 ·47** | −0 ·37** | 0 ·12 | 0 ·13 | −0 ·36** | −0 ·23* | −0 ·25* | −0 ·07 | −0 ·07 | 0 ·03 | −0 ·21 | −0 ·09 | −0 ·08 | 0 ·03 | 0 ·14 | 0 ·08 |
| BPSM | 0 ·22 | 0 ·25** | 0 ·11 | −0 ·01 | 0 ·30** | 0 ·18* | 0 ·29* | 0 ·16 | 0 ·08 | 0 ·03 | 0 ·25* | 0 ·13 | 0 ·05 | 0 ·16 | 0 ·16 | 0 ·14 |
| GS31 | 0 ·14 | 0 ·22* | −0 ·05 | −0 ·14 | 0 ·11 | 0 ·08 | 0 ·16 | −0 ·04 | −0 ·03 | −0 ·10 | 0 ·15 | 0 ·01 | 0 ·03 | 0 ·02 | 0 ·09 | −0 ·03 |
| GS39 | 0 ·00 | 0 ·06 | −0 ·13 | −0 ·09 | −0 ·11 | −0 ·03 | −0 ·04 | −0 ·09 | −0 ·14 | −0 ·11 | −0 ·01 | −0 ·04 | 0 ·09 | 0 ·01 | 0 ·09 | −0 ·06 |
| GS61 | 0 ·17 | 0 ·14 | −0 ·30** | −0 ·20* | −0 ·07 | −0 ·04 | 0 ·08 | −0 ·06 | −0 ·24* | −0 ·19* | 0 ·16 | 0 ·04 | 0 ·21 | 0 ·13 | 0 ·01 | −0 ·03 |
| GS92 | 0 ·35** | 0 ·30** | −0 ·10 | −0 ·12 | 0 ·23* | 0 ·16 | 0 ·24* | 0 ·04 | 0 ·03 | −0 ·09 | 0 ·25* | 0 ·10 | 0 ·19 | 0 ·09 | 0 ·01 | −0 ·05 |
| GS31−GS39 | −0 ·14 | −0 ·21* | −0 ·05 | 0 ·09 | −0 ·19 | −0 ·12 | −0 ·18 | −0 ·04 | −0 ·08 | 0 ·02 | −0 ·16 | −0 ·06 | 0 ·04 | −0 ·01 | −0 ·02 | −0 ·02 |
| GS39−GS61 | 0 ·22 | 0 ·16 | −0 ·25* | −0 ·23* | 0 ·03 | −0 ·02 | 0 ·14 | 0 ·02 | −0 ·16 | −0 ·18 | 0 ·22 | 0 ·14 | 0 ·18 | 0 ·22* | −0 ·08 | 0 ·04 |
| GS31−GS61 | 0 ·03 | −0 ·11 | −0 ·22 | −0 ·05 | −0 ·15 | −0 ·13 | −0 ·06 | −0 ·03 | −0 ·17 | −0 ·09 | 0 ·02 | 0 ·03 | 0 ·15 | 0 ·12 | −0 ·07 | 0 ·00 |
| GS61−GS92 | 0 ·34** | 0 ·34** | 0 ·01 | −0 ·07 | 0 ·30* | 0 ·23* | 0 ·24* | 0 ·08 | 0 ·14 | −0 ·03 | 0 ·23 | 0 ·12 | 0 ·14 | 0 ·06 | 0 ·01 | −0 ·05 |
| (2) Glasshouse in 2014 | ||||||||||||||||
| GPS | 0 ·27** | −0 ·01 | 0 ·23* | 0 ·06 | 0 ·12 | 0 ·00 | −0 ·02 | −0 ·09 | ||||||||
| TGW | −0 ·22* | 0 ·01 | −0 ·20* | −0 ·08 | −0 ·06 | −0 ·06 | 0 ·10 | −0 ·10 | ||||||||
Trait abbreviations: SRN, seminal root number; SRL, seminal root length; TRL, total root length; MW, maximum width; MD, maximum depth; W/D, width to depth ratio; TA, the tip angle of seminal root; EA, the emergence angle of seminal root; MHY, machine-harvested yield; GPSM, grains m–2; SPSM, spikes m–2; GPS, grains per spike; TGW, thousand grain weight; BPSM, biomass m–2; GS31, GS39, GS61 and GS92, the accumulated thermal time at the onset of stem elongation, full flag leaf emergence, anthesis and maturity, respectively.
Significant at P < 0 ·05, **significant at P < 0 ·01.
In the glasshouse experiment, seminal root number and total root length also showed positive relationships with grains per spike, but negative relationships with TGW, concurring with the results found in the field experiments.
Phenotypic correlations of the root traits with phenology
Similarly, there were significant phenotypic differences in growth stages between genotypes in the RIL population (Table 1), and significant relationships of the growth stages with RSA (Table 3). Seminal root number and total root length were positively associated with GS92 and GS61–GS92, indicating that a larger root system can be associated with delayed maturity and a longer grain filling period. Additionally, longer seminal roots were associated with earlier GS61 and shorter GS39–GS61. One of the root system shape parameters, maximum depth, showed a consistent negative relationship with GS61.
Performance of the top and bottom RILs categorized by total root length
To confirm the effects of total root length representing the root system size on yield and yield-related traits, the top ten (RIL-31, 63, 69, 79, 89, 101, 134, 202, 230 and 248) and bottom ten (RIL-4, 7, 21, 67, 124, 180, 185, 186, 210 and 236) lines with the highest and lowest total root length, respectively, were selected (Table 4). The top ten RILs had 44 % longer total root length than the bottom ten, showing significantly more seminal roots, and a wider and deeper root system. In addition, the top RILs exhibited significantly higher yield, more grains per m–2 and grains per spike, higher biomass m–2, delayed maturity and a longer grain filling period, but lower TGW, consistent with the above observations.
Table 4.
Performance of the top and bottom ten recombinant inbred lines (RILs) with the highest and lowest total root length
| RIL | TRL | SRN | MW | MD | MHY | GPSM | GPS | TGW | BPSM | GS92 | GS61–GS92 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Top ten (mean) | 690 | 4·5 | 105 | 186 | 6·3 | 14110 | 43 | 41·2 | 1808 | 2443 | 741 |
| Bottom ten (mean) | 480 | 3·3 | 80 | 165 | 5·7 | 12166 | 35 | 45·6 | 1639 | 2374 | 673 |
| Difference (value) | 210 | 1·2 | 25 | 21 | 0·6 | 1944 | 8 | −4·4 | 169 | 69 | 68 |
| Difference (%) | 44 | 36 | 31 | 13 | 11 | 16 | 23 | −10 | 10 | 3 | 10 |
| P-value | <0·001 | <0·001 | <0·01 | <0·01 | <0·05 | <0·01 | <0·001 | <0·01 | <0·01 | <0·01 | <0·01 |
Trait abbreviations: TRL, total root length (mm); SRN, seminal root number; MW, maximum width (mm); MD, maximum depth (mm); MHY, machine-harvested yield (t ha–1); GPSM, grains m–2; GPS, grains per spike; TGW, thousand grain weight (g); BPSM, biomass m–2 (g); GS92, the accumulated thermal time at maturity (°Cd); GS61–GS92, the duration between anthesis and maturity (°Cd).
Detection of quantitative trait loci
Quantitative trait loci for the root architectural traits.
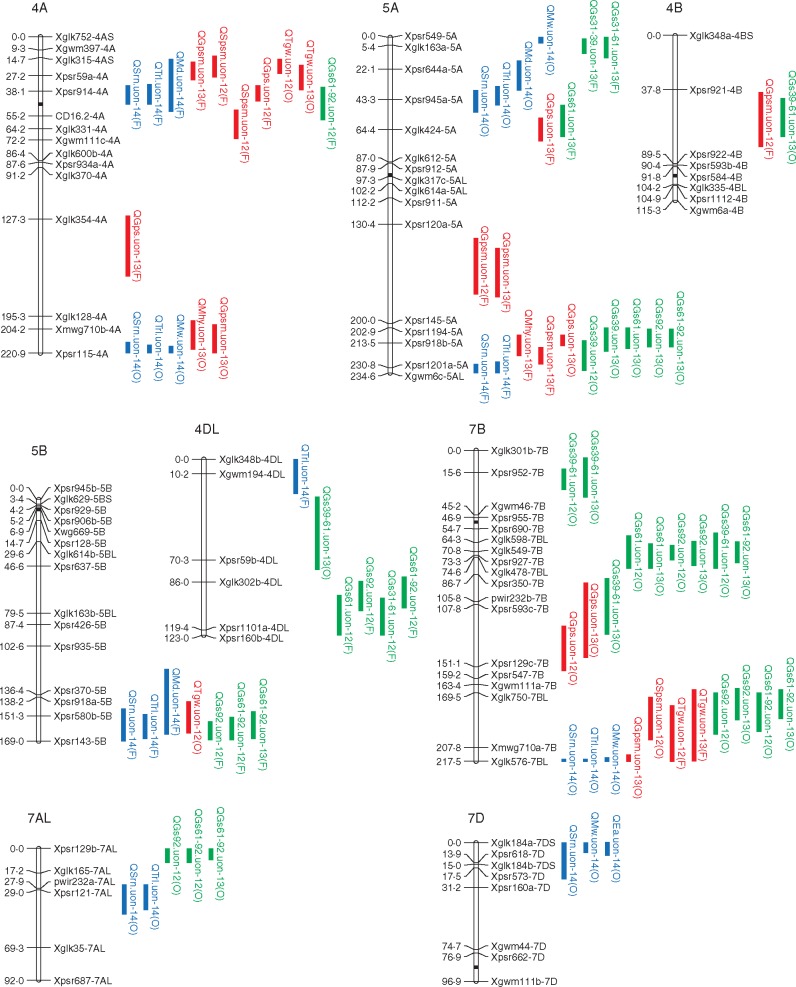
A total of 38 QTLs associated with the RSA were discovered, comprising 12 for seminal root number, two for seminal root length, 13 for total root length, five for each of root maximum width and depth, and one for the emergence angle of seminal roots (Fig. 3; Table 5). These QTLs were located on 12 chromosomes, i.e. 2A (five QTLs), 2B (one QTL), 3A (three QTLs), 3B (three QTLs), 3DL (two QTLs), 4A (six QTLs), 4DL (one QTL), 5A (six QTLs), 5B (three QTLs), 7AL (two QTLs), 7B (three QTLs) and 7D (three QTLs), individually explaining 6·3–22·3 % of the phenotypic variation observed. Forno provided 17 (45 %) increasing alleles (increasing the values of the traits) while Oberkulmer provided the remaining increasing alleles. Most of the QTLs identified (71 %) were associated with the parameters of root system size, and distributed on all 12 chromosomes where QTLs were detected.
Fig. 3.
Quantitative trait loci (QTLs) associated with seedling root traits, yield, yield components and phenology in the Forno × Oberkulmer mapping population. For each linkage group, the molecular marker identifiers are placed on the right side of the chromosome (hollow bar) and their locations are shown on the left side in centiMorgans (cM). The vertical bars on the right sides represent the 1 – LOD support intervals of significant QTLs: blue (root traits), red (yield and yield components) and green (phenology). Each QTL symbol consists of an upper case letter ‘Q’ and the abbreviations of quantitative trait, institution (uon), years when the QTL was detected (‘12’, 2012; ‘13’, 2013; ‘14’, 2014) and the parent conferring the allele with positive effect (‘F’, Forno; ‘O’, Oberkulmer). Trait abbreviations: Srn, seminal root number; Srl, seminal root length; Trl, total root length; Mw, maximum width of root system; Md, maximum depth of root system; Ea, emergence angle of seminal roots; Mhy, machine-harvested yield; Gpsm, grains m–2; Spsm, spikes m–2; Gps, grains per spike; Tgw, thousand grain weight; Gs39, the time at full flag leaf emergence; Gs61, the time at anthesis; Gs92, the time at maturity; Gs31–39, the duration from GS31 (the time at the onset of stem elongation) to GS39; Gs39–61, the duration from GS39 to GS61; Gs31–61, the duration from GS31 to GS61; Gs61–92, the duration from GS61 to GS92.
Table 5.
Quantitative trait loci (QTLs) associated with seedling root traits in the ‘pouch and wick’ system in a growth chamber as well as yield, yield components and phenology in field trials for the recombinant inbred line mapping population of Forno × Oberkulmer
| Trait/chromosome | Year | Position (cM) | LOD | R2* | Additive effect† | Closest marker |
|---|---|---|---|---|---|---|
| Seedling root traits | ||||||
| Seminal root number | ||||||
| 2A | 2014 | 24·2 | 8·7 | 18·0 | 0·18 | Xglk197-2AS |
| 2014 | 93·7 | 11·1 | 22·3 | 0·20 | Xpsr380-2A | |
| 2B | 2014 | 38·2 | 2·9 | 6·3 | –0·19 | Xglk302a-2BS |
| 3DL | 2014 | 34·2 | 3·5 | 7·7 | –0·11 | AC29-3DL |
| 4A | 2014 | 41·1 | 6·6 | 14·0 | 0·17 | Xpsr914-4A |
| 2014 | 220·8 | 5·5 | 11·8 | –0·15 | Xpsr115-4A | |
| 5A | 2014 | 43·3 | 3·1 | 6·8 | –0·11 | Xpsr945a-5A |
| 2014 | 234·0 | 4·4 | 9·5 | 0·13 | Xgwm6c-5AL | |
| 5B | 2014 | 158·3 | 3·1 | 6·7 | 0·13 | Xpsr580b-5B |
| 7AL | 2014 | 27·9 | 4·2 | 9·0 | –0·14 | pwir232a-7AL |
| 7B | 2014 | 217·4 | 8·1 | 16·8 | –0·17 | Xglk576-7BL |
| 7D | 2014 | 7·0 | 3·7 | 8·0 | –0·13 | Xpsr618-7D |
| Seminal root length (mm) | ||||||
| 3A | 2014 | 66·9 | 3·5 | 7·6 | –4 | Xprs570-3A |
| 3B | 2014 | 49·2 | 4·8 | 10·4 | 5 | Xpsr919a-3B |
| Total root length (mm) | ||||||
| 2A | 2014 | 25·5 | 4·1 | 9·0 | 22 | Xglk197-2AS |
| 2014 | 95·9 | 8·5 | 17·6 | 31 | Xpsr919b-2A | |
| 3A | 2014 | 63·9 | 5·6 | 11·9 | –30 | Xprs570-3A |
| 3B | 2014 | 51·9 | 3·3 | 7·3 | 21 | Xpsr1101b-3B |
| 3DL | 2014 | 34·2 | 3·0 | 6·6 | –19 | AC29-3DL |
| 4A | 2014 | 42·1 | 8·5 | 17·6 | 35 | Xpsr914-4A |
| 2014 | 220·8 | 6·4 | 13·6 | –28 | Xpsr115-4A | |
| 4DL | 2014 | 3·0 | 3·3 | 7·2 | 23 | Xglk348b-4DL |
| 5A | 2014 | 43·3 | 4·5 | 9·8 | –23 | Xpsr945a-5A |
| 2014 | 232·8 | 4·7 | 10·2 | 25 | Xpsr1201a-5A | |
| 5B | 2014 | 160·3 | 5·2 | 11·1 | 29 | Xpsr143-5B |
| 7AL | 2014 | 28·9 | 4·1 | 8·8 | –24 | Xpsr121-7AL |
| 7B | 2014 | 217·4 | 10·1 | 20·6 | –34 | Xglk576-7BL |
| Maximum width (mm) | ||||||
| 2A | 2014 | 97·9 | 3·3 | 7·3 | 5 | Xglk293b-2AL |
| 4A | 2014 | 220·9 | 4·5 | 9·8 | –5 | Xpsr115-4A |
| 5A | 2014 | 0·1 | 3·6 | 7·9 | –5 | Xpsr549-5A |
| 7B | 2014 | 217·4 | 4·0 | 8·6 | –5 | Xglk576-7BL |
| 7D | 2014 | 0·1 | 3·1 | 6·9 | –4 | Xglk184a-7DS |
| Maximum depth (mm) | ||||||
| 3A | 2014 | 65·9 | 4·1 | 9·0 | –6 | Xprs570-3A |
| 3B | 2014 | 40·9 | 4·1 | 9·0 | 5 | Xpsr902-3B |
| 4A | 2014 | 32·2 | 3·0 | 6·6 | 5 | Xpsr59a-4A |
| 5A | 2014 | 28·1 | 3·5 | 7·6 | –5 | Xpsr644a-5A |
| 5B | 2014 | 134·6 | 3·2 | 6·9 | 5 | Xpsr370-5B |
| Emergence angle of seminal root (°) | ||||||
| 7D | 2014 | 2·0 | 3·4 | 7·5 | –0·9 | Xglk184b-7DS |
| Yield traits | ||||||
| Machine-harvested yield (t ha–1) | ||||||
| 1DS | 2012 | 2·0 | 3·2 | 7·1 | 0·26 | Xpsr168-1DS |
| 2B | 2012 | 198·6 | 3·4 | 20·1 | –0·36 | Xpsr956a-2B |
| 3A | 2012 | 67·9 | 3·5 | 20·6 | –0·42 | Xprs570-3A |
| 2013 | 39·9 | 6·1 | 13·1 | –0·74 | Xpsr598-3A | |
| 3B | 2012 | 42·1 | 2·8 | 6·3 | 0·23 | Xpsr902-3B |
| 4A | 2013 | 210·2 | 3·8 | 8·4 | –0·42 | Xmwg710b-4A |
| 5A | 2013 | 218·5 | 9·3 | 19·2 | 0·68 | Xpsr918b-5A |
| Grains m–2 | ||||||
| 3A | 2012 | 37·9 | 4·1 | 9·0 | –1202 | Xpsr598-3A |
| 2012 | 106·4 | 5·0 | 11·0 | –847 | Xglk118-3AL | |
| 2013 | 43·9 | 6·0 | 12·9 | –1520 | Xpsr598-3A | |
| 2013 | 107·4 | 3·5 | 7·7 | –838 | Xglk577-3AL | |
| 4A | 2013 | 23·7 | 4·6 | 10·0 | 1018 | Xpsr59a-4A |
| 2013 | 212·2 | 2·8 | 6·3 | –826 | Xmwg710b-4A | |
| 4B | 2012 | 61·8 | 3·2 | 7·1 | 1186 | Xpsr921-4B |
| 5A | 2012 | 163·4 | 4·1 | 9·1 | 1562 | Xpsr120a-5A |
| 2013 | 165·4 | 4·6 | 10·1 | 1955 | Xpsr145-5A | |
| 2013 | 222·5 | 6·2 | 13·3 | 1284 | Xpsr1201a-5A | |
| 7B | 2013 | 217·4 | 3·9 | 8·5 | –811 | Xglk576-7BL |
| Spikes m–2 | ||||||
| 3A | 2013 | 50·9 | 3·2 | 12·7 | –27 | Xpsr598-3A |
| 4A | 2012 | 20·7 | 3·7 | 21·4 | 45 | Xglk315-4AS |
| 2012 | 60·2 | 3·6 | 21·0 | 42 | Xglk331-4A | |
| 7B | 2012 | 188·5 | 3·7 | 21·5 | –58 | Xglk750-7BL |
| Grains per spike | ||||||
| 2B | 2013 | 94·7 | 3·1 | 12·0 | –2·2 | Xglk687b-2BS |
| 2D | 2013 | 44·9 | 4·7 | 17·8 | –6·0 | Xpsr933b-2D |
| 3B | 2013 | 8·1 | 3·7 | 14·4 | –2·7 | Lrk10c-3BS |
| 4A | 2012 | 38·1 | 3·5 | 20·5 | 2·5 | Xpsr914-4A |
| 2013 | 151·3 | 3·6 | 14·0 | 5·5 | Xglk354-4A | |
| 5A | 2013 | 64·4 | 4·5 | 17·3 | 2·9 | Xglk424-5A |
| 2013 | 213·4 | 5·1 | 19·2 | –2·9 | Xpsr918b-5A | |
| 7B | 2012 | 137·8 | 3·2 | 18·7 | –3·6 | Xpsr129c-7B |
| 2013 | 127·8 | 3·1 | 12·2 | –3·4 | Xpsr593c-7B | |
| Thousand grain weight (g) | ||||||
| 2A | 2012 | 94·9 | 3·5 | 20·6 | –1·74 | Xpsr919b-2A |
| 2013 | 133·7 | 3·5 | 13·8 | –2·00 | Xglk699b-2AL | |
| 3B | 2012 | 80·5 | 4·2 | 23·9 | 1·95 | Xpsr1054-3B |
| 2013 | 2·9 | 3·4 | 13·4 | 1·61 | C970a-3B | |
| 2013 | 80·5 | 5·1 | 19·3 | 1·91 | Xpsr1054-3B | |
| 4A | 2012 | 20·7 | 6·6 | 35·1 | –2·65 | Xglk315-4AS |
| 2013 | 31·2 | 4·8 | 18·1 | –2·09 | Xpsr59a-4A | |
| 5B | 2012 | 154·3 | 3·0 | 17·7 | –1·80 | Xpsr580b-5B |
| 7B | 2012 | 190·5 | 5·2 | 28·9 | 3·04 | Xmwg710a-7B |
| 2013 | 187·5 | 3·2 | 12·7 | 2·24 | Xglk750-7BL | |
| Phenology | ||||||
| GS39 (full flag leaf emergence, °Cd) | ||||||
| 5A | 2012 | 222·4 | 2·9 | 17·5 | –10·3 | Xpsr1201a-5A |
| 2013 | 209·9 | 4·0 | 15·2 | –10·3 | Xpsr918b-5A | |
| GS61 (anthesis, °Cd) | ||||||
| 2D | 2013 | 45·9 | 3·7 | 14·3 | –24·3 | Xpsr933b-2D |
| 4DL | 2012 | 107·0 | 3·1 | 18·4 | 15·1 | Xpsr1101a-4DL |
| 5A | 2013 | 58·3 | 3·8 | 14·8 | 13·3 | Xglk424-5A |
| 2013 | 209·9 | 3·9 | 15·1 | –12·0 | Xpsr918b-5A | |
| 7B | 2012 | 77·6 | 5·0 | 27·9 | –15·8 | Xglk478-7BL |
| 2013 | 77·6 | 4·5 | 17·1 | –12·9 | Xglk478-7BL | |
| GS92 (maturity, °Cd) | ||||||
| 2A | 2012 | 94·9 | 5·1 | 28·6 | 39·9 | Xpsr919b-2A |
| 2013 | 94·9 | 4·2 | 16·1 | 32·8 | Xpsr919b-2A | |
| 2B | 2012 | 91·7 | 3·8 | 22·0 | –38·0 | Xpsr961-2B |
| 2013 | 81·7 | 4·3 | 16·4 | –40·5 | Xpsr961-2B | |
| 2D | 2013 | 56·9 | 4·4 | 17·0 | –81·1 | Xpsr335-2D |
| 4DL | 2012 | 97·0 | 4·4 | 25·2 | 50·5 | Xglk302b-4DL |
| 5A | 2013 | 210·9 | 3·8 | 14·8 | –36·2 | Xpsr918b-5A |
| 5B | 2012 | 162·2 | 3·7 | 21·7 | 41·9 | Xpsr143-5B |
| 7AL | 2012 | 0·1 | 4·1 | 23·7 | –39·1 | Xpsr129b-7AL |
| 7B | 2012 | 72·7 | 5·2 | 29·1 | –42·3 | Xpsr927-7B |
| 2012 | 185·5 | 3·8 | 22·2 | –51·2 | Xglk750-7BL | |
| 2013 | 73·3 | 5·3 | 19·7 | –38·6 | Xpsr927-7B | |
| 2013 | 173·5 | 3·7 | 14·3 | –35·9 | Xglk750-7BL | |
| GS31 (onset of stem elongation)−GS39 (°Cd) | ||||||
| 5A | 2013 | 5·4 | 3·4 | 13·2 | 8·6 | Xglk163a-5A |
| GS39−GS61 (°Cd) | ||||||
| 2D | 2013 | 33·9 | 3·1 | 12·2 | –10·1 | Xpsr933b-2D |
| 3B | 2013 | 0·1 | 3·3 | 12·9 | –5·4 | Xglk683-3BS |
| 2013 | 35·7 | 3·8 | 14·6 | –6·1 | Xglk538-3BS | |
| 4B | 2013 | 59·8 | 5·4 | 20·3 | –12·0 | Xpsr921-4B |
| 4DL | 2013 | 45·2 | 3·3 | 13·0 | –10·0 | Xpsr59b-4DL |
| 7B | 2012 | 18·6 | 5·3 | 29·6 | –13·7 | Xpsr952-7B |
| 2012 | 61·7 | 4·7 | 26·4 | –11·6 | Xglk598-7BL | |
| 2013 | 11·0 | 4·6 | 17·6 | –7·6 | Xpsr952-7B | |
| 2013 | 99·7 | 3·5 | 13·6 | –6·4 | pwir232b-7B | |
| GS31−GS61 (°Cd) | ||||||
| 1DS | 2013 | 0·1 | 3·2 | 12·5 | –8·8 | Xpsr168-1DS |
| 4DL | 2012 | 108·0 | 3·7 | 21·4 | 18·9 | Xpsr1101a-4DL |
| 5A | 2013 | 3·0 | 3·3 | 12·9 | 9·2 | Xpsr549-5A |
| GS61−GS92 (°Cd) | ||||||
| 2A | 2012 | 94·9 | 5·2 | 28·8 | 34·6 | Xpsr919b-2A |
| 2013 | 94·9 | 5·6 | 20·7 | 28·0 | Xpsr919b-2A | |
| 2B | 2012 | 92·7 | 4·1 | 23·6 | –34·1 | Xglk687b-2BS |
| 2013 | 81·7 | 4·2 | 16·2 | –30·3 | Xpsr961-2B | |
| 2D | 2013 | 61·9 | 4·0 | 15·4 | –54·8 | Xpsr335-2D |
| 4A | 2012 | 47·1 | 3·4 | 19·9 | 33·4 | CD16·2-4A |
| 4DL | 2012 | 92·0 | 3·3 | 19·3 | 33·7 | Xglk302b-4DL |
| 5A | 2013 | 211·9 | 3·0 | 11·9 | –24·0 | Xpsr918b-5A |
| 5B | 2012 | 161·2 | 3·3 | 19·4 | 34·7 | Xpsr143-5B |
| 2013 | 157·2 | 2·9 | 11·6 | 24·8 | Xpsr580b-5B | |
| 7AL | 2012 | 0·1 | 4·4 | 25·3 | –34·9 | Xpsr129b-7AL |
| 2013 | 0·1 | 3·4 | 13·2 | –23·3 | Xpsr129b-7AL | |
| 7B | 2012 | 185·5 | 4·5 | 25·5 | –47·4 | Xglk750-7BL |
| 2013 | 73·3 | 4·7 | 17·9 | –27·7 | Xpsr927-7B | |
| 2013 | 174·5 | 4·9 | 18·5 | –31·7 | Xglk750-7BL |
R2, the proportion of phenotypic variation explained by individual QTLs.
Positive and negative additive effects indicate that the alleles from Forno and Oberkulmer increase the values of the traits, respectively.
Twelve QTLs for total root length co-located with 12 QTLs for seminal root number and length, with the positive effects coming from the same parents (either Forno or Oberkulmer) (Fig. 3), which is in line with their positive phenotypic relationships (Table 2). Likewise, eight QTLs for total root length co-located with three QTLs for root maximum width and five for maximum depth on seven chromosomes; one QTL for the emergence angle of seminal roots co-located with one QTL for root maximum width. The increasing alleles of these coincident QTLs were from the same parents.
Quantitative trait locus coincidence between the root and yield traits.
For yield and yield components, 41 QTLs were detected on 11 chromosomes, comprising seven for machine-harvested yield, 11 for grains m–2, four for spikes m–2, nine for grains per spike and ten for TGW (Fig. 3; Table 5). These QTLs explained phenotypic variation between 6·3 and 35·1 %.
Quantitative trait locus coincidence between root and yield traits was frequently observed (Fig. 3). For the size parameters of the root system, 2–4 QTLs for seminal root number were coincident with two QTLs for yield (on 4A and 5A), four for grains m–2 (on 4A, 5A and 7B), three for spikes m–2 (on 4A and 7B), two for grains per spike (on 4A and 5A) and six for TGW (on 2A, 4A, 5B and 7B). Two QTLs for seminal root length were coincident with three QTLs for yield, two for grains m–2 and one for spikes m–2 on 3A and 3B. A similar result was found for total root length: 2–5 QTLs were coincident with five QTLs for yield (on 3A, 3B, 4A and 5A), six for grains m–2 (on 3A, 4A, 5A and 7B), four for spikes m–2 (on 3A, 4A and 7B), two for grains per spike (on 4A and 5A) and six for TGW (on 2A, 4A, 5B and 7B). For the shape parameters of the root system, one and two QTLs for root maximum width and depth, respectively, co-located with those for yield. In addition, two QTLs for root maximum width also co-located with two QTLs for grains m–2 and one for spikes m–2.
All coincident QTLs between the RSA and yield traits had the increasing alleles conferred by the same parents. The only exceptions were the coincident QTLs between the RSA and TGW, and between the RSA and grains per spike detected on 5AL, with the increasing alleles originating from the opposite parents. The results indicate tightly linked genes or pleiotropy, consistent with the correlation analysis between the RSA and yield traits.
Quantitative trait locus coincidence between the root traits and phasic development.
For the growth stages, a total of 49 QTLs were identified, individually explaining 11·6–29·6 % of the phenotypic variation (Fig. 3; Table 5). The spelt Oberkulmer provided 34 (69 %) alleles delaying phenology.
Analysis of the QTL coincidence between the RSA and phasic development showed that six QTLs for each of seminal root number and total root length co-located with seven QTLs for GS92 and ten for GS61–GS92, with the increasing alleles from the same parents except for those on 5AL (Fig. 3). One QTL for seminal root length co-located with one QTL for GS39–GS61 on 3B, with the positive effects coming from Forno and Oberkulmer, respectively. The nature of tight gene linkages or pleiotropy is in line with the positive phenotypic correlations between root system size, delayed maturity and extended grain filling period, as well as the negative correlation between seminal root length and the duration of flag leaf emergence to anthesis.
DISCUSSION
Significant links of the seedling RSA with yield and yield components
Using a germination paper-based ‘pouch and wick’ phenotyping pipeline, the seedling root architectural traits of the RILs derived from Forno and Oberkulmer were measured in a fast, cost-effective way. Although the seedling roots growing in the pouches may not develop exactly as those of plants growing in the field, this technique allowed us to characterize the differences in RSA between genotypes. Comparing the indoor seedling RSA traits with field agronomical performance has been reported in a number of studies, for instance in bread wheat (Liu et al., 2013; Atkinson et al., 2015), durum wheat (Triticum durum) (Cane et al., 2014), maize (Zea mays) (Li et al., 2015) and oilseed rape (Brassica napus) (Thomas et al., 2016). The present study identifies significant links between the seedling RSA, and field yield and yield-related traits, suggesting that the RSA of 13-day-old seedlings grown in the growth pouches could be predictive of field performance of these agronomic traits.
The root system size of wheat seedlings, as represented by seminal root number and total root length, was positively associated with grain yield, in line with a previous report by Liu et al. (2013). The positive correlations between the two root traits and yield were not found in 2012, probably due to the relatively heavy rainfall and consequent high disease levels (e.g. glume blotch that affects grain development) during grain filling in June and July of this year (Fig. S1). The bread wheat parent Forno is highly susceptible to glume blotch, whereas the spelt parent Oberkulmer has a high level of resistance (Messmer et al., 1999), which can at least partly explain the low TGW of Forno in 2012. Furthermore, the high disease pressure, rather than resource capture by roots, might limit the yields of many RILs in this year (sink limited), so the correlations between the root system size and yield could not be seen. An increase in yield as a result of a larger root system was mainly due to increased grain number rather than individual grain weight in the present study. Grains per spike, the major component of grain number, is substantially determined during stem elongation before anthesis (González et al., 2011). During this period, the spikes and stems are both growing rapidly, resulting in an intensive intra-plant competition for available assimilates (source limited). The growth of spikes, which is a key determinant of spike fertility and, in turn, grains per spike, may be limited in such a case (Fischer, 1985, 2011; González et al., 2011). There is evidence that vigorous seedling roots are beneficial to take up more soil resources (e.g. N and P) for early plant growth (An et al., 2006; Zhu et al., 2006; Cao et al., 2014), leading to a release of the intra-plant competition to a great extent. The resultant availability of more resources for young spike growth would increase grains per spike and yield. However, total root length and seminal root number had negative effects on TGW, probably because increased grain number is often associated with decreased individual grain weight (Slafer and Andrade, 1993; Miralles and Slafer, 1995; Griffiths et al., 2015). Grain size contributes to end-use quality of wheat (Gegas et al., 2010), so care must be taken that a larger root system should be selected for the purpose of improving grain yield, whereas a smaller one may improve grain quality but be accompanied by a penalty for yield.
Seedling root architectural traits also influence the phasic development. Notably, a larger root system is associated with delayed maturity and, in turn, extended grain filling, probably because of improved water and nutrient uptake for post-anthesis photosynthesis (Pinto and Reynolds, 2015). A longer period of grain filling was thought to enlarge final grain size, which contrasts with the result that the TGW is actually reduced. A further analysis showed that GS61–GS92 was positively correlated with grains per spike (r = 0·43, P < 0·001 in 2012; r = 0·60, P < 0·001 in 2013), indicating that more grains defined before anthesis as a consequence of a larger early root system require a longer period to fill thereafter. At the same time, more grains per spike are associated with smaller grains, as discussed above, which leads to an apparent negative correlation between grain size and grain filling period.
Genetic basis of the seedling RSA and its links with yield traits
A total of 25 QTLs for seminal root number and total root length were identified in this study, clustering on seven chromosomes (2A, 3DL, 4A, 5A, 5B, 7AL and 7B). Two clusters for the two traits were found on each of the short and long arms of 4A, similar to the findings of Christopher et al. (2013), and the clusters on 2AS and 5AS could correspond to those of Kabir et al. (2015) and Hamada et al. (2012). There was also a QTL identified in the distal region of 7DS for seminal root number, where the QTLs for total length of all roots and of seminal roots as well as grain yield have been detected previously (Atkinson et al., 2015). In addition, a QTL for total root length was mapped around the centromere of 3B in the present study, concurring with the reports by Bai et al. (2013) and Liu et al. (2013). The favourable QTLs identified here can be transferred into elite wheat cultivars for RSA improvement after fine mapping. Furthermore, the top ten RILs selected for their highest total root length, each of which carries mostly all the positive alleles underlying the above QTLs, will be utilized as the intermediate materials for introgression.
The phenotypic correlations of seminal root number and total root length with yield traits were confirmed by genetic analysis, showing tightly linked genes or pleiotropy. Recently, the QTL coincidence between the two root traits, yield and TGW was also found in tetraploid wheat (Maccaferri et al., 2016). qTaLRO-B1, a major 2BS QTL controlling the root length of wheat seedlings, has been used to develop near-isogenic lines with long roots and efficient phosphate uptake and biomass accumulation (Cao et al., 2014). In rice (Oryza sativa), a QTL increasing root length by 9·6 cm has been introgressed into an upland cultivar, giving rise to an increase of 0·2 t ha–1 in grain yield (Steele et al., 2013). These examples suggest the potential of introgressed coincident QTLs to improve both RSA and agronomic traits.
Spelt as a useful genetic resource for the RSA improvement in bread wheat
The significant variation in RSA in the present study is largely attributed to the use of spelt. Spelt is an ancient crop and has been cultivated since 5000 BC. As an old-world grain, spelt is adapted to high precipitation, heavy and infertile soils, and cool temperatures, and is still a minor crop grown on a considerable acreage in marginal areas at present (Campbell, 1997). Additionally, previous studies found that spelt has a faster early crop growth rate, and accumulates more biomass and N than bread wheat (Koutroubas et al., 2012). These aspects of the performance of spelt may depend greatly on its vigorous root system. In this study, the spelt Oberkulmer showed a large but narrow seminal root system, represented by more seminal roots, longer seminal and total roots, less root maximum width and smaller tip and emergence angles. A narrow but strong root system could help to penetrate the heavy soil for foraging and anchoring, while a large root system would enhance its ability for nutrient acquisition to support plant growth. In another experiment, spelt (three cultivars: ‘Oberkulmer’, ‘Tauro’ and ‘SB’) had much more root biomass at all soil layers than bread wheat (two cultivars: ‘Xi-19’ and ‘Forno’), and this is also true for root length density (root length per unit soil volume) (unpubl. res.), which, at the depth of 80 cm, is still above the threshold of approx. 1 cm cm–3 for potential water and nitrate capture (Foulkes et al., 2011). Genetically, the current study presents 14 and seven favourable alleles originating from the spelt Oberkulmer for the size and shape of the root system, respectively. Taken together, spelt is a critical genetic resource for the root system improvement in bread wheat, and its nature as a hexaploid will allow efficient hybridization with bread wheat to transfer these traits.
Conclusions
This study demonstrated that vigorous root growth at the early seedling stage affects yield formation in wheat. Seminal root number and total root length have positive effects on grain number, above-ground biomass and grain yield in the field, accompanied by a reduction of individual grain weight. A wider root system is also associated with higher grain number and yield. Delayed physiological maturity and a longer period of grain filling, as a consequence of a larger root system, may result from more grains defined before anthesis. These relationships are consistent with analysis of the QTL coincidence, indicating tight gene linkages or pleiotropy. The results suggest that the root architectural traits, particularly seminal root number and total root length, are favourable to improve yield potential, and should be incorporated into wheat ideotypes. The alleles with positive effects would permit marker-assisted selection, and the top ten RILs, which carry these alleles and also perform well in terms of yield, could be utilized as intermediate materials in breeding. In addition, spelt has a large root system, and may be a useful genetic resource for the RSA enhancement in bread wheat.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: mean air temperature, rainfall and solar radiation at University of Nottingham Farm in the 2011 − 2012 and 2012 − 2013 seasons.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the UK Commonwealth Scholarship Commission, China Scholarship Council and University of Nottingham. We thank Beat Keller (University of Zurich, Switzerland) and Monika Messmer (Research Institute of Organic Agriculture, Switzerland) for providing the seeds and marker data of the Forno × Oberkulmer population. We also thank Chuong Nguyen, Jonathan A. Atkinson, John Alcock, Matthew Tovey, Mark Meacham and Fiona Wilkinson (University of Nottingham) for their technical assistance in the field, glasshouse and laboratory work.
LITERATURE CITED
- An D, Su J, Liu Q, et al. 2006. Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant and Soil 284: 73–84. [Google Scholar]
- Atkinson JA, Wingen LU, Griffiths M, et al. 2015. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. Journal of Experimental Botany 66: 2283–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew H, Ma X, Yan G.. 2015. Screening wheat (Triticum spp.) genotypes for root length under contrasting water regimes: potential sources of variability for drought resistance breeding. Journal of Agronomy and Crop Science 201: 189–194. [Google Scholar]
- Bai C, Liang Y, Hawkesford MJ.. 2013. Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. Journal of Experimental Botany 64: 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonser AM, Lynch J, Snapp S.. 1996. Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytologist 132: 281–288. [DOI] [PubMed] [Google Scholar]
- Campbell KG. 1997. Spelt: agronomy, genetics, and breeding In: Janick J, ed. Plant breeding reviews. Oxford: John Wiley & Sons, 187–213. [Google Scholar]
- Cane MA, Maccaferri M, Nazemi G, et al. 2014. Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Molecular Breeding 34: 1629–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Ren Y, Zhang K, et al. 2014. Further genetic analysis of a major quantitative trait locus controlling root length and related traits in common wheat. Molecular Breeding 33: 975–985. [Google Scholar]
- Christopher J, Christopher M, Jennings R, et al. 2013. QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theoretical and Applied Genetics 126: 1563–1574. [DOI] [PubMed] [Google Scholar]
- Fischer RA. 1985. Number of kernels in wheat crops and the influence of solar radiation and temperature. Journal of Agricultural Science 105: 447–461. [Google Scholar]
- Fischer RA. 2011. Wheat physiology: a review of recent developments. Crop and Pasture Science 62: 95–114. [Google Scholar]
- Foulkes MJ, Slafer GA, Davies WJ, et al. 2011. Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. Journal of Experimental Botany 62: 469–486. [DOI] [PubMed] [Google Scholar]
- Gaju O, Allard V, Martre P, et al. 2014. Nitrogen partitioning and remobilization in relation to leaf senescence, grain yield and grain nitrogen concentration in wheat cultivars. Field Crops Research 155: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegas VC, Nazari A, Griffiths S, et al. 2010. A genetic framework for grain size and shape variation in wheat. The Plant Cell 22: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González FG, Miralles DJ, Slafer GA.. 2011. Wheat floret survival as related to pre-anthesis spike growth. Journal of Experimental Botany 62: 4889–4901. [DOI] [PubMed] [Google Scholar]
- Gregory PJ, Bengough AG, Grinev D, et al. 2009. Root phenomics of crops: opportunities and challenges. Functional Plant Biology 36: 922–929. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Wingen L, Pietragalla J, et al. 2015. Genetic dissection of grain size and grain number trade-offs in CIMMYT wheat germplasm. PLoS One 10: e0118847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada A, Nitta M, Nasuda S, et al. 2012. Novel QTLs for growth angle of seminal roots in wheat (Triticum aestivum L.). Plant and Soil 354: 395–405. [Google Scholar]
- Kabir MR, Liu G, Guan P, et al. 2015. Mapping QTLs associated with root traits using two different populations in wheat (Triticum aestivum L.). Euphytica 206: 175–190. [Google Scholar]
- Kirby EJM. 2002. Botany of the wheat plant In: Curtis BC, Rajaram S, Gómez Macpherson H, eds. Bread wheat improvement and production. Rome: Food and Agriculture Organization of the United Nations, 19–38. [Google Scholar]
- Koutroubas SD, Fotiadis S, Damalas CA.. 2012. Biomass and nitrogen accumulation and translocation in spelt (Triticum spelta) grown in a Mediterranean area. Field Crops Research 127: 1–8. [Google Scholar]
- Kutman UB, Kutman BY, Ceylan Y, Ova EA, Cakmak I.. 2012. Contributions of root uptake and remobilization to grain zinc accumulation in wheat depending on post-anthesis zinc availability and nitrogen nutrition. Plant and Soil 361: 177–187. [Google Scholar]
- Li P, Chen J, Wu P, et al. 2011. Quantitative trait loci analysis for the effect of Rht-b1 dwarfing gene on coleoptile length and seedling root length and number of bread wheat. Crop Science 51: 2561–2568. [Google Scholar]
- Li P, Chen F, Cai H, et al. 2015. A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. Journal of Experimental Botany 66: 3175–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li R, Chang X, Jing R.. 2013. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 189: 51–66. [Google Scholar]
- Lynch JP. 2007. Roots of the second Green Revolution. Australian Journal of Botany 55: 493–512. [Google Scholar]
- Maccaferri M, El-Feki W, Nazemi G, et al. 2016. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. Journal of Experimental Botany 67: 1161–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manschadi AM, Hammer GL, Christopher JT, deVoil P.. 2008. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant and Soil 303: 115–129. [Google Scholar]
- Messmer MM, Keller M, Zanetti S, Keller B.. 1999. Genetic linkage map of a wheat × spelt cross. Theoretical and Applied Genetics 98: 1163–1170. [Google Scholar]
- Miralles DJ, Slafer GA.. 1995. Individual grain weight responses to genetic reduction in culm length in wheat as affected by source–sink manipulations. Field Crops Research 43: 55–66. [Google Scholar]
- Pinto RS, Reynolds MP.. 2015. Common genetic basis for canopy temperature depression under heat and drought stress associated with optimized root distribution in bread wheat. Theoretical and Applied Genetics 128: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound MP, French AP, Atkinson JA, Wells DM, Bennett MJ, Pridmore T.. 2013. RootNav: navigating images of complex root architectures. Plant Physiology 162: 1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, He X, Liu D, et al. 2012. Major quantitative trait loci for seminal root morphology of wheat seedlings. Molecular Breeding 30: 139–148. [Google Scholar]
- Richard CAI, Hickey LT, Fletcher S, Jennings R, Chenu K, Christopher JT.. 2015. High-throughput phenotyping of seminal root traits in wheat. Plant Methods 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BM, Waines JG, Gill BS.. 1979. Genetic variability for seedling root numbers in wild and domesticated wheats. Crop Science 19: 843–847. [Google Scholar]
- Slafer GA, Andrade FH.. 1993. Physiological attributes related to the generation of grain yield in bread wheat cultivars released at different eras. Field Crops Research 31: 351–367. [Google Scholar]
- Smith S, De Smet I.. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society B: Biological Sciences 367: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele KA, Price AH, Witcombe JR, et al. 2013. QTLs associated with root traits increase yield in upland rice when transferred through marker-assisted selection. Theoretical and Applied Genetics 126: 101–108. [DOI] [PubMed] [Google Scholar]
- Sylvester-Bradley R, Berry P, Blake J, et al. 2008. The wheat growth guide .London: HGCA. [Google Scholar]
- Thomas CL, Graham NS, Hayden R, et al. 2016. High-throughput phenotyping (HTP) identifies seedling root traits linked to variation in seed yield and nutrient capture in field-grown oilseed rape (Brassica napus L.). Annals of Botany 118: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottman DR, Broad H.. 1987. The decimal code for the growth stages of cereals, with illustrations. Annals of Applied Biology 110: 441–454. [Google Scholar]
- Van Ooijen JW. 2009. MapQTL® 6, software for the mapping of quantitative traits loci in experimental populations of diploid species .Wageningen: Kyazma BV. [Google Scholar]
- Voorrips RE. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Waines JG, Ehdaie B.. 2007. Domestication and crop physiology: roots of Green-Revolution wheat. Annals of Botany 100: 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CA, Sylvester-Bradley R, Berry PM.. 2015. Root length densities of UK wheat and oilseed rape crops with implications for water capture and yield. Journal of Experimental Botany 66: 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, George TS, Gregory PJ, Bengough AG, Hallett PD, McKenzie BM.. 2013. Matching roots to their environment. Annals of Botany 112: 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mickelson SM, Kaeppler SM, Lynch JP.. 2006. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theoretical and Applied Genetics 113: 1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.