Abstract
We have demonstrated a novel drug delivery system to improve the selectivity of the current chemotherapy by pH-responsive, polymeric micelle carriers. The micelle carriers were prepared by the self-assembly of copolymers containing the polybasic poly(2-(diethylamino) ethyl methacrylate) (PDEAEMA) block. The mixed copolymers exhibited a comparatively low critical micelle concentration (CMC; 1.95–5.25 mg/L). The resultant mixed micelles were found to be <100 nm and were used to encapsulate the anticancer drug doxorubicin (DOX) with pretty good drug-loading content (24%) and entrapment efficiency (55%). Most importantly, the micelle carrier exhibited a pH-dependent conformational conversion and promoted the DOX release at the tumorous pH. Our in vitro studies demonstrated the comparable level of DOX-loaded mixed micelle delivery into tumor cells with the free DOX (80% of the tumor cells were killed after 48 h incubation). The DOX-loaded mixed micelles were effective to inhibit the proliferation of tumor cells after prolonged incubation. Overall, the pH-responsive mixed micelle system provided desirable potential in the controlled release of anticancer therapeutics.
Keywords: PDEAEMA, copolymers, pH-responsive, mixed micelle, DOX, targeting delivery
Introduction
Chemotherapy is one of the most widely used cancer treatment method. However, chemotherapy is far from ideal because of the heterogeneity and multidrug resistance of the tumor. The effectiveness of chemotherapy is also governed by the physical and chemical properties of the anticancer drugs, with the drug efficacy compromised by the poor water solubility, uneven in vivo distribution and weak stability.1–3
Over the past few decades, a series of novel drug delivery carriers have been extensively studied to improve the efficacy of chemotherapy, and the polymeric micelles system was considered to be one of the most promising candidates.4,5 The polymeric micelles have many prominent properties to serve as drug delivery carrier, including the enhanced solubilization of hydrophobic drugs, the reduced phagocytosis by reticuloendothelial system (RES), the enhanced permeability and retention (EPR) effect by passive targeting and the minimization of the drug toxicity on normal tissues.6,7 In addition, the micellar drug carriers can be readily prepared with the size,8–10 drug-loading capacity and the stability easily regulated by variation of the experimental conditions.11–14 Among all the polymeric micelles, the pH-responsive micelles have great potential for the controlled-release system in chemotherapy, because of the distinctive variation between the tumorous pH (4.5–5.0) and the normal physiological pH (7.4). By the conformation change in the micellar drug carriers at the tumorous pH, the drug can be released to the tumor with great precision.
The conventional strategies to prepare polymeric micelles are through the self-assembling of the graft copolymers, hyperbranched copolymers, amphiphilic multi-block copolymers or nonlinear copolymers.15–18 In particular, pH-responsive polymeric micelles are self-assembled by triblock or multi-block copolymers containing hydrophilic blocks, hydrophobic blocks and functional response blocks. A substantial proportion of hydrophilic/hydrophobic block in the micelles structure improved the in vivo micelle stability and drug-loading capability but at the price of the reduced micellar pH responsiveness. A fine-tuning of the ratio between the functional response block and the hydrophilic/hydrophobic block is pivotal to balance the micellar pH responsiveness and the micellar stability. Lang prepared a mixed micelle system by a star block copolymer S(polycaprolactone [PCL]-b-PDEAEMA) and a linear block copolymer poly(ethylene glycol) methyl ether (mPEG)-b-PCL. With the increase in mPEG-b-PCL content, the micelle size gradually decreased.19 Wu used a linear block copolymer AP-b-PEG-b-polylactide (PLA) and mPEG-b-poly(β-amino ester) (PAE) to prepare a mixed micelle system, which was used to study the anticancer drug doxorubicin (DOX) loading and control release.20 Compared to the mPEG-b-PAE micelles, the mixed micelle system showed a higher DOX-loading efficiency and better anticancer therapeutic efficacy both in vitro and in vivo.
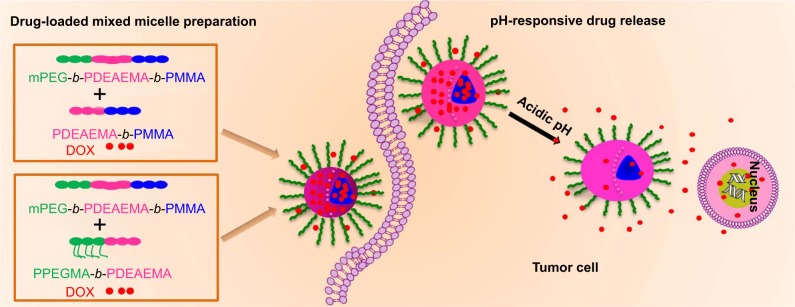
Herein, we reported a novel pH-responsive micellar system for selective targeting of the tumor cells. The pH responsiveness was originated from poly(2-(diethylamino)ethyl methacrylate) (PDEAEMA), a cationic polybasic polymer with a pKb of 6.9. In the acidic environment, the protonation of the pendant amine groups in the polymer greatly enhances the solubilization, whereas the solubilization is reduced at neutral or basic pH; thus, it serves as an excellent functional component to constitute the pH-responsive micelles for targeting tumor.21–23 In addition, the cationic tertiary amino groups in the polymeric micelles in the acidic-to-neutral environment assisted the micellar cellular uptake through the negatively charged cellular membranes.24,25 We synthesized pH-responsive triblock copolymer mPEG-b-PDEAEMA-b-poly(methyl methacrylate) (PMMA) (polymer A) and diblock copolymers PDEAEMA-b-PMMA (polymer B) and poly(poly(ethylene glycol) methyl ether methacrylate) (PPEGMA)-b-PDEAEMA (polymer C). Mixed micelles prepared by polymer A/B and polymer A/C were used as anticancer drug carriers. The PDEAEMA block improved the pH responsiveness of the mixed micelles. The hydrophilic block (mPEG and PEGMA) formed the hydrophilic shell and stabilized the structure of mixed micelles.26,27 PMMA formed the hydrophobic core of the mixed micelles to entrap the hydrophobic anticancer drug.28 The swelling mixed micelles in the acidic environment promoted the release of the loaded DOX. Compared to the single-component micelles, the mixed micelle showed a remarkable enhancement in drug-loading capability and drug release performance. The preparation of DOX-loaded mixed micelles and the pH-responsive drug release inside the tumor cell are shown in Scheme 1.
Scheme 1.
The preparation of DOX-loaded mixed micelles and the pH-responsive drug release inside the tumor cell.
Abbreviations: DOX, doxorubicin; mPEG, poly(ethylene glycol) methyl ether; PDEAEMA, poly(2-(diethylamino)ethyl methacrylate); PMMA, poly(methyl methacrylate); PPEGMA, poly(poly(ethylene glycol) methyl ether methacrylate).
Materials and methods
Synthesis of the copolymers
The triblock copolymer mPEG-b-PDEAEMA-b-PMMA (polymer A) was synthesized by activators regenerated by electron transfer atom transfer radical polymerization (ARGET ATRP) of 2-(diethylamino) ethyl methacrylate (DEAEMA) and methyl methacrylate (MMA) using brominated mPEG–Br as an initiator. The diblock copolymers PDEAEMA-b-PMMA (polymer B) and PPEGMA-b-PDEAEMA (polymer C) were synthesized by ARGET ATRP of DEAEMA and MMA or PEGMA, respectively, with ethyl 2-bromobutyrate as an initiator. All the synthesis routes and detail procedures are shown in the Supplementary materials.
Determination of the critical micelle concentration (CMC)
Fluorescence probe technique was used to determine the CMC using pyrene as the fluorescence probe.29–31 Briefly, polymer A, polymer A/polymer B (weight ratio 1:1, polymer A/B) and polymer A/polymer C (weight ratio 1:1, polymer A/C) solutions at the concentration of 0.1 mg/mL were prepared. The pyrene solution (12×10−7 M) and the polymer solution were mixed to obtain polymer concentrations ranging from 0.0001 to 0.1 mg/mL. The combined solution of pyrene and polymer was placed in dark at room temperature for 24 h to reach equilibrium before measurement. The emission intensity ratio of I338/I336 versus the logarithm of polymer concentrations was plotted. The CMC values were calculated from the intersection of the two linear fitting curves.
Study of the micelles pH responsiveness
Polymeric micelles were prepared by the solvent evaporation method. In all, 50 mg of polymer A, polymer A/B and polymer A/C were dissolved in 20 mL of acetone. The polymer solution was added into 50 mL deionized water dropwise, and the emulsion was concentrated to a 1 mg/mL micelle solution by stirring overnight. The micellar solution’s pH was adjusted by the addition of NaOH or HCl (0.01 M) solution. Dynamic light scattering (DLS) was used to measure the particle size and zeta potential of the micelles at different pH values. The base dissociation constant (pKb) buffering region of the mixed copolymers was measured. This method was reported in our previous works,21,32 and the details are shown in the Supplementary materials.
Preparation of DOX-loaded mixed micelles
The DOX-loaded mixed micelles were prepared by the dialysis method and purified by the diafiltration method. In brief, 30 mg of copolymer was dissolved in 20 mL of dimethyl sulfoxide (DMSO). A total of 15 mg of DOX⋅HCl was dissolved in another 20 mL of DMSO with 0.05 mmol of triethylamine to complex HCl. The copolymer solution and DOX solution were mixed by stirring for 4 h. Subsequently, the mixtures were transferred into a dialysis membrane (molecular weight cutoff [MWCO]: 3.5 kDa) and dialyzed with deionized water at room temperature for 48 h to purify the micelles. The deionized water was changed every 2 h for the first 12 h and then replaced every 6 h. After filtering through a 0.45 µm microporous membrane to remove the large-sized particles, the solutions were frozen and lyophilized to yield dried micelles. The dried micelles were stored at −20°C until further experiments. The method to determine the drug-loading content (LC) and entrapment efficiency (EE) was reported in our previous works,17,18 and the detail procedure is shown in the Supplementary materials.
In vitro DOX release study
The in vitro drug release study of the DOX-loaded micelles was conducted in pH 5.0, 6.5 and 7.4 buffer solutions. The experimental details and the cumulative drug release (%) calculation method are shown in the Supplementary materials.
In vitro cytotoxicity assay
The standard MTT assay was used to evaluate the cytotoxicity of the free DOX and mixed micelles II and III to HepG2 cells. The HepG2 cells were incubated in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/mL) and streptomycin (100 µg/mL) at 37°C for 3 days in a 5% CO2 atmosphere until confluence. After that, the cells were incubated in a culture medium supplemented with various concentrations of free DOX and micelle. The cells treated with an equal volume of DMEM were used as a control. All the cytotoxicity assays were performed in sextuplicate. The MTT assay procedure is shown in the Supplementary materials.
In vitro cellular uptake and endocytosis study
Confocal laser scanning microscopy (CLSM; TCS SP8; Leica Microsystems, Wetzlar, Germany) was used to study the cellular uptake of DOX. HepG2 cells were seeded on a coverslip in a 24-well plate at a density of 5×104 cells/well. The cells were cultured in a 5% CO2 atmosphere for 24 h at 37°C. Then, the culture medium was replaced with a DMEM containing 50 mg/L free DOX or DOX-loaded mixed micelles. The cells were incubated at 37°C for various times. The culture media was discarded, and the slides were immersed in PBS with gentle shaking to remove the extracellular DOX-loaded micelles. The washing step was repeated two more times. Subsequently, the cells were fixed with 4% (w/v) paraformaldehyde in PBS for 30 min at room temperature. The slides were rinsed with PBS (2 min ×3). Finally, the samples were stained with Hoechst 33324 (5 mg/mL in PBS) for 15 min at room temperature, and the slides were washed with PBS (2 min ×3) prior to the CLSM imaging.
Results and discussion
Characterization of the copolymers
In this study, all copolymers were synthesized by ARGET ATRP.33,34 mPEG–Br (Figures S1 and S2) was used as an macroinitiator to synthesize the triblock copolymer mPEG-b-PDEAEMA-b-PMMA (polymer A). The diblock copolymers PDEAEMA-b-PMMA (polymer B) and PPEGMA-b-PDEAEMA (polymer C) were polymerized with ethyl 2-bromobutyrate as the initiator. All the synthetic routes are shown in Schemes S1–S4. The molecular weight and the corresponding molecular weight distribution of the copolymers were determined by gel permeation chromatography (GPC) and proton nuclear magnetic resonance (1H NMR).
As shown in Table 1 and Figure 1, the molecular weight determined by GPC was reasonably identical to the theoretical molecular weight within the uncertainty. The unimodal symmetric distribution of the GPC traces indicated the excellent uniformity of the copolymers via ARGET ATRP. The Mw/Mn of all the copolymers was <1.4, indicating a narrow molecular weight distribution. The robustness of this synthesis method ensured the consistent performance of the prepared polymeric micelles.
Table 1.
GPC and 1H NMR data of the block copolymers
| Matrix | Polymer | Mn, Tha | Mn, GPCb | Mw/Mnc | Mn, NMRd |
|---|---|---|---|---|---|
| mPEG-Br | Initiator | 5,000 | 5,204 | 1.13 | 5,108 |
| mPEG-b-PDEAEMA21-b-PMMA17 | Polymer A | 10,437 | 10,151 | 1.18 | 10,664 |
| PDEAEMA21-b-PMMA34 | Polymer B | 8,000 | 6,835 | 1.28 | 7,335 |
| PPEGMA9-b-PDEAEMA25 | Polymer C | 10,000 | 9,231 | 1.36 | 9,685 |
Notes:
Calculated by theoretical analysis from the feed ratio of monomers to the initiator.
Measured by GPC in THF.
Calculated by the integration ratio of 1H NMR.
Abbreviations: GPC, gel permeation chromatography; 1H NMR, proton nuclear magnetic resonance; mPEG, poly(ethylene glycol) methyl ether; PDEAEMA, poly(2-(diethylamino)ethyl methacrylate); PMMA, poly(methyl methacrylate); PPEGMA, poly(poly(ethylene glycol) methyl ether methacrylate); THF, tetrahydrofuran.
Figure 1.
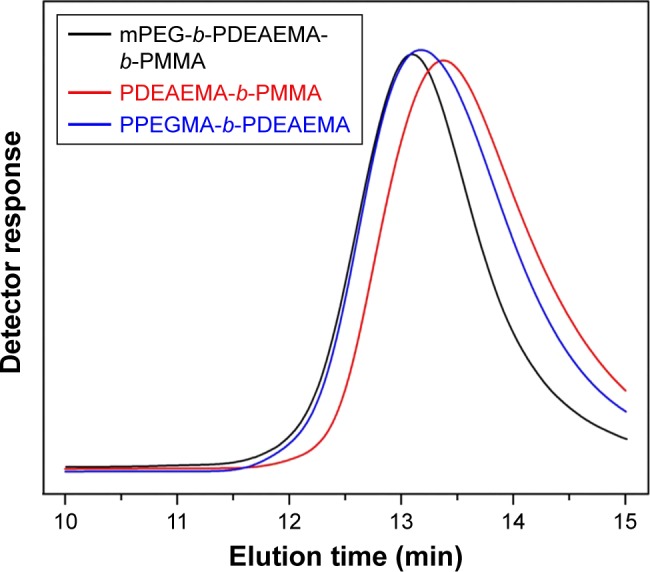
GPC traces of the block copolymers.
Abbreviations: GPC, gel permeation chromatography; mPEG, poly(ethylene glycol) methyl ether; PDEAEMA, poly(2-(diethylamino)ethyl methacrylate); PMMA, poly(methyl methacrylate); PPEGMA, poly(poly(ethylene glycol) methyl ether methacrylate).
Moreover, the composition and structure of the copolymers were confirmed by 1H NMR. The 1H NMR spectra of the polymers A, B and C are illustrated in Figure 2. For PPEGMA-b-PDEAEMA (polymer C), the peaks at 3.65, 4.09 and 1.90 ppm represented the protons of −CH2CH3 and −CH3 in the ethyl 2-bromobutyrate. The peaks at 0.98 and 1.82 ppm represented −CCH3 and −CH2− of the methacrylate main chain. The peaks at 2.71, 3.99, 0.89 and 2.57 ppm represented −CH2CH2− and −CH2CH3 in the DEAEMA block, and the peaks at 3.55 and 3.66 ppm were attributed to −CH2− and −CH3 in the PEGMA unit. For PDEAEMA-b-PMMA (polymer B), due to the variation of PMMA unit, the peaks at 3.63 ppm represented the terminal −CH3 in the MMA unit. Compared to PDEAEMA-b-PMMA, the triblock copolymer mPEG-b-PDEAEMA-b-PMMA (polymer A) contained an mPEG block. The peaks at 3.32 and 3.52 ppm of −CH2CH2− and −CH3 represented mPEG block instead of the characteristic peaks of ethyl 2-bromobutyrate.
Figure 2.
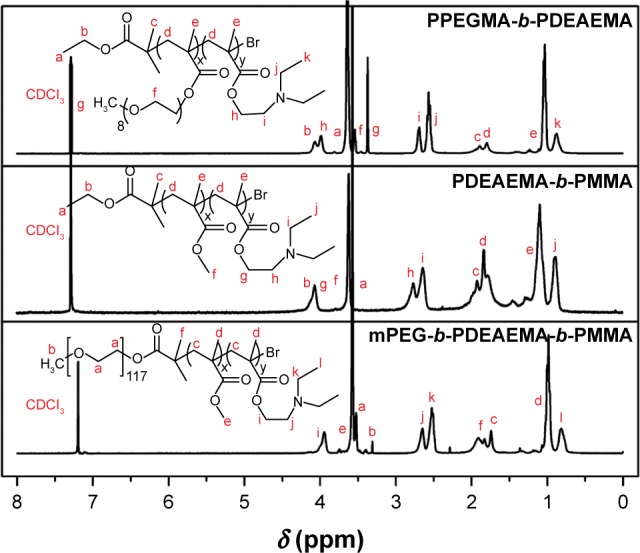
1H NMR spectra of the polymers A, B and C acquired with CDCl3 as a solvent.
Abbreviations: 1H NMR, proton nuclear magnetic resonance; mPEG, poly(ethylene glycol) methyl ether; PDEAEMA, poly(2-(diethylamino)ethyl methacrylate); PMMA, poly(methyl methacrylate); PPEGMA, poly(poly(ethylene glycol) methyl ether methacrylate).
Study of the pH-responsive micelles
Determination of the CMC was achieved by fluorescence probe technique. With the increase in the copolymer concentration, the emission of pyrene probe was shifted from 336 to 338 nm.35 The intensity ratio of I338/I336 of the polymer in various concentrations is shown in Figure 3. The CMC value can be determined from the intersection of the two linear fitting curves. Using this method, the CMC values of polymer A (mPEG-b-PDEAEMA-b-PMMA), polymer A/B (mPEG-b-PDEAEMA-b-PMMA/PDEAEMA-b-PMMA, weight ratio 1:1) and polymer A/C (mPEG-b-PDEAEMA-b-PMMA/PPEGMA-b-PDEAEMA, weight ratio 1:1) were determined to be 5.25, 1.95 and 4.18 mg/L, respectively.
Figure 3.
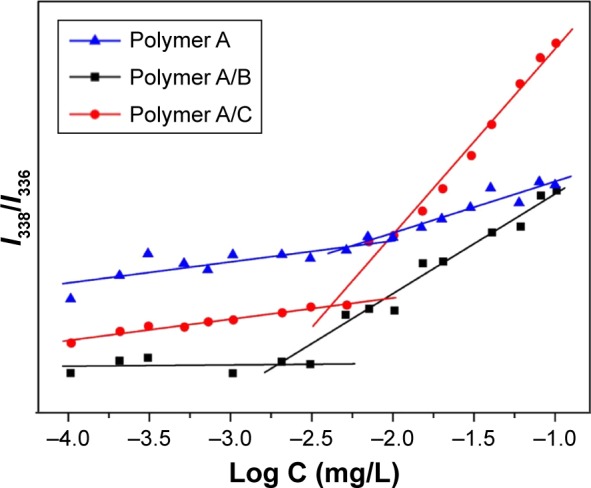
Determination of the CMC from the emission intensity ratios (I338/I336) as a function of logarithm of polymer A, polymer A/B and polymer A/C concentrations.
Abbreviation: CMC, critical micelle concentration.
To study the pKb buffer range of the copolymers in aqueous solution, acid–base titration was conducted with an automatic titrator (Hanon T-860; Hanon Instruments, Jinan, China). The PDEAEMA block in the copolymers was a weak polybase, resulting in a pH buffering capability of the copolymers. The pH buffering ranges of all the copolymers were within pH 6.5–7.4, which is close to the pH of tumorous cytoplasm. The pH buffering capability was dependent on the chain length of PDEAEMA. Enlarging the proportion of PDEAEMA block improved the overall pH buffering capability. As shown in the titration curves in Figure 4, an improved pH buffering capability was observed for polymer A/B and polymer A/C against the addition of sodium hydroxide solution.
Figure 4.
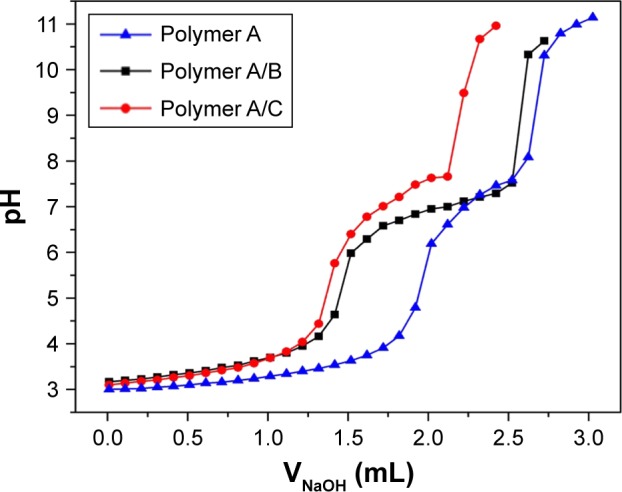
Titration curves of block copolymer solutions with NaOH.
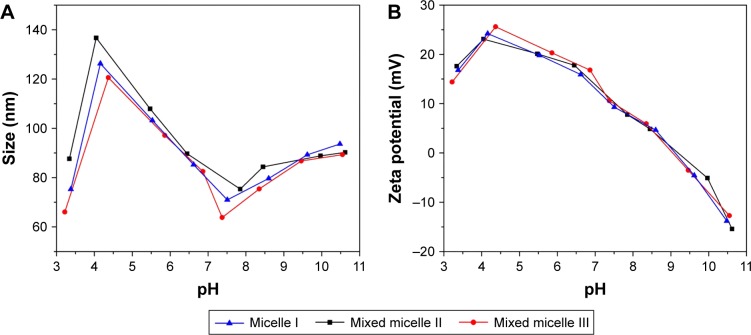
The protonation or deprotonation of the tertiary amino groups at the side chain of PDEAEMA occurred when the environmental pH was out of the pKb range. The interconversion between the protonated micelles and the deprotonated micelles resulted in the conversion of polymer polarity, which significantly impacted the size and surface charge of the micelles, as shown in Figure 5.
Figure 5.
Effects of environmental pH on the micelle sizes (A) and zeta potentials (B).
Figure 5A shows the variation of micelles sizes in aqueous solution from pH 3 to 11. Significantly, the sizes of the micelles I (self-assembled by polymer A), II (self-assembled by polymer A/B) and III (self-assembled by polymer A/C) were all <140 nm, which is critical to be used as an anticancer drug carrier.29 A similar trend of pH response was observed for the size variation of the micelles I, II and III. When the pH value was <4.5, the micellar size decreased. Decrease of the aggregation number of the polymers or even slight dissociation of the micelle structures was caused by the protonation of the PDEAEMA block in the micelles, which imparting a hydrophilic characteristic and the comparatively strong electrostatic repulsion between polymer chains. In the range of pH 4.5–7.0, the PDEAEMA block in the micelle was partially protonated. The graduate deprotonation of the tertiary amino groups increases micellar hydrophobicity and shrinks micellar structure toward the hydrophobic PMMA core. When pH >7.5, the PDEAEMA block was completely deprotonated to be the hydrophobic polymer. The increased tendency of micellar aggregation led to the further enlargement of the micelles.
The micellar zeta potential was also correlated with the pH variation, as shown in Figure 5B. The micelles I, II and III have the same tendency of reduced zeta potential with the pH increase. The zeta potential increased slightly from pH 3.0 to 4.5, presumably correlated to the higher degree of micellar aggregation.18 The zeta potential reached a maximum of 25 mV at pH 4.5. Further pH increased gradually deprotonated the DEAEMA segment, leading to the decrease in the micellar zeta potential. The zeta potential reached 0 mV at pH 9.0. In basic condition, the excess OH− resulted in the formation of the negatively charged micellar surface with negative zeta potential. Our results confirmed the formation of cationic micelles at tumorous pH (6.5–7.0). Therefore, we anticipated the promotion of the micelles’ uptake in cancer cells by the enhanced association between the micelles and the negatively charged cell membrane.25
Study of the DOX-loaded micelles
The DOX-loaded micelles I, II and III were prepared by mixing 1:2 (w/w) DOX/copolymer followed by dialysis at pH 7.4 solution. The micellar sizes, zeta potentials, drug LC and drug EE of the micelles were determined by DLS and ultraviolet (UV)–Vis analysis, and the results are summarized in Table 2. The particle size of all the DOX-free micelles was <70 nm with a narrow polydispersity index (PDI) of <0.17. The large proportion of hydrophobic PMMA block in the mixed micelle II resulted in the higher tendency of micelle aggregation and the relatively large measured micelle size. Noteworthy, the tertiary amino groups in PDEAEMA side chain are largely deprotonated at pH 7.4. Thus, the higher proportion of PDEAEMA block led to the increase in the core volume and the micelle size.36,37
Table 2.
Physicochemical properties of the blank and DOX-loaded mixed micelles
| Micelle | DOX/polymer (mg/mg) | LC (%) | EE (%) | Size (nm) | PDI | Zeta potential (mV) |
|---|---|---|---|---|---|---|
| Micelle I | 15/30 | 18.51±0.45 | 41.68±1.01 | 40.3±4.6 | 0.124±0.018 | 5.7±1.6 |
| Micelle I | 0/30 | – | – | 35.6±3.9 | 0.109±0.026 | 6.3±2.1 |
| Mixed micelle II | 15/30 | 21.72±0.39 | 48.90±0.88 | 94.3±7.2 | 0.135±0.034 | 13.6±1.8 |
| Mixed micelle II | 0/30 | – | – | 65.5±5.7 | 0.161±0.012 | 15.7±1.4 |
| Mixed micelle III | 15/30 | 23.98±0.52 | 53.99±1.17 | 85.7±6.8 | 0.118±0.051 | 9.8±1.5 |
| Mixed micelle III | 0/30 | – | – | 53.5±5.3 | 0.108±0.063 | 11.9±2.0 |
Note: The error bar represents the standard deviation of three parallel experiments (n=3). ‘–’ indicates no data.
Abbreviations: DOX, doxorubicin; LC, loading content; EE, entrapment efficiency; PDI, polydispersity index.
As shown in Table 2, the mixed micelles II and III exhibited higher DOX loading than the single-component micelle I. The high DOX loading in micelle II was the result of the formation of large micelle core by the higher proportion of the hydrophobic PMMA block. However, the highest DOX loading was obtained from the mixed micelle III. The presence of the hydrophilic PEGMA block preserved the encapsulated DOX in the micelle with a smaller micelle core. Based on our results, there are two important considerations to maximize the loading of hydrophobic drug in micelle: 1) enlarging the micelle core to generate the internal cavity for drug encapsulation and 2) fortifying the hydrophilic shell to reduce the diffusion of the encapsulated drug.38
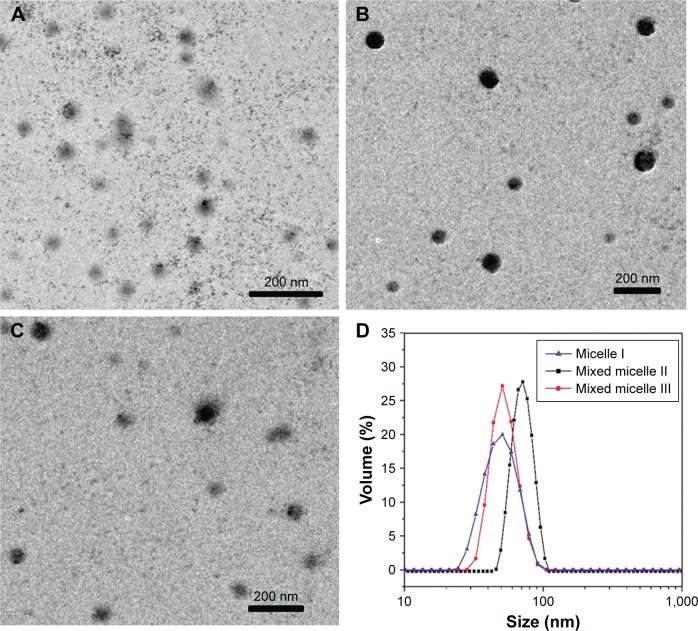
As shown in the transmission electron microscopy (TEM) images of Figure 6A–C, the DOX-loaded micelles were in spherical morphology, with the size of <100 nm. Figure 6D shows comparatively narrow size distribution of the micelles.
Figure 6.
TEM images of the block copolymer DOX-loaded micelle I (A), mixed micelle II (B) and mixed micelle III (C) and size distribution of the DOX-loaded micelles (D).
Note: Magnification ×20,000.
Abbreviations: TEM, transmission electron microscopy; DOX, doxorubicin.
In vitro release of DOX from the micelles
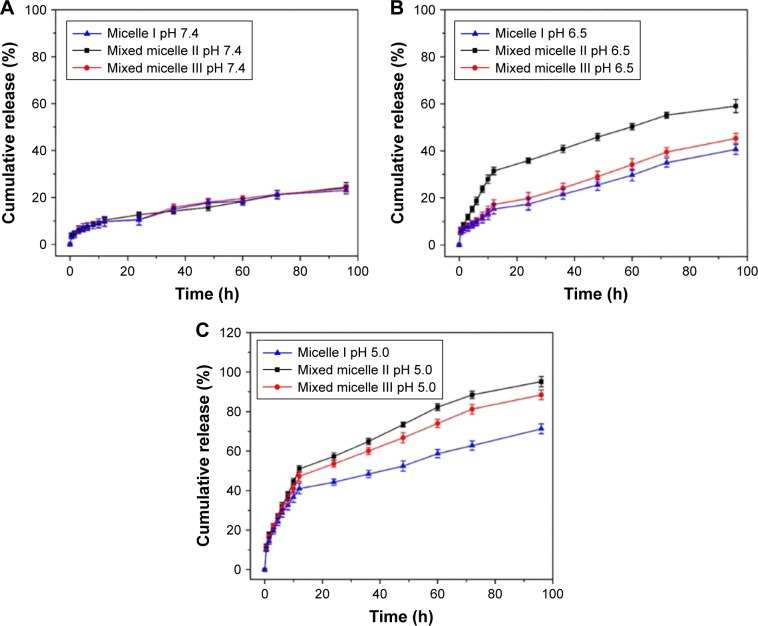
We have conducted a systematic investigation to study the pH-responsive DOX release in PBS solution. The pH 7.4 PBS solution was used to mimic the normal physiological environment. The pH 6.5 and 5.0 PBS solutions were used to mimic the extracellular tumor tissue and intracellular tumor tissue environment, respectively. The cumulative DOX release from micelles at pH 7.4, 6.5 and 5.0 is shown in Figure 7.
Figure 7.
In vitro DOX release profiles of DOX-loaded block copolymer micelles at pH 7.4 (A), pH 6.5 (B) and pH 5.0 (C).
Note: The error bar represents the standard deviation of three parallel experiments (n=3).
Abbreviation: DOX, doxorubicin.
Remarkably, the DOX release increased significantly with the pH changed from 7.4 to 5.0. In normal physiological environment (pH 7.4, Figure 7A), the encapsulated DOX was released at a constant rate with 10% release in 12 h and <25% cumulative release in 96 h. It indicated that the DOX was well retained inside the micellar core at physiological pH. It potentially minimized the DOX release into the normal tissues. In pH 6.5 PBS solution (Figure 7B), the cumulative DOX release in 12 h was 15%, 30% and 17% for micelle I, mixed micelle II and mixed micelle III, respectively. The cumulative DOX release in 96 h was 40%, 59% and 45% for micelle I, mixed micelle II and mixed micelle III, respectively. The release rate was in the following order: mixed micelle II > mixed micelle III > single-component micelle I. Compared with the other two systems, the mixed micelle II had a larger proportion of PDEAEMA, which led to a significantly faster release rate. The enhanced DOX release was contributed by the partial protonation of PDEAEMA block and the conversion to the partially swelled micelles. In the more acidic (pH 5.0) solution mimicking the cancer cell intracellular environment (Figure 7C), the DOX release was significantly accelerated. The cumulative DOX release in 12 h was 41%, 51% and 47% for micelle I, mixed micelle II and mixed micelle III, respectively. Remarkably, the cumulative release in 96 h reached 95% and 88% for mixed micelles II and III, respectively, surpassing the 70% DOX cumulative release of the single-component micelle I. The tertiary amino groups in the PDEAEMA block were completely protonated at pH 5.0. The conversion to the hydrophilic micelles was accompanied by the increased electrostatic repulsion between PDEAEMA segments. The fully swelled micellar structure potentially led to dissociation of micelles, which further accelerated the DOX release. The PDEAEMA block in the micelles served as a pH-sensitive on-and-off switch to control the release of the encapsulated DOX. In acidic conditions, the cumulative DOX release from mixed micelle II was the highest among all micelles, because of the thinnest hydrophilic shell in the micellar structure. Noteworthy, the protonation of DOX in the acidic condition also enhanced the water solubility, which accelerated the release of the encapsulated DOX.39
Therefore, the mixed micelles formed by triblock copolymer and diblock copolymers with PDEAEMA incorporation increased the drug LC. The release of the micellar-encapsulated DOX was controlled by the environmental pH. We have demonstrated the reduced drug release in normal physiological condition and the promoted drug release in tumorous condition. The pH-responsive micelle system has the great potential to be used as a drug carrier for cancer therapeutics.
Study of the DOX release mechanism
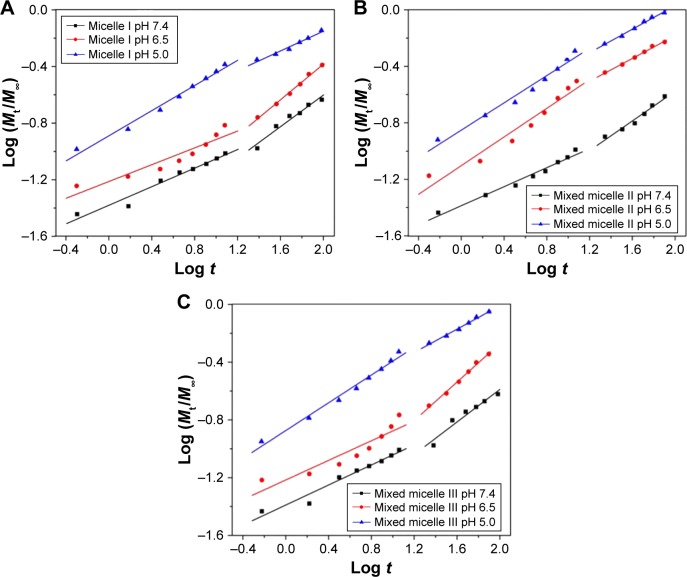
A semiempirical equation established by Ritger and Peppas was used to study the mechanism of DOX release from the polymeric micelles, as the method was validated in our previous works.17,18,40–43 Equation 1 was used to fit the drug release profiles, with n indicating the drug release mechanism and k reflecting the rate constant of the DOX release. The release process of the three DOX-loaded micelles can be divided into two stages, with the first stage from 0 to 12 h and the second stage between 12 and 96 h. The linear fitting curves of micelle I, mixed micelle II and mixed micelle III are shown in Figure 8A–C, respectively, and the fitting parameters are listed in Table 3.
| (1) |
Figure 8.
Plots of log (Mt/M∞) against log t for DOX release from micelle I (A), mixed micelle II (B) and mixed micelle III (C) at pH 7.4, 6.5 and 5.0.
Abbreviation: DOX, doxorubicin.
Table 3.
Fitting parameters of DOX release data from drug-loaded micelles at pH7.4, 6.5 and 5.0
| pH | Matrix | n1a | k1a | n2b | k2b |
|---|---|---|---|---|---|
| 7.4 | Micelle I | 0.33 | 0.04 | 0.56 | 0.02 |
| Mixed micelle II | 0.31 | 0.04 | 0.49 | 0.03 | |
| Mixed micelle III | 0.32 | 0.04 | 0.56 | 0.02 | |
| 6.5 | Micelle I | 0.30 | 0.06 | 0.63 | 0.02 |
| Mixed micelle II | 0.51 | 0.08 | 0.37 | 0.11 | |
| Mixed micelle III | 0.32 | 0.06 | 0.62 | 0.03 | |
| 5.0 | Micelle I | 0.45 | 0.13 | 0.28 | 0.14 |
| Mixed micelle II | 0.45 | 0.15 | 0.36 | 0.17 | |
| Mixed micelle III | 0.45 | 0.14 | 0.38 | 0.16 |
Notes:
The first stage 0–12 h.
The second stage 12–96 h.
Abbreviation: DOX, doxorubicin.
At pH 7.4, the n values of the three micelles were all <0.43, indicating that the release of DOX was dominated by the combination of diffusion and erosion control in the first 12 h.44 Owing to the deprotonation of PDEAEMA and the more compact micellar structure, it was difficult to diffuse out the drug from the micelle core. In the second stage, the n values were all in the range of 0.43–0.85, corresponding to the anomalous transport mechanism. The consistency of k value indicated that the DOX release rate was comparatively slow in neutral environment. At pH 6.5, the tertiary amino groups were partially protonated and the micelles structure was slightly swollen. In the first stage, the n values of the single-component micelle I and mixed micelle III were <0.43, but that of the mixed micelle II was between 0.43 and 0.85. It suggested the reduced retention of the encapsulated DOX in the micelles. In the second stage, the release mechanism of all the three micelles was reversed. Compared to the k value at pH 7.4, the higher k value at pH 6.5 indicated the release of acceleration under weak acidic conditions. However, for the single-component micelle I and mixed micelle III, the k value decreased in the second stage because of the DOX encapsulation by the hydrophilic shell. In the more acidic (pH 5.0) environment, the complete protonation of tertiary amino groups resulted in a higher degree of swelling of the micellar core. The n values of all the three micelles in the first stage were 0.45; thus, the DOX release mechanism was dominated by Fick diffusion. In the second stage, the n values of all the three micelles were <0.43, indicating the combination of diffusion and erosion control mechanism. It was the result of the micelles dissociation after full swelling. The k value of the second stage was much higher than that of pH 7.4 and 6.5, indicating the higher release rate in the acidic environment.
The abovementioned studies demonstrated the superior performance of the mixed micelles in pH-responsive DOX release. Thus, our subsequent study was focused on investigating the in vitro cytotoxicity and the cellular uptake of the mixed micelles.
In vitro cytotoxicity of mixed micelles
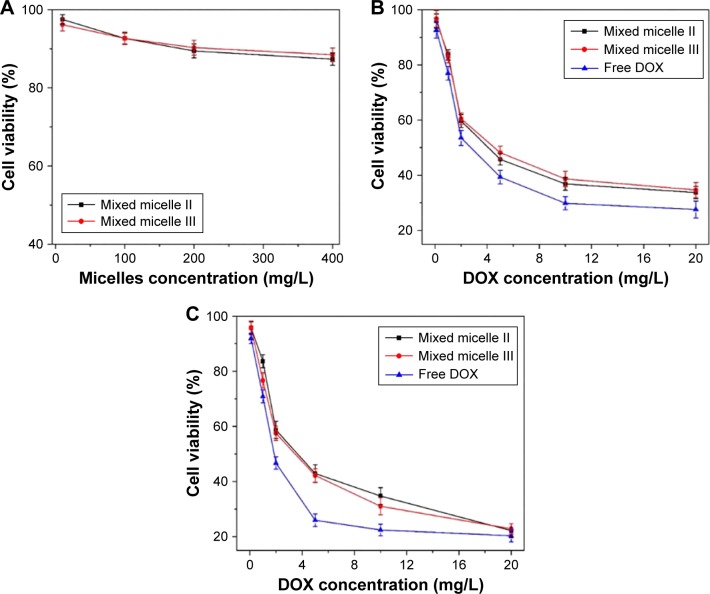
MTT assay was used to evaluate the in vitro cytotoxicity of the DOX-loaded mixed micelles on our model tumor cells HepG2. The HepG2 cell line is isolated from liver tumor, whose pH is ~5.0, and it was widely used as the in vitro experiment to study the activity of anticancer therapeutics.45,46 The DOX-free mixed micelles and DOX (0–20 mg/L) were used as negative and positive controls, respectively. No apparent toxicity was observed from the DOX-free mixed micelles, as shown in Figure 9A. The cell viability was still maintained at 85% after 48 h incubation at a comparatively high concentration (400 mg/L). The results indicated that the mixed micelles exhibited excellent biocompatibility and low toxicity.
Figure 9.
The viability of HepG2 after treating with DOX-free mixed micelles for 48 h incubation at various polymer doses (A), free DOX or DOX-loaded mixed micelles for 24 h (B) and 48 h (C) incubation at various DOX doses.
Note: The error bar represents the standard deviation of six parallel experiments (n=6).
Abbreviation: DOX, doxorubicin.
The HepG2 viability after 24 and 48 h treatment with free DOX and DOX-loaded mixed micelles is shown in Figure 9B and C, respectively. Regardless of the treatment time, the antitumor activity of the DOX-loaded mixed micelles was slightly lower than that of the free DOX. The DOX released from the micelles was a continuous process taking >96 h to complete, as shown in the in vitro release profiles (Figure 7). The cellular uptake efficiency of the mixed micelles II and III was almost the same, since they had similar charge loading. However, at 24 h, the viability of the micelle II-treated HepG2 was slightly lower than that of the mixed micelle III-treated cells, mainly because of the reduced hydrophilic shell in the mixed micelle II to retain the encapsulated DOX. As the incubation time prolonged to 48 h with 20 mg/L DOX, the cytotoxicity of both DOX-loaded mixed micelles was at a similar level, suggesting the similar DOX concentration in the cell culture. Thus, the cumulative DOX amount released from both mixed micelle systems was at a comparable level with the release time extended to >48 h. Overall, the polymeric micelles exhibited minimal cytotoxicity, but the DOX-loaded micelles were effective to inhibit the proliferation of tumor cells. Therefore, the mixed micelles are biocompatible and effective control release systems for cancer therapeutics.
In vitro cellular uptake
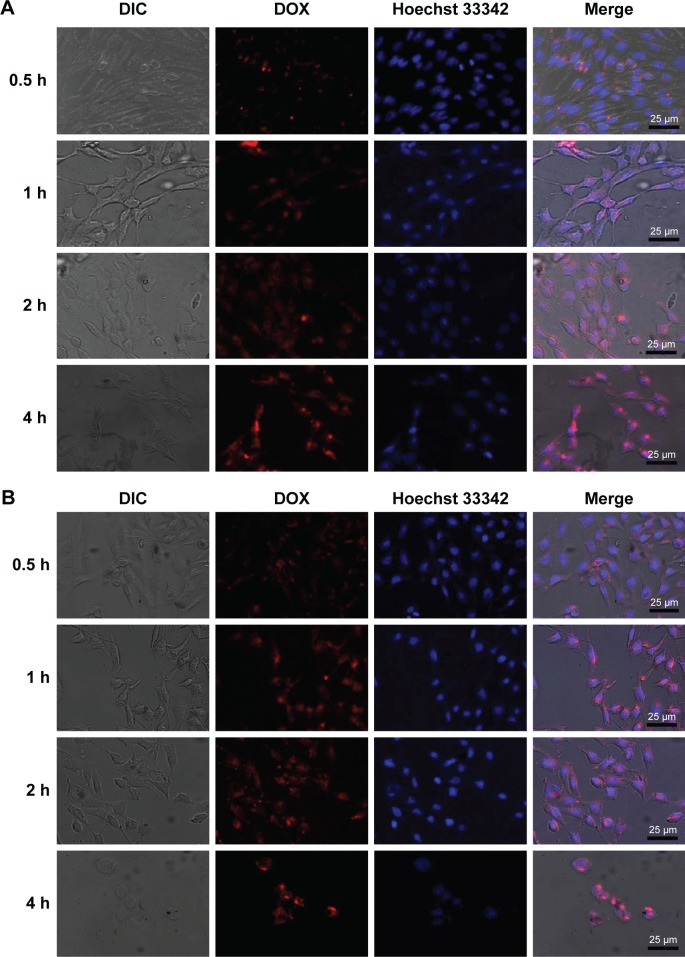
CLSM was used to image the cellular uptake and distribution of DOX in HepG2 cell. DOX emits red fluorescence and can be selectively visualized by fluorescent microscopy. The cell nucleus was stained with Hoechst 33342 for colocalization. The CLSM images of the cells treated with the DOX-loaded mixed micelles II and III for varying times are shown in Figure 10A and B, respectively, and the CLSM images of DOX-treated cells are shown in Figure S4.
Figure 10.
CLSM images of HepG2 cells incubated with mixed micelle II (A) and mixed micelle III (B) for 0.5, 1, 2, and 4 h at 37°C (red: DOX and blue: Hoechst 33342).
Note: Magnification ×40.
Abbreviations: CLSM, confocal laser scanning microscopy; DIC, differential interference contrast; DOX, doxorubicin.
The internalization of DOX into HepG2 cells was greatly promoted through passive diffusion. As shown in Figure S4, the free DOX was enriched in the cytoplasm for 0.5–1 h incubation. With the incubation time further extended, DOX started to be colocalized with the nucleus stain, suggesting the internalization of DOX into the nucleus. The intensity of the DOX red fluorescence was also increased significantly with prolonged incubation time.
As shown in Figure 10A and B, by incubating the HepG2 cells with the DOX micelles for 0.5 h, the red fluorescence from DOX was mainly distributed in the cytoplasmic region, indicating the micelle enrichment in the cytoplasm. Prolonging the incubation time to 1 h, the DOX fluorescence was observed in both the cell nucleus and the cytoplasm. The red DOX fluorescence at the cell nucleus was gradually increased by extending the incubation time to 2–4 h, indicating an enhanced amount of DOX localized in the nucleus. The results demonstrated the efficient internalization of the DOX-loaded mixed micelles with 30 min incubation. In particular, the DOX enriched in the nucleus with the prolongation of incubation time. The gradual release of DOX in the nucleus effectively inhibited the proliferation of cancer cells.
Conclusion
In the present study, we incorporated the polybasic PDEAEMA block to synthesize several pH-responsive copolymers, including a triblock copolymer mPEG-b-PDEAEMA-b-PMMA and two diblock copolymers PDEAEMA-b-PMMA and PPEGMA-b-PDEAEMA. The mixed copolymers were self-assembled to form two pH-responsive mixed micelle systems. The size of all the micelles was <100 nm, and the drug LC and EE of the mixed micelle were 24% and 55%, respectively. Most importantly, all the mixed micelles showed good pH responsiveness and promoted the DOX release at the tumorous pH. Compared to the single-component micelle I, the mixed micelles excelled in the pH responsiveness, CMC and DOX release performance. The DOX-loaded mixed micelles could be delivered into cancer cells effectively, and the anticancer activity of mixed DOX-loaded micelles was comparable to that of the free DOX. Therefore, these novel pH-responsive mixed micelles have tremendous potential to be used for the controlled release of the anticancer therapeutics.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No 91434125) and Team Project of Natural Science Foundation of Guangdong Province, China (No S2011030001366).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Du J, Lane LA, Nie S. Stimuli-responsive nanoparticles for targeting the tumor microenvironment. J Control Release. 2015;219:205–214. doi: 10.1016/j.jconrel.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Kim JY, Huh KM, Acharya G, Park K. Hydrotropic polymer micelles containing acrylic acid moieties for oral delivery of paclitaxel. J Control Release. 2008;132(3):222–229. doi: 10.1016/j.jconrel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Qian Y, Liu T, et al. Amphiphilic multiarm star block copolymer-based multifunctional unimolecular micelles for cancer targeted drug delivery and MR imaging. Biomaterials. 2011;32(27):6595–6605. doi: 10.1016/j.biomaterials.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 4.Prabhu RH, Patravale VB, Joshi MD. Polymeric nanoparticles for targeted treatment in oncology: current insights. Int J Nanomed. 2015;10:1001. doi: 10.2147/IJN.S56932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steichen SD, Caldorera-Moore M, Peppas NA. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur J Pharm Sci. 2013;48(3):416–427. doi: 10.1016/j.ejps.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rösler A, Vandermeulen GW, Klok H-A. Advanced drug delivery devices via self-assembly of amphiphilic block copolymers. Adv Drug Deliv Rev. 2012;64:270–279. doi: 10.1016/s0169-409x(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 7.Wei H, Zhuo R-X, Zhang X-Z. Design and development of polymeric micelles with cleavable links for intracellular drug delivery. Prog Polym Sci. 2013;38(3):503–535. [Google Scholar]
- 8.Attia ABE, Ong ZY, Hedrick JL, et al. Mixed micelles self-assembled from block copolymers for drug delivery. Curr Opin Colloid Interface Sci. 2011;16(3):182–194. [Google Scholar]
- 9.Huang X, Liao W, Zhang G, Kang S, Zhang CY. pH-sensitive micelles self-assembled from polymer brush (PAE-g-cholesterol)-b-PEG-b-(PAE-g-cholesterol) for anticancer drug delivery and controlled release. Int J Nanomed. 2017;12:2215–2226. doi: 10.2147/IJN.S130037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butt AM, Mohd Amin MC, Katas H. Synergistic effect of pH-responsive folate-functionalized poloxamer 407-TPGS-mixed micelles on targeted delivery of anticancer drugs. Int J Nanomed. 2015;10:1321–1334. doi: 10.2147/IJN.S78438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C, Pan P, Shan G, Bao Y, Fujita M, Maeda M. Core–shell structure, biodegradation, and drug release behavior of poly (lactic acid)/poly (ethylene glycol) block copolymer micelles tuned by macromolecular stereostructure. Langmuir. 2015;31(4):1527–1536. doi: 10.1021/la503869d. [DOI] [PubMed] [Google Scholar]
- 12.Bastakoti BP, Li Y, Imura M, et al. Polymeric micelle assembly with inorganic nanosheets for construction of mesoporous architectures with crystallized walls. Angew Chem Int Ed Engl. 2015;127(14):4296–4299. doi: 10.1002/anie.201410942. [DOI] [PubMed] [Google Scholar]
- 13.Yoo HS, Park TG. Folate receptor targeted biodegradable polymeric doxorubicin micelles. J Control Release. 2004;96(2):273–283. doi: 10.1016/j.jconrel.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Huang CK, Lo CL, Chen HH, Hsiue GH. Multifunctional micelles for cancer cell targeting, distribution imaging, and anticancer drug delivery. Adv Funct Mater. 2007;17(14):2291–2297. [Google Scholar]
- 15.Teramoto K, Kimata S, Harada T, Chuman M, Sakohara S. Separation of bisphenol-a by unimolecular micelles formed by pH-responsive polymer grafted onto polypropylene nonwoven fabrics. Chem Eng J. 2014;236:490–497. [Google Scholar]
- 16.Yao N, Lin W, Zhang X, Gu H, Zhang L. Amphiphilic β-cyclodextrin-based star-like block copolymer unimolecular micelles for facile in situ preparation of gold nanoparticles. J Polym Sci A Polym Chem. 2016;54(1):186–196. [Google Scholar]
- 17.Yang YQ, Zhao B, Li ZD, et al. pH-sensitive micelles self-assembled from multi-arm star triblock copolymers poly (ε-caprolactone)-b-poly (2-(diethylamino) ethyl methacrylate)-b-poly (poly (ethylene glycol) methyl ether methacrylate) for controlled anticancer drug delivery. Acta Biomater. 2013;9(8):7679–7690. doi: 10.1016/j.actbio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Lin W, Nie S, Zhong Q, et al. Amphiphilic miktoarm star copolymer (PCL)3-(PDEAEMA-b-PPEGMA)3 as pH-sensitive micelles in the delivery of anticancer drug. J Mater Chem B. 2014;2(25):4008–4020. doi: 10.1039/c3tb21694b. [DOI] [PubMed] [Google Scholar]
- 19.Huang X, Xiao Y, Lang M. Self-assembly of pH-sensitive mixed micelles based on linear and star copolymers for drug delivery. J Colloid Interface Sci. 2011;364(1):92–99. doi: 10.1016/j.jcis.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 20.Wu XL, Kim JH, Koo H, et al. Tumor-targeting peptide conjugated pH-responsive micelles as a potential drug carrier for cancer therapy. Bioconjug Chem. 2010;21(2):208–213. doi: 10.1021/bc9005283. [DOI] [PubMed] [Google Scholar]
- 21.Chen Q, Lin W, Wang H, Wang J, Zhang L. PDEAEMA-based pH-sensitive amphiphilic pentablock copolymers for controlled anticancer drug delivery. RSC Adv. 2016;6(72):68018–68027. [Google Scholar]
- 22.Zhao Z, Zhu F, Qu X, et al. pH-responsive polymeric Janus containers for controlled drug delivery. Polym Chem. 2015;6(22):4144–4153. [Google Scholar]
- 23.Feng C, Zhu C, Yao W, et al. Constructing semi-fluorinated PDEAEMA-b-PBTFVBP-b-PDEAEMA amphiphilic triblock copolymer via suc-cessive thermal step-growth cycloaddition polymerization and ATRP. Polym Chem. 2015;6(45):7881–7892. [Google Scholar]
- 24.Zhang Y, Ang CY, Li M, et al. Polymer-coated hollow mesoporous silica nanoparticles for triple-responsive drug delivery. ACS Appl Mater Interfaces. 2015;7(32):18179–18187. doi: 10.1021/acsami.5b05893. [DOI] [PubMed] [Google Scholar]
- 25.Chen T, Wu W, Xiao H, Chen Y, Chen M, Li J. Intelligent drug delivery system based on mesoporous silica nanoparticles coated with an ultra-pH-sensitive gatekeeper and poly (ethylene glycol) ACS Macro Lett. 2015;5(1):55–58. doi: 10.1021/acsmacrolett.5b00765. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Li L, Liu Y, et al. Treatment with mPEG-SPA improves the survival of corneal grafts in rats by immune camouflage. Biomaterials. 2015;43:13–22. doi: 10.1016/j.biomaterials.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Jeon H, Kim J, Lee YM, et al. Poly-paclitaxel/cyclodextrin-spion nano-assembly for magnetically guided drug delivery system. J Control Release. 2016;231:68–76. doi: 10.1016/j.jconrel.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Yang YQ, Zheng LS, Guo XD, Qian Y, Zhang LJ. pH-sensitive micelles self-assembled from amphiphilic copolymer brush for delivery of poorly water-soluble drugs. Biomacromolecules. 2010;12(1):116–122. doi: 10.1021/bm101058w. [DOI] [PubMed] [Google Scholar]
- 29.Chen C-Y, Kim TH, Wu W-C, et al. pH-dependent, thermosensitive polymeric nanocarriers for drug delivery to solid tumors. Biomaterials. 2013;34(18):4501–4509. doi: 10.1016/j.biomaterials.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Guo R, Yang M, Jiang X, Liu B. Thermo and pH dual-responsive nanoparticles for anti-cancer drug delivery. Adv Mater. 2007;19(19):2988–2992. [Google Scholar]
- 31.Loh XJ, Ong SJ, Tung YT, Choo HT. Dual responsive micelles based on poly [(r)-3-hydroxybutyrate] and poly (2-(di-methylamino) ethyl methacrylate) for effective doxorubicin delivery. Polym Chem. 2013;4(8):2564–2574. [Google Scholar]
- 32.Zhang CY, Xiong D, Sun Y, Zhao B, Lin WJ, Zhang LJ. Self-assembled micelles based on pH-sensitive PAE-g-mPEG-cholesterol block copolymer for anticancer drug delivery. Int J Nanomed. 2014;9:4923–4933. doi: 10.2147/IJN.S69493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar B, Negi YS. Synthesis and characterization of poly (potassium 1-hydroxy acrylate) and its derivatives by using different efficient catalysts. Polymer. 2015;80:159–170. [Google Scholar]
- 34.Matyjaszewski K. Atom transfer radical polymerization (ATRP): current status and future perspectives. Macromolecules. 2012;45(10):4015–4039. [Google Scholar]
- 35.Yang YQ, Guo XD, Lin WJ, Zhang LJ, Zhang CY, Qian Y. Amphiphilic copolymer brush with random ph-sensitive/hydrophobic structure: synthesis and self-assembled micelles for sustained drug delivery. Soft Matter. 2012;8(2):454–464. [Google Scholar]
- 36.Guo XD, Tandiono F, Wiradharma N, et al. Cationic micelles self-assembled from cholesterol-conjugated oligopeptides as an efficient gene delivery vector. Biomaterials. 2008;29(36):4838–4846. doi: 10.1016/j.biomaterials.2008.07.053. [DOI] [PubMed] [Google Scholar]
- 37.Guo XD, Zhang LJ, Chen Y, Qian Y. Core/shell pH-sensitive micelles self-assembled from cholesterol conjugated oligopeptides for anticancer drug delivery. AIChE J. 2010;56(7):1922–1931. [Google Scholar]
- 38.Sant VP, Smith D, Leroux J-C. Novel pH-sensitive supramolecular assemblies for oral delivery of poorly water soluble drugs: preparation and characterization. J Control Release. 2004;97(2):301–312. doi: 10.1016/j.jconrel.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Sun X, Nakayama-Ratchford N, Dai H. Supramolecular chemistry on water-soluble carbon nanotubes for drug loading and delivery. ACS Nano. 2007;1(1):50–56. doi: 10.1021/nn700040t. [DOI] [PubMed] [Google Scholar]
- 40.Young CR, Dietzsch C, Cerea M, et al. Physicochemical characterization and mechanisms of release of theophylline from melt-extruded dosage forms based on a methacrylic acid copolymer. Int J Pharm. 2005;301(1):112–120. doi: 10.1016/j.ijpharm.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Ritger PL, Peppas NA. A simple equation for description of solute release ii. Fickian and anomalous release from swellable devices. J Control Release. 1987;5(1):37–42. [PubMed] [Google Scholar]
- 42.Siepmann J, Peppas N. Modeling of drug release from delivery systems based on hydroxypropyl methylcellulose (HPMC) Adv Drug Deliv Rev. 2012;64:163–174. doi: 10.1016/s0169-409x(01)00112-0. [DOI] [PubMed] [Google Scholar]
- 43.Siepmann J, Göpferich A. Mathematical modeling of bioerodible, polymeric drug delivery systems. Adv Drug Deliv Rev. 2001;48(2):229–247. doi: 10.1016/s0169-409x(01)00116-8. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Li H, Wei M, Lu J, Jin L. pH-responsive composite based on prednisone-block copolymer micelle intercalated inorganic layered matrix: structure and in vitro drug release. Chem Eng J. 2009;151(1):359–366. [Google Scholar]
- 45.Zhao D, Yi X, Xu J, Yuan G, Zhuo R, Li F. Design and construction of self-hidden and pH-reversed targeting drug delivery nanovehicles via noncovalent interactions to overcome drug resistance. J Mater Chem B. 2017;5(15):2823–2831. doi: 10.1039/c6tb03211g. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Tang Z, Gao Y, Sun H, Zhou S. A bio-inspired rod-shaped nanoplatform for strongly infecting tumor cells and enhancing the delivery efficiency of anticancer drugs. Adv Funct Mater. 2016;26(1):66–79. [Google Scholar]